Abstract
Patients on multiple medications are at increased risk for adverse drug events. While physicians can reduce this risk by regularly reviewing the side-effect profiles of their patients’ medications, this process can be time-consuming. We created a decision support system designed to expedite reviewing potential adverse reactions through information visualization. The system includes a database containing 16,340 unique drug and side-effect pairs, representing 250 common medications. A numeric score is assigned to each pair reflecting the strength of association between drug and effect. Based on these scores, the system generates graphical adverse reaction maps for any user-selected combination of drugs. A study comparing speed and accuracy of retrieving side-effect data using this tool versus UpToDate® demonstrated a 60% reduction in time to complete a query (61 seconds vs. 155 seconds, p<0.0001) with no decrease in accuracy. These findings suggest that information visualization can significantly expedite review of potential adverse drug events.
Keywords: adverse drug events, side-effects, information visualization, polypharmacy
Introduction
Adverse drug events are a well-recognized cause of patient morbidity and increased health care costs in the United States [1,2]. Patients taking multiple medications are at increased risk for such events [3,4]. This risk can be reduced, however, if physicians regularly review the adverse reaction profiles of their patients’ medications [5,6]. As the number of drugs increases, however, so too does the time required to review all of the potential side-effects [7,8].
Multiple studies have shown that the expected time to complete a query is a major factor in physicians’ failure to pursue answers to clinical questions [9–11]. Even when information resources are readily available, clinicians may forgo searches that are expected to require more than two minutes to complete [12]. Some have even suggested that in the busy hospital setting this time is closer to 30 seconds [13]. Thus, patients on complex drug regimens are doubly disadvantaged. They are at increased risk for an adverse event, yet their physicians may be less likely to perform a thorough review of their medications due to the lengthy time required. As the number of medications grows, this inverse relationship is further exacerbated. Clearly a need exists to reduce the time and effort required of physicians to review the adverse reaction profiles of their patients’ drugs. Ideally, the physician effort-curve would be flat, where increasing the number of drugs would not significantly affect the time required to complete a medication review. We are unaware of any solutions to date that have achieved this goal.
Current drug information resources have several limitations when it comes to retrieving adverse reaction data. First, they are designed primarily for the sequential lookup of medications, where only one drug can be reviewed at a time. Secondly, the presentation of the results is often complex, requiring scanning and searching of text and tables to find items of importance [14]. While some applications, such as Epocrates®, have condensed side-effect results to include only the most serious and common effects, this convenience comes at the expense of losing important details regarding the frequency of specific reactions. Finally, once all the information about each drug has been retrieved and interpreted, physicians must rely on their memory (or scribbled notes) to organize the collective findings into a coherent answer to their original question. Whether searching for a particular reaction or just reviewing the most common effects of a drug regimen, the doctor must iterate through these time-consuming steps before making an informed clinical decision.
In this paper, we introduce Rxplore, a decision support system specifically tailored to the evaluation of adverse reactions in patients on multiple medications. Rxplore aims to address the above limitations and reduce the time and effort required to review complex drug regimens. We describe the system’s goals, key components, and development process. We then present a study comparing its speed and accuracy to that of a more traditional electronic drug information resource. We conclude with a discussion of the study’s implications and the applicability of the Rxplore model to other clinical domains.
Methods
Rxplore Development
Our primary goals for the system were to 1) allow the simultaneous lookup of adverse reaction profiles for multiple medications; 2) provide results in an easily interpretable fashion without sacrificing detail; and 3) reduce demands on physician memory by presenting data in a consolidated format.
Multiple medication lookup is not a new concept. It has been employed in numerous online applications (e.g., Epocrates®, Medscape®) for the purpose of identifying drug-drug interactions. Users enter a list of medications, and the system returns any pairs of drugs with known interactions. Creating such systems is straightforward because the data model for drug interactions is simple: a one-to-one relationship between two medications causes a particular effect.
