Abstract
A computational study was undertaken to understand the nature of binding and the structural features that play a significant role in the binding of arylacetylene molecules to cytochrome P450 enzymes 1A1, 1A2, 2A6 and 2B1. Nine polycyclic arylacetylenes determined to be mechanism-based P450 enzyme inhibitors were studied. The lack of polar substituents in these compounds causes them to be incapable of hydrogen bonding to the polar protein residues. The four P450 enzymes of interest all have phenylalanine residues in the binding pocket for potential π–π interactions with the aromatic rings of the inhibitors. The inhibition potency of these arylacetylenes toward P450s 1A1 and 2B1 showed a dependence on the proximity of the inhibitor’s triple bond to the prosthetic heme Fe of the enzyme. In P450 enzyme 1A2, the inhibitor’s potency showed more dependence on the π–π interactions of the inhibitor’s ring systems with the phenylalanine residues of the protein; with proximity of inhibitor triple bond to the heme Fe weighing in as the second most important factor. The results suggest that maximizing the π–π interactions with phenylalanine residues in the binding pocket and optimum proximity of the acetylene moiety to the heme Fe will provide for a substantial increase in the potency of the polyaromatic hydrocarbon mechanism-based inhibitors. A fine balance of these two aspects of binding coupled with attention to supplementing hydrophobic interactions could address potency and selectivity issues for these inhibitors.
Introduction
Cytochrome P450 enzymes constitute a large superfamily of heme proteins known to catalyze oxidation reactions of an expansive array of endogenous and exogenous compounds including drugs, sterols, fatty acids, carcinogens, vitamins and steroids (1–3). There are 57 known human P450 enzymes, including a few pseudogenes which are classified into families (sequence identity >40%) and subfamilies (sequence identity ≥55%), (see http://drnelson.utmem.edu/CytochromeP450.html). Some of these enzymes are polymorphically expressed, resulting in different metabolic activity that could influence the overall toxic effect, including carcinogenesis induced by environmental chemicals (4). The P450 enzymes 1A1, 1A2, 2A6, and 2B1 have been shown to oxidize polycyclic aromatic hydrocarbons to produce carcinogenic agents that can bind to DNA causing cancer (5–8). Targeting these enzymes for inhibition with small molecules offers new avenues in cancer prevention and therapy (9). Many acetylenic compounds are shown to irreversibly inactivate P450 enzymes by time-dependent destruction of heme via alkylation (6, 10–15). Ortiz de Montellano and coworkers have established the mechanism involving the formation of an intermediate by P450-dependent oxidation of acetylenic moiety followed by covalent binding of the unstable intermediate to a heme nitrogen (16, 17).
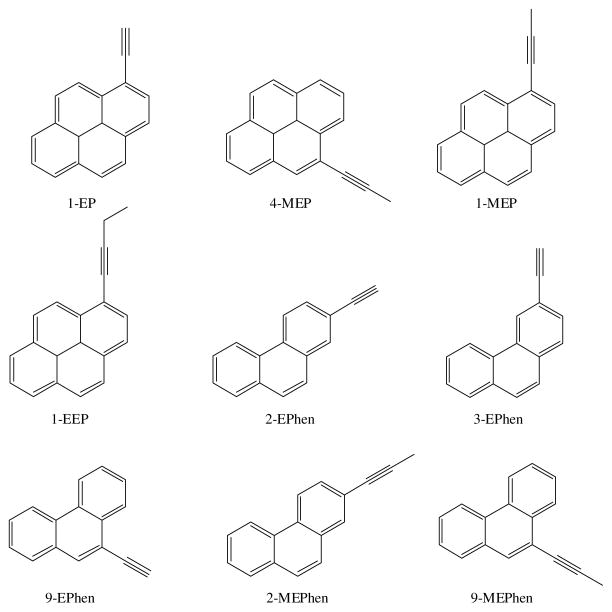
In our present study, we show that arylacetylenes, such as, 1-ethynylpyrene (1EP), 1-(1-propynyl)pyrene (1MEP), 4-(1-propynyl)pyrene (4-MEP), 1-(1-butynyl)pyrene (1-EEP), 2-ethynylphenanthrene (2-EPhen), 3-ethynylphenanthrene (3-EPhen), 9-ethynylphenanthrene (9-EPhen), 2-(1-propynyl)phenanthrene (2-MEPhen), and 9-(1-propynyl)phenanthrene (9-MEPhen) act as mechanism-based inhibitors of P450s 1A1 and 1A2, with many of them additionally inhibiting P450 2B1. None of these compounds showed significant inhibition of P450 2A6. The unique aspect of these compounds is the lack of any polar structural moiety capable of hydrogen bonding with polar protein residues. Our goal was to understand the structural basis of the interactions that these molecules have with the P450 enzymes 1A1, 1A2, and 2B1 through molecular modeling studies. We were also intrigued by the fact that none of these compounds inhibited P450 2A6. Reported X-ray crystal structures were utilized for the enzymes 1A2 and 2A6, while homology models were built for P450s 1A1 and 2B1. Docking studies of the arylacetylenes with the four P450 enzymes 1A1, 1A2, 2A6 and 2B1 were performed and the results were analyzed to obtain the consensus binding posture for each of the molecules of interest. Better understanding of the molecular nature of binding interactions will direct us in the design of more effective and selective inhibitors for these P450 enzymes.
Methods
Assays
Rat CYP2B1 supersomes (rat CYP2B1 + P450 reductase + cytochrome b5), and human CYP1A1, 1A2, and 2A6 supersomes (human CYP enzymes + P450 reductase) were purchased from B.D. Biosciences Corporation (Woburn, MA, USA). All other chemicals were purchased from Sigma Aldrich Company (Milwaukee, WI).
The P450 1A1, 1A2, and 2B1 dependent activities were assayed in 7-alkoxyresorufin dealkylation assays using ethoxyresorufin, methoxyresorufin, and pentoxyresorufin fluorescent substrates, respectively (18). P450 2A6 dependent 7-hydroxylation of coumarin was used in a similar assay with minor differences as described below for measuring 2A6 activity (19, 20).
