Abstract
Although most folding intermediates escape detection, their characterization is crucial to the elucidation of folding mechanisms. Here we outline a powerful strategy to populate partially unfolded intermediates: A buried aliphatic residue is substituted with a charged residue (e.g., Leu→Glu−) to destabilize and unfold a specific region of the protein. We apply this strategy to Ubiquitin, reversibly trapping a folding intermediate in which the β5 strand is unfolded. The intermediate refolds to a native-like structure upon charge neutralization under mildly acidic conditions. Characterization of the trapped intermediate using NMR and hydrogen exchange methods identifies a second folding intermediate and reveals the order and free energies of the two major folding events on the native side of the rate-limiting step. This general strategy may be combined with other methods and have broad applications in the study of protein folding and other reactions that require trapping of high energy states.
Keywords: Psi-analysis, ubiquitin, protein engineering, native-state hydrogen exchange, small angle X-ray scattering, NMR
To fully delineate protein folding mechanisms, multiple points on the free energy surface must be structurally and thermodynamically characterized. However, this task is extremely challenging, in particular for proteins that fold in an apparent two-state manner. Folding intermediates tend to be transient and weakly populated in both kinetic and equilibrium studies. Two major strategies are used to address this challenge. The most straightforward and information-rich method is direct trapping followed by structural characterization. However, trapping often involves the use of unnatural conditions (e.g., low pH, cosolvents),1; 2 or relies on protein-specific mutagenesis.3; 4; 5; 6; 7; 8
The other major strategy involves the detection of sparsely populated intermediates, for example, using NMR relaxation dispersion,7; 9 native state hydrogen exchange (NSHX),10; 11 chemical modification12; 13 or proteolysis.14; 15 Relaxation dispersion (RD) methods can characterize excited states that are populated as little as 0.5%, and provide interconversion rates, stability and the chemical shifts of the excited state.9 RD methods have characterized intermediates that reside either before or after the rate-limiting TS for a number of proteins.16; 17; 18
Native state hydrogen exchange (NSHX) can detect partially unfolded intermediates by examining the denaturant dependence of the HX rates. The regions of the protein that unfold cooperatively in subglobal openings are termed foldons. Foldons typically are associated with elements of secondary structure. Trapped intermediates have been created by selectively destabilizing or deleting a foldon.6; 19; 20; 21; 22 The resulting intermediates generally are on-pathway, located before or after the rate-limiting step in two-state or multi-state folding, respectively.
RD and NSHX methods are powerful but have certain limitations. RD requires appropriate interconversion rates and chemical shift differences, and the intermediate must comprise at least ~0.5% of the total population. NSHX requires that the hydrogen bonds broken in the subglobal opening are not hidden by faster exchange processes including smaller scale (local) openings (Supplementary Fig. 1). In addition, the free energy and the size of the subglobal opening must be such that its HX “isotherm” is distinct from those of global and local unfolding processes. These requirements may explain why NSHX studies on mammalian Ubiquitin (Ub) did not reveal any subglobal openings (Sidhu, N.S. and Robertson, A.D., master thesis, Univ. of Iowa).
These issues call for new strategies for trapping folding intermediates. Here, we explore a strategy that involves reversibly destabilizing a foldon. The destabilization is accomplished by replacing a buried aliphatic residue with a nearly isosteric but charged acidic residue (e.g. Leu→Glu−).23; 24 At neutral pH, the energetic penalty associated with the burial of the acidic group destabilizes the foldon, thereby creating an equilibrium intermediate amenable for detailed characterization. We apply this strategy to Ub to reversibly populate an intermediate that had escaped detection by NSHX. Kinetic folding experiments indicate that it is a late folding intermediate on the native side of the rate-limiting barrier. The application of NSHX to the trapped folding intermediate identifies a second unfolding intermediate. These results reveal a hierarchical unfolding pathway beginning with the loss of the β5 strand followed by the disruption of a small 310 helix.
Results
Rationally populating intermediates
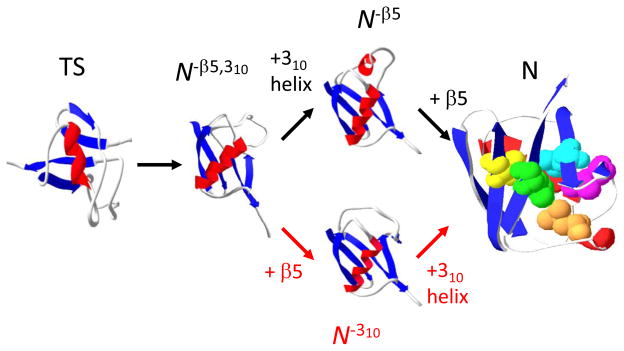
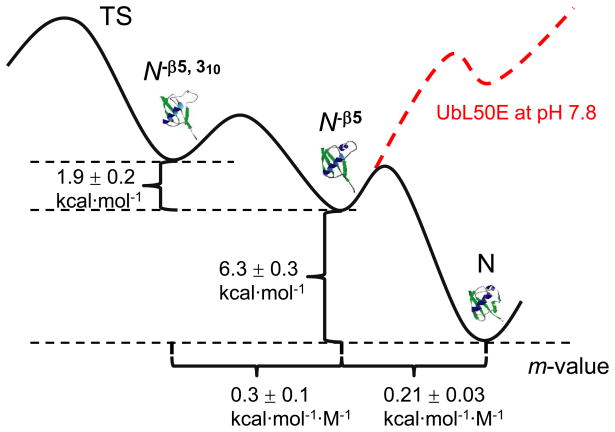
We have previously proposed a detailed folding pathway for Ub using a combination of results including HX1 and ψ-analysis, our bihistidine metal binding method which identifies inter-residue contacts in the transition state ensemble (TSE).25; 26; 27 Each major pathway step both on the way to and from the TSE is the addition or consolidation of regions of secondary structure. These events occur with a commensurate level of hydrogen bond formation and surface burial, as indicated by our kinetic isotope effect studies.28; 29 The only secondary structures which form after the TS are the small 310 helix and the β5 strand (Fig. 1). This identification suggests the presence of one or two post-TS, kinetically silent intermediates lacking one or both of these structural elements. Such intermediates are likely to be unfolding intermediates whose thermodynamics may be characterized using HX. An earlier HX study 1 indicated that hydrogen bonds between β4–β5 generally are ~ 1 kcal mol−1 less stable than bonds in the 310 helix. This difference suggests that the β5 strand unfolds prior to the helix on the unfolding pathway (N → N−β5 → N−β5,310 → TS, Fig. 1).
Figure 1. Post-TS event in Ub folding.
The present study indicates that the folding of the 310 helix and β5 strand preferentially occur along the upper pathway. Mutations employed in the present study strategy are highlighted (V5, yellow; L43, blue; L50, magenta; L56, orange; L67, green), except I30E on the interior face of the helix. The three dimensional models of the folding intermediates are created by changing the φ,ψ angles of the disrupted regions while leaving the remainder of the protein unchanged.
