Abstract
Objective
To determine the role of Dkkl1 on mouse development, viability and fertility
Design
Prospective experimental study
Setting
Government research institution
Animals
Mice of C57BL/6 and 129X1/SvJ strains, as well as transgenic mice of mixed C57BL/6 and 129X1/SvJ strains were used for the studies.
Intervention(s)
Mice were constructed that lacked a functional Dkkl1 gene.
Main Outcome Measure(s)
Deletion of the gene was confirmed by DNA, RNA, and protein analyses; in vivo fertility was examined by continuous mating scheme.
Result(s)
Previous studies have shown that Dkkl1, a gene unique to mammals, is expressed predominantly, if not exclusively, in developing spermatocytes, and the DKKL1 protein accumulates in the acrosome of mature sperm. Subsequent studies (reported in the accompanying paper) demonstrate that Dkkl1 also is expressed in the trophectoderm/placental lineage. Taken together, these results strongly suggested that DKKL1 protein is required for terminal differentiation either of trophoblast giant cells or of sperm, both of which are directly involved in fertility. To challenge this hypothesis, conditional targeted mutagenesis was used to ablate the Dkkl1 gene in mice. Surprisingly, Dkkl1 nullizygous embryos developed into viable, fertile adults, despite the fact that they failed to produce any portion of the DKKL1 protein.
Conclusion(s)
DKKL1 is a mammalian-specific acrosomal protein that is not essential either for development or fertility.
Keywords: Fertilization, Acrosome, Sperm, DKKL1, conditional knock-out
INTRODUCTION
Dickkopf-Like 1 (Dkkl1) was identified independently as a distant homologue to the Dickkopf (DKK) family of proteins that modulate WNT/β-catenin signaling (1), and as a gene located 3.8 kb upstream of the transcription factor Tead2 [Fig. 1A; (2)]. Tead2 RNA is present in oocytes and eggs, but Dkkl1 RNA is not. Both are detectable soon after the onset of zygotic gene expression, but then the expression patterns of these two genes diverge. At the blastocyst stage, Dkkl1 expression is localized to the trophectoderm whereas Tead2 expression occurs at similar levels in both the trophectoderm and inner cell mass (3), suggesting that DKKL1 functions in placental development. In adult mice, Dkkl1 RNA is expressed abundantly and almost exclusively in developing spermatocytes where DKKL1 accumulates first in developing acrosomes and then in the acrosome of mature sperm (2, 4). DKKL1 is an N-glycosylated protein that is processed during maturation of spermatocytes into sperm (4), raising the possibility that DKKL1 exists in multiple active forms.
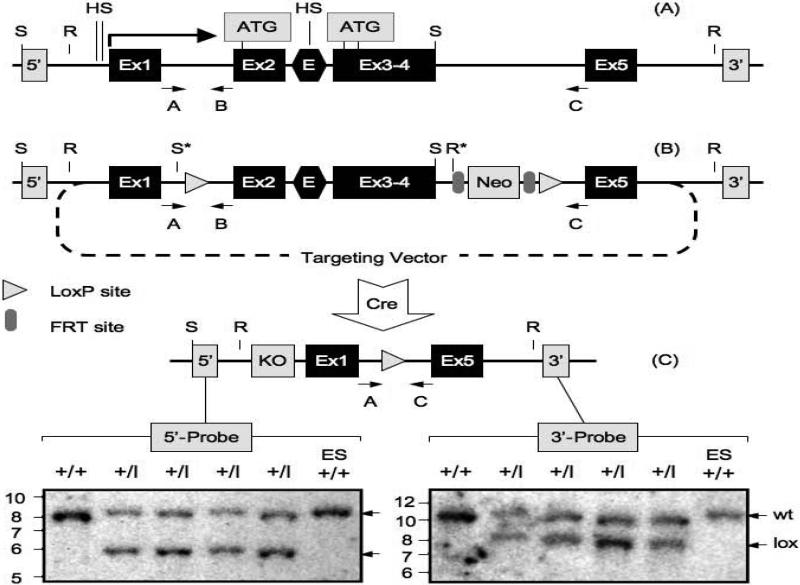
Figure 1.
