Abstract
Mutations in the genes for isocitrate dehydrogenase 1 (IDH1) and isocitrate dehydrogenase 2 (IDH2) have been recently identified in glioblastoma. In the present study, we investigated IDH1 and IDH2 mutations in follicular thyroid cancer (FTC) and anaplastic thyroid cancer (ATC), with the latter, like glioblastoma, having a rapidly aggressive and lethal clinical course. By direct genomic DNA sequencing, we analyzed exon 4 of the IDH1 and IDH2 genes that harbored the mutation hot spots codon 132 and 172 of the two genes in glioblastoma, respectively, in 12 thyroid cancer cell lines, 20 FTC and 18 ATC tumor samples. A novel homozygous G367A IDH1 mutation, resulting in a G123R amino acid change in codon 123, was identified in a case of ATC. A previously described IDH1 V71I mutation was found in a case of FTC and a case of ATC and no mutations were found in the cell lines. The overall prevalence of mutations was thus 1/20 (5%) in FTC and 2/18 (11%) in ATC. We did not find mutation in the IDH2 gene in these thyroid cancer cell lines and tumor samples. Sequence alignment analysis of 16 species revealed that the novel IDH1 G123R mutation was located in a highly conserved region, raising the possibility of a serious functional consequence as could also be predicted by the occurrence of a positively charged amino acid from this mutation. To test this, we created a G123R mutant by site-directed mutagenesis and demonstrated a decreased enzymatic activity of IDH1, similar to the expected reduction in the enzymatic activity of the previously described R132H IDH1 mutant measured as a control. Thus, functionally relevant IDH1 mutations can also occur in thyroid cancer, particularly ATC, suggesting a potential tumorigenic role of the IDH1 system that could represent a new therapeutic target for thyroid cancer.
Keywords: IDH1mutation, thyroid cancer, anaplastic thyroid cancer, follicular thyroid cancer, genetics, isocitrate dehydrogenase
Introduction
Mutations in the genes for the enzymes isocitrate dehydrogenase 1 (IDH1) and isocitrate dehydrogenase 2 (IDH2) have been recently identified in brain glioblastomas, particularly secondary glioblastomas, with a high frequency (> 70%) particularly in the IDH1 gene [1–4]. IDH is a key player in the tricarboxylic acid cycle, or Krebs cycle, and catalyzes the oxidative decarboxylation of isocitrate to produce alpha-ketoglutarate (α-KG). The activity of IDH is dependent on nicotinamide adenine dinucleotide phosphate (NADP+) and the biochemical reaction catalyzed by IDH leads to the production of NADPH, which plays an important role in the cellular control of oxidative damage [5]. IDH1 mutations were found to be exclusively in codon 132 [1]. Several different IDH1 mutations have been identified, all affecting the amino acid arginine at codon 132 and resulting in various amino acid substitutions (R132H, R132S, R132C and R132G). IDH1 mutations were relatively uncommon in other cancers, with 8% in acute myeloid leukemia [6], 2.7% in prostate cancer, and 1.7% in B-acute lymphoblastic leukemia [7]. All the IDH2 mutations were found in codon R172 and tumors without mutations in IDH1 often had mutations affecting the analogous amino acid (R172) of the IDH2 gene [3]. The R132 residue is highly conserved over evolution and is localized in the substrate binding site of IDH1 where hydrophilic interactions between R132 and the α- and β-carboxylates of isocitrates occur [8]. Codon 132 IDH1 mutants have been shown to be associated with reduced catalytic activity of the IDH1 enzyme [3, 9, 10].
Recently, the IDH1 gene was examined for mutations in medullary thyroid cancer (MTC) and papillary thyroid cancer (PTC), but no mutations were found in these types of thyroid cancer [4]. Thyroid cancer is the most common endocrine malignancy, which, in addition to MTC and PTC, also includes follicular thyroid cancer (FTC) and anaplastic thyroid cancer (ATC). These two latter thyroid cancers, particularly ATC, are more aggressive. In fact, ATC, like glioblastoma, is an extremely aggressive and rapidly lethal human cancer [11]. Given the common IDH1 and IDH2 mutations in glioblastoma and its somehow similar aggressiveness with ATC, it remains an interesting question whether IDH1 and IDH2 mutations also occur in ATC. In the present study, we analyzed the IDH1 and IDH2 mutation in both ATC and FTC since they have not been examined for mutations in the two genes.
