Abstract
Like other alpha-herpesviruses, Herpes Simplex Virus Type 1 (HSV-1) possesses the ability to establish latency in sensory ganglia as a non-integrated, nucleosome-associated episome in the host cell nucleus. Transcription of the genome is limited to the Latency-Associated Transcript (LAT), while the lytic genes are maintained in a transcriptionally-repressed state. This partitioning of the genome into areas of active and inactive transcription suggests epigenetic control of HSV-1 latent gene expression.
During latency viral transcription is not regulated by DNA methylation but likely by post-translational histone modifications. The LAT region is the only region of the genome enriched in marks indicative of transcriptional permissiveness, specifically dimethyl H3 K4 and acetyl H3 K9, K14, while the lytic genes appear under-enriched in those same marks. In addition, facultative heterochromatin marks, specifically trimethyl H3 K27 and the histone variant macroH2A, are enriched on lytic genes during latency. The distinct epigenetic domains of the LAT and the lytic genes appear to be separated by chromatin insulators. Binding of CTCF, a protein that binds to all known vertebrate insulators, to sites within the HSV-1 genome likely prevents heterochromatic spreading and blocks enhancer activity. When the latent viral genome undergoes stress-induced reactivation, it is possible that CTCF binding and insulator function are abrogated, enabling lytic gene transcription to ensue.
In this review we summarize our current understanding of latent HSV-1 epigenetic regulation as it pertains to infections in both the rabbit and mouse models. CTCF insulator function and regulation of histone tail modifications will be discussed. We will also present a current model of how the latent genome is carefully controlled at the epigenetic level and how stress-induced changes to it may trigger reactivation.
I. Introduction to HSV-1 biology and latency
The herpesviruses are large enveloped viruses that infect a wide spectrum of invertebrate and vertebrate species ranging from oysters to man. All herpesviruses share the defining trait of establishing a life-long latent infection within their hosts. The latent infection is characterized by a shutdown of virus replicative functions and the inability to detect infectious virus. Periodic reactivation of this latent infection allows for subsequent infection of other hosts. During latency the 120–300 kb double-stranded DNA genomes of these viruses are maintained as multiple copies of circular episomes within the nuclei of the cells in which they became latent. The division of herpesviruses into three sub-families, alpha, beta and gamma, is based largely on their sites of latency. This review will focus on the latency of the alpha-herpesviruses, which maintain a latent infection within neurons. Specifically, we will discuss the current understanding of epigenetic factors that regulate Herpes Simplex Virus (HSV) latent gene expression within neurons.
HSV establishes a life-long latent infection within sensory nerve ganglia
There are two species of HSV: HSV-1, which is associated most commonly with clinical features in the orofacial region such as cold sores or fever blisters, and HSV-2, which is the primary cause of genital herpes infections. While some features of their clinical diseases differ, both viruses establish a latent infection within sensory nerve ganglia. Since HSV-1 and HSV-2 are genetically very similar, the prototype of the group, HSV-1, will be the primary focus of this review.
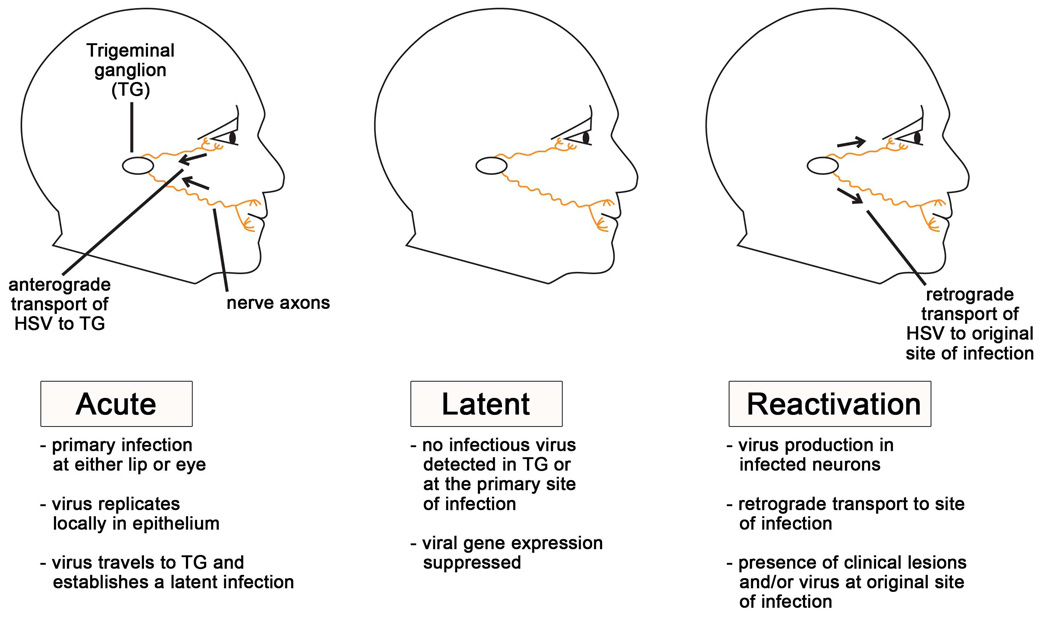
As outlined in Figure 1, HSV-1 infections are generally acquired by direct contact on the surface of the oral mucosa. The virus then replicates within the mucosal epithelial cells and enters the nerve termini of the sensory neurons which innervate the site of the primary infection. The HSV virion travels to the cell bodies of these neurons, located within the trigeminal ganglia, via fast-axonal transport. Once inside the neuron, one of two processes is initiated: either lytic replication or repression of lytic genes and establishment of latency (Figure 2). During lytic gene replication, ordered gene expression occurs, with the products of each gene class being necessary for initiation of expression of the subsequent class. The first gene class to be transcribed and translated is that of the immediate-early (IE) genes, which in addition to providing transactivation and export/transport functions, are necessary for transcription of the early genes; the early (E) genes, which are important in viral DNA replication, are prerequisite to the structural and capsid-associated late (L) genes. If the lytic genes are not expressed, the viral genome enters a nonreplicating nucleosome-associated latent state in the sensory ganglia; the virus can remain in this state until a stress stimulus causes some still unknown changes that can trigger transcription of the lytic genes, ultimately leading to reactivation. While there have been reports of HSV genomes present after resolution of acute infections in the skin or cornea, it is likely that these genomes represent residual recent reactivation events; sensory neurons are generally viewed as the principal reservoir of HSV latency [1, 2].
Figure 1. Stages of an HSV-1 infection in vivo.
Humans typically acquire HSV-1 as a primary infection of the mucosa of the lips or eye. During the initial acute infection, the virus replicates locally in the mucosal epithelium and gains access to the sensory nerve termini that underlie the skin’s surface. The virus then travels to the nerve cell bodies in the trigeminal ganglia where the virus will replicate in some cells and become latent in others. During the latent infection, the virus remains dormant in the trigeminal ganglion. During this period no infectious virus is detected, and viral gene expression is suppressed, except for the latency-associated transcript (LAT). Periodically, stress causes the virus to reactivate. During reactivation viral lytic gene transcription and DNA replication initiates in some neurons, and virions are transported back down the axons to the primary site of infection. This results in infectious virus at the site of initial infection, and in some instances, clinical lesions such as cold sores will result.
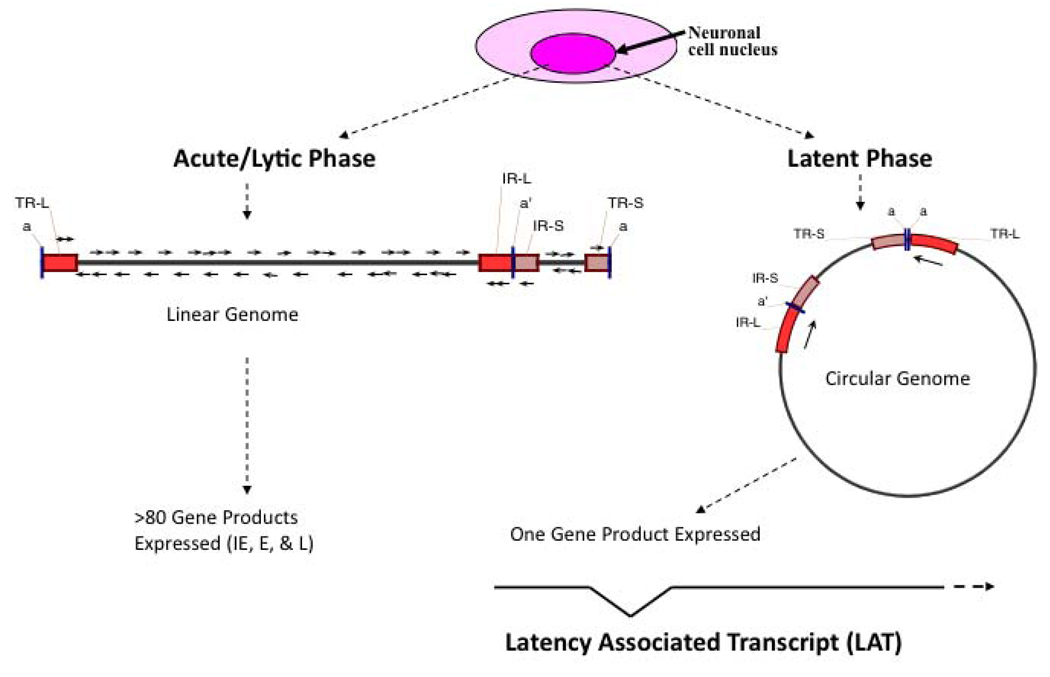
Figure 2. Molecular features of HSV-1 lytic and latent infections.