7-Ethoxyresorufin O-deethylation (EROD), 7-methoxyresorufin O-demethylation (MROD), 7-pentoxyresorufin O-depentylation (PROD), and Coumarin 7-Hydroxylation Assays
Potassium phosphate buffer (1750 μL of a 0.1 M solution, pH 7.6) was placed in a 1.0 cm quartz cuvette, and 10 μL of a 1.0 M MgCl2 solution, 15 μL of a 1.0 mM corresponding resorufin or coumarin substrate solution in Me2SO (DMSO), 10 μL of the microsomal P450 protein, and 15 μL of an inhibitor in DMSO were added. For the controls, 15 μL of pure DMSO was added in place of the inhibitor solution. The reaction was initiated by the addition of 200 μL of a NADPH regenerating solution. The regenerating solution was prepared by combining 797 μL of a 0.10 M potassium phosphate buffer solution (pH 7.6), 67 μL of a 15 mM NADP+ solution in buffer, 67 μL of a 67.5 mM glucose 6-phosphate solution in buffer, and 67 μL of a 45 mM MgCl2 solution, and incubating the mixture for 5 minutes at 37°C before the addition of 3 units of glucose 6-phosphate dehydrogenase/mL and a final 5 minute incubation at 37°C. The final assay volume was 2.0 mL. The production of 7-hydroxyresorufin anion was monitored by a spectrofluorimeter (OLIS DM 45 Spectrofluorimetry System) at 530 nm excitation and 585 nm emission, with a slit width of 2 nm. The production of 7-hydroxycoumarin was monitored at 338 nm excitation and 458 nm emission, with a slit width of 2 nm. The reactions were performed at 37°C. For each inhibitor, a number of assay runs were performed using varying inhibitor concentrations.
Data Analysis (14)
The data obtained from these assays were analyzed by a computer analysis method of the reaction progress curve in the presence of various inhibitor concentrations and in the absence of the inhibitor as the control run. Results are tabulated in Table 1. A second order curve describing product formation with respect to reaction time in seconds was obtained for each inhibitor concentration and the control. The Microsoft Excel Program was used to fit the data (Fluorescence intensity vs. Time) obtaining the parameters of the best-fit second order curves (y=ax2+bx+c). Using the parameters (Coefficient b in the second order equation is enzymatic activity) obtained from the above, activities were calculated using first order derivatives. Dixon plots were used (by plotting the reciprocals of enzymatic activity (1/v) vs. inhibitor concentrations [I]) in order to determine IC50 values (x-intercepts) for the inhibitors.
Table 1.
Inhibition data in supersomes for arylacetylenes measured using human P450 enzymes (1A1, 1A2, 2A6) and rat P450 enzyme (2B1).
| Compound | IC50 (μM) | |||
|---|---|---|---|---|
| 1A1 | 1A2 | 2B1 | 2A6 | |
| 1-EEP | 0.72 | 0.43 | >10 | >10 |
| 1-EP | 0.18 | 0.32 | 0.04 | >10 |
| 1-MEP | 0.02 | 0.06 | 0.83 | >10 |
| 4-MEP | 0.37 | 0.45 | 2.26 | >10 |
| 2-Ephen | 0.61 | 0.34 | 0.17 | >10 |
| 3-Ephen | 1.03 | 0.28 | 0.48 | >10 |
| 9-Ephen | 0.62 | 0.4 | 0.07 | >10 |
| 2-MEPhen | 0.16 | 0.6 | >10 | >10 |
| 9-MEPhen | 2.63 | 0.67 | >10 | >10 |
Pre-incubation Assays in the Presence and Absence of NADPH
To confirm mechanism-based inhibition, pre-incubation assays were performed as follows. All assay solution components had the same concentrations as in the above assays. For pre-incubation assays in the presence of NADPH, potassium phosphate buffer (1550 μL of a 0.1 M solution, pH 7.6) was placed in a 1.0 cm quartz cuvette followed by 10 μL of a 1.0 M MgCl2 solution, 10 μL of the microsomal P450 protein, 15 μL of an inhibitor in DMSO (for the control, 15 μL of pure DMSO was added in the place of the inhibitor solution), and 200 μL of a NADPH regenerating solution. The assay mixture was incubated for five minutes at 37°C, before reaction initiation by the addition of 200 μL of buffer and 15 μL of the corresponding substrate solution. The final assay volume was 2.0 mL. The production of 7-hydroxyresorufin anion was monitored at 530 nm excitation and 585 nm emission. The production of 7-hydroxycoumarin was monitored at 338 nm excitation and 458 nm emission. The reactions were performed at 37°C. For each inhibitor, a number of assay runs were performed using varying inhibitor concentrations. For the pre-incubation assays in the absence of NADPH, potassium phosphate buffer (1750 μL of a 0.1 M solution, pH 7.6) was placed in a 1.0 cm quartz cuvette followed by 10 μL of a 1.0 M MgCl2 solution, 10 μL of the microsomal P450 protein, and 15 μL of an inhibitor in DMSO (for the control, 15 μL of pure DMSO was added in the place of the inhibitor solution). The assay mixture was incubated for five minutes at 37°C, before reaction initiation by the addition of 200 μL of the NADPH regenerating solution and 15 μL of the corresponding substrate solution. The final assay volume was 2.0 mL. The production of P450-dependent reaction products were monitored as previously described (10). The reactions were performed at 37°C. For each inhibitor, a number of assay runs were performed using varying inhibitor concentrations.
Protein Crystal Structures and Homology Modeling of P450s 1A1 and 2B1
All of our in silico studies were carried out using the Molecular Operating Environment (MOE) Program (Chemical Computing Group, Montreal, Canada). The coordinates of the template P450 enzymes 1A2 (PDB ID: 2HI4) and 2A6 (PDB ID: 1Z10) were taken from the Protein Data Bank (http://www.rcsb.org). Oxygen atoms representing water were removed and hydrogen atoms were added to the template proteins using Amber99 force field.