Materials and Methods
Cell lines and tumor samples
A total of 50 samples, including 12 thyroid cancer cell lines (K1, K5, OCUT-1, OCUT-2, FB-1, SW1736, BCPAP, HTh7, HTh74, KAT 18, FTC133 and C643), 20 FTC tumors, and 18 ATC tumors were used for mutational analysis of the IDH1 and IDH2 genes as shown in Table 1. Cell lines were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine, streptomycin (100 μg/mL) and penicillin (100 U/mL). Tumor samples were from an IRB-approved study [12]. Genomic DNA from cell lines and tumors was isolated by standard phenol-chloroform extraction and ethanol precipitation procedures, using MaXtract high density gel tubes (Qiagen, Valencia, CA).
Table 1.
Mutational analysis of IDH1 and IDH2 genes in thyroid cancers
| IDH1 | IDH2 | |||
|---|---|---|---|---|
| Samples | Analyzed samples | Mutations | Analyzed samples | Mutations |
| Cell lines | ||||
| FTC | 2 | 0 | 2 | 0 |
| ATC | 8 | 0 | 8 | 0 |
| PTC | 2 | 0 | 2 | 0 |
| Tumors | ||||
| FTC | 20 | 1 (5%) | 20 | 0 |
| ATC | 18 | 2 (11%) | 18 | 0 |
PCR amplification and sequencing
PCR amplification of exon 4 of IDH1, which was the hot spot of IDH1 mutation in other cancers, was performed using the primers and PCR conditions as described previously [4]. In addition, we used a reverse primer (5′- CATGCAAAATCACATTATTGCC -3′) to sequence the amplicon for reverse orientation. The amplification of exon 4 of IDH2 was performed using the forward primer IDH2-4F 5′- TGCACTCTAGACTCTACTGCC -3′ and reverse primer IDH2-4R 5′- ACAAAGTCTGTGGCCTTGTAC -3′ with the annealing temperature at 60ºC. The PCR amplified products were directly sequenced using a Big Dye terminator v3.1 cycle sequencing ready reaction kit (Applied Biosystems) and ABI PRISM 3730 automated next generation genetic analyzer (Applied Biosystems). The gene bank accession numbers of IDH1 and IDH2 are NM_005896.2 and NM_002168.2, respectively.
Multiple amino acid sequence alignment
Amino acid sequences of IDH1 of various species were obtained from NCBI database (http://www.ncbi.nlm.nih.gov/protein/) as follows: H_sapiens (NP_005887.2), P_troglodytes (XP_001142197), C_lupus (XP_536047.2), B_taurus (NP_851355.2), M_musculus (NP_034627.2), R_norvegicus (NP_113698.1), G_gallus (XP_421965.2), D_rerio (NP_958907.1), D_melanogaster (NP_652044.1), A_gambiae (XP_001688948), O_sativa (NP_001043749), A_thaliana (NP_176768.1), K_lactis (XP_451683.1), E_gossypii (NP_984921.1), N_crassa (XP_323176.1) and S_pombe (NP_594105.1). These sequences were compared using a computer based multiple sequence alignment program (http://pir.georgetown.edu/cgi-bin/multialn.pln).
Expression vector construction
The wild-type clone of IDH1 (Catalog No: SC322129) was obtained from OriGene Technologies, Inc. (Rockville, MD). The DNA fragment containing the entire open reading frame of IDH1 was PCR amplified using a forward primer containing BamH1 and Kozak consensus sequences (IDH1 WT-F1 5′-CGCGGATCCGCCACCATGTCCAAAAAAATCAGT-3′) and a reverse primer containing Not1 sequence (IDH1 WT-R1 5′- CGTGCGGCCGCCAAGTTTGGCCTGAGCTAG- 3′). The amplified fragment was digested with BamH1 and Not1 and cloned into the pcDNA4TOmyc-His(B)™ (Invitrogen, CA). The IDH1 ORF was re-amplified from the pcDNA4TOmyc-His(B)™ using the forward primer (IDH1 WT-F1) and reverse primer containing EcoR1 (IDH1 R2 -5′- GCTGAATTCTCAATGGTGATGGTGATG-3′). The resultant fragment was digested with BamH1 and EcoR1 and subsequently cloned into a mammalian expression vector pcDNA3.1 (+)™ (Invitrogen, CA) between the BamH1 and EcoR1 sites, which produced a myc tag fused to the COOH terminus of the IDH1gene. The integrity of the plasmid was verified by restriction enzyme digestion and sequencing.