Once HSV-1 enters a sensory neuron, it can either initiate lytic replication or establish a latent infection. The acute or lytic phase is characterized by expression of all three kinetic gene classes (immediate early, IE; early, E; or late, L) and replication of the viral genome through a linear mechanism. In contrast, the latent infection is characterized by circularization of the viral genome and suppression of the lytic genes. Only one gene product, the non-coding LAT, is abundantly transcribed during latency. This figure also illustrates the four different regions of the HSV-1 genome: the unique long (UL) region, the unique short (US) region, and the two sets of repeats, the repeat long (RL) and the repeat short (RS). Each of the repeats is present in two copies, differentiated as either the terminal (T) or internal (I) segment.
In addition to the restriction of latency to neurons, evidence suggests that HSV-1 latency may be restricted to defined sub-types of sensory neurons as well. Of the 5–20 % of sensory neurons in a ganglion that harbor latent genomes, the majority of these neurons express a specific marker recognized by antibody A5 [3]. While the molecular basis and biological significance of this restricted tropism for latency is not known, it is likely that neuronal-specific proteins interact with elements within the HSV genome to direct this process. This is currently an area of vigorous study and a recent study suggests that elements within the LAT region may play a central role [4, 5].
For reasons that are not understood, a true latent state cannot be established in vitro. While a quiescent infection can be established using inhibitors of HSV replication in human foreskin fibroblasts [6], rat pheochromocytoma (PC12) cells [7, 8], or primary dorsal root ganglia neurons [6, 9, 10], these cultures do not completely parallel the latent state that exists in vivo. While they are not naturally infected by HSV, the primary models used to study HSV latency are the mouse and the rabbit, which have provided the bulk of the field’s current understanding of latency and reactivation.
The mouse represents the most commonly used model in which to study HSV latency. Following inoculation of the virus on hind footpads, flank, eyes, ear pinnae or snout, the virus replicates locally in the epithelium and travels to the sensory nerve ganglia whose afferents project to the inoculated region. The virus will productively replicate in some neurons of the sensory ganglia, while establishing latency in others during the acute phase of the infection, which typically lasts up to 21 days after infection. At 28 days post-infection, infectious virus is generally not detectable and at this point the virus is said to be latent. This makes the mouse a useful model for the study of a number of aspects of viral latency, including latent-phase transcription, epigenetic modifications, and limited aspects of reactivation. Virus can be reactivated from the latent ganglia by a procedure called “explantation-induced reactivation,” in which the ganglia are dissected from the mouse and incubated in tissue culture medium. Infectious virus can first be detected approximately 12 hours after explant [11, 12]. Although the mouse has been used to study a number of aspects of HSV latency and explant-induced reactivation, in vivo reactivation in the mouse does not result in clinical reactivation of the virus at the site of infection, which is unlike what occurs in humans and rabbits.
In the rabbit model, HSV-1 latency is generally established in the trigeminal ganglia (TG) following ocular infection. In contrast to the mouse, virus can be efficiently induced to produce clinical reactivation at the primary site of infection. A number of reactivation stimuli have been employed, but adrenergic agents such as epinephrine are the most commonly used. Following either topical or iontophoretic application of epinephrine to the eye, corneal lesions and infectious virus can be detected in the tears beginning 2 days post treatment, and continuing for up to a week. In addition to the ability to induce clinical reactivation, the efficiency of reactivation typically approaches 100% of the induced eyes, and therefore, this model has been extremely useful in studying the genetic determinants of HSV latency and reactivation.
The mouse and rabbit models have been invaluable in defining the transcriptional control, genome configuration and epigenetic control of latent gene expression, and the details of these processes described below were established using these models. Unless otherwise stated, the details apply to both the mouse and rabbit systems.
HSV Latent Transcription
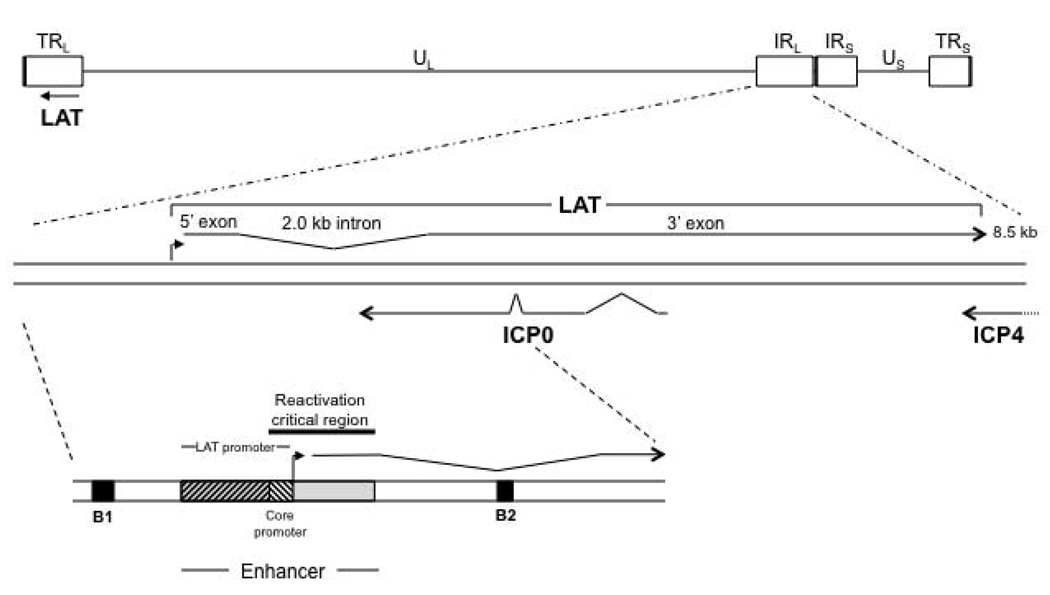
During latency in the mouse, the lytic viral genes persist in a transcriptionally silenced state, while only the Latency-Associated Transcript (LAT) is transcribed abundantly within approximately one-third of latently-infected neurons [13, 14]. The LAT is a polyadenylated non-coding RNA approximately 8.3–8.5 kb in length [15–17]. The primary LAT can be spliced to yield a very stable 2.0-kb intron, and this intron can be further spliced in some neurons to yield a 1.5-kb intron [18]. Though the long 24 hour half-life and abundance of the 2.0-kb intron in cell culture implies importance, the functions of the LAT introns remain undetermined [19]. Features of the HSV-1 LAT are highlighted in Figure 3. A number of phenotypes have been associated with the HSV-1 LAT including increasing the efficiency of establishment of latency [20, 21], reactivation [16, 17], and blocking apoptosis [22].
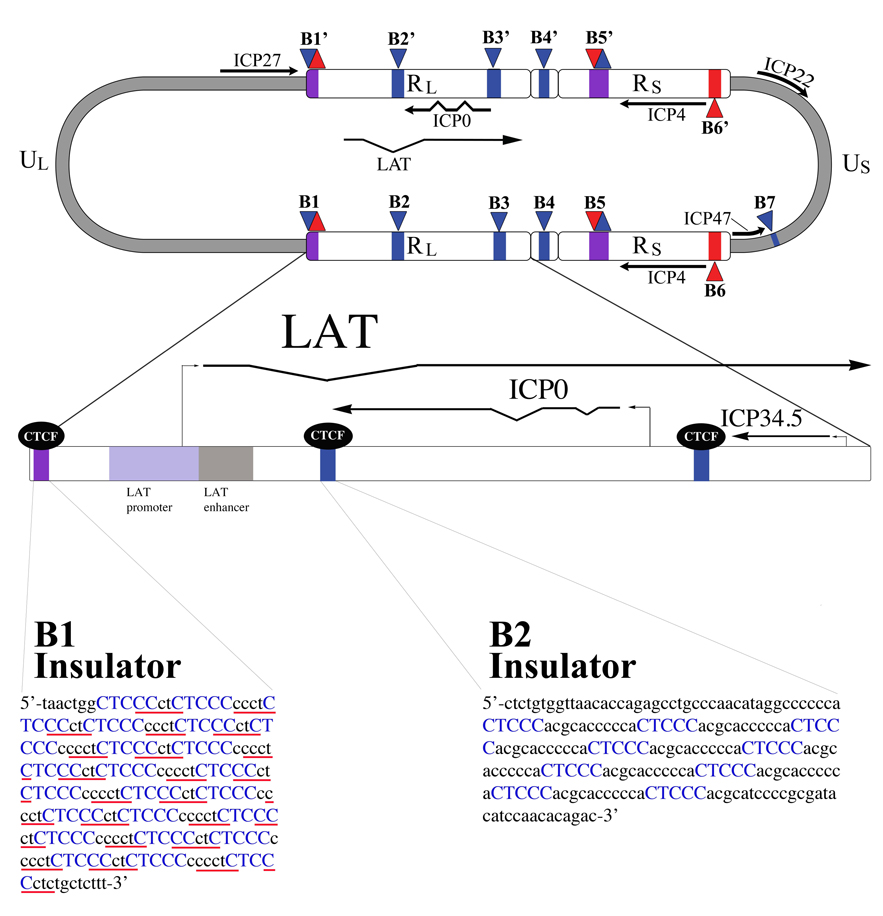
Figure 3. Features of the LAT region.