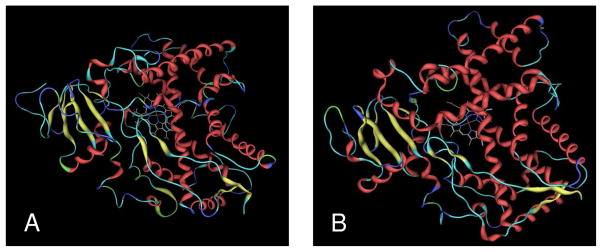
A homology model of P450 1A1 (Figure 1A) was built based on the protein crystal structure of P450 1A2 (2HI4) taken from the Protein Data Bank using the MOE-Homology program (Tripos SYBYL which was used for building 2B1 homology model lacked access to some modules at the time of 1A1 homology modeling and hence MOE was used). The target sequence of human P450 1A1 was obtained from the SwissProt database (21) (P04798). Target sequence was aligned with the sequence of 2HI4 using MOE’s multiple sequence and structural alignment algorithm (22) with the structural alignment tool enabled and the blosum62 substitution matrix. The three-dimensional model building of P450 1A1 was performed using the MOE homology program based on a segment matching procedure and a best intermediate algorithm with the option to refine each individual structure enabled. Ten intermediate models were generated and the final model was taken as the cartesian average of all the intermediate models. The heme residue was positioned using the same coordinates as in the template and the complex model was energy minimized. The minimized homology model was validated with PROCHECK(23) and ProsaII (24).
Figure 1.
3D structures of P450s 1A1 (A) and 2B1 (B) obtained from homology modeling. The protein is shown as a ribbon model and the Heme residue is depicted as stick model.
A homology model of P450 2B1 (Figure 1B) was built based on the protein crystal structures of P450 2B4 (1SUO- mammalian and 2BDM- microsomal) using the molecular modeling package SYBYL (version 7.1, Tripos, St. Louis, MO). The sequence of rat P450 2B1 was used as the target which was obtained from the SwissProt database (21) (P00176). The sequence identity was 73%. The homology model of P450 2B1 was built using Composer in the Biopolymer module of SYBYL. Conformations of loop sequences were automatically generated by the program. The heme residue was positioned using the same coordinates as in the template. Hydrogen atoms were added to the structure and the initial model was refined by energy minimization by the steepest descent method followed by the conjugate gradient method until the root mean square gradient of the potential energy was <0.05 kcal.mol−1A−1. The minimized homology model was validated with PROCHECK and ProsaII.
Binding Modes by Docking Simulation
The structures of the 9 molecules used in this study are given in Figure 2. The 3D structures of the molecules were built using the Tripos SYBYL 7.3 Program. Initial geometric optimizations of the ligands were carried out using the standard MMFF94 force field, with a 0.001 kcal/mol energy gradient convergence criterion and a distance-dependent dielectric constant employing Gasteiger and Marsili charges. Additional geometric optimizations were performed using the semi-empirical method molecular orbital package (MOPAC). The compounds were docked into the binding pockets of P450 enzymes using three programs, GOLD (Cambridge Crystallographic Data Centre), MOE and FlexX (Tripos). The consensus binding postures of the inhibitors were obtained by visual inspection of the docking simulations and their docking scores. The docked complexes were then minimized in three steps using MOE Energy Minimize application. Amber ff99 was used for standard residues of the protein and partial charges were calculated as required for this force field. The non-standard forcefield parameters for heme and the cysteine-iron bond were taken from the literature (25). MOE Energy minimization consists of finding a set of atomic coordinates that correspond to a local minimum of the molecular energy function (we used the potential energy model) by applying large scale non-linear optimization techniques to calculate a conformation (near to the starting geometry) for which the forces on the atoms are zero(26). MOE uses a success of three methods to effect an energy minimization: Steepest Descent (SD), Conjugate Gradient (SG) and Truncated Newton (TN) controlled by the root mean square (RMS) gradient of energy falling below 0.1 kcal A−1. In addition, atoms may be fixed during the calculation. First, hydrogen atom positions were relaxed by holding other atoms fixed. This was followed by allowing all side chain atoms to relax while holding the backbone atoms fixed. Finally, the entire structure was relaxed until root mean square (RMS) gradient of energy was less than 0.1 kcal A−1. Through all of the minimization steps, the planar structure of the heme residue was fixed. The distances between the external carbon of the inhibitor’s triple bonds and the heme Fe were determined. The centroids of the phenyl rings of the phenylalanine residues and those of the inhibitor molecules were calculated using the Scientific Vector Language (SVL) module of MOE.
Figure 2.
Structures of arylacetylene inhibitors used for the study
Results and Discussion
Cytochrome P450s 1A1, 1A2, 2A6 and 2B1 are known to metabolize xenobiotics and ubiquitous environmental carcinogens such as polycyclic aromatic hydrocarbons. Known X-ray crystal structures of P450s 1A2 (PDB ID: 2HI4) (27) and 2A6 (PDB ID: 1Z10) (28) were used in this study. P450s 1A1 (human) and 1A2 (human) show high sequence identity with 73% alignment score. P450s 2B4 (mammalian) and 2B1 (rat) also show high sequence identity with 70% alignment score. Based on the sequence identities of 1A1 with 1A2, and 2B1 with 2B4; homology models for P450s 1A1 and 2B1 were built using the X-ray structures 2HI4.pdb (human) (for 1A2) and 1SUO.pdb (mammalian) and 2BDM.pdb (microsomal) (for 2B1) respectively, as templates. The homology models of P450 1A1 and 2B1 are shown in Figure 1. P450 enzymes share a common overall fold and topology with twelve helices A-L and four β-sheets 1–4, despite less than 20% sequence identity across the gene superfamily. The binding site is defined by the helices E, I, J, K, and L as well as portions of β-sheet 1. The prosthetic heme group is confined between the distal I helix and proximal L helix and bound to the adjacent cysteine in the loop containing the P450 signature amino acid sequence FxxGx(H/R)xCxG. The absolutely conserved cysteine is the proximal or “fifth” ligand to the heme iron (29). The nine inhibitors (Figure 2) used in this study are unique in that they all have 3–4 fused benzene ring systems (polycyclic aromatic hydrocarbons- PAHs), incorporated with an acetylenic moiety in order to achieve mechanistic inhibition of P450 enzymes while lacking any polar group capable of hydrogen bonding with protein residues. The inhibition data for the nine inhibitors measured using human P450 enzymes 1A1, 1A2, and 2A6, and rat enzyme 2B1 are given in Table 1.