Site-directed mutagenesis
The expression vector pcDNA3.1 (+)™ carrying IDH1_myc was used to generate the IDH1 mutants, G123R and R132H, with a Quick Change XL Site-Directed mutagenesis kit according to the instruction of the manual. The primers were designed using template specific mutagenic primer design program. The primer sequences are as follows: G123R: sense, IDH1-G367A_F 5′- CCCCCGGCTTGTGAGTAGATGGGTAAAACCTAT-3′; antisense, IDH1-G367A_R 5′- ATAGGTTTTACCCATCTACTCACAAGCCGGGGG-3′; R132H: sense, IDH1- G395A-F 5′- GTAAAACCTATCATCATAGGTCATCATGCTTATGGGGATCAATAC -3′; antisense, IDH1- G395A-R 5′- GTATTGATCCCCATAAGCATGATGACCTATGATGATAGGTTTTAC -3′. All the mutations were confirmed by sequencing with primer IDH1-VEC_F 5′- ATGCAAGGAGATGAAATGACACT-3′. Plasmid DNAs for the transfection experiments were purified using an Invitrogen mini prep kit (K2100–11).
Cell transfection
HEK293T cells were grown at 37ºC in Dulbecco’s Modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum, 100 U/ml of penicillin and 100 μg/ml of streptomycin (GIBCO). Cells were transiently transfected with an empty vector, wild-type or each of mutant IDH1 expression vectors using the Lipofectamine 2000 transfection reagent per manufacturer’s instructions (Invitrogen Life Technologies, CA). Cells were harvested 48 hours after transfection and subjected to lysis for Western blotting and IDH1 enzymatic assay.
Western blotting
Western blotting was performed as described [13]. Briefly, about 20 μg of cell lysate proteins prepared for IDH1 enzymatic assay was separated on 10% SDS/PAGE and transferred to PVDF membrane (Millipore Co., Bedford, MA). After transfer, the membrane was blocked with 5% skim-milk/PBS containing 0.1% Tween-20 (PBST) for 1 hour at room temperature, incubated overnight at 4º C with anti-Myc antibody (SC-40). After washing 4 times with PBST the blots were incubated with anti-mouse HRP conjugated antibody (SC-2005) for 1 hour at room temperature. After washing with PBST, protein bands on the membrane were detected with enhancement chemiluminescence (ECL) reaction and exposure to X-ray film. To ensure the equality of protein loading, the same membrane blot was stripped off and re-probed with an anti-β-actin antibody (SC-1616-R).
IDH1 enzymatic assay
The IDH1 enzymatic assay was performed as described previously [3, 14, 15]. Briefly, cells were collected 48 hrs after transfection and centrifuged at 1000 × g for 10 minutes at 4ºC. After washing with cold phosphate- buffered saline, cells were lysed in mammalian cell lysis buffer containing 0.1% TritonX-100. Cells were then disrupted by ultra sonication and incubated on ice for 30 minutes. Cell lysates were then centrifuged at 12,000 × g for 10 minutes at 4ºC. The supernatants were collected and protein concentration was measured using a DC protein assay kit (Bio-Rad laboratories, Hercules, CA). For each enzymatic reaction, an equal volume of cell lysate containing the same amount of protein was added to the assay solution containing 33 mM Tris buffer (pH 8.0), 0.33 mM EDTA, 0.1 mM NADP+ (Sigma Cat. No. N-0505), 1.33 mM manganese chloride and 1.3 mM isocitrate (Sigma Cat. No. I-1252). Isocitrate dehydrogenase activity was measured spectrophotometrically at 27ºC by monitoring the formation of NADP+ to NADPH at 340 nm.
