Abstract
Cajal bodies (CBs) are subnuclear domains that participate in spliceosomal small nuclear ribonucleoprotein (snRNP) biogenesis and play a part in the assembly of the spliceosomal complex. The CB marker protein, coilin, interacts with survival of motor neuron (SMN) and Sm proteins. Several coilin phosphoresidues have been identified by mass spectrometric analysis. Phosphorylation of coilin affects its self-interaction and localization in the nucleus. We hypothesize that coilin phosphorylation also impacts its binding to SMN and Sm proteins. In vitro binding studies with a C-terminal fragment of coilin and corresponding phosphomimics show that SMN binds preferentially to dephosphorylated analogs and that SmB′ binds preferentially to phosphomimetic constructs. Bacterially expressed full-length coilin binds more SMN and SmB′ than does the C-terminal fragment. Co-immunoprecipitation and phosphatase experiments show that SMN also binds dephosphorylated coilin in vivo. These data show that phosphorylation of coilin influences interaction with its target proteins and, thus, may be significant in managing the flow of snRNPs through the CB.
INTRODUCTION
Cajal bodies (CBs) are subnuclear domains that participate in spliceosomal small nuclear ribonucleoprotein (snRNP) biogenesis (Jady et al., 2003) and play a part in the assembly of the spliceosomal complex (Stanek et al., 2008). Canonical CBs comprise basal transcription factors, cell-cycle factors, SMN, U snRNPS, and coilin (for reviews, see (Matera, 1999; Morris, 2008)).
Human coilin is a 576-amino acid protein found enriched in CBs, but is also found in large amounts in the nucleoplasm (Lam et al., 2002). Several important regions in coilin have been mapped and a schematic is presented in Figure 1. The highly conserved N-terminus (Shpargel et al., 2003) has been shown to mediate coilin self-interaction (Hebert and Matera, 2000); nuclear and nucleolar localization signals are found between residues 111 and 200 (Bohmann et al., 1995); the so-called RG box beginning at residue 392 is critical for SMN binding (Hebert et al., 2001); Sm proteins bind near the C-terminus of coilin (Xu et al., 2005); and, the final 10 residues of the most distal end of the C-terminus are implicated in controlling availability of the N-terminus for self-interaction, CB formation (Shpargel et al., 2003), and coilin localization (Hearst et al., 2009). Throughout the cell-cycle coilin levels remain constant (Andrade et al., 1993). This pool is available for CB formation in early- to late-G1 and is presumed to be recycled as CBs disassemble during mitosis.
Fig. 1.
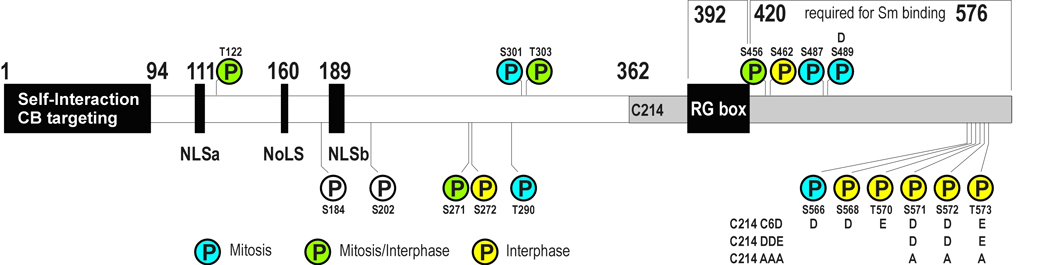
Schematic representation of the binding domains of coilin. Phosphoresidues identified by MS/MS are indicated. Residues identified from mitotic samples by MS/MS are colored blue, interphase samples are colored yellow, and residues found phosphorylated in both mitotic and interphase samples are green. S184 and S202 were identified by mutagenesis studies. Residues replaced in coilin mutants are indicated
Coilin interacts directly with SMN and Sm proteins and the regions responsible in coilin are separable (Xu et al., 2005). SMN binding is mediated by the RG box in coilin, a region rich in arginine and glycine repeats (Hebert et al., 2001). Symmetrical dimethylation of these arginine residues increases SMN/coilin interaction (Hebert et al., 2002) putatively through contact with conserved aromatic residues in the binding-pocket of the SMN Tudor domain (Sprangers et al., 2003). Sm protein and U snRNP binding require the C-terminal 156 residues of coilin (Xu et al., 2005). Whereas Sm protein binding to SMN is mediated by Sm RG-tails and is increased by their dimethylation (Brahms et al., 2001; Friesen and Dreyfuss, 2000), Sm proteins bind coilin through interaction with their Sm-fold region and binding is not affected by dimethylation (Xu et al., 2005).
Coilin is a constitutive phosphoprotein that is hyperphosphorylated during mitosis (Carmo-Fonseca et al., 1993) and it is thought that hyperphosphorylation triggers CB disassembly during cell replication. High-throughput tandem MS/MS experiments have identified 16 phosphorylated residues in coilin, five of which have been found primarily in only mitotic cell preparations and eight in only interphase samples (Fig. 1 and Table 1). Two additional residues, S184 and S202, have been identified by site-directed mutagenesis studies and demonstrate the importance of both phosphorylation and dephosphorylation to coilin localization. When serine-184 is replaced with alanine and is no longer available for phosphorylation, coilin localizes to the nucleolus and CBs in approximately 30% of transfected cells (Hebert and Matera, 2000). However, treatment with phosphatase inhibitors or mutation of an important serine to aspartate (S202D) results in snRNP and coilin localization in nucleoli (Lyon et al., 1997). Self-interaction and localization have been shown to depend on the phosphorylation state of coilin and support the hypothesis that coilin hyperphosphorylation mediates CB disassembly while hypophosphorylated coilin aids in CB assembly by increasing coilin-coilin ineteraction. GFP-coilin immunoprecipitates less endogenous coilin from mitotic lysates than from interphase lysates (Hebert and Matera, 2000). GFP-coilin in which most of the phosphoresidues are converted to alanine more strongly interacts with endogenous coilin than does a GFP-coilin with the same residues converted to aspartate or glutamate (Hearst et al., 2009); and, coilin mutants S202D, S184A, and C6A partially localize to the nucleolus in immunofluorescence microscopy studies (Hearst et al., 2009; Hebert and Matera, 2000; Lyon et al., 1997).
Table 1.
Summary of known phosphorylated residues in coilin
| Residue | Reference | Peptide | Notes |
|---|---|---|---|
| Thr 122 | (Beausoleil et al., 2006; Dephoure et al., 2008; Nousiainen et al., 2006; Olsen et al., 2006) | AFQLEEGEET*EPDCK RAFQLEEGEET*EPDCK |
Nocodozole-arrested HeLa (Beausoleil et al., 2006); Human mitotic spindle preparation (Nousiainen et al., 2006); Seen in both G- and M-phase (Dephoure et al., 2008) |
| Ser 184 | (Hebert and Matera, 2000) | S184A mutant localizes to nucleolus; S184D mutant had no effect | |
| Ser 202 | (Lyon et al., 1997; Sleeman et al., 1998) | S202D mutant localizes to nucleolus | |
| Ser 271 | (Dephoure et al., 2008) | NS*SEKLPTELSK | Seen in both G1- and M-phase (Dephoure et al., 2008) |
| Ser 272 | (Olsen et al., 2006) | VTLEARNSS*EKLPTELSK | R-X-X-pS/pT(CaMK2) |
| Thr 290 | (Dephoure et al., 2008) | NTT*ADKLAIK | M-phase (Dephoure et al., 2008) |
| Ser 301 | (Dephoure et al., 2008) | LGFS*LT*PSK | M-phase (Dephoure et al., 2008) |
| Thr 303 | (Beausoleil et al., 2004) | LGFSLT*PSK LGFS*LT*PSK |
Nuclear fraction of HeLa lysate; possible CDK1, CDK2 motifs; Seen in both G1- and M-phase (Dephoure et al., 2008) |
| Ser 456 | (Dephoure et al., 2008) | NSSTIIQNPVET*PKKDYSLLPLLAAAPQVGEK | Seen in both G1- and M-phase (Dephoure et al., 2008) |
| Ser 462 | (Dephoure et al., 2008) | NSSTIIQNPVETPKKDYS*LLPLLAAAPQVGEK | G1-phase (Dephoure et al., 2008) |
| Ser 487 | (Dephoure et al., 2008) | LLELTSS*YSPDVSDYKEGR | M-phase (Dephoure et al., 2008) |
| Ser 489 | (Beausoleil et al., 2006) | LLELTSSYS*PDVSDYK | Nocodozole-arrested HeLa |
| Ser 566 | (Nousiainen et al., 2006) | LIIES*PSNTSSTEPA | Human mitotic spindle preparation |
| Ser 568 | www.Phosida.com | ||
| Thr 570 | www.Phosida.com | ||
| Thr 571/573 | (Olsen et al., 2006) | LIIESPSNTS*ST*EPA LIIESPSNTSST*EPA |
S571: S-X-X-pS/pT(CK1);pS/pT-X-X-E(CK2) |
| Ser 572 | (Olsen et al., 2006) | LIIESPSNTSS*TEPA | S/T-X-X-X-pS(CK1); |
Knockdown of PPM1G, a Mg2+/Mn2+-dependent nuclear phosphatase, reduces dephosphorylation of SMN and abrogates SMN incorporation into CBs, but has no effect on U snRNP localization in the CB (Petri et al., 2007). Although there is not yet any evidence that coilin is modified by PPM1G in the cell, coilin is an in vitro substrate for PPM1G (Hearst et al., 2009). We hypothesized that coilin phosphorylation may also contribute to SMN/Sm protein incorporation into the CB and, therefore, that hyperphosphorylated coilin would bind Sm proteins preferentially while coilin-SMN interactions would be increased by dephosphorylation of coilin.