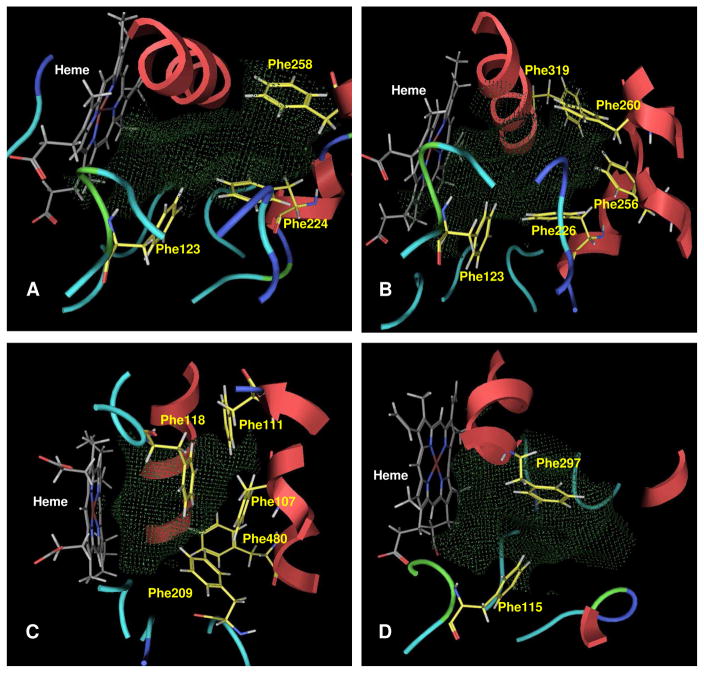
The binding sites of the four P450s of interest were studied to understand the nature of the residues defining the site (Table 2). Aromatic residues were of great interest to us as these residues would interact favorably with the PAH rings of our inhibitors through π–π interactions. Among the four P450 enzymes in our study, P450 1A2 was found to have the largest binding pocket, and P450 2B1 was found to have the smallest binding pocket. All of these enzymes have phenylalanine residues in their binding pockets, with many clustered together (highlighted in bold in Table 2 and shown in Figure 3), capable of making π–π interactions with aromatic rings of the ligand.
Table 2.
Properties of the P450 enzyme binding pocket
| Protein | Binding cavity Volume Å3* | Phenylalanine residues that define the proposed binding cavity |
|---|---|---|
| 1A1 | 82 | Phe123, Phe224, Phe258, Phe384 |
| 1A2 | 129 | Phe125, Phe226, Phe256, Phe260, Phe319 |
| 2A6 | 64 | Phe107, Phe111, Phe118, Phe209, Phe480 |
| 2B1 | 41 | Phe115, Phe297 |
Tripos SYBYL (SiteID Pockets) was used to identify the binding cavity by initiating grid-based algorithm for locating pockets. This works well for finding narrow and/or solvent inaccessible pockets. The volume of the binding cavity is also given by this algorithm.
Figure 3.
Depicts binding pockets of P450 enzymes 1A1 (A), 1A2 (B), 2A6 (C) and 2B1 (D) with heme and Phenylalanine residues as stick models showing Gaussian contact molecular surfaces in green dots.
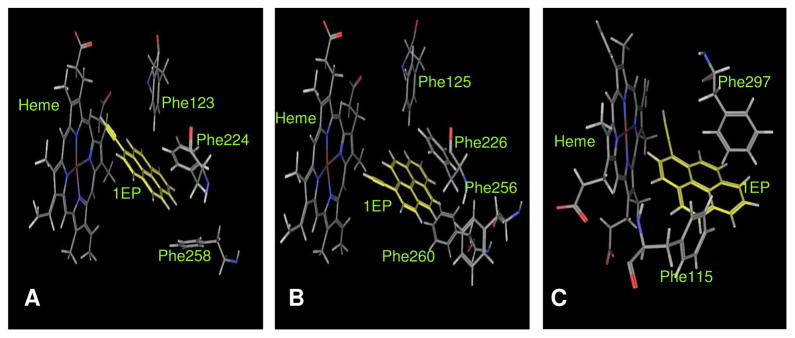
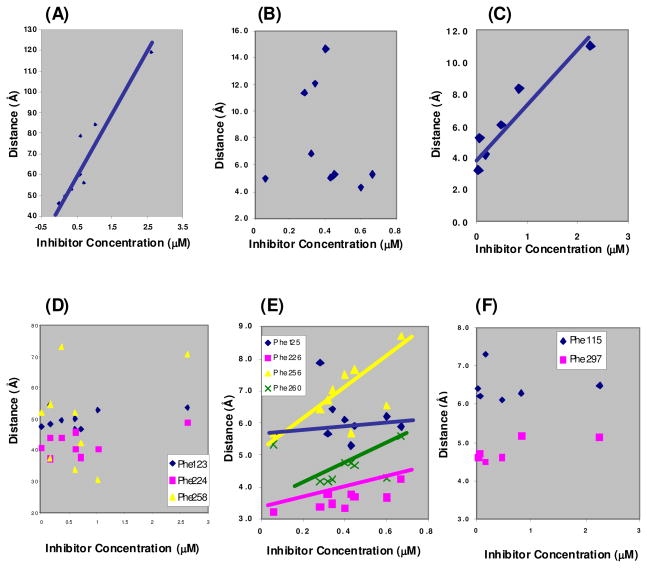
Docking studies were carried out using three different docking programs, GOLD, MOE and FlexX. We were greatly interested in the binding orientation of the molecules that exhibited significant inhibition (i.e., IC50 <10 μM) of the P450 enzymes 1A1, 1A2 and 2B1. Figure 4 depicts the binding of 1EP to P450s 1A1, 1A2 and 2B1 as an example. Two aspects of the ligand docking postures were considered. As the present inhibitors are mechanism-based inhibitors, the position of the triple bond with respect to the Fe atom of the prosthetic heme group of the P450 enzyme is crucial in the oxidation of the triple bond. The distance between the Fe atom of the heme residue and the external carbon of the triple bond was the first aspect for consideration in our docking studies. The second aspect was the effective π stacking of the inhibitors with the various aromatic rings of phenylalanine residues in the active site. The docking results revealed that all of the inhibitors in our study have π–π interactions with the phenylalanine residues in the binding pocket. The distances between the centroids of phenylalanine residue’s aromatic rings and the nearest aromatic ring of the ligands were calculated. The distances between the docked ligands and the heme-Fe and phenylalanine aromatic rings for the P450 enzymes 1A1, 1A2, and 2B1 are given in Tables 3A, 3B and 3C respectively.