The LAT exists in duplicate in the genome, since it is located in the long repeat (RL) regions. The 8.5 kb LAT primary transcript is shown, with the exons and 2.0 kb stable intron indicated. Additionally, the lytic ICP0 and ICP4 transcripts are designated. The lower blown-up section gives additional key features of the LAT that are relevant to this review, specifically those in the region upstream of the LAT through the LAT intron. The B1 and B2 insulators are indicated by black boxes; the LAT promoter, core promoter, enhancer, and reactivation critical region are also shown. The LAT transcriptional start site is designated by a filled black arrow.
While the LAT is the only transcript abundantly transcribed during latency, sensitive RT-PCR analyses have demonstrated that relatively low amounts of some HSV lytic gene transcripts can be detected during latency [23, 24]. It is not clear whether these transcripts reflect an occasional neuron that has recently reactivated from latency (spontaneous reactivation) or represent a general “leakiness” of transcription from the latent genome [25, 26].
As described earlier in this review, HSV latency can be studied in the mouse or rabbit models, and it appears that the choice of model may determine the effect of the LAT on the latent infection. In the mouse model infected with viral recombinants that do not express the LAT, explant reactivation is dramatically reduced [16], and the transcriptional silencing of the lytic genes is leaky [16, 26–28]. In fact, accumulation of the lytic genes ICP4 (a major lytic phase transactivator representative of the IE gene class) and thymidine kinase (tk) (a member of the E gene class) is approximately 5- to greater than 10-fold higher, respectively, than the background levels observed for the latent wild-type virus [28]. This suggests a potential role for the LAT in silencing the viral genome during latency in the mouse.
In the latently-infected rabbit, the LAT may play a different role than what is seen for the mouse. When rabbits are latently-infected with a recombinant that does not express the LAT, accumulation of the lytic transcripts ICP4, tk, and glycoprotein C (gC, L gene class) is between 3- and 154-fold less than what is observed for wild-type [29] (Table 1). These findings suggest that the LAT may have a role in maintaining the latent viral genome in a more activated form, which would allow it to more readily reactivate. Alternatively, these apparent model-specific effects of the LAT on transcription may be due to the strain of HSV-1 studied rather than an effect of the model. These issues will be further discussed later in this review.
Table 1.
Fold-difference in transcript abundance in LAT positive vs. LAT negative virus latent infections of the mouse and rabbit trigeminal ganglia.
| Transcript | Fold difference in transcript abundance* | |
|---|---|---|
| Mouse1 | Rabbit2 | |
| ICP4 | ~5 more | 3 less |
| tk | >10 more | 35 less |
| gC | ND3 | >154 less |
Fold-difference in transcript abundance calculated from reported data for number of transcripts per viral genomes
Chen et al., 1997.
Giordani et al., 2008.
ND = not determined
Regulation of HSV lytic gene repression does not involve DNA methylation
While it has been shown that in members of the beta- and gamma-herpesvirus families, methylation of specific CpG nucleotides plays a role in the regulation of the promoters of lytic genes, both HpaII/MspI and bisulfite sequence analyses of HSV latent genomes has failed to detect any CpG methylation [30–32]. This apparent difference in the usage of CpG methylation as a transcriptional control mechanism between alpha-herpesviruses and the beta- and gamma-herpesviruses may reflect a requirement for different transcriptional control mechanisms in the cells where these viruses establish latency: neurons in the case of alpha-herpesviruses, and lymphocytes in the case of beta- and gamma-herpesviruses. Reversing CpG methylation within terminally differentiated, non-replicating neurons might result in an insurmountable barrier to reactivation in this cell type. It does appear, however, that HSV makes extensive use of epigenetic modifications of histones as a mechanism of transcriptional control during latency, as summarized in the next section.
II. Epigenetic modification of the latent HSV-1 genome
Since latent gene expression does not appear to be controlled by DNA methylation, other mechanisms of epigenetic control were investigated, specifically chromatin formation. When naked DNA is condensed to form chromatin, it wraps around an octamer of histone proteins to form a nucleosome (for a review see [32]). The N-termini of these proteins are subject to posttranslational modifications, the patterns of which are reflective of the transcriptional status of a gene. Chromatin may be divided into two categories: that associated with transcriptionally permissive genes (euchromatin) or that associated with transcriptionally repressed genes (heterochromatin).
It was demonstrated that during latency, HSV-1 DNA associates with nucleosomes [33]. Furthermore, histone posttranslational modifications (PTMs) representative of either euchromatin or heterochromatin are found on HSV-1 genes during latency, indicating that control of latent gene expression exists at the level of chromatin.
Survey of epigenetic modifications on the HSV-1 genome during latency
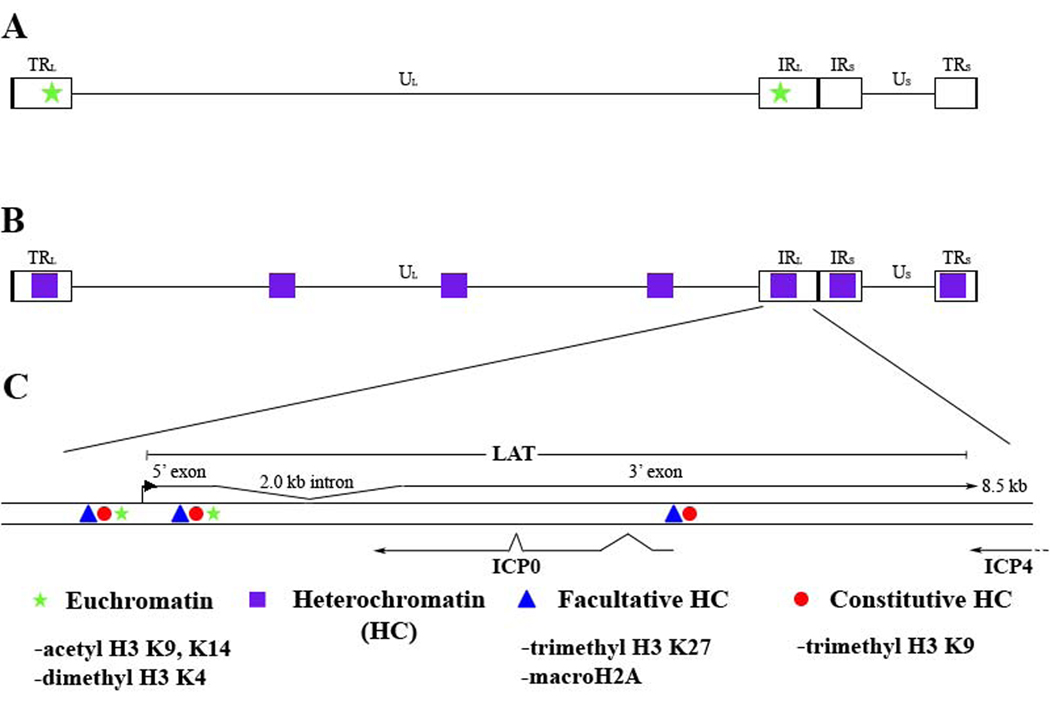
Two modifications that commonly define areas of euchromatin are acetylation of histone H3 on lysines 9 and 14 (H3 K9,K14) and dimethylation of histone H3 on lysine 4 (H3 K4). Studies looking at the deposition of these euchromatic marks found them to be enriched at the LAT locus relative to lytic targets examined [30, 34, 35] (Figure 4). This observation corresponds with data showing that the LAT is abundant during latency, signifying that the virus utilizes histone modifications in a manner similar to that of the host cell. Additionally, this suggests that transcriptional control of the latent episome might occur at the complex and dynamic level of chromatin organization.
Figure 4. Chromatin profile of the latent HSV-1 genome.
A) Regions in which euchromatic histone marks are enriched are denoted by green stars on the HSV-1 genome. B) Regions in which heterochromatic histone marks are enriched are marked in purple boxes on the HSV-1 genome. C) The enlarged LAT region displays the areas where euchromatic (acetyl H3 K9, K14 and dimethyl H3 K4) and heterochromatic (trimethyl H3 K27 and trimethyl H3 K9) marks were identified.
