Abstract
The putative structure of the Tissue Factor/Factor VIIa/Factor Xa (TF/FVIIa/FXa) ternary complex is reconsidered. Two independently derived docking models proposed in 2003 (one for our laboratory: CHeA and one from the Scripps laboratory: Ss) are dynamically equilibrated for over 10 ns in an electrically neutral solution using all-atom molecular dynamics. Although the dynamical models (CHeB and Se) differ in atomic detail, there are similarities in that TF is found to interact with the γ-carboxyglutamic acid (Gla) and Epidermal Growth Factor-like 1 (EGF-1) domains of FXa, and FVIIa is found to interact with the Gla, EGF-2 and serine protease (SP) domains of FXa in both models. FVIIa does not interact with the FXa EGF-1 domain in Se and the EGF domains of FVIIa do not interact with FXa in the CHeB. Both models are consistent with experimentally suggested contacts between the SP domain of FVIIa with the EGF-2 and SP domains of FXa.
Keywords: Factor VIIa, Factor Xa, ternary complex, Molecular Dynamics simulation
Introduction
In both the conventional blood coagulation cascade [1] and in the more recent cell-based cascade [2,3], the ternary complex of FVIIa/TF/FXa is thought to play an important role in providing free FXa for the formation of prothrombinase (Factor Va, FXa, negatively charged phospholipids and calcium ions). While the structure of FVIIa/sTF has been determined in several laboratories and under several different conditions [4–6], the structure of the ternary complex remains unsolved. A static docking model [7] and a solution-equilibrated model [8] were generated in 2003; both models were derived independently and while globally similar, showed differences in the details of the structures. Significant gains in computational power have been realized since 2003, as well as additional experiments that probe the possible structure of the complex are now available [9]. Thus we wished to revisit these two models and provide a more extensive, current “best-guess” solution-equilibrated model.
Methods
We will use the following labels to describe the various models: Ss =Scripps static, Se=Scripps solution-equilibrated (14.2 ns), CHeA=original Chapel Hill solution-equilibrated (3.6 ns) and CHeB=current Chapel Hill solution-equilibrated for 10.5 ns. The Se model derives from the Scripps static model (PDB code: 1NL8) [7] with several modifications: Glu39 was modified to a Gla39 residue, residues 159–162 in TF were added, and calcium ions were placed on Gla (γ-carboxyglutamic acids) residues Gla32 and Gla39 to be consistent with our prior simulation (CHeA) [8]. The CHeB model derives from the starting docking model (the chirality of several residues was corrected) that led to CHeA [9], but which has undergone a simulation time sufficient for equilibration.
The details of the setup for the solvent equilibration of the ternary complex models (CHeB and Se) are given in the Supplementary Information of Ref. 10. The essentials are that the docked complexes are surrounded with layers of water using periodic boundary condition so that images in surrounding boxes do not interact, the systems are carefully equilibrated at the starting conformation and the particle mesh Ewald (PME) method [11] is used to compute the electrostatic interactions. The AMBER9 [12] program was employed along with the ff99SB force field, the TIP3P water model [13] and the dynamics code PMEMD9. The importance of using the PME method for macromolecular simulations has been discussed [14]. Both CHeB and Se were simulated for sufficient times (>10 ns) that the overall RMSD of the simulation to the starting structures was relatively constant with time.
Results and Discussion
A fair question is “How good is molecular dynamics (MD) for predicting the structures of macromolecular complexes in solution?” An optimistic view of the usefulness of MD to drug design has been given recently [15]. Likewise, the improvement of docked protease-inhibitor binding energies (experimental to predicted) for a large number of HIV-1-inhibtor complexes was improved considerably by use of molecular dynamics as opposed to protein-rigid docking without dynamics [16]. Significant overlap between essential spaces of proteins defined by NMR ensembles and molecular dynamics has also been shown [17]. In our hands, we were able to refine an NMR structure of the factor IX Gla domain obtained with significant denaturing agent present and locate the ω-loop (residues 1–13) and positions of the calcium ions to obtain a refined whole Gla domain that compared closely to similar domains of prothrombin and factor VIIa [18].