Figure 4.
Illustrates the π–π interactions between the protein and the ligand and the proximity of the triple bond to the heme Fe atom for the representative molecule 1EP docked in the binding pockets of P45 enzymes 1A1 (A), 1A2 (B) and 2B1 (C). The residues and inhibitor are shown as stick models.
Table 3.
Distances between Heme Fe, Phe residues and the ligand moieties for P450 1A1 (A), 1A2 (B) and 2B1 (C).
| (A) | |||||
|---|---|---|---|---|---|
| Distance in Å | |||||
| Ligand | Inhibition (μM) | Heme-C1 | Phe1232 | Phe2242 | Phe2582 |
| 1EEP | 0.72 | 5.30 | 4.7 | 3.8 | 4.2 |
| 1EP | 0.18 | 4.80 | 4.8 | 4.4 | 5.5 |
| 1MEP | 0.02 | 4.60 | 4.8 | 4.1 | 5.2 |
| 4MEP | 0.37 | 5.30 | 5.0 | 4.4 | 7.3 |
| 2EPHEN | 0.61 | 6.01 | 4.7 | 4.6 | 5.2 |
| 3EPHEN | 1.03 | 8.41 | 5.3 | 4.0 | 3.1 |
| 9EPHEN | 0.62 | 7.89 | 5.0 | 4.0 | 3.4 |
| 2MEPHEN | 0.16 | 4.95 | 5.5 | 3.7 | 3.7 |
| 9MEPHEN | 2.63 | 11.90 | 5.3 | 4.9 | 7.1 |
| (B) | ||||||
|---|---|---|---|---|---|---|
| Distance in Å | ||||||
| Ligand | Inhibition (μM) | Heme-C1 | Phe1252 | Phe2262 | Phe2562 | Phe2602 |
| 1EEP | 0.43 | 5.07 | 5.3 | 3.8 | 5.7 | 4.7 |
| 1EP | 0.32 | 6.79 | 5.7 | 3.8 | 6.7 | 4.2 |
| 1MEP | 0.06 | 4.93 | 5.7 | 3.2 | 5.7 | 5.3 |
| 4MEP | 0.45 | 5.31 | 5.9 | 3.7 | 7.7 | 4.7 |
| 2EPHEN | 0.34 | 12.08 | 6.4 | 3.5 | 7.1 | 4.2 |
| 3EPHEN | 0.28 | 11.39 | 7.9 | 3.4 | 6.4 | 4.2 |
| 9EPHEN | 0.4 | 14.64 | 6.1 | 3.3 | 7.5 | 4.7 |
| 2MEPHEN | 0.6 | 4.31 | 6.2 | 3.7 | 6.5 | 4.3 |
| 9MEPHEN | 0.67 | 5.27 | 5.9 | 4.2 | 8.7 | 5.6 |
| (C) | ||||
|---|---|---|---|---|
| Distance in Å | ||||
| Ligand | Inhibition (μM) | Heme-C1 | Phe1152 | Phe2972 |
| 1EP | 0.04 | 3.25 | 6.4 | 4.6 |
| 1MEP | 0.83 | 8.40 | 6.3 | 5.2 |
| 4MEP | 2.26 | 11.00 | 6.5 | 5.1 |
| 2EPHEN | 0.17 | 4.20 | 7.3 | 4.5 |
| 3EPHEN | 0.48 | 6.00 | 6.1 | 4.6 |
| 9EPHEN | 0.07 | 5.20 | 6.2 | 4.7 |
Heme-C distances represent the distance between the Heme Fe and the external carbon of the triple bond.
Distance between the centroids of the Phenyl ring of Phenylalanine residues and the nearest aromatic ring of the inhibitor.
It has been shown that terminal acetylenes are metabolized by cytochrome P450 to form a reactive ketene intermediate by a 1,2-hydrogen shift. The ketene intermediate may react with the P450 heme moiety becoming covalently bound to heme nitrogen resulting in a time-dependent destruction of the heme chromophore leading to loss of P450 enzymatic activity (16, 30, 31). The putative ketene intermediate can also react with nucleophilic residues of the protein resulting in covalent binding to the protein with concomitant loss of P450 enzymatic activity (10, 32, 33). Alternatively, excess ketene generated in the active site of the enzyme may react with water to form a carboxylic acid metabolite (16, 30). These mechanistic studies indicate the relevance of proximity of the acetylenic moiety of the inhibitors to the heme Fe atom which is the first aspect of our study.