Studies linking euchromatin to expression of LAT concluded that the composition of the chromatin may play a role in latent gene expression. In order to understand repression of the lytic genes, the type of heterochromatin present on the latent genome was investigated. While heterochromatin is generally associated with transcriptionally inactive genes, it exists in two forms. The tightly condensed 30 nm structure of chromatin is called constitutive heterochromatin and is often considered irreversible, as it will not revert back to euchromatin [36]. This type of repression is most often seen at the telomeres and centromeres in mammalian cells. Alternatively, a form of repression that is referred to as reversible exists in a variable state of compaction and is called facultative heterochromatin. This type of repression is seen at developmentally controlled genes (i.e. the HOX locus) or the inactive X chromosome. These two types of repression can be characterized by the presence of certain PTMs that are established by different groups of proteins; thus, proteins catalyzing PTMs in facultative heterochromatin are not the same as those catalyzing PTMs in constitutive heterochromatin. Histone PTMs that differentiate constitutive vs. facultative heterochromatin are summarized in Figure 4.
Thus far, studies examining heterochromatin during HSV latency have been limited to histone modifications indicative of constitutive heterochromatin [37]. Wang et al.’s analysis of H3 K9 dimethylation over the course of establishment demonstrated that the lytic genes become enriched over time [38]. Due to this mark’s association with both actively transcribed and inactive genes, it is more suggestive of heterochromatin than reflective of it [39]. To verify this observation, several histone modifications representative of heterochromatin were investigated. It was found that lytic genes are in fact enriched in heterochromatin during latency. Interestingly, histone modifications signifying both types of heterochromatin were present on the latent genome.
The facultative heterochromatin PTM trimethyl H3 K27 (triMe K27) [40, 41] and deposition of macroH2A [40], a repressive histone H2A variant, are enriched on the latent genome with no particular bias toward gene class [40]. Additionally, trimethyl H3 K9 (triMe K9), a PTM of constitutive heterochromatin is also enriched on lytic genes of all three kinetic classes [40, 41]. Since these PTMs generally indicate different types of gene repression, it suggests that in a pool of the three mouse DRG or TG that make up a single ChIP sample, some genomes have one type of heterochromatin associated with them, while other genomes have a different type. Reasons for why this may occur are discussed later in this review. To add to this complex situation, the LAT region, previously characterized as euchromatic, was found to be enriched in the same heterochromatic marks (triMe H3K27, macroH2A, triMe H3K9) that were observed on lytic genes [41]. This supports the hypothesis that not all HSV-1 genomes are created equally in terms of PTMs, since differences in LAT expression, or the transcriptional profile of the entire genome, could lead to an altered chromatin profile. As above, these findings suggest that some genomes within a ChIP sample have an actively transcribed LAT locus associated with euchromatin, while other genomes are not transcribing LAT and are associated with heterochromatic marks. This phenomenon is discussed in detail later on.
To assess if the expression of the LAT affects the chromatin status of the latent genome, the LAT negative mutant, 17ΔPst, in which the 202 bp core LAT promoter was deleted, was examined for heterochromatic marks. The viral genes investigated in 17ΔPst (the LAT 5’ exon, ICP0 and ICP4) had a significant enrichment of facultative heterochromatin (trimethyl H3 K27, macroH2A) relative to wild-type 17syn+ [40]. However, no difference in accumulation was observed in the constitutive heterochromatin PTM, triMe H3K9, within the rabbit model [29]. Therefore, it appears that in 17syn+ the LAT plays a role specifically in limiting the deposition of facultative heterochromatin marks. Alternatively the effect of the LAT in HSV-1 strain KOS seems to play a role in enhancing the deposition of heterochromatic marks. When the same LAT promoter deletion was made in this strain and analyzed for triMe H3 K27 deposition, a decrease relative to the rescue virus was observed [41]. It should be noted, however, that some of these studies compared LAT promoter deletions with wild-type HSV-1 instead of a rescuant virus. None-the-less, these observations highlight potential differences in the regulation of latent gene expression and the function of the LAT between HSV-1 strains. It is tempting to speculate that these strain-specific differences in heterochromatin formation could explain the variation in reactivation potential of 17syn+ and KOS.
Organization of the HSV genome into chromatin domains by CTCF insulators
The transcriptional and chromatin profile of the latent virus makes it clear that there must be barriers in place to keep these domains separate. Unless a physical barrier like a chromatin insulator is in place within the genome, there would be no way of preventing the spread of heterochromatin into areas of euchromatin. Recently the CCCTC-binding protein (CTCF), known to bind to all vertebrate insulators, was found to have several binding regions around the latent HSV genome [42, 43]. The ubiquitously expressed CTCF is an eleven zinc-finger DNA-binding protein with phosphorylation-dependent transcriptional regulatory activity [44, 45]. While a single DNA binding motif has been shown to be sufficient for binding, the binding motifs are often present as clusters that afford higher binding affinity [46]. CTCF-binding enables the formation and regulation of chromatin insulators through interactions with chromatin-modifying proteins; further, through the formation of chromatin loops, CTCF may segregate chromatin into spatially separated domains [45]. When CTCF is bound in HSV, the resulting insulators may act as physical barriers to block the spread of heterochromatin. CTCF has also been recently shown to play a role in regulating latent transcription in other herpesviruses, specifically EBV [47].
Analysis of the HSV genome for the presence of CTCF-binding sites revealed two large clusters surrounding the LAT promoter/enhancer, as well as five other clusters of motifs throughout the genome (Figure 6, [42]). Chromatin immunoprecipitation (ChIP) analysis revealed that all seven of these motifs are occupied during HSV-1 latency. Additionally, transient enhancer-blocking analysis of the insulator located within the LAT intron region (B2/CTRL2 insulator) has revealed that this insulator possesses enhancer-blocking and silencing activities [42]. This finding suggests that the CTCF insulators in the HSV-1 genome function analogously to cellular insulator elements. In fact, HSV-1’s B2 (CTRL2) insulator can act to block enhancers and protect against positional effects in Drosophila melanogaster embryos and eyes, respectively [43]. In addition, the insulators may play a role in partitioning the HSV-1 genome into functional chromatin domains and may recruit specific chromatin-modifying proteins that direct the transcriptional activity of these domains. Therefore, CTCF may play an integral role in the transcriptional regulation of lytic genes during latency through the formation of chromatin insulators. Since much of the analysis to date has been performed using transient insulator assays, future work is necessary to fine-map the chromatin boundaries around these insulator elements in the context of the viral genome, as well as to determine the effects of deleting these insulator elements on the ability of the virus to establish and maintain latency in vivo.
Figure 6. Location of CTCF binding sites in the HSV-1 genome.
The HSV-1 genome contains 7 unique clusters of CTCF binding sites. CTCF-binding sites are often present in clusters, and the cluster CTRL2 (of the B2 insulator) located 3’ to the LAT enhancer contains 9 copies of the consensus, whereas the cluster CTRL1 upstream of the LAT promoter (of the B1 insulator) contains 46 copies, in total, of two separate consensus motifs, CTCCC (blue), and CCCTC (underlined in red). These CTCF binding sites have been shown to bind the cellular insulator protein CTCF during latency. The B1 and B2 insulators flank the only region of the genome that is marked with transcriptionally permissive histone modifications during latency. It is speculated that these two insulators act as boundary elements to segregate the transcriptionally active LAT promoter regions from the repressed regions of the genome, as well as to prevent the LAT enhancer from acting on the surrounding lytic genes.