Results
Relatively common IDH1 mutations in thyroid cancer
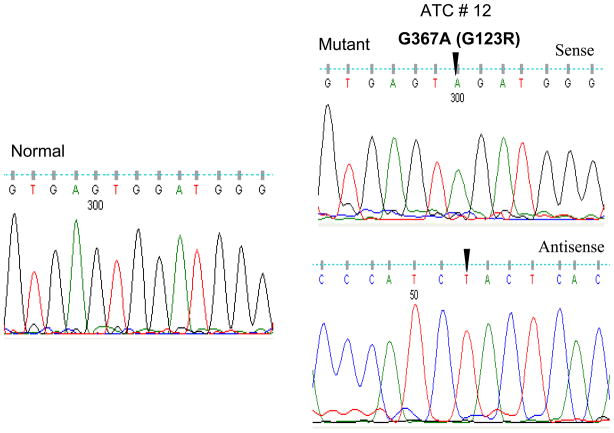
We analyzed exon 4 of the IDH1 and IDH2 genes for mutations in FTC and ATC. This exon was chosen to be analyzed because it contains the hot spots most commonly carrying somatic mutations in glioblastomas. In total, 50 samples were examined, which included 12 thyroid cancer cell lines, 20 FTC and 18 ATC tumor samples. We did not find any mutation at codon 132 of IDH1 and at codon 172 of IDH2 in any of the thyroid cancer cell lines and thyroid cancer samples. However, we found a novel missense mutation of IDH1 at codon 123 in an ATC sample. As shown in Fig 1, this point mutation represented a G-to-A transition in nucleotide position 367 and was a homozygous mutation. Codon 123 was changed by this mutation from GGA to AGA, resulting in the amino acid change of glycine to arginine (G123R). This G367A mutation represents a novel IDH1 mutation in human cancer that has not been described previously. We also found a mutation in codon 71, causing the codon change from GTC to ATC, resulting in the amino acid change of V71I of the IDH1 protein. This mutation was seen in one FTC sample and one ATC sample. The V71I mutation was previously reported in the myeloma cell line RPMI-8226 [4]. The IDH1 mutation rates were therefore 1/20 (5%) in FTC and 2/18 (11.11%) in ATC. Table 1 and 2 summarizes the prevalences of the IDH1 mutations in various types of thyroid cancers.
Figure 1. Identification of the G367A IDH1 mutation in codon 123 on sequence analysis of exon 4 of the IDH1 gene.
Sequencing chromatogram of sense and antisense strands reveals the presence of a homozygous G-to-A mutation at nucleotide position 367 in codon 123 in exon 4 of the IDH1 gene from a anaplastic thyroid cancer. The nucleotide number, amino acid alteration and tumor number are indicated above the arrow. Nucleotide number refers to the position within the coding sequence, where position 1 corresponds to the first nucleotide of the initiation codon. The left panel shows a fragment of wild-type DNA sequence corresponding to the region where the G367A mutation occurred.
Table 2.
Mutations and single nucleotide polymorphism of the IDH1 gene in thyroid cancers
| Sample | Exon | Nucleotide | Codon | Amino acid | Type of mutation |
|---|---|---|---|---|---|
| Cell line | |||||
| C643 | 4 | G255A | GAG-GAA | E85E | Silent |
| BCPAP | 4 | C315T | GGC-GGT | G105G | Silent |
| Tumor | |||||
| FTC | |||||
| # 10 | 4 | G211A | GTC-ATC | V71I | Missense |
| # 10 | 4 | C315T | GGC-GGT | G105G | Silent |
| # 27 | 4 | C315T | GGC-GGT | G105G | Silent |
| ATC | |||||
| #6 | 4 | G211A | GTC-ATC | V71I | Missense |
| #6 | 4 | C315T | GGC-GGT | G105G | Silent |
| #12 | 4 | G367A | GGA-AGA | G123R | Missense |
Common single nucleotide polymorphisms (SNP) of the IDH1 gene in thyroid cancers
In addition to these mutations, as shown in Table 2, we also identified several single nucleotide polymorphisms (SNP) in two thyroid cancer cell lines (C643 and BCPAP), two FTC samples and one ATC sample. They were located at codons 85 (E85E) and 105 (G105G). The SNP E85E was not previously reported while the SNP G105G (rs11554137) was reported in the NCBI and ENSEMBL SNP data bases (http://www.ncbi.nlm.nih.gov/projects/SNP/ and http://www.ensembl.org/index.html). The biological relevance of these SNPs remains to be determined.