In contrast, adverse reactions present a more complex data model for several reasons. First, a single drug can cause hundreds of effects at varying frequencies and levels of severity. Secondly, similar effects may be described using different terms (e.g., sedation, somnolence, sleepiness). Thirdly, the data themselves may come from numerous sources, including clinical trials and post-marketing reports. Finally, descriptions of adverse reaction frequency may be quantitative (“occurs in 12% of patients”), semi-quantitative (“occurs frequently”), or qualitative (“was reported”). These highly variable and non-specific descriptions result in a complex set of information for even a single drug, much less an entire regimen. Thus, simultaneously retrieving adverse reactions for multiple medications offers no intrinsic value unless the complexity of the data is also addressed.
Creation of a Standardized Adverse Reaction Knowledge Base
In developing Rxplore, we sought to harmonize the side-effect data model. We first selected 250 commonly used medications, based on national and local dispensing patterns. We then retrieved the Food and Drug Administration-approved Structured Product Labels (SPL’s) for these medications from the DailyMed website [15]. For each SPL, we extracted adverse event data using a combination of manual and natural language processing techniques.
We began by isolating the text found in the "Adverse Reactions" section of each SPL using XML section identifiers (specifically, code "34084-4" of the LOINC code system). We then manually divided this section into tables and paragraphs. Tables were converted to tab-delimited format and column order was adjusted so that placebo data was consistently located in the final column. These tables were then processed using regular expressions to extract side-effect name, drug frequency, and placebo frequency. Each side-effect was then mapped to a corresponding preferred term in the Medical Dictionary of Regulatory Activities (MedDRA) Version 11.1. This use of preferred terms provided consistency of adverse event naming across multiple labels, so for example the terms “hyponatremia,” “decreased serum sodium,” and “blood sodium decreased” would be all be represented by the preferred term “hyponatremia.” The majority of these mappings were accomplished by exact term matching or by stemming of terms. Manual mapping was required in approximately 15% of cases.
Data in paragraph format required additional manual curation. Paragraphs were first grouped as either organ system-based side-effect lists or unspecified side-effect lists. Those arranged by organ system followed a generally consistent format, displaying a category name followed by frequent, infrequent, and rare events. Based on this pattern, we used regular expressions to parse these paragraphs into side-effect terms and corresponding frequencies. Unspecified lists were more challenging in that they comprised a wide variety of formats. For example, one paragraph began, "The following events occurred in >2% of patients. Events shown in italics occurred in >5% of patients." In this case, we had to separate normal from italicized text and manually assign the appropriate frequency values. Given such idiosyncratic formats, careful label review and data entry was essential. This manual effort resulted in a lengthy extraction process, with an average of 1.25 hours per label.
Development of a Frequency Scoring System
In order to standardize the extracted frequencies from multiple labels, we devised a scoring system by which the relationship between each drug and associated side-effect could be represented using a single numeric value. This value was known as the Rxplore Score and was calculated using algorithms developed by the authors and based upon expert consensus and review of term use across multiple labels (examples shown in Table 1). The algorithms covered a broad range of frequency reporting formats, ranging from precise drug versus placebo incidence rates to vague phrases such as "may sometimes occur." In total, 39 different algorithms were required for the describe all the frequency relationships found in our set of 250 labels. Our algorithms required iterative modification as new labels were introduced. Thus, the original text-based frequency descriptors (e.g., "occurred rarely") were also retained in the database so that scores could be recalculated in the event of an algorithm modification.
Table 1.
Example algorithms for Rxplore Scores
| Frequency Descriptor | Algorithm | Algorithm |
|---|---|---|
| Seen in 27% of patients on drug and 12% on placebo |
Drug % - Placebo % |
27 − 12 = 15 |
| Occurred in 3% (Low) to 9% (High) of patients |
Low + (High−Low)/2 |
3+(9-6) / 2 = 6 |
| Occurred “rarely” | Expert consensus |
0.3 |
A common challenge in implementing our scoring system was the presence of reactions which were listed multiple times in a single label. For example, the label of a selective serotonin reuptake inhibitor might include clinical trials for both depression and anxiety, but report very different rates of nausea in one population than the other. Alternatively, a given side-effect might occur in a high percentage of patients during a controlled trial but be reported as "rare" in post-marketing reports. To ensure consistency in our system, we used the following preferences to prioritize which report type would serve as the source for the Rxplore Score calculation: placebo-controlled clinical trial over post-marketing report; larger trial over smaller trial; longer trial over shorter trial; and trial for primary indication over trial for secondary indication.