In order to investigate the sequential order of unfolding events, we set out to trap one or both these intermediates. The buried leucine on β5 is replaced with a glutamic acid (UbL50E). The energetic penalty for burying the charged Glu−, or more precisely, the energy required to shift acid’s pKa so that it remains neutral at a pH above its intrinsic pKa,24 is anticipated to unfold the β strand, thereby populating the N−β5 conformation. This unfolding event would be analogous to the disruption of the LOV2 protein’s signaling helix upon a I→E substitution 23, and the partial unfolding of a helix in SNase;30 but possibly different than other results with this protein as nearly all substitutions are well tolerated.24 For UbL50E at pH below glutamic acid’s intrinsic pKa, the nearly isosteric Gluo may adequately mimic the Leu allowing the β strand to refold, and generate the native-like N* conformation. If successful, this strategy provides a system whereby pH can be used to reversibly convert Ub from a native-like state to a late folding intermediate, which can be characterized using equilibrium methods.
Thermodynamic and kinetic properties
Structural, kinetic and thermodynamic measurements are performed on UbL50E to investigate the ability of the charge burial strategy to trap a partially unfolded state. A pH titration of UbL50E at 4° C, monitoring near-UV circular dichroism (CD) indicates the presence of a minor unfolding transition with a midpoint near pH 6 (Fig. 2a, a similar transition is observed by NMR following the chemical shift of the E50 amide proton, see below). At pH 4, the protein has approximately the wild-type [θ]230 nm value while the high pH species has lost structure (Δ[θ] 230 nm = 2,700 deg cm2 dmol−1). The observed pH transition midpoint is 1.7 units above the intrinsic pKaGlu because β5’s folding energy helps stabilize the folding transition by 2 kcal mol−1. This amount is 4 kcal mol−1 less than the free energy for opening β5 in the F45W pseudo-wildtype Ub (pWTUb, see HX results below), presumably because the Gluo does not fully mimic the Leu30 in the low pH, N* species.
Figure 2. Equilibrium and kinetic folding behavior of UbL50E.
a) Upon acidification, the protein shifts from an intermediate to a native-like state. b) At both high and low pH, the chevron plot of UbL50E and pWTUb have nearly overlapping folding arms but distinct unfolding arms. At high pH, the UbL50E unfolding arm is shallower, suggesting disruption of β5 strand in the intermediate state. Illustrative folding energy landscapes are shown at right. Chevron values for the pWTUb have been shifted by the amount indicated by the arrows to account for the addition of 2 or 2.3 M guanidine hydrochloride required to unfold the pWTUb.
To further characterize the low and high pH conformations of UbL50E, their folding rates are measured as a function of denaturant concentration using chevron analysis and stopped-flow methods following the F45W fluorescence (Fig. 2). At pH 7.8, the folding rates of UbL50E and pWTUb are statistically identical (ΔΔG‡ = −0.25 ± 0.32 kcal mol−1) in spite of the considerable destabilization imparted by the substitution of the acidic residue (ΔΔGeqL50E ~ 6.4 ± 0.4 kcal mol−1). At pH 4, the folding rate of UbL50E is mildly slower than for pWTUB. In energetic terms, however, the difference (ΔΔG‡ = 1.1 ± 0.2 kcal mol−1) remains much smaller than the destabilization (ΔΔGeqL50E = 6.8 ± 0.3 kcal mol−1). The resulting mutational φL50E values are 0.16 ± 0.03 and 0.04 ± 0.05 at pH 4 and 7.8, respectively. These low values are consistent with our identification of β5 as a structure formed after the TS along the folding pathways of pWTUB and UbL50E.
At both low and high pH, the folding rates of the two proteins have the same urea dependence (mf-value, Table 1). This correspondence indicates a comparable amount of urea sensitive surface area is buried in their TSs (relative to their denatured states). This result along with the similarity of their folding rates and the lack of β5 in their TS suggests that the two proteins have very similar TS structures at both pH conditions.
Table 1.
Kinetic parameters obtained from chevron analysis1
| Protein | Solvent condition | mf (kcal mol−1 M−1) | mu (kcal mol−1 M−1) | ΔGf‡(0) (kcal mol−1) | ΔGo(kcal mol−1) |
|---|---|---|---|---|---|
| pWTUb (Fig. 2) | N-state pH 7.8 | 0.60 ± 0.04 | 0.35 ± 0.02 | 3.97 ± 0.24 | −8.64 ± 0.35 |
| pWTUb (Fig. 2) | N-state pH 4.0 | 0.64 ± 0.03 | 0.36 ± 0.02 | 4.62 ± 0.21 | −9.18 ± 0.28 |
| L50E (Fig. 2) | I-state pH 7.8 | 1.08 ± 0.15 | 0.14 ± 0.02 | 4.22 ± 0.21 | −2.29 ± 0.17 |
| L50E (Fig. 2) | N*-state pH 4.0 | 0.97 ± 0.05 | 0.32 ± 0.01 | 3.47 ± 0.06 | −2.43 ± 0.04 |
| L50E (Suppl. Fig. 3, HX condition) | I-state pDread 7.5 | 0.71 ± 0.02 | 0.18 ± 0.01 | 4.18 ± 0.05 | −2.98 ± 0.07 |
| L67E (Fig. 8) | I-state pH 7.8 | 0.63 ± 0.02 | 0.21 ± 0.01 | 4.31 ± 0.04 | −4.00 ± 0.07 |
| L67E (Fig. 8) | N*-state pH 4.0 | 0.68 ± 0.02 | 0.28 ± 0.01 | 4.10 ± 0.08 | −4.39 ± 0.07 |
Although the folding rate of UbL50E is largely unchanged, its unfolding rate at both pH 4 and 7.8 is much faster than the unfolding rate of pWTUb. This acceleration is due to the stabilizing effect of burying the acidic group in UbL50E. In addition for unfolding at higher pH, the slope of the chevron unfolding arm (mu-value) for UbL50E is shallower by ~15% of the equilibrium mo-value, as compared to pWTUb. This decrease in slope for UbL50E indicates that the starting state of its unfolding reaction has lost some structure, presumably the destabilized β5 strand. This result is suggestive that high pH conformation of UbL50E is a folding intermediate on the native side of the major folding barrier.
NMR analysis
To characterize the low and high pH forms of UbL50E in detail, a series of 1H-15N heteronuclear single quantum coherence (HSQC) spectra are acquired from pH 4 to 7.8 (Figs. 3,4). The spectra of the high and low pH states contain well dispersed, sharp peaks indicating that both states contain well structured regions. At pH 4 (Fig. 3) and slightly higher pH (Fig. 4), most of the amide resonances overlap or remain close to the pWTUb’s resonances, suggesting that the UbL50E with Glu0 retains a very native-like structure.