In this study, we investigate the implications of coilin phosphorylation on binding SMN and Sm proteins, specifically SmB′. We demonstrate that phosphorylation of a C-terminal fragment of coilin significantly affects its interaction with SMN and SmB′ in vitro and that this effect is also evident in SMN binding to full length coilin in vivo. These data demonstrate that phosphorylation is important in moderating coilin interaction with its target proteins and thus may be significant in managing the flow of snRNPs through the CB.
MATERIALS AND METHODS
Cell lines and antibodies
HeLa cells stably expressing YFP-SmB′ (Sleeman et al., 2001) were a gift from Dr. Angus Lamond (University of Dundee, Dundee, Scotland). HeLa and WI-38 cells were acquired from the American Type Culture Collection (Manassas, VA). Polyclonal antibodies: α-coilin (H300, Santa Cruz, CA), α-SMN (BD Biosciences, San Jose, CA), α-GFP (Santa Cruz Biotechnology, Santa Cruz, CA). Monoclonal antibodies: α-GST (Santa Cruz Biotechnology, Santa Cruz, CA), α-T7 (Novagen, Gibbstown, NJ), α-SMN (BD Biosciences, San Jose, CA), and α-GFP (Roche, Indianapolis, IN).
C-terminal and full-length coilin constructs
GST-C-terminal fusion protein variants were prepared by site-directed mutagenesis of GST-C214, coilin residues 362-576 (Hebert et al., 2001), using the QuikChange Mutagenesis kit (Stratagene, La Jolla, CA) and previously described mutagenesis primers (Hearst et al., 2009). GST-coilin was prepared by subcloning the His-T7-coilin cDNA from pET-28a coilin (Hebert et al., 2001). The His-coilin insert was removed from pET-28a-coilin by digestion with BamH1 and ligated into BamH1 restriction site in the pGEX-3X vector (GE Healthcare, Piscataway, NJ). Orientation was screened by EcoR1 digestion and the GST-coilin construct was confirmed by sequence analysis (SeqWright, Houston, TX). Full length GST-coilin variants were made by subcloning GFP-coilin mutants GFP-coilin ON, GFP-coilin TORSO ON, and GFP-coilin C6D (Hearst et al., 2009) by digestion with EcoR1 and insertion into EcoR1-digested pGEX-coilin. Orientation was screened by EcoR1 digest and constructs were confirmed by sequence analysis. GFP-coilin AAA (S571-572A and T573A) and GFP-coilin DDE (S571-572D and T573E) were constructed by site-directed mutagenesis of previously described construct where GFP was fused to the N-terminus of coilin (Hearst et al., 2009). GFP-coilin Δ1nt, GFP-coilin Δ3nt, and mutants were made by modifying the above GFP-coilin constructs by site-directed mutagenesis to contain change of one or three nucleotides to protect against RNAi. Primers used: Δ1nt forward 5′-GAG AAC CTG GGA AAT TCG ATT TAG TTT ATC AC-3′; Δ1nt reverse 5′- GTG ATA AAC TAA ATC GAA TTT CCC AGG TTC TC-3′; Δ3nt forward 5′-CCT TAC CTG CCT TGA GGG AAC CGG GGA AAT TCG ATT TAG TTT ATC ACA AT-3′; Δ3nt reverse 5′-ATT GTG ATA AAC TAA ATC GAA TTT CCC CGG TTC CCT CAA GGC AGG TAA GG-3′. GFP-coilin mutants were confirmed by sequence analysis. His-T7-SmB′ and His-T7-SMN were prepared as previously described (Hebert et al., 2001).
In vitro binding assays
BL21(DE3)pLysS cells (Invitrogen, Carlsbad, CA) or Rosetta pLysS cells (Novagen, Gibbstown, CA) were transformed with His- or GST-fusion constructs. Protein expression was induced with 0.5 mM IPTG for 2–4 hours and purified using either Ni-NTA Superflow beads (Qiagen, Valencia, CA) or glutathione sepharose beads (GE Healthcare, Piscataway, NJ). Purified proteins were evaluated by SDS-PAGE or western blotting. His-T7-fusion proteins and GST-fusion proteins immobilized on glutathione-sepharose beads were incubated in 1 mL of mRIPA (50 mM Tris-HCl pH 7.6; 150 mM NaCl; 1% (v/v) NP-40; 1 mM EDTA) plus 2 mM dithiothreitol (DTT) at 4°C with rocking for 1 hr. The beads were washed 3 × 1 mL mRIPA plus DTT, resuspended in 10 µL SDS loading buffer, heated at 95°C for 5 minutes, and subjected to SDS-PAGE (10% or 12%). After transfer to nitrocellulose, western blots were probed with α-GST, α-T7, or α-SMN antibodies.
Phosphatase treatment and co-immunoprecipitation
HeLa, WI-38, or YFP-SmB′ stably expressing HeLa cells were cultured in DMEM (Mediatech, Manassas, VA) supplemented with 10% FBS (Gibco, Carlsbad, CA) and penicillin/streptomycin. Cell pellets were lysed in 1 mL mRIPA followed by brief sonication. Lysates were cleared by centrifugation. Samples with volumes of 460 µL were treated with 50 U of CIP (New England Biolabs, NEB) in 1x NEB Buffer 3 in a total volume of 520 µL and incubated at 37°C for 30 min. Mock samples were identical to CIP-treated samples with the exception that 5 µL of Buffer 3 were used in place of phosphatase. The samples were precleared by rocking at 4°C with 500 µL mRIPA and 30 µL of 50% Protein G sepharose bead slurry (in PBS) for 1 hr. For experiments to determine an interaction between endogenous coilin and YFP-SmB′, untreated lysates were precleared for 2 hours. Samples were split into 500 µL aliquots and further incubated 1 hr with 500 µL mRIPA and either 2 µg anti-SMN antibody or an equivalent amount of normal mouse serum. In the case of the YFP-SmB′ stable lines, monoclonal GFP antibody was used. Samples were rocked overnight at 4°C with 30 µL of a 50% Protein G sepharose bead suspension. Proteins bound to Protein G beads were washed 3 × 1 mL mRIPA, resuspended in 10 µL SDS loading buffer, heated at 95°C for 5 minutes, and subjected to SDS-PAGE. Western Blots were probed with α-coilin, α-GFP, and α-SMN antibodies.
cDNA synthesis and quantitative real-time PCR
The iScript cDNA synthesis kit (Bio-Rad) was used to synthesize cDNA from HeLa RNA. Samples were incubated at 25°C for 5 minutes, 42°C for 30 minutes, and 85°C for 5 minutes using a PCT-200 Peltier Thermal Cycler (MJ Research). For qRT-PCR, separate reactions were conducted to detect coilin and actin. Equal amounts of cDNA were used in both reactions and primers have been described previously (Hearst et al., 2009). Primers and cDNA were added to the Brilliant SYBR green QPCR Master Mix kit (Stratagene, Ja Jolla, CA) and incubated 10 min at 95°C, followed by 40 amplification cycles (95°C for 30 seconds, 50°C for 30 seconds, 72°C for 1 minute) and a dissociation curve-analysis step on a MX3000P real-time PCR system (Stratagene, Ja Jolla, CA). Amplification rates, Ct values and dissociation curve analyses of products were determined using MxPro (version 4.01) software. Relative expression was determined using the 2(−ΔΔCt) method (Livak and Schmittgen, 2001).