Upon completion of the SPL extraction and standardization process, the knowledgebase contained information on 16,340 distinct medication and side-effect pairs. See Table 2 for a summary of the knowledgebase development process.
Table 2.
Creation of the Quantitative Adverse Reaction Knowledgebase
| Step | Process Details | Example |
|---|---|---|
| Medication Selection | Identified 250 commonly prescribed medications based on national and local pharmacy dispensing data |
Lisinopril |
| SPL Extraction | Retrieved Structure Product Label for each drug. Extracted side-effect and frequency data |
“Taste disturbances occurred rarely in patients on Lisinopril” |
| Data Standardization | Standardized side-effects to MedDRA preferred terms. Standardized frequency data to a simplified numeric value (Rxplore Score). |
“Taste Disturbance” → “Dysgeusia” “Rarely” → 0.3 |
| Storage in Knowledgebase | Stored drug name, coded effect, and Score for 16,340 drug-effect pairs |
Lisinopril | Dysgeusia | 0.3 |
Design of the Visualization Tool
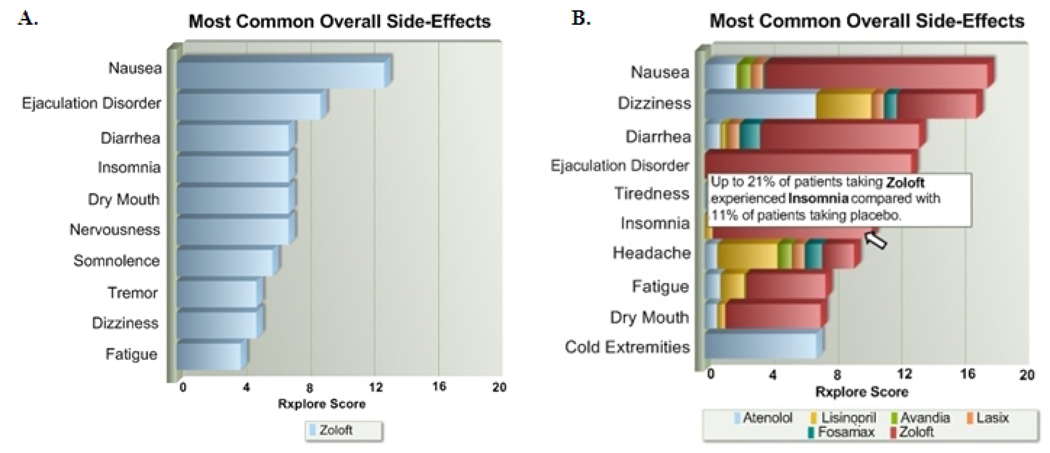
The Rxplore Score, though simplistic in nature, facilitated the development of a visual interface for representing adverse reaction data at both an overview and detailed level. Our browser-based application allows the user to enter a list of patient medications. Upon entry of the first medication, the system displays a horizontal bar graph showing the drug’s most common side effects; bar lengths are proportional to the calculated Rxplore Score for each effect (Figure 1a). With each new medication the graph is recalculated to display the most common side-effects of the collective regimen as determined by the summation of Rxplore Scores (Figure 1b). To see more detailed adverse reaction data for a particular drug and side-effect combination, the user moves the mouse to the appropriate location on the graph (Figure 1b). The results can be filtered in several ways: by organ system, by therapeutic class, or by direct search for a specific side-effect of interest. Each medication also contains a link to the original FDA label as maintained on DailyMed [15].
Fig. 1.
Results display in Rxplore. A. The most common reactions seen with Zoloft alone. B. The most common “combined” effects of a 6 drug regimen.