III. Potential mechanisms of epigenetic regulation of latent transcription
The nature of HSV-1 heterochromatin
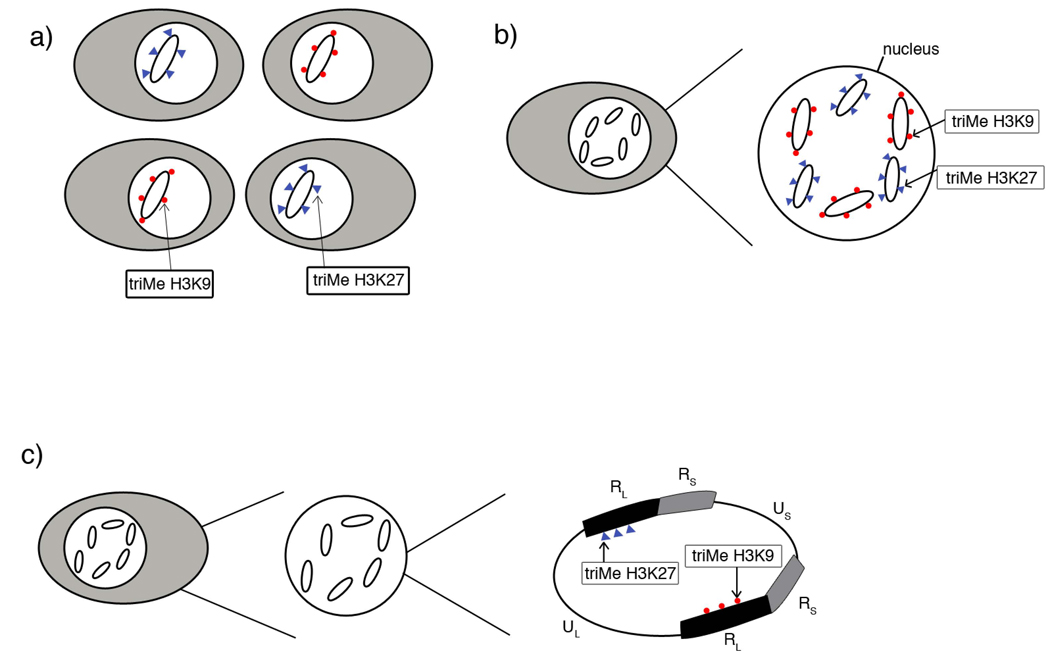
Because repressed chromatin is enriched on the latent genome, it is possible that the virus uses this as a mechanism to control expression during latency or possibly even reactivation. Interestingly, both types of heterochromatin were found in the same areas of the genome, specifically the LAT promoter and enhancer, and lytic genes ICP4, ICP0, ICP27, tk and gC. This indicates that the viral genome is interacting with the histone methyl transferases, EZH2 (trimethylates H3K27) and SUV39H (trimethylates H3K9), suggesting two independent mechanisms of gene repression in HSV-1. Since these marks are generally considered to be mutually exclusive of each other, it is likely that there are two populations of genomes: those that associate with triMe H3 K27 and those that associate with triMe H3 K9. This would explain why the ChIP experimental results suggest that these PTMs are in the same location. This assay, which utilizes pooled DRG from three mice, is not capable of distinguishing on which genomes the PTMs are present. Whether those populations consist of genomes with opposing marks in different cells, opposing marks on genomes within the same cell, or on different copies of a repeated gene within the same genome of an infected cell remains unclear (see Figure 5). Due to the tighter state of repression that is associated with triMe H3K9, one can hypothesize that genomes that are highly associated with this PTM might be less likely to be transcribed or reactivate.
Figure 5. Potential chromatin populations of latent HSV-1 genomes.
A) Genomes that associate with trimethyl H3 K27 (blue triangles) are present in different cells than genomes associated with trimethyl H3 K9 (red circles). B) Genomes enriched in either trimethyl H3K27 or trimethyl H3 K9 are present within the same cell in latent ganglia. C) Genes in the viral genome present in duplicate have differing histone post-translational marks on the two different copies of the same gene.
This idea of populations of genomes is not unprecedented. During latency there are at least two populations of neurons that harbor latent genomes: those that express LAT and those that do not. Given that HSV-1 genomes cannot be visualized by in situ hybridization within the latent host cell for unknown reasons, other techniques must be explored to assess genomes per cell. In Mehta et al., in situ PCR as well as RNA in situ hybridizations demonstrated that there were about three-fold more viral genomes detected than LAT RNA [13]. Since the number of LAT-expressing cells represents only one-third of the cells harboring latent genomes, the pattern of transcription does not appear to be universal between all genomes. Thus, genomes that express the LAT might have a different chromatin configuration than those that do not, resulting in the appearance of different populations of genomes as described above. This might explain why the LAT enhancer was shown to have both euchromatic and heterochromatic PTMs, since LAT-expressing genomes may be associated with euchromatic marks while non LAT-expressing may be enriched in heterochromatic marks.
The expression of the LAT appears to be very important in the repression of HSV-1. Though it is only expressed in 1/3 of latently-infected neurons, it has a large effect on facultative heterochromatin. A significant increase in triMe H3K27 and macroH2A deposition was noted in a LAT promoter deletion virus relative to the wild-type parent [40]. This indicates that the transcription of LAT, the act of transcribing, or cis elements in the LAT promoter play a role in chromatin formation on the latent genome. It also supports the above hypothesis that LAT-expressing genomes would have differing heterochromatin marks or different enrichment of those marks. The increased deposition of facultative heterochromatin marks might provide an explanation for the reduced reactivation phenotype associated with 17ΔPst. Perhaps the increase in facultative heterochromatin is a barrier that cannot be reversed during a reactivation stimulus. [29]
Role of polycomb proteins in regulating HSV gene expression
The presence of triMe H3 K27 on the latent genome suggests the involvement of the polycomb group proteins, a class of proteins involved in the deposition and maintenance of repressive histone marks [48]. In cellular systems, the EZH2 protein of the polycomb repressive complex (PRC) protein group, is the only protein known to catalyze the methylation of H3 K27 [49]. This strongly suggests that this cellular protein, in coordination with other PRC2 proteins, is involved in the repression of the HSV genome. ChIP analysis of the latent 17syn+ genome demonstrated that a member of the PRC1 maintenance complex, Bmi1, was enriched on the viral genes, especially the LAT enhancer [40]. It is not yet known what factor contributes to recruitment and direction of those proteins during establishment of infection nor whether this factor is a viral protein or an RNA or DNA-binding recruitment site.
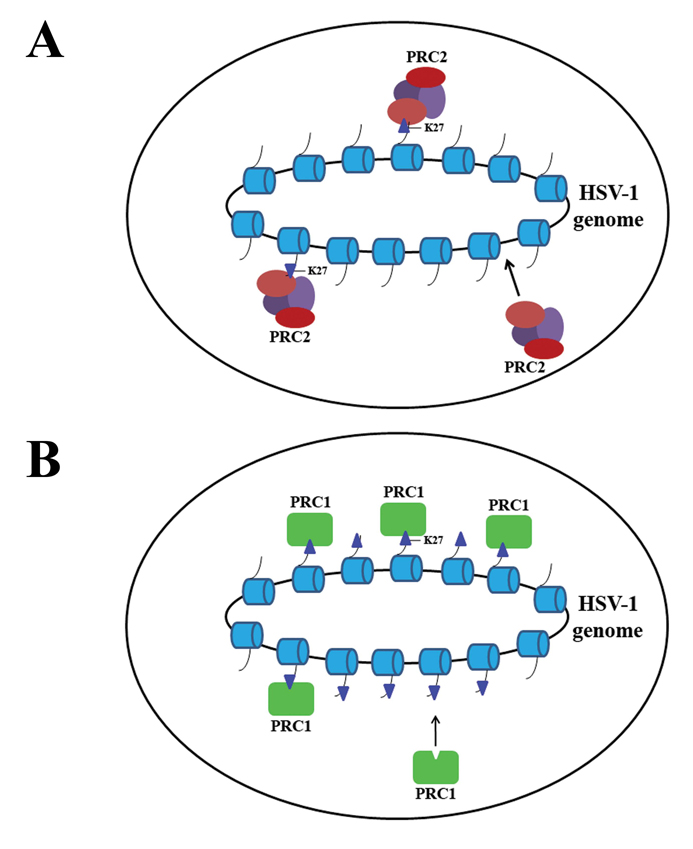
The idea that polycomb group proteins play a role in the establishment and maintenance of the latent genome is an attractive model for HSV-1 latency since it explains how the genome can be repressed and reversed in order to facilitate reactivation (Figure 7). Upon entry into the nucleus, the virus may associate with the PRC2 group of proteins containing EZH2 to catalyze the trimethylation of H3 K27 while simultaneously removing transcriptionally permissive marks. To preserve these repressive marks on the histones, PRC1 replaces the PRC2 proteins to associate with the region and maintain the PRC2-established repression. This replacement of PRC2 by the PRC1 complex on the genome is apparent by the presence of Bmi1 during latency [40]. Thus, in the latent viral context, once latency has been established and the genome has been repressed, only the proteins of the PRC1 complex remain associated with the virus. A stress stimulus could induce reactivation by displacing the PRC1 complexes and allowing for demethylation of H3 K27 and subsequent transcriptional activation.
Figure 7. Model of polycomb-mediated repression on the latent HSV-1 genome.
A) PRC2 protein complexes are recruited to the genome and trimethylate H3K27. B) PRC1 replaces PRC2 on the genome to maintain repressive histone marks to keep the latent genome in a transcriptionally inhibited state.
Potential role of CTCF in regulating HSV-1 gene expression
There are several plausible mechanisms through which CTCF could influence epigenetic regulation of HSV latent gene expression. The first is through its activity as an insulator protein. As mentioned in the section above, CTCF insulators play key roles as barriers for separating functionally disparate chromatin domains. Since the B2 (CTRL2) insulator element of HSV possesses enhancer-blocking activity and the B1 (CTRL1) and B2 insulators demarcate regions of transcriptionally permissive vs. non-permissive chromatin, it appears that these two insulators act as classic barriers [42, 43].
It is important to note that a second well-known role of CTCF is as a transcriptional co-repressor with other silencer proteins. Given that the HSV B2 insulator appears to possess both transcriptional silencing as well as enhancer-blocking activities, it is possible that when binding to the B2 insulator, CTCF interacts with a silencer protein to repress the surrounding lytic regions.