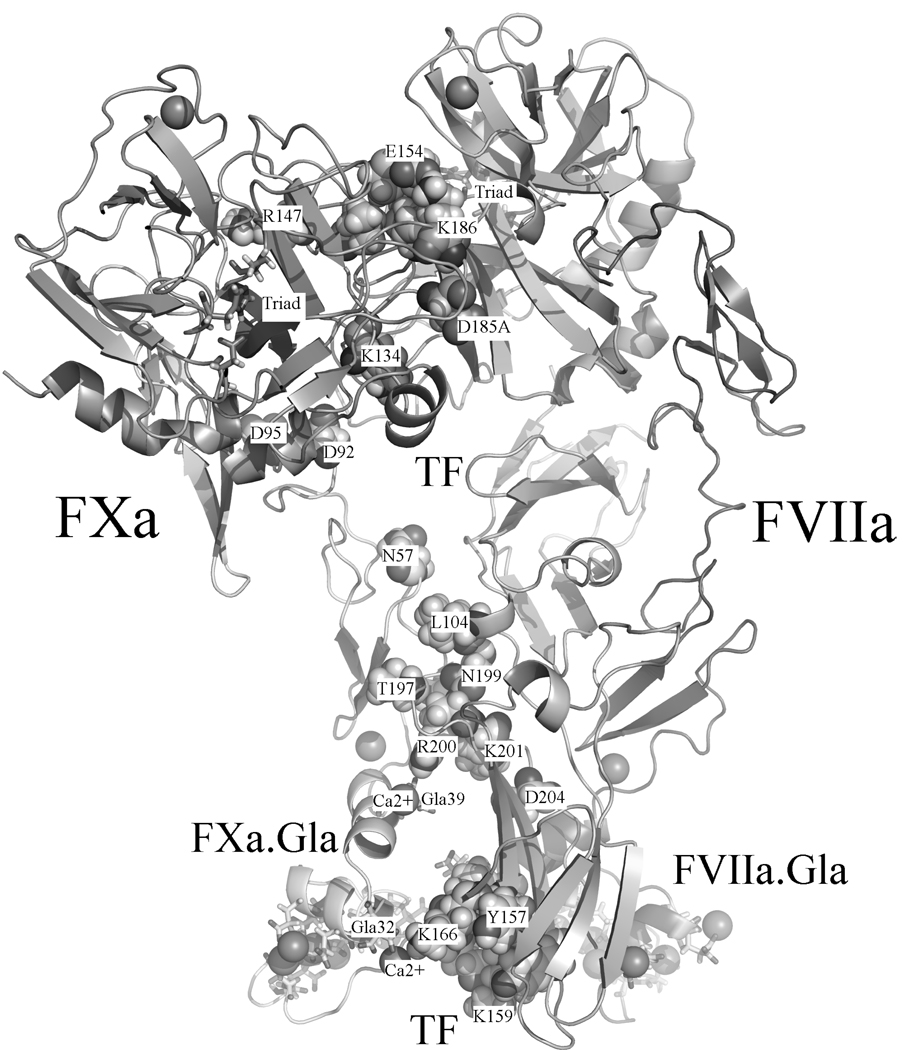
The goal of this work was to solution-refine two independently derived docking models for the FVIIa/TF/FXa complex and thereby arrive at an up-to-date estimates of the solution structure. The same force field, water model, integration method and treatment of electrostatic forces were employed for both models. Fig. 1 shows the Se model for orientation purposes. Table 1 provides a comparison of the contacts derived, TF/FVIIa with FXa, from the simulations (10.5 ns for CHeB and 14.2 ns for Se) and also gives a comparison to the experimental contacts [20–32]. Eight of the contacts are similar in both models, of these five have partial agreement with experiment. Both models bury about the same area in binding FXa, 4150 (CHeB) and 3921(Se) Å2. The TF/FVIIa units have an RMSD (backbone) of 2.92 Å between the models. For comparison, Se is 2.72 Å and CHeB is 2.78 Å RMSD to the X-ray crystal structure of TF/FVIIa in Ref. 4. On the other hand, the TF/FVIIa/FXa (des-Gla) units have an RMSD of 5.53 Å. Both models are consistent with experimentally suggested contacts between the SP domain of FVIIa with the EGF-2 and SP domains of FXa. FVIIa does not, however, interact with the FXa EGF-1 domain in the Se model and the EGF domains of FVIIa do not interact with FXa in the CHeB model. Overall, the differences in the models appear to trace back to the relative orientation of the EGF-1 in the original docking models. These models, which can be obtained on request, should be useful for comparison for new experiments.
Fig. 1.