Evaluation Study
We conducted a pilot study comparing the speed and accuracy of Rxplore in retrieving adverse reaction data to that of UpToDate®, a popular clinical information resource. The subjects were 24 physicians in residency training in internal medicine or family practice at Indiana University. All subjects had prior experience using UpToDate.
Subjects were given a 3-minute orientation to both Rxplore and UpToDate. For each system, the retrieval of adverse reaction data for two drugs was demonstrated. Following this orientation, the subjects were presented with two sample clinical tasks consisting of a patient description, a medication list, and 4 hypothetical adverse reactions (Table 3). For each reaction, the subjects were asked one of the following: which of the patient’s drugs are known to cause this reaction; or which one drug is most likely to cause this reaction. To answer these questions, the subjects were randomly assigned to use either Rxplore or UpToDate. They were allowed to use any method of searching or filtering for information provided by these sites. The resource was switched between tasks so that all subjects answered questions using both systems. The order of tasks and order of questions within each task were randomized.
Table 3.
Clinical scenarios used in Rxplore Study
| Patient | 70 year-old male with hypertension, congestive heart failure, and hyperlipidemia |
| Medications | atorvastatin, amlodipine, atenolol, furosemide, fluoxetine |
| Adverse Reactions |
alopecia, tremor, flushing, erythema multiforme |
| Patient | 64 year-old female with diabetes, GERD, osteoporosis, and mild cognitive impairment |
| Medications | rosiglitazone, glimepiride, alendronate, donepezil, pantoprazole |
| Adverse Reactions |
pruritus, hyponatremia, flatulence, photosensitivity |
For each question, the subject’s answer and time to completion were recorded. Data on task order, question order, and resource order were also stored. The authors verified that answers to all questions were available and identical in both resources. After finishing the tasks, subjects completed an online survey regarding their experience using Rxplore. The survey consisted of six questions generated by the authors addressing the clinical utility and speed of Rxplore as well as overall user satisfaction.
Our null hypothesis was that there would be no difference in the average task-wise time between the two software tools, and our alternative hypothesis was that Rxplore would entail less time to complete a task than UpToDate. Study subjects were blinded to this hypothesis and were unaware that measurements of time and accuracy were being recorded. In order to account for intra-subject correlation (i.e., the relationship between an individual subject’s performance on each of the two tasks), we analyzed time-to-answer using mixed models and we used generalized estimating equations to evaluate answer accuracy.