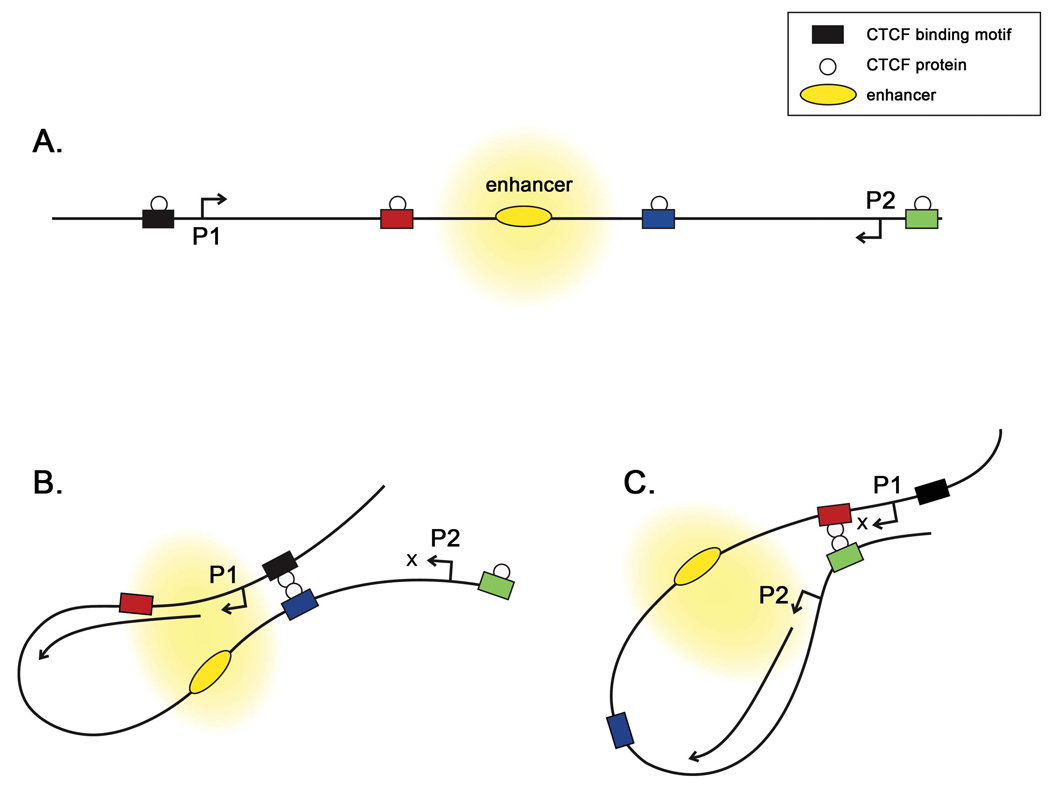
Finally, a third intriguing possible role of the HSV-1 genome’s CTCF binding sites is the potential to form loop domains, which could provide transcriptional activation of different regions of the genome depending on interactions defined by the looping partners (Figure 8). Examples of CTCF-mediated looping domains that regulate transcription are the Igf2 and gypsy insulator domains [50, 51]. In these examples, differential occupancy of CTCF sites promotes loop formation to bring an enhancer element in proximity with different promoters, depending on the loop domains formed. One attractive model is that differential occupancy of the B2 insulator could bring the LAT enhancer in proximity with a key lytic gene promoter, such as ICP4, which could promote transcriptional activation. Given the number of CTCF insulators present within the HSV genome, there are a number of possible interactions; the specific functions and interactions of the HSV insulators are currently under investigation.
Figure 8. CTCF insulators can interact to regulate transcription through the formation of loop domains that can direct the formation of transcriptional hubs.
A) Chromatin insulators, with CTCF bound, can act as enhancer-blockers to prevent the enhancer from activating genes located distal to the insulators. B) Interactions between the CTCF proteins bound to the blue and black insulators form a loop domain that allows the P1 promoter to be influenced by the enhancer. C) Interaction between the CTCFs on the green and red insulator elements excludes the P1 promoter from activation, but now allows the P2 promoter to be activated by the enhancer.
Potential Mechanisms of Epigenetic Regulation of Latent Transcription – the LAT
As discussed earlier, the LAT may play a role in maintaining or establishing the viral genome in either a repressed state in the mouse or an activated state in the rabbit. The LAT may act similarly to other transcriptionally regulative ncRNAs, which exist in a wide range of organisms. These regulatory ncRNAs include Xist, which is important in X-inactivation in placental mammals, and the roX RNAs, which regulate dosage-compensation in Drosophila. These examples play complex roles in repression and activation, respectively, of the genes upon which they act.
In mammalian cells, the non-coding Xist RNA acts in cis to silence the X chromosome that will remain inactive, associating with it and essentially “painting” the chromosome into a silenced state [52]. Xist is negatively regulated by Tsix, an antisense transcript which is necessary for silencing Xist on the active X chromosome (reviewed in [53]).
During HSV-1 latency in the mouse, transcription of the LAT appears to maintain the HSV-1 lytic genes in a tightly repressed state [28, 38], perhaps playing a role similar to Xist in X-inactivation [54]. Additionally, only a subpopulation of latently-infected neurons expresses the LAT [13, 14], suggesting that there could be a negative regulatory mechanism at play in some cells, which prevents LAT expression. A transcript antisense to the LAT (TAL) was recently identified during latency in both the mouse and the rabbit, suggesting that the transcript could be a regulatory element of the LAT, potentially playing a negative regulatory role of the LAT in a manner similar to that of Tsix and its regulation of Xist (unpublished data). It was recently shown that Tsix is critical for maintaining repressive chromatin at the Xist promoter, implicating it in Xist silencing; specifically, truncation of Tsix results in increased levels of diMe H3 K4, a marker of transcriptional permissiveness [55]. Because HSV-1 appears to be regulated at the level of histone modifications, it is possible that the TAL may play a similar role in maintaining a repressive chromatin profile of the LAT. Experiments are ongoing to determine whether this is the case and if the regulation of HSV-1 latency in the mouse is driven by a mechanism of repression similar to that of Xist, in which the ncRNA is responsible for silencing.
The roX RNAs in Drosophila provide a model for what could be occurring during HSV-1 latency in the rabbit. The Drosophila roX RNAs participate in X chromosome dosage-compensation and act to form an RNA-protein complex, which causes transcriptional activation of X-linked target genes (reviewed in [56] [57]). In the rabbit, the LAT appears to maintain the virus in a more transcriptionally permissive and activated state, a role similar to that of Drosophila’s roX non-coding RNAs [29, 56]. The two opposing roles played by the LAT, repressive like Xist in the mouse and activating like roX in the rabbit, may be what enables the virus to reactivate efficiently from latency in the rabbit and not in the mouse.
IV. A model of latency and reactivation
From this review it should be clear that the regulation of HSV gene expression during latency is quite complex, with multiple regulatory elements acting to maintain the finely tuned switch between viral latency and reactivation.
Control of HSV latency appears to be regulated at an epigenetic level, exclusive of DNA methylation. Specifically, one mechanism in the establishment of latency appears to be the recruitment of the PRC2 protein family members to lytic gene promoters. The PRC2 proteins likely deacetylate and trimethylate the histones associated with those promoters, transforming them from a euchromatic state to a reversible facultative heterochromatic state. Once these changes have occurred, the PRC1 proteins would interact with the repressed genome to maintain latency. There may also be other classes of cellular proteins acting upon the genome to assist in the establishment of latency, such as in the deposition of triMe H3 K9 to act as a repressor of gene transcription.
Data suggest that HSV latency may be also regulated by the LAT. While it appears that there may be mechanistic differences in the LAT’s function in the rabbit vs. the mouse or between viral strains, there are clear differences in the suppression of latent-phase transcription of the lytic genes between LAT-negative and wild-type viruses regardless of the model. This raises the possibility that the LAT may be recruiting or interacting with cellular proteins, such as transcriptional machinery or histone modification enzymes, to maintain or relieve repression of the lytic genes. How this is accomplished is still under investigation, but it likely involves LAT RNA-protein interactions that are absent in LAT-negative viruses, which may explain both the differences in latent-phase transcription of lytic genes and the ability to reactivate.