Residues involved in experimental contacts are mapped onto the Se model. TF: (Lys165, Lys166) [20–25], (Tyr157, Lys159, Ser163, Gly164) [23,25], Tyr185 [24,25], (Asn199, Asp204) [24], Arg200 [9,24,26,27], Lys201 [9,27], (Leu104, Thr197) [27]; FVIIa: Arg 36 [28], SP((Val21, Glu154, Met156)[29,30], Leu144 [30] Ala152 [30,31], Arg147 [32]); FXa [7]: Glu51, Asn57, Asp92, Asp95, SP(Lys134, Asp185A, Lys186).
Table 1.
Comparison of predicted and experimental contacts from the Se and CHeB models. Interactions were computed with default setting in the Protein Interactions calculator server [19]. Chymotrypsin numbering system as in 1NL8 (pdb of Ss) is used for SP domains of FVIIa and FXa. Bold residues correspond to experimental contacts [20–32]. Abbreviations: HI, hydrophobic; HB, hydrogen bond; MS, main chain-side chain; SS, side chain-side chain; LC, light chain of FVIIa and FXa for residues over EGF-2 domain in light chain. Italic residues designate residues adjacent to residues involved in experimental contact.
| CHeB | Se | |
|---|---|---|
| TF/fXa | Glu24TF@OE1,OE2 ∷ Gly66fXa.EGF-1@O (MS-HB) | |
| Lys41TF ∷ Glu77fXa.EGF-1 (Ionic interaction) | ||
| Thr70TF@OG1 ∷ Gln58fXa.EGF-1@OE1 (SS-HB) | ||
| Glu99TF@N ∷ Glu77fXa.EGF-1@OE1 (MS-HB) | Glu99TF@OE1∷ Gln58fXa.EGF-1@NE2(SS-HB) | |
| Glu105TF ∷ Lys79fXa.EGF-1 (Ionic interaction) | ||
| Glu105TF@N ∷ Lys79fXa.EGF-1@OE2 (MS-HB) | ||
| Lys165TF@NZ ∷ Asp35fXa.Gla@OD2 (SS-HB) | ||
| Lys165TF ∷ Asp35fXa.Gla (Ionic interaction) | ||
| Lys166TF ∷ Gla32fXa.Gla (Ionic interaction) | ||
| Thr197TF@OG1∷ Cys61fXa.EGF-1@N (MS-HB) | ||
| Val198TF ∷ Pro54fXa.EGF-1 (HI) | ||
| Asn199TF@OD1∷ Ser53fXa.EGF-1@OG (SS-HB) | ||
| Arg200TF ∷ Gla39fXa.Gla (Ionic interaction) | ||
| Arg200TF ∷ Asp48fXa.EGF-1 (Ionic interaction) | ||
| Lys201TF@NZ ∷ Glu51fXa.EGF-1@OE2 (SS-HB) | ||
| Lys201TF ∷ Glu51fXa.EGF-1 (Ionic interaction) | ||
| Asp204TF ∷ Lys43fXa.Gla (Ionic interaction) | ||
| Asp204TF@OD1,OD2 ∷ Lys43fXa.Gla@NZ (SS-HB) | ||
| fVIIa/fXa | Ala34fVIIa.Gla ∷ Met18fXa.Gla (HI) | |
| Gla35fVIIa.Gla ∷ Lys36fXa.Gla (Ionic interaction) | ||
| Gla35fVIIa.Gla@N ∷ Met18fXa.Gla@SD (MS-HB) | ||
| Arg36fVIIa.Gla ∷ Gla14fXa.Gla (Ionic interaction) | ||
| Arg36fVIIa.Gla@NH1,NH2 ∷ Gla14fXa.Gla@OE1,OE2 (SS-HB) | ||
| Leu39fVIIa.Gla ∷ Phe31fXa.Gla (HI) | ||