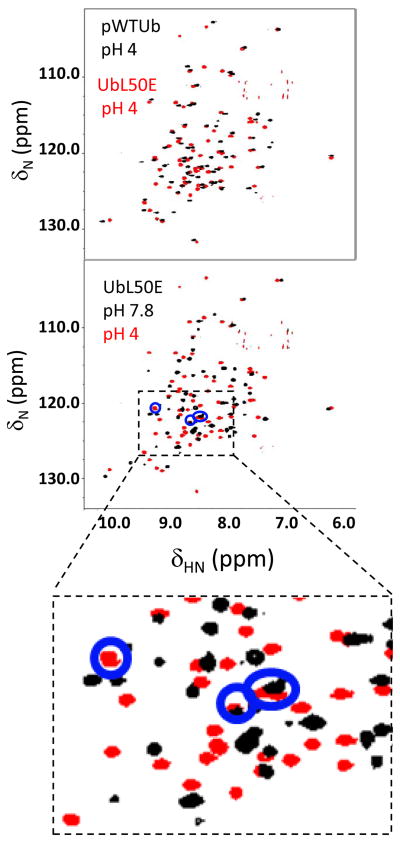
Figure 3. NMR analysis.
Overlay of 1H-15N HSQC spectra of pWTUb and UbL50E at pH 4 (upper panel). The extensive peak overlap indicates that the two proteins have similar conformations. At pH 7.8, UbL50E has sharp and well-dispersed peaks, although with fewer resonances than at pH 4 (lower panel). The four peaks (blue circles) observable at pH 4 but not pH 5, are attributed to the four unassigned residues on β5.
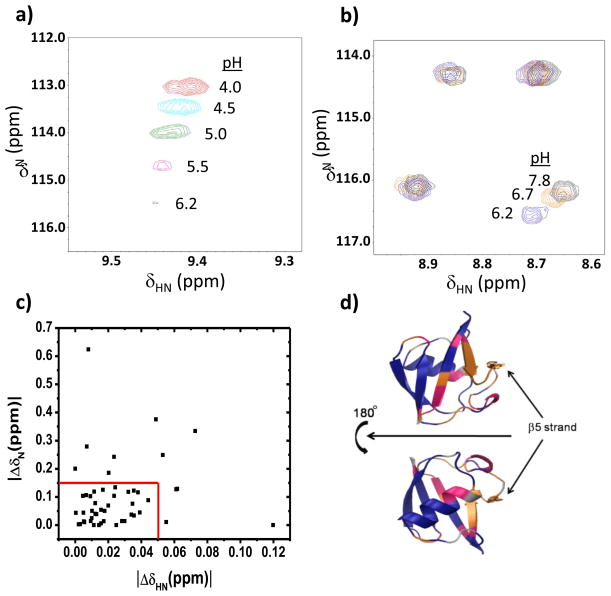
Figure 4. pH titration of UbL50E from pH 4 to pH 7.8.
a) Representative resonance which shifts and then disappears due to exchange broadening. b) In the pH 6.2 to 7.8 range, some resonances move while others do not (right panel). c) Resonances classified as non-moving if their chemical shift remained unchanged from pH 6.2 to 7.8 (red box). d) The three classes of behavior of UbL50E peaks are mapped onto the structure: unchanged (blue), changed (magenta), and chemical shift changed with peak disappearing at neutral pH due to exchange broadening (orange).
Resonances are assigned using triple resonance HNCACB and CBCACONH measurements in conjunction with the pWTUb assignments (Massi, F., unpublished). To obtain a concentrated protein sample for the 3D measurements, we used a solubility enhancing buffer (22) (15 mM sodium phosphate, 225 mM sodium chloride, 50 mM Glutamic acid, 50 mM Arginine). The enhancement is best near neutral pH, but the NMR spectra gradually deteriorate from pH 5 to pH 7.8. Accordingly, assignments are conducted at pH 5.3 where all NH resonances are assignable except for residues 48-51 on the β5 strand. These four resonances are not observed presumably due to exchange broadening (pH 5.3 is in the middle of the pH-induced folding transition). At pH 4, four additional peaks appear in the region appropriate for amide NH resonances. Because the other amide resonances are accounted for, we associate the new resonances with the four missing β5 resonances (Fig. 3). The assignment of peaks at other solvent conditions is accomplished by tracking peak movement during the pH titration (Fig. 4a,b).
In the pH titration, all the NH peaks shift and some start to disappear near pH 6. The shifts reflect both the folding transition as well as the less interesting ionization of the surface exposed Glu and Asp groups. Fortunately, their ionization equilibria are ~2 pH units below the midpoint of the CD-monitored folding transition. Consequently, the surface Glu and Asp side chains are nearly fully ionized by pH 6 and hence, the remaining chemical shift movement at higher pH’s reflects the partial unfolding transition.
Across the pH 6.2 – 7.8 range, 48, 28 and 24% of the observable NH resonances either remain unchanged (|ΔδHN| < 0.05 ppm and |ΔδN| < 0.15), shift, or shift and then disappear, respectively (Fig. 4). These three classes are interpreted as minimal or significant backbone conformational change, which also may be accompanied by extreme exchange broadening due to millisecond time scale dynamics. The residues composing each class map onto the native structure of Ub’s according to their distance from the β5 strand (Fig. 4d) and ψ-values 25. The resonances for the β1–β2 hairpin and carboxy terminus of the α-helix, the most distal regions, are the most invariant while the resonances β4, 310 helix, located adjacent to β5, are the most perturbed. The exchanged broadened peaks do not reappear as sharp peaks with random coil chemical shifts. Hence, the β5 strand is not behaving as a completely unfolded loop. Presumably, the loop interacts dynamically with the rest of the protein.
Hydrogen exchange
HX measurements are conducted on the intermediate form of UbL50E to characterize its structure, thermodynamics, and potentially identify other partially unfolded states. HX probes thermally activated fluctuations that break backbone hydrogen bonds. Once the bond is broken, HX between amide and solvent protons occurs according to:31; 32
| (1) |
where kopen and kclose are the opening and closing rates of the fluctuation, and kint is the intrinsic HX rate, which depends upon the amino acid sequence and solvent conditions.33; 34 For a stable protein in the EX2 limit where kclose≫kint, the protein establishes an equilibrium between the ground and excited states. The observed HX rate reduces to the fraction of time the protein is in the excited state multiplied by the intrinsic rate of exchange, kHX = kint/(1+kclose/kopen) = kint/(1+1/Keq). The free energy of the excited state is calculated according to ΔGHX = − RT ln (Keq) = − RT ln (kHX/(kint−kHX)), where RT is the product of the gas constant and temperature.