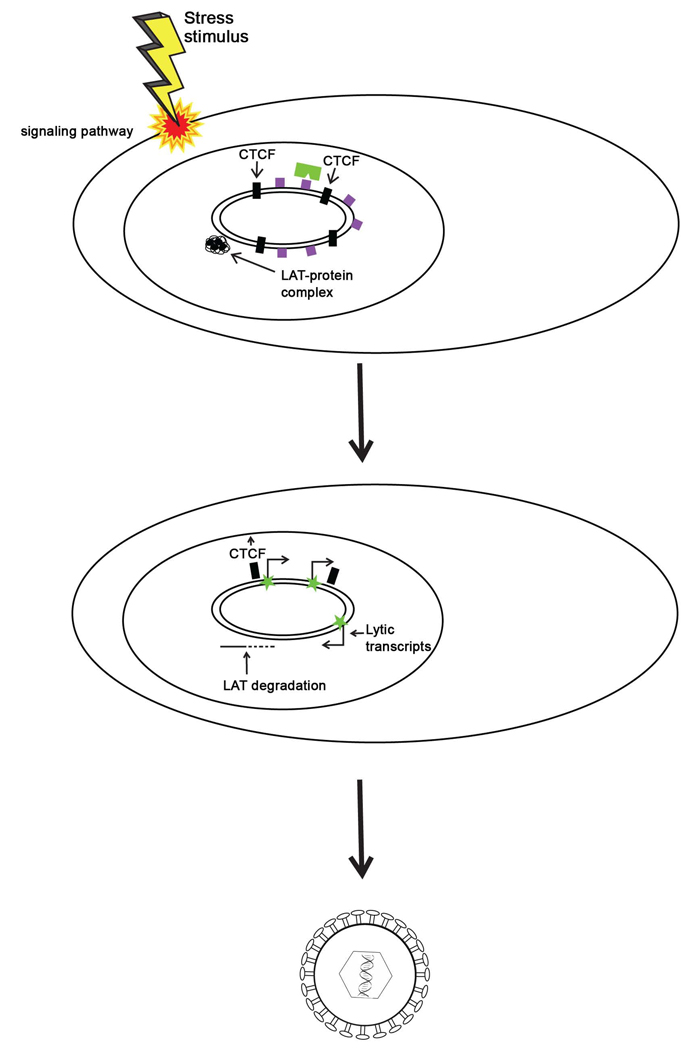
Since the rabbit model closely parallels clinical reactivation in humans, one could argue that in human HSV infections, the LAT would play a similar role as it does in the rabbit. Thus, from recent evidence, it seems that the LAT maintains the lytic genes in a repressed yet reversible state to keep them poised for reactivation. Additionally, the CTCF motifs are likely responsible for not only blocking the LAT enhancer from acting upon lytic gene promoters, but also for maintaining the lytic genes in distinct domains that help to ensure that rogue transcription from one gene does not trigger a cascade of transcription from other lytic genes to result in premature reactivation. Following a stress stimulus, there is evidence that chromatin remodeling around the LAT region and surrounding lytic genes likely occurs [35, 58]. Eviction of CTCF proteins from the insulators in response to stress-induced phosphorylation, as well as eviction of the polycomb protein complexes to allow euchromatin to replace triMe H3 K27 on lytic genes could permit the genes to be accessible for transcription, leading to productive expression of the lytic genes and eventually, reactivation of HSV (Figure 9).
Figure 9. Model for reversal of epigenetic repression of lytic genes during HSV reactivation.
During latency, the LAT acts to balance transcriptionally repressive (but reversible) facultative heterochromatin marks (purple boxes) on the lytic gene regions of the genome. These heterochromatic marks are established and maintained by polycomb group proteins (green rectangle). The CTCF insulators (black boxes) flanking that LAT promoter/enhancer block the LAT enhancer from activating ICP0 and other lytic genes. Stressors and other reactivation mediators cause a transient decrease in the LAT, and possibly a loss of insulator occupancy at the B2 and/or B1 insulators. This allows the LAT enhancer to activate lytic genes, resulting in viral replication and the production of infectious virus.
While the mechanistic details surrounding the epigenetic control of HSV latency are incomplete, it is becoming clearer that the virus has manipulated cellular epigenetic processes to allow a reversible suppression of its lytic gene program. A deeper understanding of these mechanisms will not only provide insight into how HSV maintains its latent state but may also provide new insight into details of cellular epigenetic processes.
ACKNOWLEDGEMENTS
The authors would like to acknowledge C. Lilly, L. Watson, R. Johns and D. Jacobs for helpful comments on the manuscript, and R. Eson for help with the illustrations. Finally, we have to give some credit to B. Loco for getting us through the morning meetings.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Cook SD, Ophth FC, Hill JH. Herpes simplex virus: molecular biology and the possibility of corneal latency. Survey of Ophthalmol. 1991;36(2):140–148. doi: 10.1016/0039-6257(91)90127-2. [DOI] [PubMed] [Google Scholar]
- 2.Clements GB, Jamieson FE. Reactivation of latent herpes simplex virus-1 (HSV) from mouse footpad cells demonstrated by in situ hybridization. Arch Virol. 1989;104(1–2):95–106. doi: 10.1007/BF01313811. [DOI] [PubMed] [Google Scholar]
- 3.Yang L, Voytek CC, Margolis TP. Immunohistochemical analysis of primary sensory neurons latently infected with herpes simplex virus type 1. J. Virol. 2000;74(1):209–217. doi: 10.1128/jvi.74.1.209-217.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Margolis TP, et al. Herpes simplex virus type 2 (HSV-2) establishes latent infection in a different population of ganglionic neurons than HSV-1: role of latency-associated transcripts. J Virol. 2007;81(4):1872–1878. doi: 10.1128/JVI.02110-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertke AS, et al. Latency-associated transcript (LAT) exon 1 controls herpes simplex virus species-specific phenotypes: reactivation in the guinea pig genital model and neuron subtype-specific latent expression of LAT. J Virol. 2009;83(19):10007–10015. doi: 10.1128/JVI.00559-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jamieson DR, et al. Quiescent viral genomes in human fibroblasts after infection with herpes simplex virus type 1 Vmw65 mutants. J Gen Virol. 1995;76(Pt 6):1417–1431. doi: 10.1099/0022-1317-76-6-1417. [DOI] [PubMed] [Google Scholar]
- 7.Danaher RJ, Jacob RJ, Miller CS. Establishment of a quiescent hertpes simplex virus type 1 infection in neurally-differentiated PC12 cells. J Neurovirol. 1999;5(3):258–267. doi: 10.3109/13550289909015812. [DOI] [PubMed] [Google Scholar]
- 8.Su YH, et al. Human corneal cells and other fibroblasts can stimulate the appearance of herpes simplex virus from quiescently infected PC12 cells. J. Virol. 1999;73(5):4171–4180. doi: 10.1128/jvi.73.5.4171-4180.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arthur JL, et al. Herpes simplex virus type 1 promoter activity during latency establishment, maintenance, and reactivation in primary dorsal root neurons in vitro. J Virol. 2001;75(8):3885–3895. doi: 10.1128/JVI.75.8.3885-3895.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilcox CL, Johnson EM. Characterization of nerve growth factor-dependent Herpes Simplex Latency in neurons in vitro. J. Virol. 1988;62(2):393–399. doi: 10.1128/jvi.62.2.393-399.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stevens JG, Cook ML. Latent herpes simplex virus in spinal ganglia of mice. Science. 1971;173(999):843–845. doi: 10.1126/science.173.3999.843. [DOI] [PubMed] [Google Scholar]
- 12.Pesola JM, et al. Herpes Simplex Virus 1 Immediate-Early and Early Gene Expression during Reactivation from Latency under Conditions That Prevent Infectious Virus Production. J Virol. 2005;79(23):14516–14525. doi: 10.1128/JVI.79.23.14516-14525.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mehta A, et al. In situ DNA PCR and RNA hybridization of herpes simplex virus sequences in trigeminal ganglia of latently infected mice. Virology. 1995;206:633–640. doi: 10.1016/s0042-6822(95)80080-8. [DOI] [PubMed] [Google Scholar]
- 14.Gressens P, Martin JR. In situ polymerase chain reaction: localization of HSV-2 DNA sequences in infections of the nervous system. J. Virol. Methods. 1994;46:61–83. doi: 10.1016/0166-0934(94)90017-5. [DOI] [PubMed] [Google Scholar]
- 15.Stevens JG, et al. RNA complementary to a herpesvirus alpha gene mRNA is prominent in latently infected neurons. Science. 1987;235(4792):1056–1059. doi: 10.1126/science.2434993. [DOI] [PubMed] [Google Scholar]
- 16.Leib DA, et al. A deletion mutant of the latency-associated transcript of herpes simplex virus type 1 reactivates from the latent state with reduced frequency. J Virol. 1989;63(7):2893–2900. doi: 10.1128/jvi.63.7.2893-2900.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hill JM, et al. Herpes simplex virus latent phase transcription facilitates in vivo reactivation. Virology. 1990;174(1):117–125. doi: 10.1016/0042-6822(90)90060-5. [DOI] [PubMed] [Google Scholar]
- 18.Farrell MJ, Dobson AT, Feldman LT. Herpes simplex virus latency-associated transcript is a stable intron. Proc. Natl. Acad. Sci. U.S.A. 1991;88(3):790–794. doi: 10.1073/pnas.88.3.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomas DL, et al. The 2-kilobase intron of the herpes simplex virus type 1 latency-associated transcript has a half-life of approximately 24 hours in SY5Y and COS-1 cells. J Virol. 2002;76(2):532–540. doi: 10.1128/JVI.76.2.532-540.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sawtell NM. Comprehensive quantification of herpes simplex virus latency at the single-cell level. J. Virol. 1997;71(7):5423–5431. doi: 10.1128/jvi.71.7.5423-5431.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson RL, Sawtell NM. Herpes simplex virus type 1 latency-associated transcript gene promotes neuronal survival. J Virol. 2001;75(14):6660–6675. doi: 10.1128/JVI.75.14.6660-6675.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perng GC, et al. Virus-induced neuronal apoptosis blocked by the herpes simplex virus latency-associated transcript. Science. 2000;287(5457):1500–1503. doi: 10.1126/science.287.5457.1500. [DOI] [PubMed] [Google Scholar]