High conservativeness of G123 among different species and functional consequence of G123R mutation in the IDH1 gene
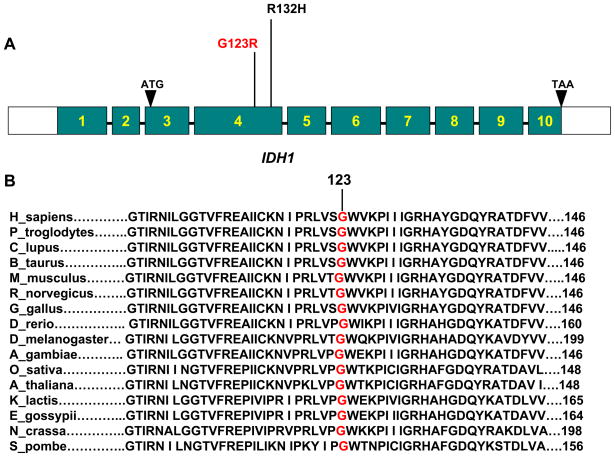
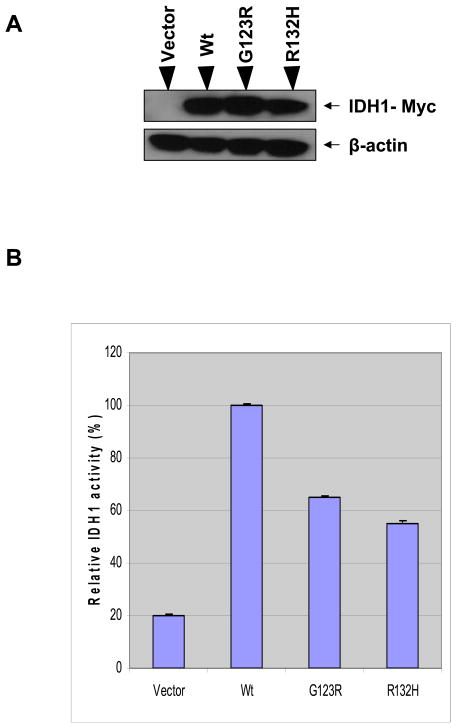
Alignment comparison of amino acid sequences in IDH1 proteins from different species revealed that the novel G123R mutation found in this study occurred at the highly conserved amino acid residue G123 of IDH1 protein that was evolutionarily conserved among sixteen different species analyzed (Fig 2). Previous studies have demonstrated that expression of IDH1 mutants disturb the function of endogenous IDH1 through a negative dominant mechanism [3, 9, 10]. As the G123R represents the change from an electrically neutral amino acid to a positively charged amino acid, the function of IDH1 is likely affected. The fact that this change occurred at an amino acid that was otherwise widely conserved among different species also raised a strong possibility that this novel G123R IDH1 mutation might have a serious functional consequence. We therefore next examined the function of the G123R mutation. To this end, we generated the G123R IDH1 mutant by site-directed in vitro mutagenesis. We similarly generated the previously characterized R132H IDH1 mutant as a positive control as this mutant, highly prevalent in glioblostoma, was known to reduce the enzymatic activity of the IDH1 enzyme. Our constructed mammalian expression vectors included Myc-tag-fused IDH1 (wild-type) and Myc-tag-fused IDH1 G123R and R132H. These vectors, as well as the empty control vector, were transiently transfected in the HEK 293T human embryonic kidney cell line. The supernatants from these transfectants were measured for the enzymatic activity of IDH1 (reduction of NADP+ to NADPH) of IDH1. Fig 3 shows that exogenous expression of the wild-type IDH1 in HEK 293T cells significantly increased the production of NADPH, whereas, in comparison with the wild-type of IDH1, reduced IDH1 activities were observed in cells that were transfected with the novel IDH1 mutant G123R. Similarly and as expected, the control IDH1 mutant R132H was also associated with reduced IDH1 activities.
Figure 2. Schematic diagram and amino-acid sequence alignment of IDH1.
A. Schematic diagram of the IDH1 gene showing the relative positions of the new IDH1 mutation G123R and the previously characterized mutation R132H in exon 4. The IDH1 gene is located on chromosome 2q33.3 and contains 10 exons with intervening sequences. B. Amino acid sequence alignment of IDH1 from 16 species showing that G123 is a highly and widely conserved residue among different species. Numbers indicate amino acid or codon positions. Amino-acid sequences are numbered with the initiation codon (methionine) of each protein as No 1.
Figure 3. Reduced enzymatic activities of the IDH1 protein with mutations.