Results
Accuracy of Answers Independent of Time
Physicians using Rxplore answered 78% (95% CI [65%, 87%]) questions correctly, compared with 70% (95% CI [57%, 80%]) questions correct by those using UpToDate (p=0.27). The order of the clinical tasks and order of individual questions within each task did not affect accuracy.
Time to Answer
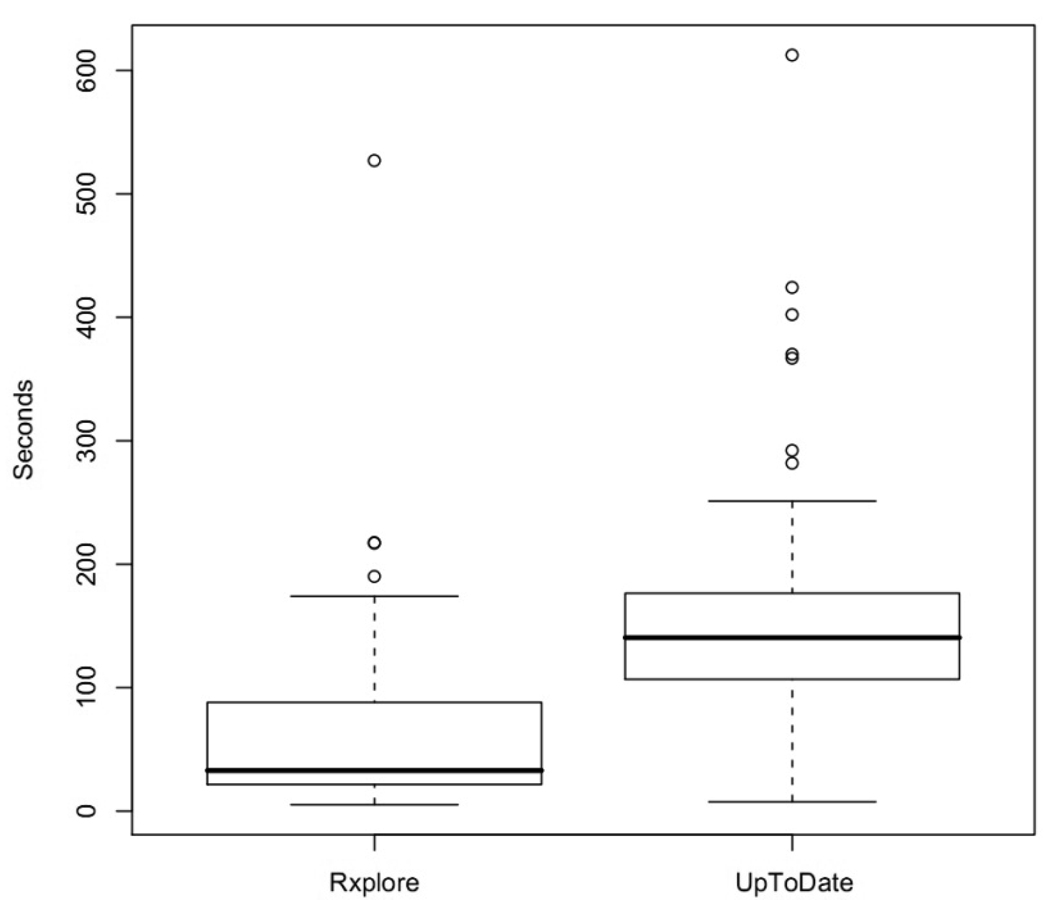
When using Rxplore, clinicians answered questions in significantly less time than with UpToDate. The mean time per question regardless of accuracy was 61 seconds for Rxplore (95% CI [46, 77]) versus 155 seconds (95% CI[138, 171]) (p<0.0001) for UpToDate (Figure 2). The mean time to complete a four-question task using Rxplore was 239 seconds (95% CI [174,304]) compared with 625 seconds (95% CI [553,698]) using UpToDate (p<0.001).
Fig. 2.
Average time to assess a 5 drug regimen for possible causes of an adverse reaction
Correct answers were also obtained in significantly less time using Rxplore. The average time to a correct answer was 62 seconds (95% CI [46,79]) for Rxplore and 160 seconds (95% CI [142,179]) for UpToDate (p<0.0001).
As evident in Figure 2, outliers requiring 200 seconds or more to answer a question were found in both groups. To assess the impact of these outliers, we repeated answer time calculations using median rather than mean times. Statistical trends remained consistent. The median time per question regardless of accuracy was 39 seconds (95% CI [31, 49]) for Rxplore versus 142 seconds (95% CI [122, 163]) for UpToDate. Correct answers were reached in a median time of 37 seconds (95% CI [29, 47]) with Rxplore compared with 159 seconds (95% CI [137, 206]) for UpToDate. Median four-question task time was 228 seconds for Rxplore (95% CI [193, 262]) versus with 602 seconds (95% CI [534,678]) for UpToDate.
Time to Answer By Question Order
For both systems, question order had a statistically significant impact on time to answer. Subjects required more time to complete the first question in a given task, regardless of the question’s content (p<0.0001 for Rxplore, p=0.003 for UpToDate). For Rxplore, finding a correct answer took an average of 145 seconds for the first question, while questions 2, 3, and 4 were answered correctly in 42, 33, and 30 seconds respectively. With UpToDate, this differential was less pronounced but still present. Correct answers took 212 seconds for question one, followed by 143, 133, and 155 seconds for the final three questions in a task.
Survey Results
23/24 subjects completed the survey. Respondents showed a very favorable impression of Rxplore, with 100% describing their overall experience as good or excellent, and 100% stating they would be likely or very likely to use Rxplore again. All respondents rated Rxplore as faster than or much faster than their current drug information resource.