Construction of a Dkkl1 conditional knock-out mouse. (A) The Dkkl1 gene contains five exons (Ex) that encode 230 amino acids. The transcription start site is indicated by a long arrow (2), while three in-frame translation start sites (ATGs) are located in exons 2 and 3. A putative Dkkl1 gene enhancer (E) is located within intron 2. Two DNase I hypersensitive sites (HS) are located in the Dkkl1 promoter and one in the enhancer (3). Also indicated are EcoRV (R) and SacI (S) restriction endonuclease sites, 5’- and 3’-sequence specific DNA probes used in Southern blotting-hybridization analysis (shaded 5’ and 3’ boxes), and PCR primers A, B and C (see Materials and Methods) used for multiplex genomic DNA PCR assays. (B) The ‘floxed gene’ within the targeting vector was used to introduce a mutated Dkkl1 locus by homologous recombination, and spans the region from just upstream of the Dkkl1 promoter (EcoRV site) through exon 5. Lox P sites were inserted within introns 1 and 4. A neomycin resistance gene (Neo), driven by the phosphoglycerate kinase (PGK) gene promoter and flanked by Flip recombinase target (FRT) sites, was inserted into intron 4 just upstream of the 3’-loxP site. Newly created restriction sites are indicated with an asterisk. (C) Cre recombinase excised the region between the two lox P sites to generate the Dkkl1 knock-out allele. The probe (shaded KO box) used to detect DNA fragments containing this allele was just upstream of exon 1. Genomic DNA from the tails of pups born from matings between chimeric males and C57BL/6J females was digested either with SacI (5’-probe, nucleotides 6135−7514, Genbank AF274313.1) or EcoRV (3’-probe, nucleotides 190309−189131, Genbank AC149868.2) restriction endonuclease and then subjected to Southern blotting-hybridization. SacI digested DNA was detected with the 5’-probe as indicated, and EcoRV DNA was detected with the 3’-probe. SacI produced a 7.9kb DNA fragment from the wild-type allele (wt), and a 5.9kb DNA fragment from the mutant allele (lox). EcoRV produced a 10.8kb fragment from the wild-type allele (wt) and an 8.1kb fragment from the lox allele (lox). DNA from wild-type ES cells was included as a positive control. DNA from agouti pups are in lanes 2−5 (+/lox), and DNA from a black littermate (+/+) is in lane 1.
The most direct approach to understanding gene function is to mutate the gene using gene targeting methodology (5). Given the possibility that deletion of Dkkl1 may result in lethality and/or sterility, a conditional Dkkl1 knockout mouse was constructed so that Dkkl1 could be ablated either in all mouse tissues or in selected tissues. Subsequent analysis of the Dkkl1 gene locus (RNA and protein products) confirmed the complete absence of DKKL1 protein in mice nullizygous for the Dkkl1 gene. Surprisingly, Dkkl1−/− mice were as fertile as wild-type mice. Therefore, Dkkl1 is not essential for development, viability or fertility.
MATERIALS AND METHODS
Construction of conditional knockout allele and RNA/protein analyses
The conditional Dkkl1 knockout allele was constructed in a manner similar to that described for a Tead2 conditional knockout allele (6). The targeting vector (Fig. 1A) was constructed using recombinant DNA methods [(7); described at http://recombineering.ncifcrf.gov/] using BAC clone 75.6i [Genome Systems (2)] as template for all PCR reactions]. Briefly, gap repaired plasmid was made by PCR amplification using primers D/E (see below) and restriction endonucleases NotI/HindIII as well as PCR amplification using primers F/G and HindIII/SpeI. The two fragments were cloned into NotI/SpeI-digested pL253. This plasmid was digested with HindIII, electroporated into heat-induced EL350 E. coli containing BAC 75.6i, and selected for ampicillin resistance. LoxP site was introduced into intron 1 (Fig. 1B) in the gap repaired plasmid as described (6). The “right-arm” was constructed by PCR using primers H/I and digested with NotI/EcoRI. The “left-arm” was constructed by PCR using primers J/K and digested with EcoRI/XhoI. The two arms were mixed and ligated to NotI/SalI digested pBluescript (Stratagene). LoxP flanked PGK-Neo cassette was ligated into EcoRI/BamHI sites of the above mini-targeting vector. This mini-targeting cassette was excised from the vector by digestion with NotI/Acc65I and along with gap repaired plasmid, was electroporated into heated-induced EL350 E. coli. Neo cassette was removed from resulting plasmid (leaving a single loxP site within intron 1) as described (6). A second mini-targeting vector was constructed as follows. The ‘left arm’ was constructed by PCR using primers L/M and digested with NotI/EcoRI. The ‘right arm’ was constructed by PCR using primers N/O and digested with SalI/EcoRI. The two fragments were ligated into NotI/SaI digested pBluescript. The FRT flanked loxP/PGK-Neo cassette (digested with EcoRI/BamHI) was then ligated into the EcoRI/BglII-digested mini-targeting vector. NotI/SalI digested mini-targeting cassette along with gap repaired plasmid containing the loxP in intron 1 was electroporated into heat-treated EL350. The final targeting vector was linearized by NotI and electroporated into the GSI-1 ES line (Genome Systems). ES selection and injection of appropriately cloned ES lines into blastocysts were done exactly as previously described (6).