Coilin RNAi and co-immunoprecipitation
Endogenous coilin was depleted by transfecting HeLa cells with duplex coilin or control siRNA (Whittom et al., 2008) using Lipofectamine (Invitrogen) according to the manufacturer’s protocol. The cells were harvested after 72 hrs of RNAi. The coilin duplex siRNA is targeted to the coilin sequence 5′-GAG AGA ACC TGG GAA ATT T-3′. Coilin knockdown was verified by Western blotting and RT-PCR. For transfections of DNA after RNAi treatment, cells were exposed to the siRNAs for 48 hrs, and then transfected with GFP-coilin, GFP-coilin Δ1nt or GFP-coilin Δ3nt constructs using Superfect (Qiagen, Valencia, CA) or FuGENE 6 (Roche) following the manufacturer’s instructions. To determine the in vivo interaction of SMN with various coilin proteins, HeLa cells were transfected with full length, unmodified GFP-coilin or mutants where S571-572 and T573 were replaced with either alanine (GFP-coilin AAA) or phosphomimetic residues (GFP-colin DDE). Thus, 48 hrs post control- or coilin-siRNA treatment, HeLa cells were transfected with GFP-coilin Δ3nt, GFP-coilin AAA Δ3nt, GFP-coilin DDE Δ3nt, or GFP-C3 control (Clontech, Mountain View, CA) and harvested 24 hrs later. HeLa cells for CoIP experiments, cultured in T25 flasks, were harvested and pellets frozen in liquid nitrogen, followed by resuspension in 1 mL mRIPA. The lysates were incubated 5 min on ice, and further lysed by 28g syringe passage. Cleared lysates were precleared with 30 µL of Protein G sepharose beads (50% slurry in PBS) for 30 min. Proteins in the samples were coimmunoprecipitated with 3 µg α-GFP polyclonal and 30 µL of Protein G for 4 hrs at 4°C. Proteins bound to Protein G beads were washed 3 × 1 mL mRIPA, resuspended in 10 µL SDS loading buffer, heated to 95°C for 5 minutes, and subjected to SDS-PAGE. Western blots were probed with α-SMN and α-GFP monoclonal antibodies.
RESULTS
SmB′ shows increased binding to the phosphomimetic C-terminus of coilin in vitro
Coilin/Sm protein interactions are mediated through the C-terminus of coilin as seen in pulldown experiments with the C214 fragment, the final 214 residues of coilin comprising amino acids 362-576 (Xu et al., 2005). In order to investigate the effects of phosphorylation on the C-terminus of coilin, we were interested to see if phosphorylation of C214 would lead to changes in Sm protein binding. We first generated a series of GST-fused C214 phosphomimics (summarized in Table 2) in which up to 6 residues (identified by MS/MS) were replaced by aspartate or glutamate to mimic phosphorylation. Modifications were made in two regions of coilin where groups of phosphoresidues have been reported (Fig. 1): the very C-terminus comprising the last 11 amino acids (565 –576) and S489 which is found central to the C214 fragment and is conserved as serine or threonine across 12 species. GST pulldown experiments were conducted with bacterially expressed recombinant T7-fusion SmB′ protein. We have previously shown that the C214 fragment of coilin can interact with SmB′, SmD3, SmD1 and the Lsm proteins Lsm4, Lsm10 and Lsm11 (Xu et al., 2005), and thus believe it is appropriate to use SmB′ as a representative of these proteins. Western blots were probed with α-T7 antibodies to detect T7-tagged SmB′. The conversion of phosphoresidues with analogous acidic residues in GST-C214 lead to increased SmB′ binding relative to unmodified GST-C214 (Fig. 2a). The same blot was probed with α-GST antibody to confirm that quantities of GST-proteins were approximately equal. These data indicate that Sm proteins have a higher affinity for coilin when the C-terminus is phosphorylated.
Table 2.
Summary of GST-C214 construct and mutants
| Construct Name | Description |
|---|---|
| C214 | Coilin residues 362–576 |
| C214 S489D | S489D |
| C214 DDE | S571-572D, T573E |
| C214 S489D + DDE | S489D, S571-572D, T573E |
| C214 C6D | S566D, S568D, T570E, S571-572D, T573E |
Fig. 2.
SmB′ and SMN binding to C-terminal coilin fragments is affected by phosphorylation. a SmB′ shows increased binding to the phosphomimic C-terminal coilin fragment (GST-C214). Equal amounts of GST, GST-fused C-terminal coilin fragment, or C-terminal coilin phosphomimic fragments immobilized on glutathione beads were incubated with bacterially produced T7-tagged full length His-SmB′. Western blots were probed with antibodies to the T7 tag. The membrane was reprobed with anti-GST antibodies to demonstrate that approximately equal amounts of fusion proteins were used. Bands of different molecular weights from the same reprobed blot are outlined and aligned for ease of comparison. b SMN demonstrates reduced binding to phosphomimics. Equal amounts of immobilized GST, GST-C214, and GST-C214 phosphomimic fragments were incubated with His-SMN. Western blots were probed with antibodies to SMN and GST. The input lanes in both a and b contain 10% of the amount used in the binding reactions. c Histogram of densitometric analysis showing that His-SMN has significantly higher avidity for C214 than the DDE phosphomimic (p-value = 0.022). Error bars represent the standard deviation from the mean
SMN shows decreased binding to phosphomimetic C214 in vitro
SMN binding to coilin is mediated through the so-called RG box, coilin residues 392 to 420 (Hebert et al., 2001), and enhanced when these arginine residues are dimethylated (Hebert et al., 2002). Pulldown experiments as above were carried out to assess SMN interaction with the coilin C214 fragment and phosphomimics. In contrast to SmB′, SMN binds preferentially to the dephosphorylated form of the protein; SMN binding is reduced with phosphomimetic protein fragments (Fig. 2b). In order to compensate for slight deviations in the amounts of GST-protein in each lane, SMN levels were normalized to the amounts of GST-protein and densitometric analysis of three separate experiments confirmed that the amounts of His-SMN pulled down by unmodified GST-C214 and GST-C214 DDE differed significantly (Fig. 2c, p-value = 0.022).
Full length coilin pulls down more SmB′ and SMN than the C214 fragment
GST-fusion full length coilin was expressed in E. coli, purified, and used in GST pulldown experiments to confirm relevance of the fragment data. SmB′ and SMN binding to unmodified GST-214 and GST-coilin were examined and in both cases significantly more protein was pulled down by full length coilin than by the C214 fragment (Fig. 3). Blots were probed with anti-GST to verify that equal amounts of GST-proteins were present and that proteins did not bind significantly to GST alone (Fig. 3, lane 2 for both panels). We next assessed whether phosphorylation of the coilin C-terminus affected SMN and SmB′ interaction in the full-length coilin protein context. A GST-coilin DDE construct was designed, analogous to the C214 DDE fragment, where three of the last 6 amino acid residues (S571-572 and T573) were replaced by aspartic or glutamic acid residues. The difference in the amount of SMN or SmB′ recovered by the two full-length GST-coilin proteins was not as distinctive as with the fragments, so we chose to probe the interaction with titration experiments. Supplemental Fig. 1 shows representative pulldown experiments where GST-coilin and GST-coilin DDE were incubated with increasing amounts of either His-SMN or His-SmB′ (1.5 µL to 15 µL) in separate reactions. After densitometric analysis, we concluded that there was no significant difference in the amount of SMN or SmB′ bound by the bacterially expressed full-length phosphomimic or wild-type protein.