Discussion
In this paper, we described the development of a decision support system for the evaluation of adverse reactions in patients on multiple medications. Use of this system resulted in a 60% reduction in the time required to research medication side-effects when compared with a traditional drug information resource. This increased speed was achieved with no decrease in accuracy. Furthermore, physicians rated the application very highly in terms of usability and overall satisfaction.
Reducing adverse drug events is a national mandate, yet physicians face increasing time pressures that limit their ability to perform thorough medication reviews [1,11]. Thus, technological innovations are necessary to maximize patient safety while maintaining physician efficiency. Rxplore, which significantly decreases the time required to review adverse drug reactions, may be an example of such innovation, and it is worth considering which particular design elements were most responsible for its success.
Key System Design Features
Simultaneous lookup of multiple medications is an essential component of Rxplore. As discussed earlier, multi-drug lookup has been used in numerous applications for the identification of drug-drug interactions. Yet it had not previously been extended to other types of drug information. In this study, we demonstrated its effectiveness in evaluating adverse reactions. We believe it could also be successfully applied to drug contraindications, pregnancy safety categories, formulary status, and other commonly cross-referenced information. The volume of data produced by multiple medication lookup can be significant, however, and effective filters are essential. In Rxplore, we provide the user with a means to view side-effects by overall frequency, by organ system, or by specific name. These filters may prevent the user from being overwhelmed by the data and facilitate rapid retrieval of the effects of interest.
Another principle design feature of Rxplore is the use of information visualization to present the query results. Studies have shown that visualization can reduce physician cognitive load and support the extraction of useful information from complex medical data [14,16,17]. By shifting workload from the cognitive to perceptual system, the subject is able to interact with the data at a higher level, expending less effort on remembering discrete bits of information than on interpreting their meaning [18]. In Rxplore, all results are shown on a single page, meaning that the user does not need to write down or remember information in order to make comparisons between drugs. Furthermore, by using standard bar graphs and tooltips for detailed data, we have created visualizations that are intuitive to use and require no prior training.
In sum, the increased efficiency seen with Rxplore is likely attributable to a combination of simultaneous multi-data retrieval, rapid filtering, and intuitive visual results. These elements, when taken together, form a process which may offer benefit in other clinical information tasks, such as review of laboratory results, selection of diagnostic tests, and narrowing of differential diagnoses. The common factor in these cognitive processes is that they require the physician to consider and compare multiple data points, retain the acquired knowledge for at least several minutes, and make a decision once all elements have been reviewed. They are thus susceptible to the inefficiencies of looking up multiple items and to gaps in physician concentration and working memory. Our results with Rxplore suggest that these limitations can be ameliorated by 1) initial retrieval of a large dataset containing all or most of the elements to be reviewed (e.g. all patient laboratory tests); 2) filtering the data by a user-selected dimension of interest (e.g. test frequency); 3) visualization showing the relationship among the multiple elements according to the stated parameter (e.g., word cloud of patient labs sized by frequency). A prerequisite to these steps is data model standardization. In the example of a laboratory results viewer, all test names and units would need to be harmonized in advance (preferably using a terminology such as LOINC) in order for the system to deliver data in a consolidated form. While of course each application requires its own customization and choice of data model, this three-step process, if applied correctly, has the potential to yield efficiency gains.
Speed Variability based on Question Order
A notable finding of our study was the significant difference between time to answer the first question in a patient scenario and time to answer subsequent questions. This pattern was seen with both Rxplore and UpToDate, and may in part be explained by increasing familiarity with these systems over the course of answering four questions. But with Rxplore, a likely additional factor is that users must first type in each of the patients’ medications in order to answer the initial question in a task. But for follow-up questions on the same patient, they can simply filter the existing results. With answer times averaging over 100 seconds faster for the latter questions, we suspect that medication entry is a significant bottleneck in Rxplore. Thus, Rxplore’s speed gains may be improved even further through automatic pre-population of the medication list from an electronic medical record via web service or standards-based CDS integration approach such as InfoButtons [19]. This step would eliminate the need for manual entry of drugs. More importantly, it would potentially allow physicians to answer any adverse reaction query—regardless of number of drugs involved—in less than forty seconds. We would thus achieve our goal of bringing the time required to perform a medication review down to a level acceptable to physicians and practicable in busy clinical settings.