Under our experimental conditions (pDread 7.5, 4° C), intrinsic exchange rates are from seconds to minutes, which is faster than standard HSQC acquisition times. Accordingly, a pulse-labeling strategy is employed to measure HX rates. HX is initiated with a 1:4 dilution with D2O to pDread 7.5. After a specified time, H-to-D exchange is quenched by reducing the pDread to 2.9 with DCl. In addition to quenching HX, acidification also generates the N* state where the exchange-broadened resonances in the intermediate state become sharp and assignable. Hence, the HX rates for otherwise NMR invisible residues in the intermediate can be determined using this protocol. HX likely occurs via the EX2 mechanism as kint is much slower than global refolding rates. Hence, the stability of the hydrogen bonds can be determined for ~ 80% of the intra-molecular hydrogen bonds observable in the crystal structure.
In the intermediate, significant HX protection is observed for all of the native secondary structures except for the single measured hydrogen bond between the β4 and β5 strands (NHW45- - O=CK48). For this bond, kHX/kint = 0.3 ± 0.3 s−1 / 0.3 s−1. Hence the protection fact essentially is unity. The four unassigned peak associated with β5 also have kHX/kint ~ 1 (observed HX is complete within 3 seconds, kint ~ 0.5 – 3 s−1). Hence, the hydrogen bonds between β4–β5 are broken most of the time. For the remaining hydrogen bonds, ΔGHX is between 1.7 and 3.5 kcal mol−1 (Fig. 5, Table 2, Supplementary Fig. 2).
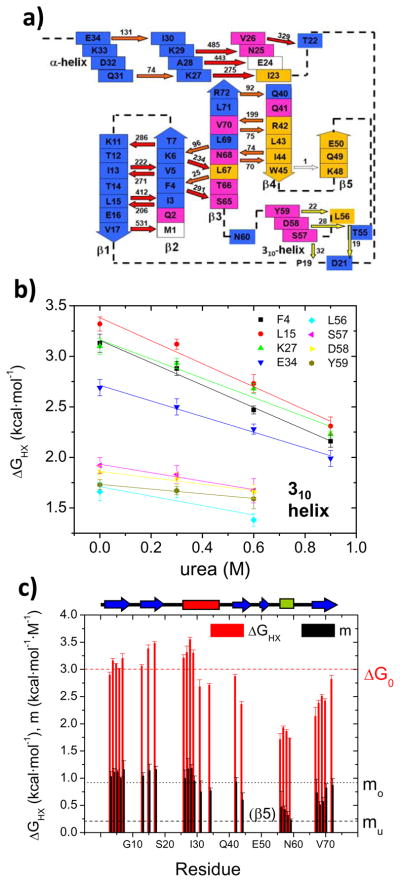
Figure 5. Native state hydrogen exchange on UbL50E I state at pDread 7.5.
a) Hydrogen bond network with direction of bond (NH→ O=C) and Keq noted. Arrows are color-coded according to their stability (red = global, yellow= 310 opening, orange = other, white = absent) while boxes are color-coded according to the type of chemical shift movement during the pH titration (adapted from Fig. 4). b) Representative NSHX denaturant dependent isotherms. Isotherms for protons on the 310 helix have lower DGHX and m values compared to the rest of the isotherms. c) Histogram of ΔGHX and m-values. The positions of the strands (blue), α- (red) and 310 helices (green) are noted at the top.
Table 2.
Native State Hydrogen Exchange on UbL50E intermediate, pDread 7.51
| 2nd structure of H-bond donor | H-bond donor (-NH) | H-bond acceptor (-C=O) | ΔGHX (kcal mol−1) | m (kcal mol−1 M−1) |
|---|---|---|---|---|
| β2 strand | I3 | L15 | 2.90±0.05 | 1.03±0.09 |
| F4 | S65 | 3.16±0.04 | 1.11±0.07 | |
| V5 | I13 | 3.10±0.01 | 1.12±0.03 | |
| K6 | L67 | 3.01±0.01 | 1.00±0.02 | |
| T7 | K11 | 3.20±0.09 | 1.16±0.16 | |
| β1 strand | I13 | V5 | 3.06±0.03 | 1.04±0.06 |
| L15 | I3 | 3.38±0.07 | 1.16±0.06 | |
| V17 | M1 | 3.48±0.03 | 1.16±0.06 | |
| α helix | V26 | T22 | 3.21±0.06 | 1.00±0.15 |
| K27 | I23 | 3.31±0.12 | 1.17±0.19 | |
| A28 | E24 | 3.55±0.04 | 1.18±0.07 | |
| K29 | N25 | 3.30±0.06 | 0.95±0.09 | |
| Q31 | K27 | 2.68±0.13 | 0.75±0.20 | |
| E34 | I30 | 2.71±0.03 | 0.77±0.05 | |
| β4 strand | R42 | V70 | 2.87±0.04 | 0.93±0.08 |
| I44 | N68 | 2.36±0.05 | 0.60±0.13 | |
| W45 | K48 | ~01 | ND2 | |
| 310 helix | L56 | D21 | 1.71±0.11 | 0.47±0.29 |
| S57 | P19 | 1.93±0.03 | 0.42±0.07 | |
| D58 | T55 | 1.86±0.03 | 0.32±0.07 | |
| Y59 | L56 | 1.73±0.01 | 0.23±0.02 | |
| β3 strand | L67 | F4 | 2.14±0.16 | 0.73±0.25 |
| N68 | I44 | 2.38±0.05 | 0.51±0.06 | |
| L69 | K6 | 2.51±0.03 | 0.57±0.08 | |
| V70 | R42 | 2.42±0.04 | 0.82±0.08 | |
| R72 | Q40 | 2.82±0.07 | 0.87±0.12 |
kHX/kint~ 1
Not determined.
Small-angle X-ray scattering
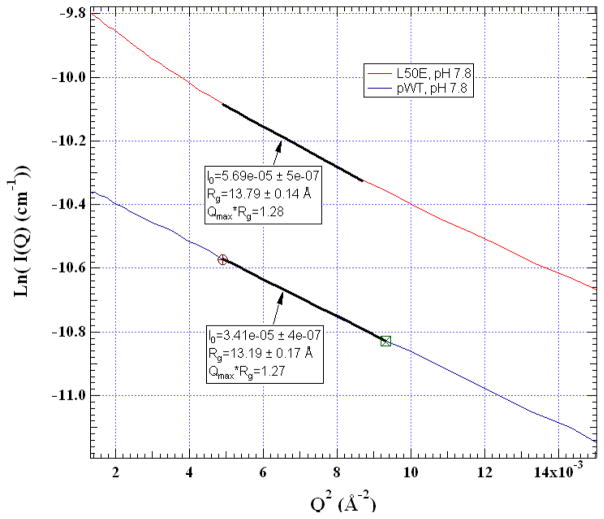
We compare the global dimensions of pWTUb to the UbL50E intermediate using SAXS (Fig. 6). At pH 7.8, the measured radius of gyration (Rg) is 13.2 ± 0.2 and 13.8 ± 0.1 Å for the two proteins, respectively. This difference matches the 0.5 Å difference between Rg’s of the native state and the model for the N−β5 intermediate but is less than the 1.6 Å difference for the N−β5,310 intermediate (Fig. 1).
Figure 6. Dimensions of pWt and L50E intermediate measured using SAXS.