Interactions between aromatic moieties also known as π–π interactions are important forces in molecular recognition, and play an important role in controlling the conformations and substrate binding properties of nucleic acids and proteins. The geometry preferences and energetic forces of these interactions have been widely studied in recent years (34, 35). Hunter and coworkers have shown that edge-to-face type orientations and offset stacked orientations are electrostatically attractive geometries while face-to-face and edge-to-edge orientations are unfavorable (36–38). Observations in proteins support the presence of such favorable geometries (37).
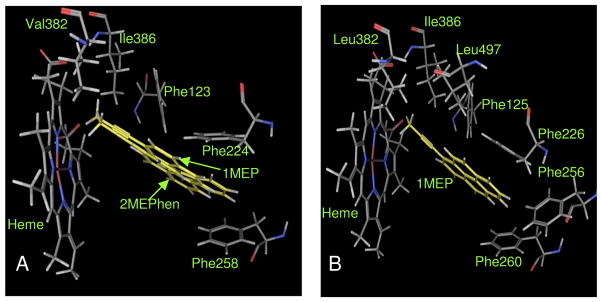
The binding pocket of cytochrome P450 1A1 incorporates four phenylalanine residues at positions 123, 224, 258 and 384. Three of them (Phe123, Phe224, and Phe258) showed favorable interactions with the ligand molecules. The distance between aromatic rings of phenylalanine residues in the native enzyme was 6.0 Å (Phe123-Phe224), 7.7 Å (Phe224-Phe258) and 11.5 Å (Phe123-Phe258). The docked molecules exhibited edge to face interactions with Phe123 and offset stacked interactions with Phe224 and Phe258. The distance between the external carbon of the triple bond and the heme Fe (Table 3A) showed a direct linear correlation with inhibitory activity (Figure 5A). The molecule 1MEP with the closest triple bond to the heme Fe (4.6 Å) showed the most inhibition with an IC50 of 0.02 μM while the molecule 9MEPhen with the farthest triple bond from the heme Fe (11.9 Å) showed the least inhibition with an IC50 of 2.26 μM. The distance between the ligands and centroids of aromatic rings of phenylalanine residues ranged from 4.7 to 5.5 Å for Phe123, 3.7 to 4.9 Å for Phe224, and 3.1 to 7.3 Å for Phe258 (Table 3A). Even though the aromatic rings of phenylalanine residues had π–π interactions with the ligand molecules, they did not show any clear correlation to the activity profile. It is clear that the cytochrome P450 1A1 inhibition potency of these molecules depends on the distance of the triple bond from the heme Fe. The π stacking of the polycyclic aromatic core of the molecules with the aromatic rings of phenylalanine residues did not contribute to the differences in the potency of these inhibitors. The terminal methyl substituted aryl acetylenes (1MEP and 2MEPhen) showed higher inhibition potency due to many favorable interactions with the protein such as shorter distance of the triple bond to the heme Fe, effective offset π stacking distance with Phe224 and Phe258, and the additional fact that the methyl group fits perfectly in the hydrophobic region of the pocket defined by amino acid residues Val382 and Ile386 (Figure 6A).
Figure 5.
(A), (B), and (C) Relationship between the inhibitor concentration and Heme-C distance for P450 enzymes 1A1, 1A2 and 2B1; (D), (E), and (F) Relationship between the inhibitor concentration and distance between centroids of aromatic rings of Phenylalanine residues and inhibitor for P450 enzymes 1A1, 1A2 and 2B1.
Figure 6.
(A) Represents binding modes of most potent inhibitors 1MEP and 2EPHEN for P450 enzyme 1A1; (B) represents binding modes of most potent inhibitor 1MEP for P450 enzyme 1A2. All interacting residues and the inhibitors are shown as stick models.
The active site of cytochrome P450 1A2 depicts the presence of five phenylalanine residues Phe125, Phe226, Phe256, Phe260 and Phe319. The aromatic rings of Phe125, Phe226, Phe256 and Phe260 were clustered together in the binding pocket with the rings pointing inwards, whereas the Phe319 aromatic ring pointed outwards. Hence the contribution to π stacking with the inhibitors could be seen only from the first four phenylalanine residues. The distances between these aromatic rings in the native 1A2 enzyme is 5.9 Å for Phe125-Phe226, 6.5 Å for Phe226-Phe256, 5.8 Å for Phe256-260 and 6.9 Å for Phe226-Phe260. The distance between the external carbon of the ligand molecules and the heme Fe ranged between 4.3 Å and 14.6 Å (Table 3B). This distance did not show any clear relationship with the variation in inhibitory activity of the molecules. The distances between the centroids of the aromatic rings of the phenylalanine residues and the ligands were 5.3–7.9 Å for Phe125, 3.2–4.2 Å for Phe226, 5.7–8.7 Å for Phe256 and 4.2–5.6 Å for Phe260 (Table 3B). The variation in the ligand-phenylalanine aromatic ring distance was minimal for Phe125 and Phe226, implying that even though it can be a positive factor towards effective binding, it does not account for the differences in inhibition potency. Distances between the ligand and the phenylalanines Phe256 and Phe260 indeed show a linear relationship to their inhibition potency (Figure 5B). The data points that did not fit in the linear relationship for Phe256 and Phe260 had shorter distances to the heme Fe atom which factored into the increase in potency. These results show that a good π–π interaction of the ligand with all four of the phenylalanines increases the inhibition potency of these molecules towards P450 enzyme 1A2. This when combined with a shorter distance of the triple bond from the heme Fe should result in higher inhibition potency. 1MEP showed the highest potency among the ligands as its triple bond was one of the closest to the heme Fe, it had favorable π–π interactions with the phenylalanine residues lining the pocket, and the methyl group was positioned in the hydrophobic region of the pocket defined by protein residues Leu382, Ile386 and Leu497 (Figure 6B).