Limitations
Our system does have a few limitations. The Rxplore algorithms were designed to apply a consistent metric across a wide variety of qualitative and quantitative data, and are thus not precise measures of side-effect frequency. Validation studies are currently underway to assess and refine these algorithms through comparison of Rxplore-predicted frequencies with those reported in large pharmacosurveillance datasets such as MedWatch. Such validation efforts are necessarily limited, however, by the lack of a true "gold standard" in defining the incidence rates of adverse drug events. Nonetheless, we expect that iterative improvement of algorithms, particularly with respect to the synergistic or interacting effects of multiple drugs, will enhance both the efficiency of our system and physicians' confidence in its use.
A second issue is the difficulty of building and maintaining a comprehensive adverse reaction database based on FDA labels. A successful drug information system requires data on a broad array of commonly used medications and these data must be updated regularly. Our method of extracting adverse events, incorporating both manual and automated processes, was time-consuming and unlikely to scale well. Available vendor-based drug compendia, while comprehensive, do not contain frequency data at the level of granularity required by our system. Thus, to successfully implement Rxplore in the clinical setting we must create a more fully automated mechanism of extracting side-effect information from the Structured Product Label (SPL). Challenges to developing such a system include the SPL's lack of standard terminology for describing adverse events (e.g., MedDRA, SNOMED-CT) and inconsistencies in its tabular and text formats. Despite these challenges, we are currently underway in building such a system and creating a full-scale implementation of Rxplore.
In regards to our evaluation study, it was performed in a controlled environment and therefore may not account for the unpredictable nature of the clinical setting. Furthermore, our study used sample clinical scenarios designed by the authors and limited in scope to those drugs currently available in Rxplore. While we expect the application's efficiency to be maintained across a broad range of medications, it is possible that selecting a different combination of medications for our tasks would have yielded different results. Additionally, we chose UpToDate® as our comparator based on its popularity among practicing physicians and the level of detail provided by its drug monographs; however, other systems such as ePocrates® or DailyMed® might have yielded different results. Finally, our study’s endpoints were speed and accuracy of answers, not of medical decision-making. Thus, further studies are necessary to determine Rxplore’s potential impact on patient outcomes and adverse drug events.
Conclusion
The combination of multi-drug lookup and data visualization can reduce the time required to review potential adverse reactions. At a broader level, this process of high-volume information retrieval, filtering, and visualization may extend beyond medications to other domains of clinical decision support. To successfully apply this technique, however, the underlying data must be properly structured and standardized so that concise visualizations are possible.
Our future work includes validation and refinement of Rxplore’s scoring algorithms, expanded task analysis and assessment of alternate visualization schemes, integration into a computerized order entry system, and evaluation of the application’s impact on adverse drug events in the clinical setting.