Multiplex PCR genotyping was done as described (6) using primers A/B/C. Southern, Northern, Western, immunofluorescence and histological analyses were done as described (4, 6).
PCR Primer Sequences (5’→3’)
(A) acacattgcaggtgggtgggag, (B) ccagaactggatattaactgtccg, (C) cccttttgacacgttacact, (D) ataagcggccgcaagtggctcctatcccactta, (E) gtcaagcttctgtcccaggatagacagag, (F) gtcaagcttgcttgggtacctattaatgcgg, (G) ggactagttatcggcagctttcttctgcg, (H) ataagcggccgcagggcgtggtgagacaattag, (I) gtcgaattcgagctcggcgttctctccatttcagg, (J) ggaattcggatcctacagaatccagggggatgc, (K) ccgctcgagttggagaagcctctgaaggc, (L) ataagcggccgcttccagtttcctctctgctg, (M) gtcgaattcgagctccagatatctcaagga, (N) ggaattcagatctagtgtaacgtgtcaaaagggag, (O) acgcgtcgactttctggaacgaccaaaacgc, (P) aagtagaagcaaaagagcccc, (Q) cctgaggatgtagagaaagtgtgtc, (R) tatgcacagttccagctgcg, (S) aggtggcagacagatggtgt
RESULTS
Construction Of A Dkkl1 Conditional Knockout Allele
To determine whether or not Dkkl1 is required during either mammalian development or reproduction, a conditional knockout allele of Dkkl1 was constructed to allow inactivation by expression of Cre-recombinase in specific tissues. EIIa-Cre-mediated excision in the progeny removed exons 2 through 4, encoding 56% of the protein (Fig. 1). This deletion removed the first in-frame ATG and the putative signal peptide, both of which are encoded by exon 2. The deletion also removed the next two downstream ATGs present in exon 3 and the putative Dkkl1 transcription enhancer element in intron 2 (Kaneko K.J., Vassilev, A. and M.L. DePamphilis, unpublished results). This deletion should prevent production of full-length functional DKKL1 with a signal peptide.
The wild-type Dkkl1 allele (Fig. 1A) was mutated by homologous recombination in ES cells using a genetically altered targeting vector that contained a loxP site within intron 1 and another loxP site upstream of exon 5 (Fig. 1B). Cre-recombinase then directs excision of the intervening sequence. The targeting vector also contained the phosphoglycerate kinase promoter driving a neomycin-resistance gene (PGK-Neo) in intron 4 upstream of the loxP site to allow selection of recombinants. The Neo gene was flanked by FRT sites (the target of FLP-recombinase) to allow deletion of Neo (6, 8).
Genomic DNA prepared from tail clippings of agouti offspring (indicative of germline transmission) were screened by PCR for the presence of the mutant allele (data not shown), and the results were confirmed by Southern blotting-hybridization using either 5’-/3’-specific DNA probes (Fig. 1C). Both assays confirmed that the mutant allele was transmitted through the germline. These mice were then mated with EPEIIa-Cre+ mice (9) to delete exons 2 through 4 by Cre-dependent recombination (Fig. 1B/C). Since EIIa promoter activity in mice is restricted to oocytes and preimplantation embryos (10), mating with EIIa-Cre+ mice produced a constitutive knock-out allele.
Inactivation Of Dkkl1 Did Not Prevent Development Of Viable Adult Animals
Dkkl1+/− F1 mice were mated and their offspring genotyped both by Southern blotting-hybridization and PCR analyses (Fig. 2A). Both assays confirmed Cre-mediated deletion of the region flanked by the two loxP sites (‘floxed’ gene, Fig. 1B). The genotypic ratios of Dkkl1+/+, Dkkl1+/−, and Dkkl1−/− offspring were consistent with Mendelian segregation. Of the 173 mice produced by mating Dkkl1+/− mice, 28% were +/+, 45% were +/− and 27% were −/−. Moreover, the ratio of males to females was 1.5, 1.2 and 1.8, respectively, indicating that Dkkl1 did not influence sex determination. Furthermore, the size, morphology, and behavior of Dkkl1 nullizygous mice were indistinguishable from wild-type mice with the same mixed genetic background. Therefore, despite the strong expression of Dkkl1 in placental tissues (11), Dkkl1 is not required for development of fertilized eggs into healthy adult animals.