Fig. 3.
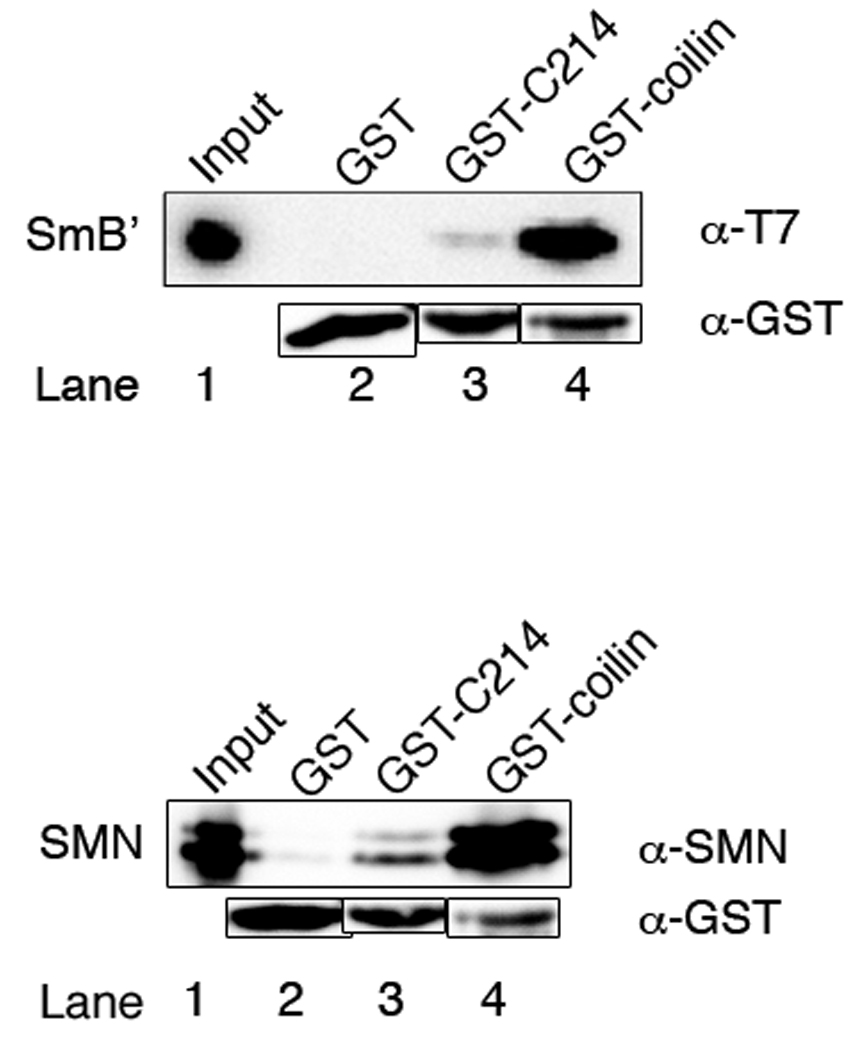
SmB′ and SMN bind more to full length GST-coilin than to GST-C214. Equal amounts of GST, GST-C214, or GST-coilin immobilized on glutathione beads were incubated with either His-SmB′ (top panel) or His-SMN (bottom panel) and analyzed by western blotting. Antibodies to the T7-tag were used to probe for His-SmB′ and antibodies to SMN were used to probe the SMN blot. Antibodies to GST were used on both blots to show that similar amounts of GST-fusion proteins were used. Input lanes represent 5% of the total protein used in the experiments
GFP-coilin AAA mutant co-immunoprecipitates more SMN than GFP-coilin DDE in HeLa cell lysate
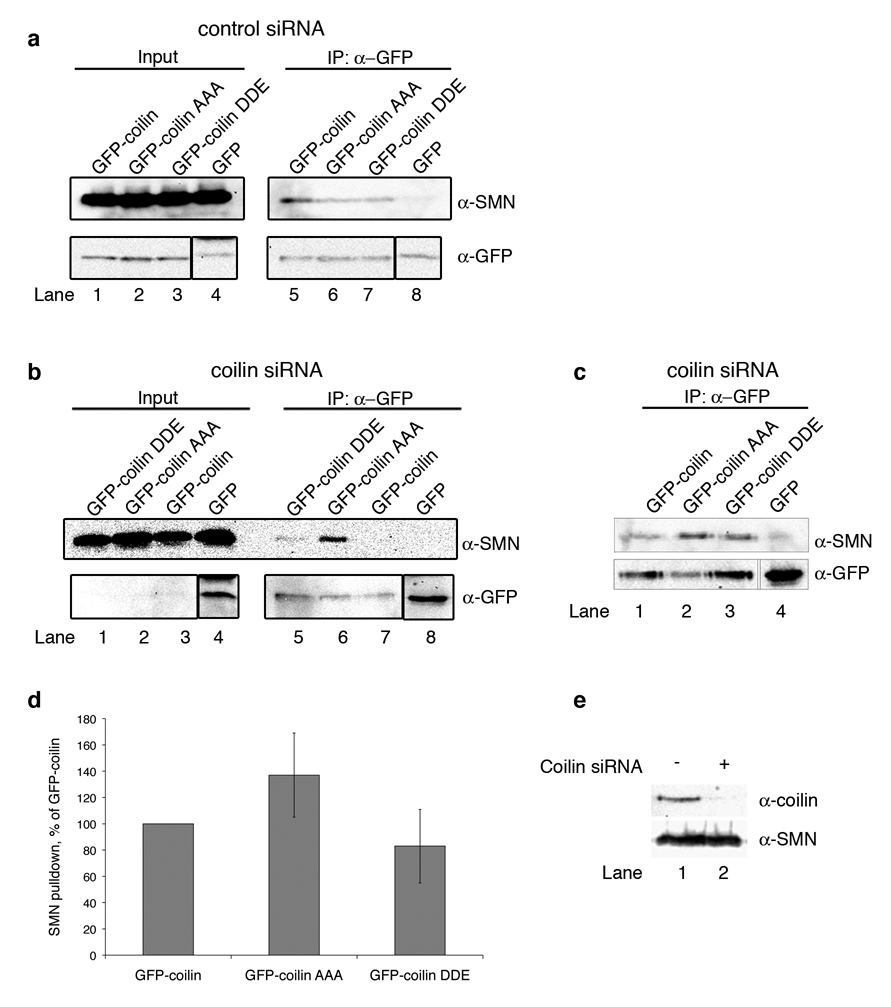
Although we did not see a difference in coilin-SMN interaction with full-length bacterial GST-coilin or phosphomimic in vitro, we wondered if further in-cell modification might be required to restore differential SMN binding. Thus, in order to test the importance of phosphorylation of the C-terminus of full-length coilin in vivo, we introduced the DDE mutation into GFP-coilin (GFP-coilin DDE). Further, to ensure that S571-572 and T573 remained in a dephosphorylated state, we also replaced those residues with alanines in a separate construct (GFP-coilin AAA). The coilin constructs also contain a three nucleotide mutation (described below). GFP-coilin, GFP-coilin AAA, GFP-coilin DDE, and GFP-C3 vector alone were transfected into HeLa cells treated with control siRNA. Lysates from these cells were co-immunoprecipitated with α-GFP antibodies, and western blots were probed with α-SMN antibodies (Fig. 4a). The input lanes all contained similar amounts of SMN (lanes 1–4), and all three coilin constructs co-immunoprecipitated SMN (lanes 5 – 7) at levels higher than the background amount recovered with GFP alone (lane 8). When comparing the amount of SMN recovered relative to the level of immunoprecipitated GFP-coilin proteins, no obvious changes in the SMN binding potential of the various coilin constructs was detected. One caveat to this experiment, however, is the presence of endogenous coilin. Given that coilin contains an N-terminal self-association domain (Hebert and Matera, 2000), it is possible that endogenous coilin, by interacting with the ectopically expressed GFP-coilin proteins, could interfere with the amount of SMN recovered by each GFP-coilin construct. We chose to deplete the level of endogenous coilin in HeLa cells by treatment with duplex coilin siRNA (Whittom et al., 2008) (Hearst et al., 2009), specific to one site on the coilin message, to ensure that the endogenous coilin would not mask any differences in the SMN binding potential of the different GFP-coilin constructs.