The binding pocket of P450 2B1 was the smallest of all the P450 enzymes studied. It had two phenylalanine residues Phe115 and Phe297. Phe297 made an edge-to-face interaction with the inhibitors whereas Phe115 made an edge-to-edge π interaction with the inhibitors. The distances between the centroid of the aromatic rings of phenylalanine residues and the nearest aromatic ring of the inhibitors ranged from 4.5–5.2 Å for Phe297, and 6.1–7.3 Å for Phe115 (Table 3C). The distance between the heme Fe and the ligand external carbon of the triple bond varied from 3.2 Å to 11 Å and showed a linear correlation to the inhibition potency (Figure 5C). The inhibitors did not show significant difference in their π stacking interactions with the phenylalanine residues. The molecule 1EP showed the highest inhibition which could be attributed to its triple bond being closest to heme Fe atom (3.2 Å).
The binding pocket of P450 enzyme 2A6 consists of many phenylalanine residues (Phe107, Phe11, Phe118, Phe209 and Phe480) that could potentially interact favorably with the inhibitors. However, the shape of the binding pocket and the orientation of many of the phenylalanine residues in the binding pocket cause distortion in the planar inhibitors when docked in the binding pockets. This is a highly unfavorable situation which is reflected by the absence of inhibitory activity of these ligands towards cytochrome P450 2A6.
Conclusion
The polyaromatic hydrocarbon inhibitors studied here do not have any polar substituents capable of hydrogen bonding with the polar residues of P450s 1A1, 1A2, 2A6 and 2B1. The binding pockets of all four target P450 enzymes contain aromatic residues that can form π–π interactions with the aromatic rings of the inhibitors. In P450s 1A1 and 2B1, the distance between the triple bond and the heme Fe is a crucial factor in determining the potency of the inhibitor. In P450 enzyme 1A2, the π–π interactions with the four phenylalanine residues play a more dominating role in determining the potency of the inhibitors. Further more favorable positioning of the hydrophobic groups of the inhibitor’s side chains positioned in the hydrophobic region of the binding pocket further increases the potency of inhibition. Through these studies, we have gained valuable insight into the factors governing the potency of the inhibition of cytochrome P450s 1A1, 1A2, 2A6 and 2B1 by polyaromatic hydrocarbons. This knowledge will help us in designing inhibitors with increased potency and selectivity for these enzymes.
Acknowledgments
We wish to acknowledge NIH grants SC1 GMO84722 and S06 GM08008 for financial support of this work.
Abbreviations
- 1EP
1-ethynylpyrene
- 1MEP
1-(1-propynyl)pyrene
- 4-MEP
4-(1-propynyl)pyrene
- 1-EEP
1-(1-butynyl)pyrene
- 2-EPhen
2-ethynylphenanthrene
- 3-EPhen
3-ethynylphenanthrene
- 9-EPhen
9-ethynylphenanthrene
- 2-MEPhen
2-(1-propynyl)phenanthrene
- 9-MEPhen
9-(1-propynyl)phenanthrene
- EROD
7-Ethoxyresorufin O-deethylation
- MROD
7-methoxyresorufin O-demethylation
- PROD
7-pentoxyresorufin O-depentylation
- MOE
Molecular Operating Environment
- MOPAC
Molecular Orbital Package
- SD
Steepest Descent
- SG
Conjugate Gradient
- TN
Truncated Newton
- RMS
Root Mean Square
- SVL
Scientific Vector Language
References
- 1.Nelson DR, Koymans L, Kamataki T, Stegeman JJ, Feyereisen R, Waxman DJ, Waterman MR, Gotoh O, Coon MJ, Estabrook RW, Gunsalus IC, Nebert DW. P450 superfamily: update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics. 1996;6:1–42. doi: 10.1097/00008571-199602000-00002. [DOI] [PubMed] [Google Scholar]
- 2.Nelson DR, Kamataki T, Waxman DJ, Guengerich FP, Estabrook RW, Feyereisen R, Gonzalez FJ, Coon MJ, Gunsalus IC, Gotoh O, et al. The P450 superfamily: update on new sequences, gene mapping, accession numbers, early trivial names of enzymes, and nomenclature. DNA Cell Biol. 1993;12:1–51. doi: 10.1089/dna.1993.12.1. [DOI] [PubMed] [Google Scholar]
- 3.Guengerich FP. Cytochrome P450: What Have We Learned and What Are the Future Issues? Drug Metabol Rev. 2004;36:159–197. doi: 10.1081/dmr-120033996. [DOI] [PubMed] [Google Scholar]
- 4.Agundez JA. Cytochrome P450 gene polymorphism and cancer. Curr Drug Metab. 2004;5:211–224. doi: 10.2174/1389200043335621. [DOI] [PubMed] [Google Scholar]
- 5.Shimada T, Oda Y, Gillam EM, Guengerich FP, Inoue K. Metabolic activation of polycyclic aromatic hydrocarbons and other procarcinogens by cytochromes P450 1A1 and P450 1B1 allelic variants and other human cytochromes P450 in Salmonella typhimurium NM2009. Drug Metab Dispos. 2001;29:1176–1182. [PubMed] [Google Scholar]
- 6.Shimada T, Guengerich FP. Inhibition of human cytochrome P450 1A1-, 1A2-, and 1B1-mediated activation of procarcinogens to genotoxic metabolites by polycyclic aromatic hydrocarbons. Chem Res Toxicol. 2006;19:288–294. doi: 10.1021/tx050291v. [DOI] [PubMed] [Google Scholar]