| Leu39fVIIa.Gla ∷ Phe40fXa.Gla (HI) | ||
| Trp41fVIIa.Gla ∷ Met18fXa.Gla (HI) | ||
| Ile42fVIIa.Gla ∷ Met18fXa.Gla (HI) | ||
| Ile42fVIIa.Gla ∷ Phe40fXa.Gla (HI) | ||
| Ile42fVIIa.Gla ∷ Tyr44fXa.Gla (HI) | ||
| Gln150fVIIa.LC@O ∷ Lys134fXa.SP@NZ(MS-HB) | ||
| Arg152fVIIa.LC@O ∷ Lys134fXa.SP@NZ (MS-HB) | ||
| Lys20fVIIa.SP@NZ ∷ Tyr162fXa.SP@O (MS-HB) | Lys20fVIIa.SP ∷ Glu159fXa.SP (Ionic interaction) | |
| Lys20fVIIa.SP@NZ ∷ Glu159fXa.SP@OE1 (MS-HB) | ||
| Val21fVIIa.SP ∷ Pro161fXa.SP (HI) | ||
| Val21fVIIa.SP ∷ Tyr185fXa.SP (HI) | Val21fVIIa.SP ∷ Tyr185fXa.SP (HI) | |
| Glu26fVIIa.SP@OE1 ∷ Thr185BfXa.SP@OG1 (SS-HB) | ||
| Asp72fVIIa.SP ∷ Lys186fXa.SP (Ionic interaction) | ||
| Glu75fVIIa.SP ∷ Lys223fXa.SP (Ionic interaction) | ||
| Leu145fVIIa.SP ∷ Ile137fXa.SP (HI) | ||
| Leu145fVIIa.SP ∷ Met157fXa.SP (HI) | ||
| Leu145fVIIa.SP ∷ Tyr207fXa.SP (HI) | ||
| Asp146fVIIa.SP ∷ Arg202fXa.SP (Ionic interaction) | ||
| Arg147fVIIa.SP@NH1,NH2 ∷ Glu138fXa.LC@OE1,OE2 (SS-HB) | ||
| Arg147fVIIa.SP ∷ Glu138fXa.LC (Ionic interaction) | ||
| Leu153fVIIa.SP@N ∷ Gln20fXa.SP@OE1 (MS-HB) | ||
| Glu154fVIIa.SP@OE1,OE2 ∷ Tyr185fXa.SP@OH (SS-HB) | ||
| Glu154fVIIa.SP ∷ Lys186fXa.SP (Ionic interaction) | Glu154fVIIa.SP ∷ Lys186fXa.SP (Ionic interaction) | |
| Glu154fVIIa.SP@OE1 ∷ Lys186fXa.SP@OH (SS-HB) | Glu154fVIIa.SP@OE2 ∷ Lys186fXa.SP@NZ (SS-HB) | |
| Lys170DfVIIa.SP ∷ Glu74fXa.EGF-1 (Ionic interaction) | ||
| Lys170DfVIIa.SP@NZ ∷ Glu74fXa.EGF-1@OE1,OE2 (SS-HB) | ||
| Lys170DfVIIa.SP ∷ Asp92fXa.EGF-2 (Ionic interaction) | Arg170CfVIIa.SP ∷ Asp92fXa.EGF-2 (Ionic interaction) | |
| Lys170DfVIIa.SP ∷ Asp95fXa.EGF-2 (Ionic interaction) | Arg170CfVIIa.SP ∷ Asp95fXa.EGF-2 (Ionic interaction) | |
| Arg170CfVIIa.SP@NH1,NH2 ∷ Asp95fXa.EGF-2@OD1 (SS-HB) | ||
| Tyr184fVIIa.SP ∷ Lys134fXa.SP (Cation-Pi interaction) | ||
| Asp186fVIIa.SP ∷ Lys134fXa.SP (Ionic interaction) | ||
| Asp186fVIIa.SP@OD1 ∷ Lys134fXa.SP@NZ (SS-HB) | ||
| Ser188AfVIIa.SP@OG ∷ Lys204fXa.SP@N (MS-HB) | Ser188AfVIIa.SP@O ∷ Arg202fXa.SP@NH2 (MS-HB) | |
Acknowlegements
This work was supported in part by NIH (HL-06350), NSF (FRG DMR-0804549) and by the Intramural Research Program of NIEHS. We are grateful for access to ITS computing resources at UNC-CH.
Footnotes
Conflict of Interest Statement
All authors declare no conflicts of interest to disclose.