Fig. 4.
SMN binds more to dephosphorylated coilin in vivo. a HeLa cells treated with control siRNA were transfected with GFP-coilin Δ3nt, GFP-coilin AAA Δ3nt, GFP-coilin DDE Δ3nt or empty GFP vector. Lysate from these cells was subjected to immunoprecipitation with polyclonal anti-GFP antibodies and SDS-PAGE. After western transfer, blots were probed with anti-SMN, followed by reprobing of the same blot with anti-GFP monoclonal antibodies. Input lanes (1–4) represent 1% of the total protein used in the Co-IP experiments (lanes 5–8). Lysate with GFP vector alone did not recover significant amounts of SMN (lane 8). b HeLa cells treated with coilin siRNA were transfected and treated as described above, except that half the amount of DNA was used. c Immunoprecipitation reactions from another experiment in which cells were treated with coilin siRNA followed by DNA transfection and processing as described in (a). d Histogram of the densitometric analysis of the data shown in (b), (c) and other results. The amount of SMN bound to GFP-coilin, GFP-coilin AAA and GFP-coilin DDE relative to the corresponding GFP signal of each protein was calculated. Amounts of SMN were normalized to the amount of SMN immunoprecipitated by GFP-coilin (p-value = 0.025; two-tailed t-test). The error bar represents standard error of the mean. e Endogenous coilin knock down with coilin siRNA. HeLa cells were transfected with duplex control (−) or coilin siRNA (+). Lane 2 clearly shows that coilin levels are depleted by treatment with the siRNA specific to one site on the coilin message, and that there is no apparent effect on SMN levels. At 72 hours post-siRNA treatment, cells were harvested and subjected to western analysis. Proteins were detected with α-coilin and α-SMN antibodies
In order to protect the transfected GFP-coilin constructs from depletion themselves, they were modified by the addition of a three nucleotide mutation (Δ3nt), introducing three silent mutations in the DNA sequence and rendering them impervious to the siRNA treatment. Initially, we tested a GFP-coilin construct with only a single nucleotide change (Δ1nt). Although this construct was somewhat resistant to the siRNA depletion, it was not as effective as the Δ3nt modification that was chosen for these experiments (Supplemental Fig. 2). Accordingly, at 48 hours post-siRNA transfection, HeLa cells were transfected with equal quantities of GFP-coilin Δ3nt, GFP-coilin AAA Δ3nt, GFP-coilin DDE Δ3nt, or empty GFP vector alone. Low levels of DNA were used in these experiments to preclude overexpression artefacts and increase the likelihood that the proteins were adequately modified. Cells were harvested 24 hours post DNA transfection (72 hours post siRNA knockdown) and proteins were co-immunoprecipitated with anti-GFP antibodies. Typically, coilin knockdown was around 65% as determined by RT-PCR and western blotting (Fig 4e). The samples were subjected to western blot analysis and probed with α-SMN monoclonal antibodies. Representative results are shown in Figure 4b and c. In Figure 4b, similar SMN levels are apparent in the input lanes (1–4), however, considerably more SMN co-immunoprecipitates with GFP-coilin AAA (lane 6) than GFP-coilin DDE (lane 5). No SMN was detected when empty GFP vector was used in the transfection and co-immunoprecipitation (lane 8). Blots were re-probed with α-GFP antibodies to confirm that transfection levels of the various GFP-coilin proteins were equal. As expected, GFP-coilin signals detected in the input lanes were low (lanes 1–3), but adequate GFP-coilin proteins were present in the immunoprecipitation reactions (lanes 5–7). Immunoprecipitation reactions from another experiment are shown in Figure 4c, and likewise demonstrate that the relative recovery of SMN is greatest with GFP-coilin AAA. Densitometric analysis of these and other data show that the amount of SMN co-immunoprecipitated with GFP-coilin AAA is significantly greater than that recovered by GFP-coilin DDE (Fig. 4d; p-value = 0.025; n = 5). There is no significant difference in the relative amount of SMN recovered by GFP-coilin AAA compared to that recovered by GFP-coilin, nor is there a significant difference between SMN recovered by GFP-coilin compared to that recovered by GFP-coilin DDE. These data clearly show that the AAA coilin construct more tightly interacts with SMN than does coilin DDE and are in vivo confirmation of our coilin C-terminal fragment data showing that SMN preferentially binds dephosphorylated coilin.
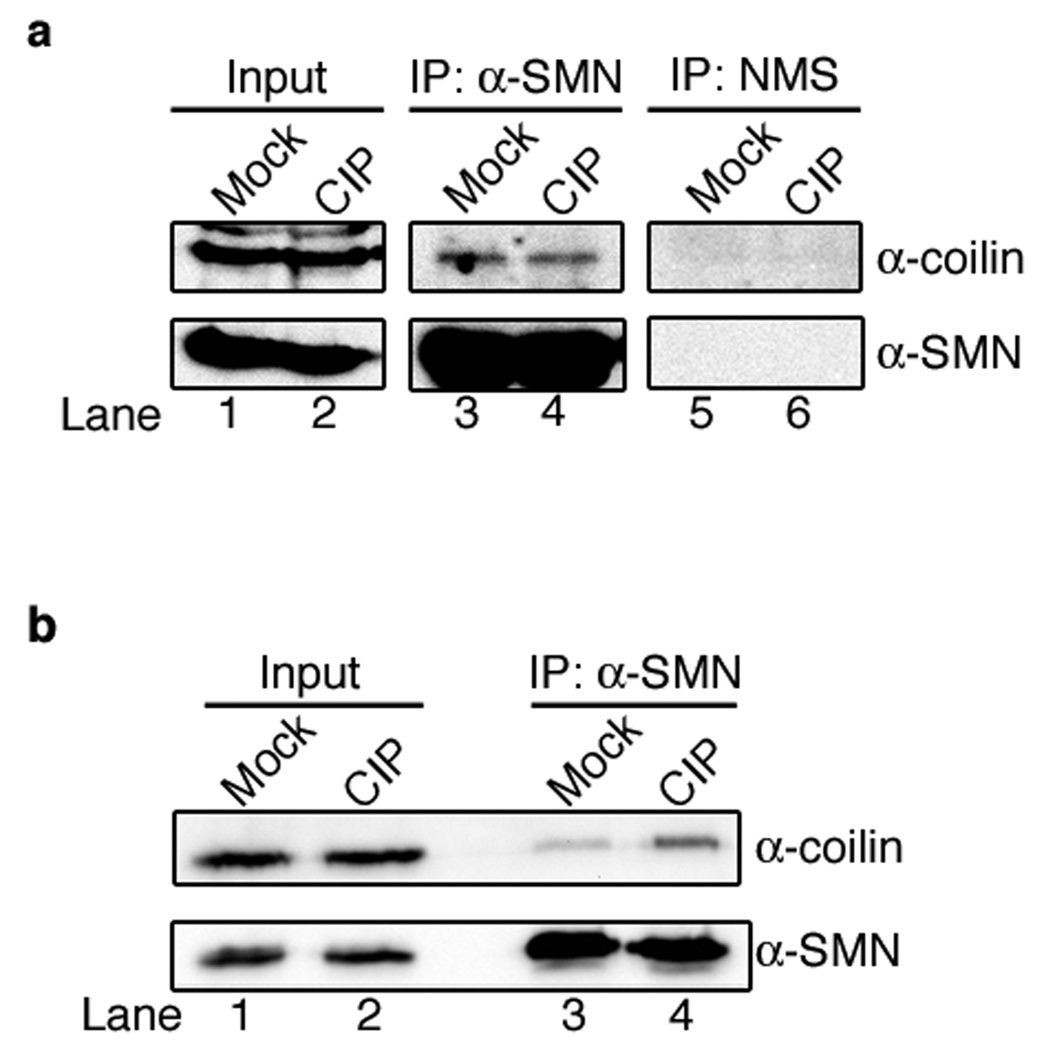
Coilin from phosphatase treated WI-38 cell lysate binds better to SMN
To investigate the in vivo effects of phosphorylation on endogenous coilin interaction with SMN, cleared HeLa lysates were mock- or calf intestinal alkaline phosphatase (CIP)-treated followed by immunoprecipitation with anti-SMN antibodies or normal mouse serum (NMS). Isoelectric focusing experiments have shown that CIP-treatment increases the pI of coilin, consistent with dephosphorylation (Hearst et al., 2009). Additionally, CIP treatment results in a very slight shift of coilin migration on SDS-PAGE (Carmo-Fonseca et al., 1993). Based on the C-terminal coilin fragment pulldown data, and the GFP-coilin phosphomimic experiments, we would expect that CIP-induced dephosphorylation would increase interaction between coilin and SMN. However, we saw no apparent difference in the amount of coilin co-immunoprecipitated with SMN between CIP- or mock-treated HeLa lysate (Fig. 5a, compare the coilin signal in lane 3 to that in lane 4). Little SMN or coilin was detected in reactions using normal mouse serum as a negative immunoprecipitation control (lanes 5 and 6).