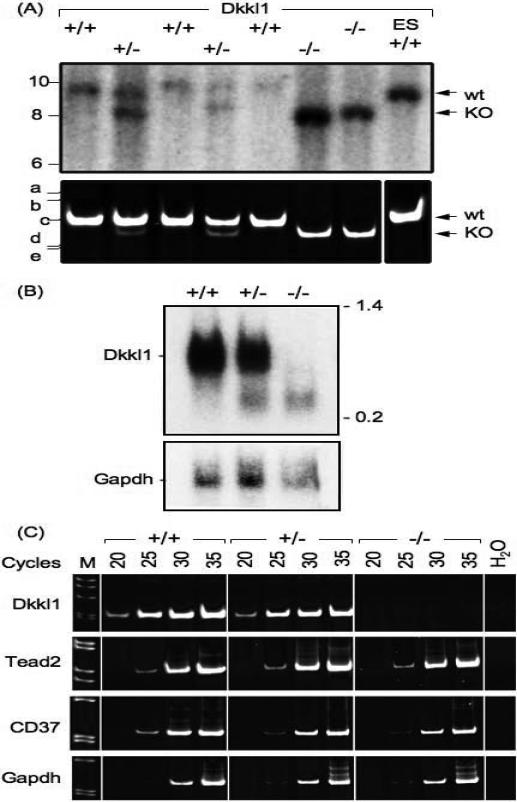
Figure 2.
Absence of Dkkl1 mRNA in Dkkl1−/− mice. Total RNA was isolated (14) and subjected to Northern blotting-hybridization and reverse transcription-polymerase chain reaction (RT-PCR) (2). (A) Mice containing the mutant allele were mated to EIIa-Cre+ mice, and resulting pups were assayed for the deleted allele (KO). Dkkl1+/− mice were then mated to each other, and their litter of seven pups was assayed for the presence of the KO allele. Top panel: DNA was digested with EcoRV, subjected to Southern blotting-hybridization, and the DNA detected with the ‘KO probe’ (nucleotides 2168−3034 Genbank AF274313.1) indicated in Figure 1C. The wild-type allele generated a 10.8kb DNA fragment (wt) whereas the deleted allele generated a 8.8kb DNA fragment (KO). Bottom panel: The same DNA samples also were subjected to multiplex PCR analysis. Primers A and B (Fig. 1A) amplified a 297bp fragment from the wild-type allele (wt), and primers A and C (Fig. 1C) amplified a 271bp fragment from the KO allele (KO). (B) Total RNA (15μg) was prepared from adult testes that were either wild-type, heterozygous, or nullizygous for Dkkl1 and subjected to hybridization-blotting as in Figure 3 using Dkkl1 32P-cDNA as a probe. Position of full-length Dkkl1 mRNA is indicated. The blot was stripped and rehybridized with Gapdh 32P-cDNA as control. The 0.24−9.5kb RNA ladder from Invitrogen was used to determine migration of RNA according to molecular size. (C) RNA from panel B was used as a template for cDNA synthesis to determine whether or not Dkkl1 exons 2 and 3 were present. RT-PCR was performed for the indicated number of cycles using Dkkl1 primers that amplified the region from exons 2 and 3, as described (2). The same aliquot of RNA also was used to amplify Tead2 and CD37 specific sequences (11). The 1kb DNA ladder from Invitrogen (M) was used to determine the size of the RT-PCR products. RT-PCR of Gapdh mRNA confirmed that equal amount of RNA samples were amplified. Water (H2O) was used as a negative control. Probes and PCR primers for Tead2, Dkkl1 and Gapdh are described in (3). Probe for Dkkl1 Exon5 was generated using PCR primers P and Q (Materials and Methods). Primers R and S were used to amplify a 617bp DNA fragment from the CD37 gene.
Functional Dkkl1 Protein Was Absent In Dkkl1 Knockout Mice
To confirm that Dkkl1 expression was indeed absent in Dkkl1−/− mice given their unexpected viability, the levels of Dkkl1 specific mRNA were examined by Northern blotting-hybridization with RNA collected from testes. The results confirmed that Dkkl1 mRNA was reduced in Dkkl1 heterozygous mice, and absent in Dkkl1−/− mice (Fig. 2B). A smaller band was generated by the deleted allele, but its level was significantly reduced compared with the wild-type allele, consistent with the fact that the Dkkl1 mutant allele retains the native Dkkl1 promoter, but it lacks the enhancer found in intron 2 (Fig. 1).