Cell lysates were prepared from HEK293T cells after transfection with equal amounts of control vector and wild-type or mutant IDH1 (G123R and R132H). A. Shown is the protein expression of wild-type and IDH1 mutants as determined by western blotting with an anti-myc antibody and the same blot was stripped off and re-probed with anti-β-tubulin antibody as the loading control. B. Shown is the enzymatic activities of the IDH1 proteins as examined by the production of NADPH. The relative IDH1 enzymatic activity is shown as a ratio to wild-type (Wt, 100). Results obtained for the enzymatic activities are mean ± S.D. of four assay measurements, representing two independent experiments.
Discussion
We report here the identification of a novel and functionally relevant IDH1 mutation, G123R, in ATC and a previously described IDH1 mutation, V71I, in FTC and ATC, with a relatively high overall prevalence (11.11%) of IDH1 mutations in ATC. We did not find mutation in the previously well described hot spots of codon 132 of the IDH1 and codon 172 of the IDH2 genes, suggesting that thyroid cancers are different than brain tumors. Several old and new SNPs were also found in the IDH1 gene, whose function and significance remain to be defined.
The IDH1 mutations have been shown to occur with a high frequency of 12–74% in glioblastoma of various types [1, 3]. The incidence of IDH1 mutations was high particularly in secondary glioblastomas while the mutation of IDH1 was shown to be a relatively rare event in primary glioblostomas [10]. The IDH1 mutation also occurred preferentially in high-grade gliomas in a tissue-specific pattern [4]. The IDH1 mutation was only rarely reported in other human cancers, including acute myeloid leukemia [6], prostate carcinomas, and B-acute lymphoblastic leukemia [7]. Therefore, thyroid cancer, particularly ATC, represents another human cancer that can sometimes harbor IDH1 mutations with a relatively high prevalence.
It is interesting that nearly all IDH1 mutations identified to date caused a single amino acid substitution at Arg 132. Our finding of a point mutation at codon 123 of the IDH1 gene is therefore an interesting exception. As this was a homozygous mutation, the IDH1 enzyme activities in the ATC tumor harboring this mutation were likely seriously affected. No IDH1 mutation was found in MTC and PTC in a previous study [4], suggesting that the IDH1 mutation was unique to FTC and ATC, particularly the latter. The mechanism of IDH1 mutation in FTC and ATC tumorigenesis remains to be defined. As it is conventionally believed that ATC can derive from FTC, it remains an interesting question whether IDH1 mutations play any role in the progression from FTC to ATC.
The novel G123R mutation is located in the catalytic domain of IDH1. More specifically, it is localized in a region between the two substrate binding sites or residues (codon 104) and (codon 132) of IDH1 [8]. Therefore, the change from an electrically neutral amino acid to a positively charged amino acid caused by this mutation in this region can be expected to alter the function of IDH1. Indeed, the G123R IHD1 mutant, like the previously well characterized R132H mutant, showed impaired enzymatic activities in comparison with the wild-type IDH1. It has recently been shown that tumor-derived IDH1 mutations impair the enzyme’s affinity for its substrate and dominantly inhibit wild-type IDH1 activities through the formation of catalytically inactive heterodimers, and structural modeling predicted that the R132H mutation would weaken the hydrogen bonding of the IDH1 enzyme [9]. It remains to be elucidated whether this is the mechanism that the novel G123R IDH1 mutation affects the enzymatic activities of the IDH1 protein. As the V71I mutation of IDH1 found in thyroid cancers in the present study did not cause a change in the electrical change of the amino acids, the functional relevance of this IDH1 mutation is not predicatively straightforward.
We have demonstrated relatively common IDH1 mutations in thyroid cancer, particularly ATC, including a novel mutation G123R that impaired the enzymatic activities of the IDH1 protein as functional consequence. This study represents the first report of IDH1 mutations in thyroid cancer, although their specific tumorigenic and pathogenic roles in thyroid cancer remain to be defined. The IDH1 pathway may represent a potentially novel therapeutic target for thyroid cancer, particularly ATC that harbors IDH1 mutations most commonly.
Acknowledgments
We thank Drs. N. E. Heldin, K. B. Ain, N. Onoda, M. Santoro, D.Wynford-Thomas, G. Brabant, A. P. Dackiw, G. J. Juillard, R. E. Schweppe, and B. R. Haugen for kindly providing us or facilitating the accessibility to the cell lines used in this study. This work was supported by National Institutes of Health Grant R01CA134225-01 (to M.X.).