Fig. 5.
SMN binds more to dephosphorylated coilin in WI-38. a HeLa lysates were CIP- or mock-treated and immunoprecipitated with either antibodies against SMN or normal mouse serum (NMS). The reactions were subjected to SDS-PAGE and western transfer, followed by the detection of coilin and SMN with appropriate antibodies. Similar amounts of coilin were co-immunoprecipitated with anti-SMN antibody from CIP- and mock-treated HeLa lysates (lanes 3 and 4). No coilin or SMN were detected in the NMS control indicating that the CoIP experiment was specific for those proteins (lanes 5 and 6). b WI-38 lysates were CIP- or mock-treated followed by immunoprecipitation with anti-SMN antibodies and further processing as described above. Note that more coilin was co-immunoprecipitated from CIP-treated WI-38 lysate (lane 4) compared to that recovered using mock-treated lysate (lane 3). Input lanes represent 6% of the total protein used in the experiments
We have previously showed that coilin is hyperphosphorylated in the primary WI-38 cell line compared to that found in transformed HeLa cells (Hearst et al., 2009). We thus reasoned that these cells might serve as a better resource to demonstrate that phosphorylation impacts coilin/SMN interaction. Towards this end, WI-38 lysate was subject to CIP- and mock-treatment followed by immunoprecipitation with anti-SMN antibodies as conducted for HeLa extract. Significantly more coilin was co-immunoprecipitated in the CIP-treated WI-38 sample (Fig. 5b, lane 4) compared to the mock-treated reaction (lane 3), yet approximately equal amounts of SMN were recovered (lanes 3 and 4, lower panel). Thus, dephosphorylation of coilin and SMN results in more complex formation of these proteins in WI-38 extract compared to that observed with normal levels of phosphorylation. These data confirm the coilin C-terminal fragment data showing that SMN interacts less with phosphomimics. It is possible that alkaline phosphatase treatment of SMN may also contribute to the coilin/SMN interaction, but we are not able to assess the contribution of specifically dephosphorylating SMN in this experiment.
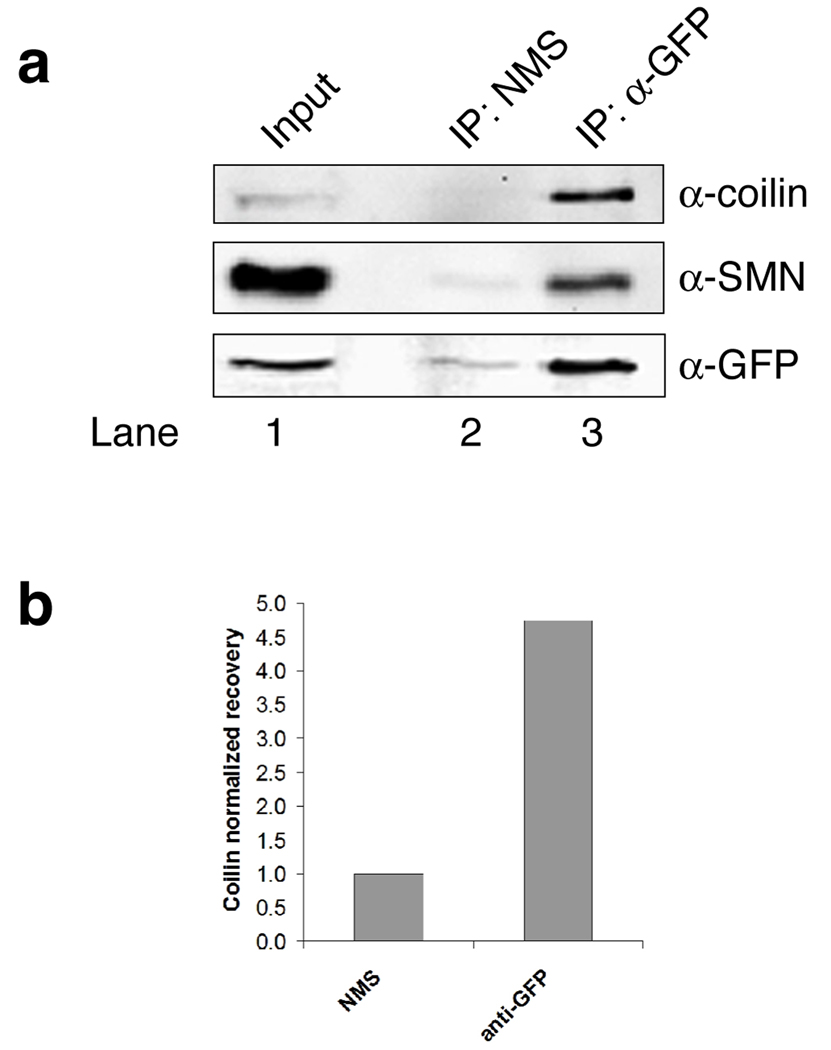
Coilin interaction with SmB′ in HeLa lysate
As with the coilin/SMN interaction described above, we were also interested in assessing the effect of phosphorylation on coilin/Sm interaction in vivo. No studies have shown an interaction between endogenous coilin and Sm proteins. Since the commonly used anti-Sm antibody Y12 also detects dimethylated coilin (Hebert et al., 2002), we chose to examine coilin/Sm interaction using a HeLa cell line stably expressing YFP-SmB′. In so doing, antibodies to the YFP tag could be used to immunoprecipitate YFP-SmB′ and the level of co-immunoprecipitating coilin could be detected. As shown in Fig. 6, more coilin is recovered when YFP-SmB′ lysate is incubated with anti-GFP antibodies compared to that found using normal mouse serum in the immunoprecipitation reaction (upper panel, compare the amount of coilin in lane 2 to that in lane 3). The amount of SMN and YFP-SmB′ recovered using normal mouse serum is greatly reduced compared to that detected when using anti-GFP antibodies (middle and lower panels, lane 3), confirming the specificity of the reaction. Thus endogenous coilin can form a complex with YFP-SmB′. To determine if phosphorylation impacts the interaction of endogenous coilin with YFP-SmB′, lysates from the HeLa YFP-SmB′ stable cell line were CIP- or mock-treated, followed by immunoprecipitation with anti-GFP antibodies. Based on our coilin C-terminal fragment pulldown data, we would expect that dephosphorylation would decrease the interaction between coilin and YFP-SmB′ and thus more coilin should be recovered from the mock-treated lysate relative to the CIP-treated lysate. We could not, however, detect any differences in the amount of co-immunoprecipitating coilin relative to the YFP-SmB′ signal between the reactions subject to CIP- or mock-treatment (our unpublished observations). Additionally, we could not detect an interaction between transiently transfected GFP- or myc-coilin with SmB/B’ or YFP-SmB′, whether or not endogenous coilin expression was reduced by RNAi (our unpublished observations). One interpretation of this finding is that transiently transfected coilin is not in the proper phosphorylation state for interaction with Sm proteins. In summary, therefore, while we could detect an interaction between endogenous coilin and YFP-SmB′ in the HeLa cell background, no obvious changes in the level of this interaction are induced upon CIP-treatment.
Fig. 6.