To confirm that Dkkl1 exons 2, 3 and 4 were deleted from any Dkkl1 mRNA that may have been produced from the residual exons in Dkkl1−/− mice, total RNA was prepared from testis and splenocytes, the only tissues in adult mice that produce detectable levels of Dkkl1 (3), and the presence of exons 2−4 was assayed for by RT-PCR, as previously described (2). As expected, Dkkl1 mRNA containing exons 2−4 was detected in both tissues either from Dkkl1 wild-type or from heterozygous mice, but exons 2−4 were absent in mRNA from both testis and splenocytes of Dkkl1 nullizygous mice (Fig. 2C; data not shown). Hence, any mRNA produced from the Dkkl1 mutant allele could not produce a functional DKKL1 protein. Of equal importance is the fact that the mutated Dkkl1 allele did not affect expression of either Tead2 or CD37 (Fig. 2C), two genes that lie in closest proximity to Dkkl1 (2).
To confirm that DKKL1 protein was absent in Dkkl1−/− mice and examine the effects of DKKL1 loss on acrosome structure, tissue sections of testis were stained either with anti-DKKL1 antiserum to detect DKKL1 protein, or with Periodic acid-Schiff (PAS) to detect glycoproteins (12). No differences were detected between testis from Dkkl1+/+ and Dkkl1−/− males stained with PAS (Fig. 3, “testis”/PAS). In contrast, DKKL1 protein was abundant in testis from wild-type males, but absent in testis from Dkkl1−/− males (Fig. 3, “testis”/DKKL1). Testis sections from Dkkl1+/− males exhibited an intermediate level of staining (data not shown).
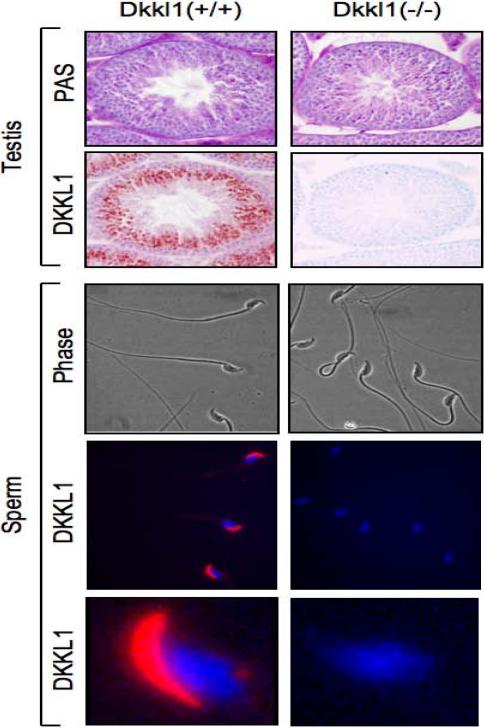
Figure 3.
DKKL1 protein was absent in Dkkl1−/− testis and sperm. Testes from wild-type and Dkkl1−/− males at six months of age were fixed overnight in Bouin's fixative, sectioned and stained with periodic Acid Schiff (PAS) to visualize glycoproteins. In both cases, sections were counterstained with hematoxylin. The same tubule is shown in both top and bottom images from neighboring sections. Sperm from cauda epididymides was collected from wild-type and Dkkl1 nullizygous males, washed several times with phosphate buffered saline and spread on glass slides. DKKL1 protein was detected in tissue sections using a goat polyclonal antibody (AF1508, R&D Systems), followed by Cy3-coupled anti-goat secondary antibody (Jackson Immunologics) or biotinylated anti-goat secondary antibody (Invitrogen) (4). PAS staining was performed according to the manufacturer's protocol (Polysciences). Sperm from wild-type and nullizygous males were stained for DKKL1 protein (red). Nuclei were visualized with DAPI (Blue). One of the sperm heads from above panel was enlarged to show DKKL1 specific staining at the base of the sperm head (bottom, white arrow). Phase contrast images corresponding to above immunostaining show normal morphology of sperm (top).