Acknowledgements
This work was performed at the Regenstrief Institute and supported in part by grant T15 LM07117 from the National Library of Medicine. Our gratitude goes to Dr. Lisa Hofler for her work in the early stages of this research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bates DW, Cullen DJ, Laird N, Petersen LA, Small SD, et al. Incidence of adverse drug events and potential adverse drug events. Implications for prevention. ADE Prevention Study Group. JAMA. 1995 Jul 5;274(1):29–34. [PubMed] [Google Scholar]
- 2.Ernst FR, Grizzle AJ. Drug-related morbidity and mortality: updating the cost-of-illness model. J Am Pharm Assoc (Wash) 41(2):192–199. doi: 10.1016/s1086-5802(16)31229-3. [DOI] [PubMed] [Google Scholar]
- 3.Hajjar ER, Cafiero AC, Hanlon JT. Polypharmacy in elderly patients. Am J Geriatr Pharm. 2007;5(4):345–351. doi: 10.1016/j.amjopharm.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 4.Nguyen JK, Fouts MM, Kotabe SE, Lo E. Polypharmacy as a risk factor for adverse drug reactions in geriatric nursing home residents. Am J Geriatr Pharm. 2006;4(1):36–41. doi: 10.1016/j.amjopharm.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 5.Zarowitz BJ, Stebelsky LA, Muma BK, Romain TM, Peterson EL. Reduction of high-risk polypharmacy drug combinations in patients in a managed care setting. Pharmacotherapy. 2005 Nov;25(11):1636–1645. doi: 10.1592/phco.2005.25.11.1636. [DOI] [PubMed] [Google Scholar]
- 6.Ballentine NH. Polypharmacy in the elderly: maximizing benefit, minimizing harm. Crit Care Nurs Q. 31(1):40–45. doi: 10.1097/01.CNQ.0000306395.86905.8b. [DOI] [PubMed] [Google Scholar]
- 7.Shelton PS, Fritsch MA, Scott MA. Assessing medication appropriateness in the elderly: a review of available measures. Drugs Aging. 2000;16(6):437–450. doi: 10.2165/00002512-200016060-00004. [DOI] [PubMed] [Google Scholar]
- 8.Hanlon JT, Schmader KE, Samsa GP, et al. A method for assessing drug therapy appropriateness. J Clin Epidemiol. 1992;45(10):1045–1051. doi: 10.1016/0895-4356(92)90144-c. [DOI] [PubMed] [Google Scholar]
- 9.Ely JW, Osheroff JA, Chambliss ML, Ebell MH, Rosenbaum ME. Answering Physicians' Clinical Questions: Obstacles and Potential Solutions. J Am Med Inform Assoc. 2005 Apr;12(2):217–224. doi: 10.1197/jamia.M1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D'Alessandro DM, Kreiter CD, Peterson MW. An evaluation of information-seeking behaviors of general pediatricians. Pediatrics. 2004;113 (1 Pt 1):64–69. doi: 10.1542/peds.113.1.64. [DOI] [PubMed] [Google Scholar]
- 11.Gorman PN, Helfand M. Information seeking in primary care: how physicians choose which clinical questions to pursue and which to leave unanswered. Med Decis Making. 15(2):113–119. doi: 10.1177/0272989X9501500203. [DOI] [PubMed] [Google Scholar]
- 12.Ramos K, Linscheid R, Schafer S. Real-time information-seeking behavior of residency physicians. Fam Med. 2003;35(4):257–260. [PubMed] [Google Scholar]
- 13.Sackett DL, Straus SE. Finding and applying evidence during clinical rounds: the "evidence cart". JAMA. 1998;280(15):1336–1338. doi: 10.1001/jama.280.15.1336. [DOI] [PubMed] [Google Scholar]
- 14.Lamy J, Venot A, Bar-Hen A, Ouvrard P, Duclos C. Design of a graphical and interactive interface for facilitating access to drug contraindications, cautions for use, interactions and adverse effects. BMC Medical Informatics and Decision Making. 2008;8(1):21. doi: 10.1186/1472-6947-8-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. [Accessed February 20, 2009];DailyMed: About DailyMed. Available at: http://dailymed.nlm.nih.gov/dailymed/about.cfm.
- 16.Workman M, Lesser MF, Kim J. An exploratory study of cognitive load in diagnosing patient conditions. Int J Qual Health Care. 2007;19(3):127–133. doi: 10.1093/intqhc/mzm007. [DOI] [PubMed] [Google Scholar]
- 17.Wenkebach U, Pollwein B, Finsterer U. Visualization of large datasets in intensive care. Proc Annu Symp Comput Appl Med Care. 1992:18–22. [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas JJ, Cook KA. Illuminating the Path. 2005:200. [Google Scholar]
- 19.Cimino JJ. Infobuttons: anticipatory passive decision support. AMIA Annu Symp Proc. 2008:1203–1204. [PubMed] [Google Scholar]