SmB′ interaction with coilin in HeLa. a HeLa YFP-SmB′ lysate was subjected to immunoprecipitation with anti-GFP antibodies or normal mouse serum (NMS) followed by SDS-PAGE and western transfer. The blot was probed with antibodies to coilin, SMN, and GFP (to detect YFP-SmB′). The input lane contains 1.5% of the protein used in the IP experiments. b Quantification of this and other results. The amount of coilin recovered in anti-GFP immunoprecipitation reactions was normalized to the amount of coilin recovered in the NMS reactions. There is approximately 4.5-fold more coilin recovered using anti-GFP relative to NMS
DISCUSSION
Despite the identification of human coilin as a phosphoprotein more than 15 years ago (Carmo-Fonseca et al., 1993), very little work has been published detailing how this modification impacts coilin function. Additionally, the signalling pathways that regulate the level of coilin phosphorylation in interphase and hyperphosphorylate coilin during mitosis have not been elucidated. Also unknown are the actual kinases and phosphatases that directly modify coilin in vivo, although the kinases CDK2-cyclinE and casein kinase 2 as well as the phosphatase PPM1G have been shown to modify coilin in vitro (Hebert and Matera, 2000) (Liu et al., 2000) (Hearst et al., 2009). Considering the important role that coilin plays in CB formation and composition, a basic understanding as to how phosphorylation influences coilin’s interaction with other proteins is needed but lacking. For example, coilin hyperphosphorylation in mitosis corresponds with CB disassembly (Carmo-Fonseca et al., 1993) and decreased self-association (Hebert and Matera, 2000). Additionally, the primary cell line WI-38 which essentially lacks CBs contains hyperphosphorylated coilin relative to that found in HeLa cells, in which CBs are plentiful (Hearst et al., 2009). Clearly, therefore, coilin phosphorylation is crucial for CB formation, yet mechanistic insight into the role of phosphorylation on coilin activity is not known. Recent proteomic analyses (Table 1) have provided a valuable resource towards investigating the function of coilin phosphorylation by defining which coilin residues are modified in specific phases of the cell cycle. In particular, these studies have demonstrated that coilin phosphorylation can take place on both serines and threonines (Table 1, Fig. 1). Moreover, mass spectrometric analysis has revealed that the C-terminus of coilin, comprising aa 362-576 and previously shown to mediate coilin interaction with both SMN and Sm proteins (Hebert et al., 2001; Xu et al., 2005), contains 10 potential phosphorylation sites. Along with phosphorylation, this region of coilin also contains symmetrical dimethylarginine modifications (Hebert et al., 2002) (Boisvert et al., 2002) within the RG box that influence coilin/SMN interaction. Upon consideration of the interactions and modifications of the coilin C-terminus, it is logical to assume that phosphorylation impacts coilin interaction with SMN and Sm proteins.
The results shown here provide evidence that coilin phosphorylation differentially affects its binding to SMN and SmB′. Replacement of phosphoresidues in the coilin C-terminal fragment (residues 362-576, C214) leads to significant changes in the binding profiles of SMN and SmB′ in vitro. Specifically, SmB′ preferentially binds C214 phosphomimics while SMN interaction with these fragments is reduced (Fig. 2). We speculate that coilin phosphorylation in the cell facilitates transfer of SmB′ from SMN to coilin. These data agree with a previous report where it was shown that SMN and SmB′ compete for coilin in binding experiments (Hebert et al., 2001). Full length bacterially expressed coilin and phosphomutants bound SMN and SmB′ to a much greater extent than did the unmodified C214 fragment (Fig. 3). The differential effect of phosphorylation on binding is diminished in experiments with the full-length recombinant proteins, however, we do describe two in vivo experiments (Fig. 4 and Fig. 5) demonstrating that the phosphorylation state of full-length coilin affects the binding of SMN. In figure 4, we show that the coilin phospho-null mutant (GFP-coilin AAA) recovers more SMN than the corresponding phosphomimic (GFP-coilin DDE). In figure 5, we demonstrate that phosphatasetreatment increases the amount of coilin/SMN interaction the primary cell line WI-38. Thus, it appears that the phosphorylation state of the C-terminus of coilin is one important factor in coilin-SMN and coilin-Sm protein interaction, but it is not the only one. In addition to post-translational modification of the phosphoresidues on the C-terminus, coilin may require additional modifications on other residues. For example, MS/MS experiments show that T122, S271, T303, and S456 are apparently constitutively phosphorylated and, thus, may be essential for the selective binding activity of coilin (Table 1, Fig. 1). It is unlikely that bacterially expressed full-length coilin contains the appropriate configuration of phosphorylated residues. Consequently, the absence of phosphorylation on these residues in our bacterial coilin constructs may be one explanation for loss of phosphorylation-dependent differential SMN and SmB′ binding in the full-length context. There are no reports of symmetrical dimethlyarginine in prokaryotes. Thus, along with the absence of phosphorylation, we likewise also do not know how the lack of symmetrical dimethlyarginine in bacterially expressed coilin will impact overall coilin structure, if it all. While the pulldown data using the coilin C-terminal fragment clearly has its limitations, corroborating data using endogenous and HeLa expressed GFP-coilin proteins support our hypothesis that coilin phosphorylation mediates SMN and Sm interaction.
SMN is hyperphosphorylated in the cytoplasm relative to the nucleoplasm and its phosphorylation state is important to its activity, i.e. recruitment of proteins to the SMN complex in the cytoplasm (Grimmler et al., 2005). Similarly, we suggest that coilin phosphorylation plays a crucial role in recruiting U snRNPs to the CB and propose the following model. First, snRNPs are imported into the nucleus as components in the SMN-complex. In the nucleus, hypophosphorylated coilin recruits the SMN complex to the CB. An as yet unidentified kinase phosphorylates the coilin C-terminus, disrupting the coilin-SMN interaction and enhancing coilin-Sm protein interaction. Thus, U snRNPs are handed off from SMN to coilin in the CB and are made available to the modification machinery in the CB. Dephosphorylation of coilin, possibly by PPM1G, then facilitates the release of U snRNPs, returning coilin to its hypophosphorylated state.
In conclusion, we demonstrate that, in addition to affecting coilin-coilin interaction and localization in the nucleus, the phosphorylation state of coilin also impacts its interaction with SMN and SmB′ and may play a role in controlling U snRNP cycling through the CB. Further work will be necessary to identify the individual phosphatases and kinases that act on coilin and to parse the signalling pathway that manages CB formation. It will also be important to determine the relationship between coilin phosphorylation and symmetrical arginine dimethylation. However, the results of this current report do extend understanding of coilin’s role in the cell and provide further evidence that phosphorylation is essential in that role.
Supplementary Material
SmB′ and SMN interaction with full length GST-coilin and GST-coilin DDE. GST-proteins were incubated with decreasing amounts of SmB′ (top panel) or SMN (lower panel). Reactions were subject to SDS-PAGE and western transfer, followed by detection of the various proteins with appropriate antibodies. There is no significant difference between SmB′ and SMN binding with full length coilin or DDE phosphomimic. GST-protein levels are shown to demonstrate that similar amounts were used throughout the experiment. Input lanes represent 5% of the maximum amount of total protein used in the CoIP experiments (lanes 2 and 6)
The GFP-coilin Δ3nt construct is resistant to siRNA treatment. Constructs were designed, including silent mutations comprising either a single or three nucleotide change, to be resistant to the siRNA mediated depletion of endogenous coilin. Endogenous coilin was knocked down by transfecting HeLa cells with duplex coilin (+) and control siRNA (−). After incubating 48 hours, cells were transfected with equal amounts of GFP-coilin, GFP-coilin Δ1nt, or GFP-coilin Δ3nt DNA. At 72 hours posts-iRNA treatment, cells were harvested and subjected to western analysis. Proteins were detected with anti-GFP antibodies. Unprotected GFP-coilin was clearly depleted (lane 1) compared to the control (lane 2). The single nucleotide modification conferred some protection (lanes 3 and 4), but replacement of three nucleotides was required for complete resistance (lanes 4 and 5)
ACKNOWLEDGMENTS
This work was supported by NIH grant R01GM081448 to MDH. We thank Dr. Angus Lamond (University of Dundee, Dundee, Scotland) for his gift of the YFP-SmB′ HeLa stable line.