- 23.Kramer MF, et al. Accumulation of viral transcripts and DNA during establishment of latency by herpes simplex virus. J. Virol. 1998;72(2):1177–1185. doi: 10.1128/jvi.72.2.1177-1185.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kramer MF, Coen DM. Quantification of transcripts from the ICP4 and thymidine kinase genes in mouse ganglia latently infected with herpes simplex virus. J. Virol. 1995;69(3):1389–1399. doi: 10.1128/jvi.69.3.1389-1399.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feldman L, et al. Spontaneous molecular reactivation of herpes simplex virus type 1 latency in mice. Proc Natl Acad Sci USA. 2002;99(2):978–983. doi: 10.1073/pnas.022301899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garber DA, Schaffer PA, Knipe DM. A LAT-associated function reduces productive-cycle gene expression during acute infection of murine sensory neurons with herpes simplex virus type 1. J. Virol. 1997;71(8):5885–5893. doi: 10.1128/jvi.71.8.5885-5893.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Devi-Rao GB, et al. Herpes simplex virus type 1 DNA replication and gene expression during explant induced reactivation of latently infected murine sensory ganglia. J. Virol. 1994;68(3):1271–1282. doi: 10.1128/jvi.68.3.1271-1282.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen SH, et al. A viral function represses accumulation of transcripts from productive-cycle genes in mouse ganglia latently infected with herpes simplex virus. J. Virol. 1997;71:5878–5884. doi: 10.1128/jvi.71.8.5878-5884.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giordani NV, et al. During HSV-1 Infection of Rabbits, the Ability to Express the LAT Increases Latent-Phase Transcription of Lytic Genes. J Virol. 2008;82(12):6056–6060. doi: 10.1128/JVI.02661-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kubat NJ, et al. Specific histone tail modification and not DNA methylation is a determinant of herpes simplex virus type 1 latent gene expression. J Virol. 2004;78(3):1139–1149. doi: 10.1128/JVI.78.3.1139-1149.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dressler GR, Rock DL, Fraser NW. Latent herpes simplex virus type 1 DNA is not extensively methylated in vivo. J Gen Virol. 1987;68:1761–1765. doi: 10.1099/0022-1317-68-6-1761. [DOI] [PubMed] [Google Scholar]
- 32.Quina AS, Buschbeck M, Di Croce L. Chromatin structure and epigenetics. Biochem Pharmacol. 2006;72(11):1563–1569. doi: 10.1016/j.bcp.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 33.Deshmane SL, Fraser NW. During latency, herpes simplex virus type 1 DNA is associated with nucleosomes in a chromatin structure. J Virol. 1989;63(2):943–947. doi: 10.1128/jvi.63.2.943-947.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kubat NJ, et al. The Herpes Simplex Virus Type 1 Latency-Associated Transcript (LAT) Enhancer/rcr Is Hyperacetylated during Latency Independently of LAT Transcription. J Virol. 2004;78(22):12508–12518. doi: 10.1128/JVI.78.22.12508-12518.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neumann DM, et al. In vivo changes in the patterns of chromatin structure associated with the latent herpes simplex virus type 1 genome in mouse trigeminal ganglia can be detected at early times after butyrate treatment. J Virol. 2007;81(23):13248–13253. doi: 10.1128/JVI.01569-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trojer P, Reinberg D. Facultative heterochromatin: is there a distinctive molecular signature? Mol Cell. 2007;28(1):1–13. doi: 10.1016/j.molcel.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 37.Knipe DM, Cliffe A. Chromatin control of herpes simplex virus lytic and latent infection. Nat Rev Microbiol. 2008;6(3):211–221. doi: 10.1038/nrmicro1794. [DOI] [PubMed] [Google Scholar]
- 38.Wang QY, et al. Herpesviral latency-associated transcript gene promotes assembly of heterochromatin on viral lytic-gene promoters in latent infection. Proc Natl Acad Sci U S A. 2005;102(44):16055–16059. doi: 10.1073/pnas.0505850102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vakoc CR, et al. Profile of histone lysine methylation across transcribed mammalian chromatin. Mol Cell Biol. 2006;26(24):9185–9195. doi: 10.1128/MCB.01529-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kwiatkowski DL, Thompson HW, Bloom DC. The polycomb group protein Bmi1 binds to the herpes simplex virus 1 latent genome and maintains repressive histone marks during latency. J. Virol. 2009;83(16):8173–8181. doi: 10.1128/JVI.00686-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cliffe AR, Garber DA, Knipe DM. Transcription of the herpes simplex virus latency-associated transcript promotes the formation of facultative heterochromatin on lytic promoters. J Virol. 2009;83(16):8182–8190. doi: 10.1128/JVI.00712-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Amelio AL, McAnany PK, Bloom DC. A Chromatin Insulator-Like Element in the Herpes Simplex Virus Type 1 Latency-Associated Transcript Region Binds CCCTC-Binding Factor and Displays Enhancer-Blocking and Silencing Activities. J Virol. 2006;80(5):2358–2368. doi: 10.1128/JVI.80.5.2358-2368.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Q, et al. CTCF-dependent chromatin boundary element between the latency-associated transcript and ICP0 promoters in the herpes simplex virus type 1 genome. J Virol. 2007;81(10):5192–5201. doi: 10.1128/JVI.02447-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.West AG, Gaszner M, Felsenfeld G. Insulators: many functions, many mechanisms. Genes Dev. 2002;16:271–288. doi: 10.1101/gad.954702. [DOI] [PubMed] [Google Scholar]
- 45.Yusufzai TM, et al. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol Cell. 2004;13(2):291–298. doi: 10.1016/s1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]
- 46.Chao W, et al. CTCF, a candidate trans-acting factor for X-inactivation choice. Science. 2002;295(5553):345–347. doi: 10.1126/science.1065982. [DOI] [PubMed] [Google Scholar]
- 47.Kang H, Lieberman PM. Cell cycle control of Kaposi's sarcoma-associated herpesvirus latency transcription by CTCF-cohesin interactions. J Virol. 2009;83(12):6199–6210. doi: 10.1128/JVI.00052-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schwartz YB, Pirrotta V. Polycomb complexes and epigenetic states. Curr Opin Cell Biol. 2008;20(3):266–273. doi: 10.1016/j.ceb.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 49.Kirmizis A, et al. Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev. 2004;18(13):1592–1605. doi: 10.1101/gad.1200204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gerasimova TI, et al. Coordinated control of dCTCF and gypsy chromatin insulators in Drosophila. Mol Cell. 2007;28(5):761–772. doi: 10.1016/j.molcel.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405(6785):482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 52.Andersen AA, Panning B. Epigenetic gene regulation by noncoding RNAs. Curr Opin Cell Biol. 2003;15(3):281–289. doi: 10.1016/s0955-0674(03)00041-3. [DOI] [PubMed] [Google Scholar]
- 53.Thorvaldsen JL, Verona RI, Bartolomei MS. X-tra! X-tra! News from the mouse X chromosome. Dev Biol. 2006;298(2):344–353. doi: 10.1016/j.ydbio.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 54.Bloom DC, et al. A 348-bp region in the latency associated transcript facilitates herpes simplex virus type 1 reactivation. J. Virol. 1996;70(4):2449–2459. doi: 10.1128/jvi.70.4.2449-2459.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ohhata T, et al. Crucial role of antisense transcription across the Xist promoter in Tsix-mediated Xist chromatin modification. Development. 2008;135(2):227–235. doi: 10.1242/dev.008490. [DOI] [PubMed] [Google Scholar]
- 56.Deng X, Meller VH. Non-coding RNA in fly dosage compensation. Trends Biochem Sci. 2006;31(9):526–532. doi: 10.1016/j.tibs.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 57.Straub T, et al. The Drosophila MSL complex activates the transcription of target genes. Genes Dev. 2005;19(19):2284–2288. doi: 10.1101/gad.1343105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Amelio AL, et al. Deacetylation of the Herpes Simplex Virus Type 1 Latency-Associated Transcript (LAT) Enhancer and a Decrease in LAT Abundance Precede an Increase in ICP0 Transcriptional Permissiveness at Early Times Postexplant. J Virol. 2006;80(4):2063–2068. doi: 10.1128/JVI.80.4.2063-2068.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]