Similarly, anti-DKKL1 antiserum revealed that DKKL1 protein in wild-type sperm localized to the acrosome (4), whereas DKKL1 protein was absent in sperm from Dkkl1−/− males (Fig. 3 “sperm”). Interestingly, a relatively weak DKKL1 signal was consistently detected at the base of the head in wild-type sperm (Fig. 3, white arrow). Visual inspection of sperm by phase contrast microscopy failed to detect any consistent morphological differences between Dkkl1+ and Dkkl1− sperm (Fig. 3; data not shown).
Although mRNA transcripts from the ablated Dkkl1 locus could not produce a normal DKKL1 protein, they might still produce a truncated protein made from the open reading frame (ORF) within exon 5, the only coding exon remaining after Cre-mediated excision (Fig. 1). Such a truncated protein might contain partial DKKL1 activity, thereby masking the phenotype of Dkkl1−/− mice. To address this possibility, we first determined whether or not the polyclonal DKKL1 antibody recognized truncated proteins.
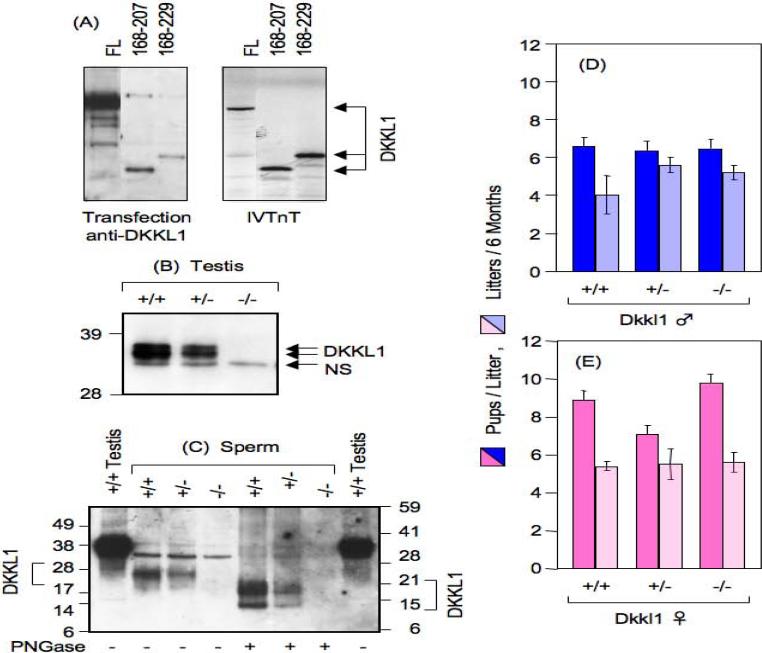
Expression vectors were constructed that contained either full-length or partial sequences of the Dkkl1 ORF (including one that contained only exon 5 with an in-frame ATG) fused to enhanced green fluorescence protein (EGFP) attached at the DKKL1 C-terminus. NIH3T3 cells were then transfected with these plasmids. All of the chimeric proteins were detected by Western immuno-blotting (Fig. 4A, left panel), and each chimeric protein produced in vivo co-migrated with [35S]methionine-labeled proteins that had been translated in vitro (Fig. 4A, right panel). Similarly, immuno-fluorescent staining detected both full-length and partial fragments of DKKL1 in EGFP expressing cells (data not shown). These results confirmed that the polyclonal DKKL1 antibody recognized epitopes throughout the DKKL1 protein, including an epitope specific to exon 5, and therefore would have detected a partial protein produced from the deleted allele, if it was present.
Figure 4.
DKKL1 protein and its effect on mouse fertility. (A) To determine whether or not polyclonal Dkkl1-specific antibody can recognize truncated DKKL1 protein, an EGFP fusion expression vector containing full-length (FL) or potential open reading frames within Exon 5 (corresponding to amino acid residues 168−207 or 168−229) were driven by a CMV promoter in vivo or by a bacteriophage T7 promoter in vitro. Plasmids were transfected into mouse NIH3T3 cells, and 48 hours later the cells were probed with the antibody by Western immuno-blotting (left panel). The antibody recognized the same size DKKL1 products as those produced from 35S-labeled in vitro translation (right panel). To determine whether or not DKKL1 was absent in Dkkl1−/− testis and sperm, whole cell extracts of testis (B) or sperm (C) prepared from mice that were wild-type (+/+), heterozygous (+/−), or nullizygous (−/−) for the Dkkl1 allele were denatured and fractionated on SDS-PAGE followed by Western immuno-blotting (4) using the same polyclonal antibody specific to DKKL1. DKKL1 products are highlighted with bracket/arrows, whereas a non-specific background band is indicated by “NS”. DKKL1 from testis migrated ∼34 kDa, as previously reported (4). Sperm samples were treated with N-glycosidase-F (PNGase) to reveal the presence of N-glycosylation (4). Sperm extracts from wild-type CD-1 males (labeled “+/+ Testis”) were used as controls in panel C. To determine the effect of DKKL1 on mouse fertility, Dkkl1+/+, or +/− or −/− males (♂) were mated with C57/B16 females (D), and Dkkl1+/+, or +/− or −/− females (♀) were mated with C57/B16 females (E). The number of pups born per litter (dark blue and red bars) and the number of litters produced per 6 months (light blue and pink bars) were recorded. Error bars indicate the SEM. N = 5 for all animals except for Dkkl1 wild-type males (N = 2) and heterozygous females (N = 4). An average of 20 litters per mating were analyzed.