REFERENCES
- Andrade LE, Tan EM, Chan EK. Immunocytochemical analysis of the coiled body in the cell cycle and during cell proliferation. Proc Natl Acad Sci U S A. 1993;Vol. 90:1947–1951. doi: 10.1073/pnas.90.5.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohmann K, Ferreira JA, Lamond AI. Mutational analysis of p80 coilin indicates a functional interaction between coiled bodies and the nucleolus. J Cell Biol. 1995;Vol. 131:817–831. doi: 10.1083/jcb.131.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert FM, Cote J, Boulanger MC, Cleroux P, Bachand F, Autexier C, Richard S. Symmetrical dimethylarginine methylation is required for the localization of SMN in Cajal bodies and pre-mRNA splicing. J Cell Biol. 2002;159:957–969. doi: 10.1083/jcb.200207028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahms H, Meheus L, de Brabandere V, Fischer U, Luhrmann R. Symmetrical dimethylation of arginine residues in spliceosomal Sm protein B/B' and the Sm-like protein LSm4, and their interaction with the SMN protein. Rna. 2001;Vol. 7:1531–1542. doi: 10.1017/s135583820101442x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M, Ferreira J, Lamond AI. Assembly of snRNP-containing coiled bodies is regulated in interphase and mitosis--evidence that the coiled body is a kinetic nuclear structure. J Cell Biol. 1993;Vol. 120:841–852. doi: 10.1083/jcb.120.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen WJ, Dreyfuss G. Specific sequences of the Sm and Sm-like (Lsm) proteins mediate their interaction with the spinal muscular atrophy disease gene product (SMN) J Biol Chem. 2000;Vol. 275:26370–26375. doi: 10.1074/jbc.M003299200. [DOI] [PubMed] [Google Scholar]
- Grimmler M, Bauer L, Nousiainen M, Korner R, Meister G, Fischer U. Phosphorylation regulates the activity of the SMN complex during assembly of spliceosomal U snRNPs. EMBO Rep. 2005;Vol. 6:70–76. doi: 10.1038/sj.embor.7400301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearst SM, Gilder AS, Negi SS, Davis MD, George EM, Whittom AA, Toyota CG, Husedzinovic A, Gruss OJ, Hebert MD. Cajal-body formation correlates with differential coilin phosphorylation in primary and transformed cell lines. J Cell Sci. 2009;Vol. 122:1872–1881. doi: 10.1242/jcs.044040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert MD, Matera AG. Self-association of coilin reveals a common theme in nuclear body localization. Mol Biol Cell. 2000;Vol. 11:4159–4171. doi: 10.1091/mbc.11.12.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert MD, Shpargel KB, Ospina JK, Tucker KE, Matera AG. Coilin methylation regulates nuclear body formation. Dev Cell. 2002;Vol. 3:329–337. doi: 10.1016/s1534-5807(02)00222-8. [DOI] [PubMed] [Google Scholar]
- Hebert MD, Szymczyk PW, Shpargel KB, Matera AG. Coilin forms the bridge between Cajal bodies and SMN, the spinal muscular atrophy protein. Genes Dev. 2001;Vol. 15:2720–2729. doi: 10.1101/gad.908401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jady BE, Darzacq X, Tucker KE, Matera AG, Bertrand E, Kiss T. Modification of Sm small nuclear RNAs occurs in the nucleoplasmic Cajal body following import from the cytoplasm. Embo J. 2003;Vol. 22:1878–1888. doi: 10.1093/emboj/cdg187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam YW, Lyon CE, Lamond AI. Large-scale isolation of Cajal bodies from HeLa cells. Mol Biol Cell. 2002;Vol. 13:2461–2473. doi: 10.1091/mbc.02-03-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Hebert MD, Ye Y, Templeton DJ, Kung H, Matera AG. Cell cycle-dependent localization of the CDK2-cyclin E complex in Cajal (coiled) bodies. J Cell Sci. 2000;113(Pt 9):1543–1552. doi: 10.1242/jcs.113.9.1543. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;Vol. 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lyon CE, Bohmann K, Sleeman J, Lamond AI. Inhibition of protein dephosphorylation results in the accumulation of splicing snRNPs and coiled bodies within the nucleolus. Exp Cell Res. 1997;Vol. 230:84–93. doi: 10.1006/excr.1996.3380. [DOI] [PubMed] [Google Scholar]
- Matera AG. Nuclear bodies: multifaceted subdomains of the interchromatin space. Trends Cell Biol. 1999;Vol. 9:302–309. doi: 10.1016/s0962-8924(99)01606-2. [DOI] [PubMed] [Google Scholar]
- Morris GE. The Cajal body. Biochim Biophys Acta. 2008 doi: 10.1016/j.bbamcr.2008.07.016. [DOI] [PubMed] [Google Scholar]
- Petri S, Grimmler M, Over S, Fischer U, Gruss OJ. Dephosphorylation of survival motor neurons (SMN) by PPM1G/PP2Cgamma governs Cajal body localization and stability of the SMN complex. J Cell Biol. 2007;Vol. 179:451–465. doi: 10.1083/jcb.200704163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleeman JE, Ajuh P, Lamond AI. snRNP protein expression enhances the formation of Cajal bodies containing p80-coilin and SMN. J Cell Sci. 2001;114:4407–4419. doi: 10.1242/jcs.114.24.4407. [DOI] [PubMed] [Google Scholar]
- Shpargel KB, Ospina JK, Tucker KE, Matera AG, Hebert MD. Control of Cajal body number is mediated by the coilin C-terminus. J Cell Sci. 2003;Vol. 116:303–312. doi: 10.1242/jcs.00211. [DOI] [PubMed] [Google Scholar]
- Sprangers R, Groves MR, Sinning I, Sattler M. High-resolution X-ray and NMR structures of the SMN Tudor domain: conformational variation in the binding site for symmetrically dimethylated arginine residues. J Mol Biol. 2003;Vol. 327:507–520. doi: 10.1016/s0022-2836(03)00148-7. [DOI] [PubMed] [Google Scholar]
- Stanek D, Pridalova-Hnilicova J, Novotny I, Huranova M, Blazikova M, Wen X, Sapra AK, Neugebauer KM. Spliceosomal small nuclear ribonucleoprotein particles repeatedly cycle through Cajal bodies. Mol Biol Cell. 2008;Vol. 19:2534–2543. doi: 10.1091/mbc.E07-12-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittom AA, Xu H, Hebert MD. Coilin levels and modifications influence artificial reporter splicing. Cell Mol Life Sci. 2008;Vol. 65:1256–1271. doi: 10.1007/s00018-008-7587-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Pillai RS, Azzouz TN, Shpargel KB, Kambach C, Hebert MD, Schumperli D, Matera AG. The C-terminal domain of coilin interacts with Sm proteins and U snRNPs. Chromosoma. 2005;Vol. 114:155–166. doi: 10.1007/s00412-005-0003-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SmB′ and SMN interaction with full length GST-coilin and GST-coilin DDE. GST-proteins were incubated with decreasing amounts of SmB′ (top panel) or SMN (lower panel). Reactions were subject to SDS-PAGE and western transfer, followed by detection of the various proteins with appropriate antibodies. There is no significant difference between SmB′ and SMN binding with full length coilin or DDE phosphomimic. GST-protein levels are shown to demonstrate that similar amounts were used throughout the experiment. Input lanes represent 5% of the maximum amount of total protein used in the CoIP experiments (lanes 2 and 6)
The GFP-coilin Δ3nt construct is resistant to siRNA treatment. Constructs were designed, including silent mutations comprising either a single or three nucleotide change, to be resistant to the siRNA mediated depletion of endogenous coilin. Endogenous coilin was knocked down by transfecting HeLa cells with duplex coilin (+) and control siRNA (−). After incubating 48 hours, cells were transfected with equal amounts of GFP-coilin, GFP-coilin Δ1nt, or GFP-coilin Δ3nt DNA. At 72 hours posts-iRNA treatment, cells were harvested and subjected to western analysis. Proteins were detected with anti-GFP antibodies. Unprotected GFP-coilin was clearly depleted (lane 1) compared to the control (lane 2). The single nucleotide modification conferred some protection (lanes 3 and 4), but replacement of three nucleotides was required for complete resistance (lanes 4 and 5)