Rg values are obtained from a Guinier analysis performed over the data range highlighted. Protein concentration is ~2 mg/ml. Solvent condition was the same as used for NMR assignments except at pH 7.8, 0% D2O.
In summary, a combination of results – the loss of secondary structure (CD, pH titration) which is formed on the native-side of the TS (shallower unfolding chevron arm), the dynamic behavior of the β5 strand relative to other regions (HSQC assignments, line broadening), the loss of hydrogen bonds only between β4–β5 (HX), and the 0.5 Å increase in Rg (SAXS) – demonstrate that the L50E mutant selectively disrupts the β5 strand at neutral pH thereby trapping the protein in the N−β5 conformation.
Denaturant dependence of HX
Next we investigated the denaturant dependence of HX on the N−β5 state of UbL50E to identify the nature of the openings leading to exchange for each amide proton (Fig. 5, Table 2, Supplementary Fig. 2). From the linear decrease in stability as a function of added urea, ΔGHX([denaturant]) = ΔGHX(0) + m[denaturant], the surface exposure of the structural fluctuation leading to exchange is determined. Nearly half of the amide protons exchange with the same ΔGHX and m-value as measured for global unfolding, 3 kcal mol−1 and 1 kcal mol−1 M−1, respectively, determined by equilibrium denaturation and kinetic measurements (Table 1, Supplementary Fig. 3). These amide protons are exchanging from the globally unfolded state, and are located in the β1–β2 hairpin and the N-terminal half of the α-helix as well as two long-range bonds between strands β2–β3 (NHK6- -O=CL67 and NHF4- - O=CS65).
The hydrogen bonds on the 310-helix stand out with their uniformly low stability (ΔGHX =1.8–2.0 kcal mol−1) and mild response to denaturant (m = 0.25–0.50 kcal mol−1 M−1). Their uniformity indicates that the small helix is a second foldon that unfolds in a concerted event, after the loss of the β5 strand. The similarity of the m-value for the 310-helix and the mu-value from the chevron analysis indicates that the amount of surface burial exposed in the unfolding of the foldon is nearly the same as that exposed in the TSE.
The hydrogen bonds between β3 – β4 and in the carboxy-terminal portion of the α helix have slightly lower ΔGHX and m-values compared to the globally exchanging hydrogen bonds. Although this difference is near our experimental error, it is observed uniformly across these regions. Potentially, the mild decrease is due to a slight amount of mixing between a global exchange process and a smaller scale event. Alternatively, the unfolded state may contain some residual structure in the β1–β3 strands and α helix, which is marginally stable only under aqueous conditions.
A free energy surface can be constructed for pWTUb and the UbL50E using the kinetic, NMR and NSHX data (Fig. 7). Structurally, the unfolding pathway goes through two PUFs, N ↔ N−β5 ↔ N−β5,310 ↔ TS. At pH 7.8, ΔGN is 0.9 kcal mol−1 less stable than Δ GN− (β 5+ 310) according to calculations based on apparent pKa from pH-induced folding titration monitored by CD (Fig. 2). Hence, the ladder of free energies for UbL50E at pH 4 and pWTUb at both pH 4 and 7.8 is ΔGN < Δ GN− β 5 < Δ GN− (β 5+ 310) whereas for UbL50E at pH 7.8, Δ GN− β 5 < Δ GN− (β 5+ 310) < ΔGN.
Figure 7. Post-TS free energy surface.
Values for the N−β5 are derived from kinetic data for UbL50E and pWTUb while values for the N−310, −β5 are obtained from NSHX studies on UbL50E intermediate. The energy of the UbL50E at pH 7.8 with the β5 strand folded is obtained from the apparent pKa of the CD-monitored pH-induced unfolding transition (pH 5.8, see Fig. 2).
Other charged substitutions
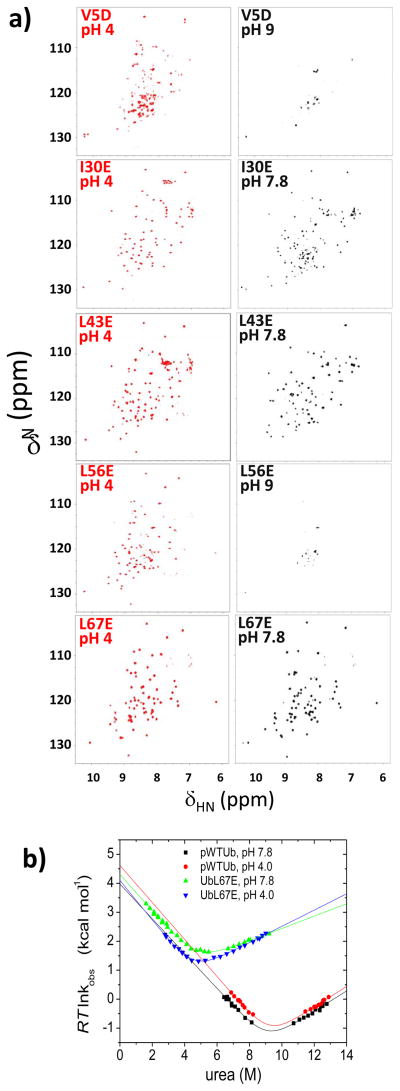
We individually introduced five other buried aliphatic-to-charge mutations into β1, the α-helix, β4, 310 helix and β3 (V5D, I30E, L43E, L56E, and L67E, Figs. 1, 8). At neutral pH, the L56E substitution on the 310 helix is anticipated by folding hierarchy to unfold both this helix and the β5 strand, thereby populating the N−β5,310 state. This species is unstable and prone to precipitate with a virtually blank HSQC spectrum presumably due to extreme line broadening. Nevertheless, this species refolds at pH 4 with an NMR spectrum having similar dispersion as the native protein. The potential loss of the 310 helix could be tested with HX labeling at neutral pH followed by NMR analysis at low pH, as done for UbL50E. Irrespective of the exact structural content, the NMR spectrum of the L56E species lacks sharp and dispersive peaks at neutral pH suggesting that the protein becomes more dynamic on the unfolding pathway back towards the TS.
Figure 8. Charge-burial partial unfolding strategy applied at other positions.
a) NMR 1H-15N HSQC spectra. b) Corresponding chevron plots of UbL67E at low and high pH compared to pWTUb.
The L67E and L43E substitutions are located on β3 and β4, respectively. These two strands are present in the TS according to ψ-analysis. Both mutants have well dispersed HSQC spectra at both pH 4 and 8. The L67E species has a native number of peaks while L43E has slightly less at pH 8. The kinetic folding properties of L67E mutant are similar to those of L50E, in particular possessing a shallower chevron unfolding arm slope at pH 8 compared to pWTUb (Fig. 8b, Table 1). However the ~10 % reduction of mu-value for L67E is insufficient to account for disruption of the entire β3 strand, which makes multiple interactions with β2 and β4 in the TS. We infer that the L67E mutation induces some local structural fraying of β3 at high pH that exposes some buried surface area, rather than the loss of the entire strand.