Footnotes
Disclosure Statement
The authors have no conflict of interest to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, Hong SM, Fu B, Lin MT, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, von Deimling A. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 2008;116:597–602. doi: 10.1007/s00401-008-0455-2. [DOI] [PubMed] [Google Scholar]
- 3.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, Reardon D, Herndon J, Kinzler KW, Velculescu VE, Vogelstein B, Bigner DD. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bleeker FE, Lamba S, Leenstra S, Troost D, Hulsebos T, Vandertop WP, Frattini M, Molinari F, Knowles M, Cerrato A, Rodolfo M, Scarpa A, Felicioni L, Buttitta F, Malatesta S, Marchetti A, Bardelli A. IDH1 mutations at residue p.R132 (IDH1(R132)) occur frequently in high-grade gliomas but not in other solid tumors. Hum Mutat. 2009;30:7–11. doi: 10.1002/humu.20937. [DOI] [PubMed] [Google Scholar]
- 5.Geisbrecht BV, Gould SJ. The human PICD gene encodes a cytoplasmic and peroxisomal NADP (+)-dependent isocitrate dehydrogenase. J Biol Chem. 1999;274:30527–30533. doi: 10.1074/jbc.274.43.30527. [DOI] [PubMed] [Google Scholar]
- 6.Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, Fulton LA, Locke DP, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Eng J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang MR, Kim MS, Oh JE, Kim YR, Song SY, Seo SI, Lee JY, Yoo NJ, Lee SH. Mutational analysis of IDH1 codon 132 in glioblastomas and other common cancers. Int J Cancer. 2009;125:353–355. doi: 10.1002/ijc.24379. [DOI] [PubMed] [Google Scholar]
- 8.Xu X, Zhao J, Xu Z, Peng B, Huang Q, Arnold E, Ding J. Structures of human cytosolic NADP-dependent isocitrate dehydrogenase reveal a novel self regulatory mechanism of activity. J Biol Chem. 2004;279:33946–33957. doi: 10.1074/jbc.M404298200. [DOI] [PubMed] [Google Scholar]
- 9.Zhao S, Lin Y, Xu W, Jiang W, Zha Z, Wang P, Yu W, Li Z, Gong L, Peng Y, Ding J, Lei Q, Guan KL, Xiong Y. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF1alpha. Science. 2009;324:261–265. doi: 10.1126/science.1170944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ichimura K, Pearson DM, Kocialkowski S, Bäcklund LM, Chan R, Jones DT, Collins VP. IDH1 mutations are present in the majority of common adult gliomas but rare in primary glioblastomas. Neuro Oncolol. 2009;11:341–347. doi: 10.1215/15228517-2009-025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neff RL, Farrar WB, Kloos RT, Burman KD. Anaplastic thyroid cancer. Endocrinol Metab Clin North Am. 2008;37:525–538. doi: 10.1016/j.ecl.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 12.Murugan AK, Dong J, Xie J, Xing M. MEK1 mutations, but not ERK2 mutations, occur in melanomas and colon carcinomas, but none in thyroid carcinomas. Cell Cycle. 2009;8:2122–2124. doi: 10.4161/cc.8.13.8710. [DOI] [PubMed] [Google Scholar]
- 13.Murugan AK, Hong NT, Fukui Y, Munirajan AK, Tsuchida N. Oncogenic mutations of the PIK3CA gene in head and neck squamous cell carcinomas. Int J Oncol. 2008;32:101–111. [PubMed] [Google Scholar]
- 14.Steen IH, Lien T, Birkeland NK. Biochemical and phylogenetic characterization of isocitrate dehydrogenase from a hyperthermophilic archaeon, Archaeoglobus fulgidus. Arch Microbiol. 1997;168:412–420. doi: 10.1007/s002030050516. [DOI] [PubMed] [Google Scholar]
- 15.Jennings GT, Minard KI, McAlister-Henn L. Expression and mutagenesis of mammalian cytosolic NADP+-specific isocitrate dehydrogenase. Biochemistry. 1997;36:13743–13747. doi: 10.1021/bi970916r. [DOI] [PubMed] [Google Scholar]