No DKKL1 protein or protein fragment detected either in testis or in sperm from either Dkkl1+/+ or Dkkl1+/− males was detected either in testes or in sperm from Dkkl1−/− males (Fig. 4B; 4C). Moreover, sperm from Dkkl1+/− males contained about half as much DKKL1 as wild-type sperm. Taken together, these results demonstrate that all forms of DKKL1 protein were effectively absent from Dkkl1 nullizygous mice.
DKKL1 Protein Was Post-Translationally Modified In Wild-Type Mice
Western immuno-blotting further demonstrated that DKKL1 protein was post-translationally modified. For example, DKKL1 from testis migrated during SDS-PAGE with a molecular weight of approximately 34 kDa, but after treatment with a glycosidase, it migrated with a molecular weight of approximately 29 kDa. This was similar in size to DKKL1 protein translated in vitro (4). Therefore, DKKL1 in testis is a glycoprotein containing predominantly, if not exclusively, N-linked carbohydrates. With increased resolution, DKKL1 in testis migrated as two bands with approximate molecular weights of 37 kDa and 35 kDa (Fig. 4B). DKKL1 in mature sperm migrates predominantly as a 22 kDa protein before treatment with glycosidase, and as an 18 kDa protein after treatment (Fig. 4C). In addition, a minor fraction migrated at 17 kDa before treatment, and 13 kDa after treatment. Since none of these proteins were detected in Dkkl1 nullizygous sperm, all of them must be forms of DKKL1. Thus, two modifications occur during sperm maturation; DKKL1 protein is N-glycosylated at multiple sites, and it is truncated.
Fertility Does Not Require DKKL1 Protein
To determine whether or not Dkkl1 expression affected fertility, wild-type C57BL/6 males and C57BL/6 females were continually mated with mice that were either Dkkl1+/+, Dkkl1+/− or Dkkl1−/−, and both the number of pups born per litter and the number of litters produced in six months were recorded. Surprisingly, ablation of the Dkkl1 gene did not affect the ability of these mice to reproduce by natural matings (Fig. 4D).
DISCUSSION
In previous studies, we have shown that Dkkl1 expression is restricted to developing sperm, trophectoderm and T-lymphocytes (2-4). In a companion study, we extended the expression pattern to the trophoblast cells that give rise to the placenta (11). To determine whether or not Dkkl1 affected mammalian development, viability or fertility, we constructed a mouse line that permitted complete ablation of the Dkkl1 gene either throughout the animal or in specific tissues. Comparisons of wild-type and Dkkl1 deficient mice revealed that between spermatogenesis in the testis and sperm maturation in the epididymis, N-glycosylated DKKL1 protein undergoes what appears to be partial deglycosylation and protease cleavage that reduces its apparent molecular weight by about 40%. Hence, the DKKL1 protein in mature sperm is a different form of the protein than that in developing spermatocytes. The biological significance of this transition remains to be elucidated.
Remarkably, mice lacking any form of the DKKL1 protein developed into what appear to be normal, healthy, viable, fertile adults. Given that the Dkkl1 gene is unique to mammals, we assume that its function, particularly in fertility, is too subtle to be revealed under normal laboratory conditions. Presumably, it imparts an evolutionary advantage to mammals in the wild. In fact, as revealed in the accompanying manuscript (11), Dkkl1 is, in fact, required for efficient sperm penetration of the zona pellucida.