The V5D and I30E mutations are located on the amino-terminal hairpin and the α helix, respectively. These two secondary structures form prior to the TS in the folding pathway and the two substituted positions are located in the core of the protein. The HSQC spectrum of V5D exhibits extensive peak broadening at pH 8.5 whereas I30E’s spectrum remains dispersive with extra sets of peaks. At pH 4, the V5D spectrum has less dispersion than I30E, which is similar to pWTUb. The V5D substitution is disruptive, indicating that charge burial can identify regions of the protein that cannot be unfolded without disrupting the rest of the protein. Such regions are likely to form early in the folding pathway.
Discussion
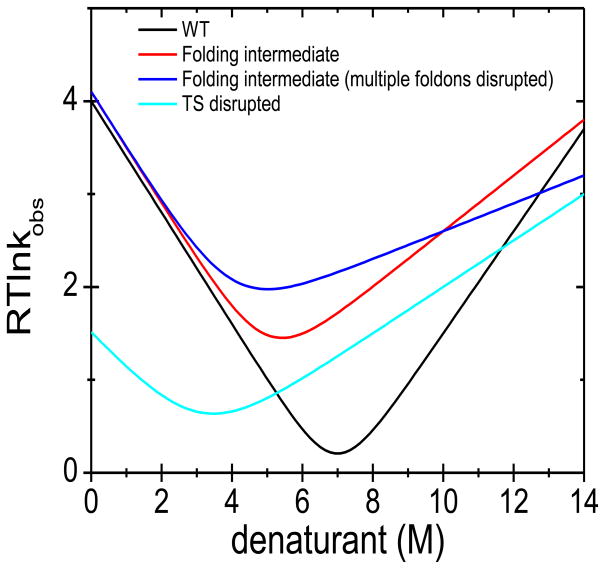
Rather than relying on serendipitously found mutations, our charge burial strategy is a rational, active and generally applicable approach for reversibly trapping partially unfolded states and delineating folding pathways. For a protein where little is known about its folding behavior, the charge burial strategy can be extremely revealing. Leu/Ile → Glu or Val → Asp substitutions can be introduced into each potential foldon. The acquisition of NMR spectra under acidic and neutral conditions can identify which parts of the protein can be selectively disrupted without unfolding the entire protein. Once a potential folding intermediate is found through such substitutions, it can be further interrogated with HX, chevron analysis (Fig. 9), and possibly structure determination. The intermediate also can be used as the new ground state to perform additional NSHX and RD measurements to “walk” up the energy surface, identify other folding intermediates and test tentative conclusions from the preliminary scan using charged residues.
Figure 9. Interpretation of possible chevrons generated by the charge-burial strategy.
For an aliphatic→ charged substitution, the chevron can have a similar folding but altered unfolding arm at neutral pH. The change in slope can be used to estimate the amount of structure lost due to the substitution. If the decrease of slope is consistent with the estimate (red), it is suggestive of the creation of a folding intermediate lacking structure near the site of the substitution that otherwise is formed after the TS in the folding of the WT protein. If decrease of slope is smaller than estimation (not shown), it may be indicative of a locally frayed conformation rather than a genuine folding intermediate. Alternatively, the decrease may be consistent with the loss of an additional foldon (blue) if folding is hierarchical, being dependent on the perturbed foldon. This possibility can be tested by further characterization, e.g. with NSHX. Changes in the folding arm indicate that the TS is destabilized by the substitution. When the slope of the folding is decreased, some structure has been lost in both the TS and the ground state (cyan). More complicated scenarios also are possible.
By introducing the L50E substitution into Ub, we trapped the N−β5 species, a late folding intermediate and characterized its structure, thermodynamics and dynamics using CD, NMR, SAXS and chevron analysis. This trapped species have very native-like regions on the opposite side of the protein (no shift in the sharp 15N-1H resonances). The disrupted β5 strand and neighboring regions, however, undergo extreme exchange broadening due to millisecond time scale dynamics although the backbone hydrogen bonds remain in the 310 helix and between strands β3–β4.
We performed NSHX measurements on the intermediate taking advantage of the well-behaved NMR spectrum of the low pH, native-like species. These measurements identified an additional unfolding intermediate lacking the 310 helix. As far as we are aware, this is the first report of a folding intermediate identified through NSHX applied to another intermediate.
Even for an equilibrium study that identifies an extensive ladder of unfolding intermediates, the kinetic connectivity can only be inferred. In the case of Ub, we have additional kinetic information. The only secondary structures that form between the TS native state are the β5 strand and the 310 helix.25 Hence, the unfolding of either one of these two elements is highly likely to be first step along the unfolding pathway. Our work demonstrates that a species lacking only the β5 strand lies at a lower free energy than the state lacking the 310 helix and that the β5 strand can be individually disrupted. Furthermore, the stability of the 310 helix responds to the L50E substitution to the same degree as does the β5 strand, thereby demonstrating that a structural hierarchy exists between these two foldons. We also found that the unfolding rate starting from the N−β5 species is faster than from N yet the pathways go through the same TS, and the trapped species is in between the native state and TS when considering urea sensitive surface burial (m-value). All these results point to the early stages of the kinetic unfolding pathway going through two intermediates, N → N −β5 → N−β5,310 → TS.
When NSHX was performed with wild-type Ub, neither of these two intermediates were readily detectable.35 Similarly, RD methods on wild-type protein would not have detected these intermediates because their populations are too low (< 0.002%). However, RD measurements could be applied to the trapped L50E intermediate. Such measurements should detect the N−β5,310 species as it now comprises 3% of the total population. This potential application illustrates the possibilities of combining the charge burial strategy with other methods to enable structural and thermodynamic characterization of folding intermediates.
Other trapping strategies and dynamics in intermediate states
Other protein engineering strategies have been used to trap intermediates. However, these methods often lack the degree of control possible with the charge burial strategy and they typically require advanced knowledge of the structural content of the intermediate. For example, Bai and coworkers used HX in conjunction with truncation and glycine substitutions to create analogs of intermediates of T. thermophilus RNase H36 and T4 lysozyme.20 They conducted NSHX on the T4 lysozyme analog, but found no additional intermediates.20 Based on HX results on E. coli RNase H, Marqusee and coworkers generated a stable mini-core,22 and a fractionally populated intermediate using a single I25A substitution.8 Other late intermediates have been created using glycine substitutions in cyt-b56221 and IM9.6
The NMR characterization of the IM9 intermediate indicated that it has a fluid core with fluctuations on the NMR timescale due to the repacking of the remaining three helices. This repacking is similar to that of the remaining helices in the cyt-b562 intermediate.21 However, this intermediate and the T. thermophilus RNase H intermediate36 remain very native-like according to NMR line widths, as do the regions distal to the perturbation in UbL50E intermediate. In contrast, the E. coli I25A intermediate has more dynamic behavior.8 We suggest that an I25D charged substitution would fully populate the E. coli intermediate and help address the apparent contradiction between the Bai and the Marqusee studies of RNase H. This example further highlights the utility of the charge burial strategy.