References
- 1.Roberts HR, Tabares AH. Overview of the coagulation reactions. In: High KA, Roberts HR, editors. Molecular Basis of Thrombosis and Haemostatis. New York: Marcel Dekker; 1995. pp. 35–50. [Google Scholar]
- 2.Hoffman M, Monroe DM. A cell-based model of hemostasis. Thrombosis and Haemostasis. 2001;85:958–965. [PubMed] [Google Scholar]
- 3.Roberts HR, Hoffman M, Monroe DM. A cell-based model of thrombin generation. Seminars in Thrombosis and Hemostasis. 2006;32:32–38. doi: 10.1055/s-2006-939552. [DOI] [PubMed] [Google Scholar]
- 4.Banner DW, Darcy A, Chene C, Winkler FK, Guha A, Konigsberg WH, Nemerson Y, Kirchhofer D. The crystal structure of the complex of blood coagulation factor VIIa with soluble tissue factor. Nature. 1996;380:41–46. doi: 10.1038/380041a0. [DOI] [PubMed] [Google Scholar]
- 5.Zhang E, St Charles R, Tulinsky A. Structure of extracellular tissue factor complexed with factor VIIa inhibited with a BPTI mutant. J Mol Biol. 1999;285:2089–2104. doi: 10.1006/jmbi.1998.2452. [DOI] [PubMed] [Google Scholar]
- 6.Bajaj SP, Schmidt AE, Agah S, Bajaj MS, Padmanabhan K. High resolution structures of p-aminobenzamidine and benzamindine-VIIa soluble tissue factor– Unpredicted conformation of the 192–193 peptide bond and mapping of Ca2+, Mg2+, Na+ and Zn2+ sites in factor VIIa. J Biol Chem. 2006;281:24873–24888. doi: 10.1074/jbc.M509971200. [DOI] [PubMed] [Google Scholar]
- 7.Norledge BV, Petrovan RJ, Ruf W, Olson AJ. The tissue factor/factor VIIa/factor Xa complex: A model built by docking and site-directed mutagenesis. Proteins: Struc Func and Gen. 2003;53:640–648. doi: 10.1002/prot.10445. [DOI] [PubMed] [Google Scholar]
- 8.Venkateswarlu D, Duke RE, Perera L, Darden T, Pedersen LG. An all-atom solution-equilibrated model for human extrinsic blood coagulation complex (sTF-VIIa-Xa): a protein-protein molecular dynamics refinement study. J Thromb Haem. 2003;1:2577–2588. doi: 10.1111/j.1538-7836.2003.00421.x. [DOI] [PubMed] [Google Scholar]
- 9.Manithody C, Yang LK, Rezaie AR. Identification of a basis region of tissue factor that interacts with the first epidermal growth factor-like domain of factor X. Biochemistry. 2007;46:3193–3199. doi: 10.1021/bi6025193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee CJ, Lin P, Chandrasekaran V, Duke RE, Everse SJ, Perera L, Pedersen LG. Proposed structural models of human factor Va and prothrombinase. J Thromb Haem. 2007;6:83–89. doi: 10.1111/j.1538-7836.2007.02821.x. [DOI] [PubMed] [Google Scholar]
- 11.Darden T, York D, Pedersen L. Particle Mesh Ewald – An N logN method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 12.Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Pearlman DA, Crowley M, Walker RC, Zhang W, Wang B, Hayik S, Roitberg A, Seabra G, Wong KF, Paesani F, Wu X, Brozell S, Tsui V, Gohlke H, Yang L, Tan C, Mongan J, hornak V, Cui G, Beroza P, mathews DH, Schafmeister C, Ross WS, Kollman PA. AMBER 9. San Francisco: University of California; 2006. [Google Scholar]
- 13.Jorgensen WL, Chadrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 14.Darden T, Perera L, Li L, Pedersen L. New tricks for modelers from the crystallographic toolkit: the particle mesh Ewald algorithm and its use in nucleic acid simulations. Structure. 1999;7:R55–R60. doi: 10.1016/s0969-2126(99)80033-1. [DOI] [PubMed] [Google Scholar]
- 15.Galeazzi R. Molecular Dynamics as a tool in rational drug design: current status and some major applications. Curr Computer-aided Drug Des. 2009;5:225–240. [Google Scholar]
- 16.Jenwitheesuk E, Samudrala R. Improved prediction of HIV-1 protease-inhibitor binding energies by molecular dynamics simulation. BMC Struct Biol. 2003;3:2. doi: 10.1186/1472-6807-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abseher R, Horstink L, Hilbers CW, Nilges M. Essential spaces defined by NMR structure ensembles and molecular dynamics simulation show significant overlap. Proteins: Struc Func and Gen. 1998;31:370–382. [PubMed] [Google Scholar]