While this manuscript was in preparation, another study appeared that also reported that Dkkl1−/− mice were fertile (13), although no analysis of fertility was included. This study concluded that DKKL1 induces apoptosis in post-pubertal spermatocytes, thereby regulating their population density. We did not detect differences in levels of apoptosis in testis sections of Dkkl1−/−males compared to wild-type or heterozygous males. One explanation for this difference may be that our studies were done on 6-month old animals, whereas the conclusions of Dakhova et al. were based on younger animals. Other possibilities arise from the fact that their Dkkl1−/− mice were created by insertional mutagenesis which is susceptible to artifacts (5). Their construction retained two downstream ATGs that may produce truncated forms of DKKL1 (2) from mRNAs that read through the polyA stop signal. Since DKKL1 exists in different forms (Fig. 4C), different forms of DKKL1 may have different functions. Their construction also contained a PGK promoter and the Dkkl1 enhancer, elements that may alter expression of other genes that are responsible for their observed phenotype.
Acknowledgments
This work was funded by the intramural program of the National Institute of Child Health and Human Development. Animals were treated according to the Guide for the Care and Use of Laboratory Animals, Institute of Laboratory Animal Research, National Academy of Sciences, USA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Capsule: Ablation of the Dkkl1 gene, which is expressed predominantly, perhaps exclusively, in developing spermatocytes and trophoblast giant cells, does not prevent production of viable, fertile mice.
LITERATURE CITATIONS
- 1.Niehrs C. Function and biological roles of the Dickkopf family of Wnt modulators. Oncogene. 2006;25:7469–81. doi: 10.1038/sj.onc.1210054. [DOI] [PubMed] [Google Scholar]
- 2.Kaneko KJ, DePamphilis ML. Soggy, a spermatocyte-specific gene, lies 3.8 kb upstream of and antipodal to TEAD-2, a transcription factor expressed at the beginning of mouse development. Nucleic Acids Res. 2000;28:3982–90. doi: 10.1093/nar/28.20.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaneko KJ, Rein T, Guo ZS, Latham K, DePamphilis ML. DNA methylation may restrict but does not determine differential gene expression at the Sgy/Tead2 locus during mouse development. Mol Cell Biol. 2004;24:1968–82. doi: 10.1128/MCB.24.5.1968-1982.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kohn MJ, Kaneko KJ, DePamphilis ML. DkkL1 (Soggy), a Dickkopf family member, localizes to the acrosome during mammalian spermatogenesis. Mol Reprod Dev. 2005;71:516–22. doi: 10.1002/mrd.20314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muller U. Ten years of gene targeting: targeted mouse mutants, from vector design to phenotype analysis. Mechanisms of development. 1999;82:3–21. doi: 10.1016/s0925-4773(99)00021-0. [DOI] [PubMed] [Google Scholar]
- 6.Kaneko KJ, Kohn MJ, Liu C, DePamphilis ML. Transcription Factor TEAD2 Is Involved In Neural Tube Closure. Genesis. 2007;45:577–87. doi: 10.1002/dvg.20330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–84. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farley FW, Soriano P, Steffen LS, Dymecki SM. Widespread recombinase expression using FLPeR (flipper) mice. Genesis. 2000;28:106–10. [PubMed] [Google Scholar]
- 9.Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, et al. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:5860–5. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dooley TP, Miranda M, Jones NC, DePamphilis ML. Transactivation of the adenovirus EIIa promoter in the absence of adenovirus E1A protein is restricted to mouse oocytes and preimplantation embryos. Development. 1989;107:945–56. doi: 10.1242/dev.107.4.945. [DOI] [PubMed] [Google Scholar]
- 11.Kohn MJ, Sztein J, Yagi R, DePamphilis ML, Kaneko KJ. The Acrosomal Protein Dickkopf-Like 1 (DKKL1) Facilitates Sperm Penetration Of The Zona Pellucida. Fertil Steril. 2009;(this issue) doi: 10.1016/j.fertnstert.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leuchtenberger C, Schrader F. The chemical nature of the acrosome in the male germ cells. Proceedings of the National Academy of Sciences of the United States of America. 1950;36:677–83. doi: 10.1073/pnas.36.11.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dakhova O, O'Day D, Kinet N, Yucer N, Wiese M, Shetty G, et al. Dkkl1 regulates post-pubertal spermatocyte apoptosis and testosterone production. Endocrinology. 2009;150:404–12. doi: 10.1210/en.2008-0673. [DOI] [PubMed] [Google Scholar]
- 14.Kaneko KJ, Cullinan EB, Latham KE, DePamphilis ML. Transcription factor mTEAD-2 is selectively expressed at the beginning of zygotic gene expression in the mouse. Development. 1997;124:1963–73. doi: 10.1242/dev.124.10.1963. [DOI] [PubMed] [Google Scholar]