Conclusion
The buried aliphatic→ charged substitution strategy is a general method for trapping and characterizing folding intermediates under equilibrium conditions. In the case of Ub, the intermediate’s stability relative to a native-like species is controllable, with acidification generating a well-dispersed NMR spectra under conditions where HX is conveniently quenched. As a result, NSHX measurements can be performed on intermediates with poor NMR spectra. The strategy is particularly powerful in combination with other methods such as NSHX and RD. Although we applied the strategy with foreknowledge of Ub’s folding behavior, we envision that even without this foreknowledge, a series of charged substitutions throughout a protein can help identify foldons and their relationship to folding pathways in other proteins.
Materials and methods
Ub L50E and other mutants, based on a pseudo WT version of Ubiquitin (pWTUb, with F45W and H68N) was prepared following ref. 25. Protein in inclusion bodies were solubilized in 8M urea buffer before HPLC purification.
Equilibrium and kinetic measurements
CD experiments were performed with a Jasco 715 spectropolarimeter with a pathlength of 1 cm in 15 mM sodium phosphate, 225 sodium chloride. Rapid mixing fluorescence experiments used a Biologic SFM-400 stopped-flow apparatus connected via a fiber optic cable to a PTI A101 arc lamp. Fluorescence spectroscopy used excitation and emission wavelengths of 280–290 nm and 300–400 nm, respectively.
NMR spectroscopy and HX chemistry
All the NMR experiments were run on a Varian Unity Inova 600 MHz spectrometer equipped with cryo-probe. To obtain resonance assignments, HNCACB and CBCACONH experiments were run on 15N, 13C-labeled UbL50E at 0.5 mM concentration in 15 mM sodium phosphate, 225 mM sodium chloride, 50 mM Glu and 50 mM Arg, pH 5.3, 5% D2O. The 2D 15N HSQC spectra were taken with 15N-labeled UbL50E at 0.1 mM protein concentration in the same buffer with an added 10 mM Glu and Arg. To initiate HX, the protein at 0.5 mM, pH 6.6 was diluted 1:4 to a final condition of 15 mM sodium phosphate, 225 mM sodium chloride, 10 mM Glu, 10 mM Arg, 80% D2O, pDread 7.5. Exchange was quenched by the addition of 3.5 μL 6 M DCl to the 500 μL protein solution, which dropped pDread to 2.9.
In order to correct for back exchange during the NMR measurement, HX rate at the quenching condition (kq) was also measured for a fully protonated sample and fit with equation:
Where To is the HSQC starting time, L is length of the HSQC measurement, VT is the NMR peak volume at To, and kq and V0 are fitting parameters. The HX results were fit with the equation:
where t is length of the labeling period at pDread = 7.5, Vt is the peak volume at time t, and kHX and V0: fitting parameters.
Small angle X-ray scattering
Data were collected at the BioCAT beamline at the Advanced Photon Source, Argonne National Laboratory. Samples were flowed through a 1.5 mm capillary at a rate of 2 μL/s and 15 exposures of ~ 1 s duration were collected at room temperature. The exposures were averaged, buffer blanks subtracted, and Guinier analysis performed using IGOR Pro.
Supplementary Material
Acknowledgments
We thank Josh Kurutz for assistance in NMR measurements, L. Guo with the SAXS experiments, and B. Garcia-Moreno, N. Kallenbach, S.W. Englander, Y. Bai, A. Robertson, F. Massi and members of the Sosnick laboratory for helpful discussions and comments on the manuscript. This work was supported by grants from the NIH. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract No. W-31-109-ENG-38. BioCAT is a National Institutes of Health-supported Research Center RR-08630. The content is solely the responsibility of the authors and does not necessarily reflect the official views of the National Center for Research Resources or the National Institutes of Health.
ABBRIEVIATIONS
- biHis
bihistidine
- CD
circular dichroism
- GdmCl
guanidinium chloride
- HSQC
heteronuclear single quantum coherence
- pWTUb
pseudo-wildtype ubiquitin
- Rg
radius of gyration
- SAXS
small angle X-ray scattering
- TSE
transition state ensemble
- Ub
mammalian ubiquitin
- UbL50E
Leu50Glu version of pseudowild-type Ub
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pan Y, Briggs MS. Hydrogen exchange in native and alcohol forms of ubiquitin. Biochemistry. 1992;31:11405–12. doi: 10.1021/bi00161a019. [DOI] [PubMed] [Google Scholar]
- 2.Jourdan M, Searle MS. Insights into the stability of native and partially folded states of ubiquitin: effects of cosolvents and denaturants on the thermodynamics of protein folding. Biochemistry. 2001;40:10317–25. doi: 10.1021/bi010767j. [DOI] [PubMed] [Google Scholar]
- 3.Vallee-Belisle A, Michnick SW. Multiple tryptophan probes reveal that ubiquitin folds via a late misfolded intermediate. J Mol Biol. 2007;374:791–805. doi: 10.1016/j.jmb.2007.09.018. [DOI] [PubMed] [Google Scholar]
- 4.Rea AM, Simpson ER, Crespo MD, Searle MS. Helix mutations stabilize a late productive intermediate on the folding pathway of ubiquitin. Biochemistry. 2008;47:8225–36. doi: 10.1021/bi800722d. [DOI] [PubMed] [Google Scholar]
- 5.Eliezer D, Yao J, Dyson HJ, Wright PE. Structural and dynamic characterization of partially folded states of apomyoglobin and implications for protein folding. Nature Struct Biol. 1998;5:148–55. doi: 10.1038/nsb0298-148. [DOI] [PubMed] [Google Scholar]
- 6.Whittaker SB, Spence GR, Gunter Grossmann J, Radford SE, Moore GR. NMR analysis of the conformational properties of the trapped on-pathway folding intermediate of the bacterial immunity protein Im7. J Mol Biol. 2007;366:1001–15. doi: 10.1016/j.jmb.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Korzhnev DM, Salvatella X, Vendruscolo M, Di Nardo AA, Davidson AR, Dobson CM, Kay LE. Low-populated folding intermediates of Fyn SH3 characterized by relaxation dispersion NMR. Nature. 2004;430:586–90. doi: 10.1038/nature02655. [DOI] [PubMed] [Google Scholar]