- 18.Li LP, Darden TA, Freedman SJ, Furie BC, Furie B, Baleja JD, Smith H, Hiskey RG, Pedersen LG. Refinement of the NMR solution structure of the gamma-carboxyglutamic acid domain of coagulation factor IX using molecular dynamics simulation with initial Ca2+ positions determined by a genetic algorithm. Biochem. 1997;36:2132–2138. doi: 10.1021/bi962250r. [DOI] [PubMed] [Google Scholar]
- 19.Tina KG, Bhadra R, Srinivasan N. PIC: Protein Interactions Calculator. Nucleic Acids Research. 2007;35:W473–W476. doi: 10.1093/nar/gkm423. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roy S, Hass PE, Bourell JH, Henzel WJ, Vehar GA. Lysine residues 165 and 166 are essential for the cofactor function of tissue factor. J Biol Chem. 1991;266:22063–22066. [PubMed] [Google Scholar]
- 21.Huang Q, Neuenschwander PF, Rezaie AR, Morrissey JH. Substrate recognition by tissue factor-factor VIIa: Evidence for interaction of residues Lys165 and Lys166 of tissue factor with the 4-carboxyglutamate-rich domain of factor X. J Biol Chem. 1996;271:21752–21757. doi: 10.1074/jbc.271.36.21752. [DOI] [PubMed] [Google Scholar]
- 22.Rao LVM, Ruf W. Tissue factor residues Lys165 and Lys166 are essential for rapid formation of the quaternary complex of tissue factor·VIIa with Xa·Tissue factor pathway inhibitor. Biochemistry. 1995;34:10867–10871. doi: 10.1021/bi00034a020. [DOI] [PubMed] [Google Scholar]
- 23.Ruf W, Miles DJ, Rehemtulla A, Edgington TS. Tissue factor residues 157–167 are required for efficient proteolytic activation of factor X and factor VII. J Biol Chem. 1992;267:22206–22210. [PubMed] [Google Scholar]
- 24.Kirchhofer D, Lipari MT, Moran P, Eigenbrot C, Kelley RF. The tissue factor region that interacts with substrates factor IX and Factor X. Biochemistry. 2000;39:7380–7387. doi: 10.1021/bi000182+. [DOI] [PubMed] [Google Scholar]
- 25.Kirchhofer D, Eigenbrot C, Lipari MT, Moran P, Peek M, Kelley RF. The tissue factor region that interacts with factor Xa in the Activation of factor VII. Biochemistry. 2001;40:675–682. doi: 10.1021/bi002013v. [DOI] [PubMed] [Google Scholar]
- 26.Wiréhn J, Carlsson K, Herland A, Persson E, Carlsson U, Svensson M, Hammarstrom P. Activity, folding, misfolding, and aggregation in vitro of the naturally occurring human tissue factor mutant R200W. Biochemistry. 2005;44:6755–6763. doi: 10.1021/bi047388l. [DOI] [PubMed] [Google Scholar]
- 27.Carlsson K, Freskgård P, Persson E, Sralsson U, Svensson M. Probing the interface between factor Xa and tissue factor in the quaternary complex tissue factor-factor VIIa-factor Xa-tissue factor pathway inhibitor. Eur J Biochem. 2003;270:2576–2582. doi: 10.1046/j.1432-1033.2003.03625.x. [DOI] [PubMed] [Google Scholar]
- 28.Ruf W, Shobe J, Rao SM, Dickinson CD, Olson A, Edgington TS. Importance of factor VIIa Gla-domain residue Arg-36 for recognition of the macromolecular substrate factor X Gla-domain. Biochemistry. 1999;38:1957–1966. doi: 10.1021/bi982254r. [DOI] [PubMed] [Google Scholar]
- 29.Persson E, Olsen OH. Assignment of molecular properties of a superactive coagulation factor VIIa variant to individual amino acid changes. Eur J Biochem. 2002;269:5950–5955. doi: 10.1046/j.1432-1033.2002.03323.x. [DOI] [PubMed] [Google Scholar]
- 30.Bjelke JR, Persson E, Rasmussen HB, Kragelund BB, Olsen OH. A loop of coagulation factor VIIa influencing macromolecular substrate specificity. FEBS Letters. 2007;581:71–76. doi: 10.1016/j.febslet.2006.11.079. [DOI] [PubMed] [Google Scholar]
- 31.Toso R, Pinotti M, High KA, Pollak ES, Bernardi F. A frequent human coagulation Factor VII mutation (A294V, c152) in loop 140s affects the interaction with activators, tissue factor and substrates. Biochem J. 2002;363:411–416. doi: 10.1042/0264-6021:3630411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolfram Ruf. Factor VIIa residue Arg290 is required for efficient activation of the macromolecular substrate factor X. Biochemistry. 1994;33:11631–11636. doi: 10.1021/bi00204a026. [DOI] [PubMed] [Google Scholar]