- 8.Connell KB, Horner GA, Marqusee S. A single mutation at residue 25 populates the folding intermediate of E. coli RNase H and reveals a highly dynamic partially folded ensemble. J Mol Biol. 2009;391:461–70. doi: 10.1016/j.jmb.2009.05.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neudecker P, Lundstrom P, Kay LE. Relaxation dispersion NMR spectroscopy as a tool for detailed studies of protein folding. Biophys J. 2009;96:2045–54. doi: 10.1016/j.bpj.2008.12.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bai Y, Sosnick TR, Mayne L, Englander SW. Protein folding intermediates: native-state hydrogen exchange. Science. 1995;269:192–7. doi: 10.1126/science.7618079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Englander SW, Mayne L, Bai Y, Sosnick TR. Hydrogen exchange: the modern legacy of Linderstrom-Lang. Protein Sci. 1997;6:1101–9. doi: 10.1002/pro.5560060517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silverman JA, Harbury PB. Rapid mapping of protein structure, interactions, and ligand binding by misincorporation proton-alkyl exchange. J Biol Chem. 2002;277:30968–75. doi: 10.1074/jbc.M203172200. [DOI] [PubMed] [Google Scholar]
- 13.Sridevi K, Udgaonkar JB. Unfolding rates of barstar determined in native and low denaturant conditions indicate the presence of intermediates. Biochemistry. 2002;41:1568–78. doi: 10.1021/bi011494v. [DOI] [PubMed] [Google Scholar]
- 14.Park C, Marqusee S. Probing the high energy states in proteins by proteolysis. J Mol Biol. 2004;343:1467–76. doi: 10.1016/j.jmb.2004.08.085. [DOI] [PubMed] [Google Scholar]
- 15.Wang L, Kallenbach NR. Proteolysis as a measure of the free energy difference between cytochrome c and its derivatives. Protein Sci. 1998;7:2460–4. doi: 10.1002/pro.5560071124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korzhnev DM, Kay LE. Probing invisible, low-populated States of protein molecules by relaxation dispersion NMR spectroscopy: an application to protein folding. Acc Chem Res. 2008;41:442–51. doi: 10.1021/ar700189y. [DOI] [PubMed] [Google Scholar]
- 17.Tang Y, Grey MJ, McKnight J, Palmer AG, 3rd, Raleigh DP. Multistate folding of the villin headpiece domain. J Mol Biol. 2006;355:1066–77. doi: 10.1016/j.jmb.2005.10.066. [DOI] [PubMed] [Google Scholar]
- 18.Grey MJ, Tang Y, Alexov E, McKnight CJ, Raleigh DP, Palmer AG., 3rd Characterizing a partially folded intermediate of the villin headpiece domain under non-denaturing conditions: contribution of His41 to the pH-dependent stability of the N-terminal subdomain. J Mol Biol. 2006;355:1078–94. doi: 10.1016/j.jmb.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 19.Bai Y, Feng H, Zhou Z. Population and structure determination of hidden folding intermediates by native-state hydrogen exchange-directed protein engineering and nuclear magnetic resonance. Methods Mol Biol. 2007;350:69–81. doi: 10.1385/1-59745-189-4:69. [DOI] [PubMed] [Google Scholar]
- 20.Kato H, Feng H, Bai Y. The folding pathway of T4 lysozyme: the high-resolution structure and folding of a hidden intermediate. J Mol Biol. 2007;365:870–80. doi: 10.1016/j.jmb.2006.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng H, Vu ND, Bai Y. Detection and structure determination of an equilibrium unfolding intermediate of Rd-apocytochrome b562: native fold with non-native hydrophobic interactions. J Mol Biol. 2004;343:1477–85. doi: 10.1016/j.jmb.2004.08.099. [DOI] [PubMed] [Google Scholar]
- 22.Chamberlain AK, Fischer KF, Reardon D, Handel TM, Marqusee AS. Folding of an isolated ribonuclease H core fragment. Protein Sci. 1999;8:2251–7. doi: 10.1110/ps.8.11.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harper SM, Christie JM, Gardner KH. Disruption of the LOV-Jalpha helix interaction activates phototropin kinase activity. Biochemistry. 2004;43:16184–92. doi: 10.1021/bi048092i. [DOI] [PubMed] [Google Scholar]
- 24.Isom DG, Cannon BR, Castaneda CA, Robinson A, Garcia-Moreno B. High tolerance for ionizable residues in the hydrophobic interior of proteins. Proc Natl Acad Sci U S A. 2008;105:17784–8. doi: 10.1073/pnas.0805113105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krantz BA, Dothager RS, Sosnick TR. Discerning the structure and energy of multiple transition states in protein folding using psi-analysis. J Mol Biol. 2004;337:463–75. doi: 10.1016/j.jmb.2004.01.018. [DOI] [PubMed] [Google Scholar]
- 26.Sosnick TR, Krantz BA, Dothager RS, Baxa M. Characterizing the Protein Folding Transition State Using psi Analysis. Chem Rev. 2006;106:1862–76. doi: 10.1021/cr040431q. [DOI] [PubMed] [Google Scholar]
- 27.Baxa MC, Freed KF, Sosnick TR. Psi-constrained simulations of protein folding transition states: implications for calculating. J Mol Biol. 2009;386:920–8. doi: 10.1016/j.jmb.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krantz BA, Moran LB, Kentsis A, Sosnick TR. D/H amide kinetic isotope effects reveal when hydrogen bonds form during protein folding. Nature Struct Biol. 2000;7:62–71. doi: 10.1038/71265. [DOI] [PubMed] [Google Scholar]
- 29.Krantz BA, Srivastava AK, Nauli S, Baker D, Sauer RT, Sosnick TR. Understanding protein hydrogen bond formation with kinetic H/D amide isotope effects. Nature Struct Biol. 2002;9:458–63. doi: 10.1038/nsb794. [DOI] [PubMed] [Google Scholar]
- 30.Karp DA, Gittis AG, Stahley MR, Fitch CA, Stites WE, Garcia-Moreno EB. High apparent dielectric constant inside a protein reflects structural reorganization coupled to the ionization of an internal Asp. Biophys J. 2007;92:2041–53. doi: 10.1529/biophysj.106.090266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Englander SW, Kallenbach NR. Hydrogen exchange and structural dynamics of proteins and nucleic-acids. Q Rev Biophys. 1984;16:521–655. doi: 10.1017/s0033583500005217. [DOI] [PubMed] [Google Scholar]
- 32.Englander SW, Mayne LC, Bai Y, Sosnick TR. Hydrogen exchange: the modern legacy of Linderstrom-Lang. Protein Sci. 1997;6:1101–1109. doi: 10.1002/pro.5560060517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Connelly GP, Bai Y, Jeng M-F, Mayne L, Englander SW. Isotope effects in peptide group hydrogen exchange. Proteins. 1993;17:87–92. doi: 10.1002/prot.340170111. [DOI] [PubMed] [Google Scholar]
- 35.Sidhu NS, Robertson AD. Master degree thesis. University of Iowa; Exploring the conformational manifold of ubiquitin by native state hydrogen exchange. (unpublished) [Google Scholar]
- 36.Zhou Z, Feng H, Ghirlando R, Bai Y. The high-resolution NMR structure of the early folding intermediate of the Thermus thermophilus ribonuclease H. J Mol Biol. 2008;384:531–9. doi: 10.1016/j.jmb.2008.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.