- 7.Guengerich FP, Shimada T. Activation of procarcinogens by human cytochrome P450 enzymes. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 1998;400:201–213. doi: 10.1016/s0027-5107(98)00037-2. [DOI] [PubMed] [Google Scholar]
- 8.Conney AH. Induction of microsomal enzymes by foreign chemicals and carcinogenesis by polycyclic aromatic hydrocarbons: G. H. A. Clowes Memorial Lecture. Cancer Res. 1982;42:4875–4917. [PubMed] [Google Scholar]
- 9.Bruno RD, Njar VC. Targeting cytochrome P450 enzymes: a new approach in anti-cancer drug development. Bioorg Med Chem. 2007;15:5047–5060. doi: 10.1016/j.bmc.2007.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foroozesh M, Primrose G, Guo Z, Bell LC, Alworth WL, Guengerich FP. Aryl acetylenes as mechanism-based inhibitors of cytochrome P450-dependent monooxygenase enzymes. Chem Res Toxicol. 1997;10:91–102. doi: 10.1021/tx960064g. [DOI] [PubMed] [Google Scholar]
- 11.Shimada T, Murajama N, Tanaka K, Takenaka S, Imai Y, Hopkins NE, Foroozesh MK, Alworth WL, Yamazaki H, Guengerich FP, Komori M. Interaction of polycyclic aromatic hydrocarbons with human cytochrome P450 1B1 in inhibiting catalytic activity. Chem Res Toxicol. 2008;21:2313–2323. doi: 10.1021/tx8002998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alexander DL, Zhang L, Foroozesh M, Alworth WL, Jefcoate CR. Metabolism-based polycyclic aromatic acetylene inhibition of CYP1B1 in 10T1/2 cells potentiates aryl hydrocarbon receptor activity. Toxicol Appl Pharmacol. 1999;161:123–139. doi: 10.1006/taap.1999.8794. [DOI] [PubMed] [Google Scholar]
- 13.Roberts ES, Hopkins NE, Foroozesh M, Alworth WL, Halpert JR, Hollenberg PF. Inactivation of cytochrome P450s 2B1, 2B4, 2B6, and 2B11 by arylalkynes. Drug Metab Dispos. 1997;25:1242–1248. [PubMed] [Google Scholar]
- 14.Hopkins NE, Foroozesh MK, Alworth WL. Suicide inhibitors of cytochrome P450 1A1 and P450 2B1. Biochem Pharmacol. 1992;44:787–796. doi: 10.1016/0006-2952(92)90417-h. [DOI] [PubMed] [Google Scholar]
- 15.Courter LA, Musafia-Jeknic T, Fischer K, Bildfell R, Giovanini J, Pereira C, Baird WM. Urban dust particulate matter alters PAH-induced carcinogenesis by inhibition of CYP1A1 and CYP1B1. Toxicol Sci. 2007;95:63–73. doi: 10.1093/toxsci/kfl137. [DOI] [PubMed] [Google Scholar]
- 16.Ortiz de Montellano PRaK, KL Self-catalyzed inactivation of hepatic cytochrome P-450 by ethynyl substrates. J Biol Chem. 1980;255:5578–5585. [PubMed] [Google Scholar]
- 17.Ortiz de Montellano PR. Mechanism-based inactivation of cytochrome P450: isolation and characterization of N-alkyl heme adducts. Methods Enzymol. 1991;206:533–540. doi: 10.1016/0076-6879(91)06123-k. [DOI] [PubMed] [Google Scholar]
- 18.Burke MD, Thompson S, Weaver RJ, Wolf CR, Mayer RT. Cytochrome P450 specificities of alkoxyresorufin O-dealkylation in human and rat liver. Biochem Pharmacol. 1994;48:923–936. doi: 10.1016/0006-2952(94)90363-8. [DOI] [PubMed] [Google Scholar]
- 19.Buters JT, Schiller CD, Chou RC. A highly sensitive tool for the assay of cytochrome P450 enzyme activity in rat, dog and man. Direct fluorescence monitoring of the deethylation of 7-ethoxy-4-trifluoromethylcoumarin. Biochem Pharmacol. 1993;46:1577–1584. doi: 10.1016/0006-2952(93)90326-r. [DOI] [PubMed] [Google Scholar]
- 20.Deluca JG, Dysaft GR, Rasnick D, Bradley MO. A Direct, Highly Sensitive Assay for Cytochrome P-450 Catalyzed O-Deethylation Using a Novel Coumarin Analog. Biochem Pharmacol. 1989;37:1731–1739. doi: 10.1016/0006-2952(88)90436-4. [DOI] [PubMed] [Google Scholar]
- 21.O’Donovan C, Martin MJ, Gattiker A, Gasteiger E, Bairoch A, Apweiler R. High-quality protein knowledge resource: SWISS-PROT and TrEMBL. Brief Bioinform. 2002;3:275–284. doi: 10.1093/bib/3.3.275. [DOI] [PubMed] [Google Scholar]
- 22.Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 23.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 24.Sippl MJ. Recognition of errors in three-dimensional structures of proteins. Proteins. 1993;17:355–362. doi: 10.1002/prot.340170404. [DOI] [PubMed] [Google Scholar]
- 25.Oda A, Yamaotsu N, Hirono S. New AMBER force field parameters of heme iron for cytochrome P450s determined by quantum chemical calculations of simplified models. J Comput Chem. 2005;26:818–826. doi: 10.1002/jcc.20221. [DOI] [PubMed] [Google Scholar]
- 26.Gill P, WM . Practical Optimization. Academic Press; London: 1981. [Google Scholar]
- 27.Sansen S, Yano JK, Reynald RL, Schoch GA, Griffin KJ, Stout CD, Johnson EF. Adaptations for the oxidation of polycyclic aromatic hydrocarbons exhibited by the structure of human P450 1A2. J Biol Chem. 2007;282:14348–14355. doi: 10.1074/jbc.M611692200. [DOI] [PubMed] [Google Scholar]
- 28.Yano JK, Hsu MH, Griffin KJ, Stout CD, Johnson EF. Structures of human microsomal cytochrome P450 2A6 complexed with coumarin and methoxsalen. Nat Struct Mol Biol. 2005;12:822–823. doi: 10.1038/nsmb971. [DOI] [PubMed] [Google Scholar]
- 29.Dawson JH, Holm RH, Trudell JR, Barth G, Linder RE, Bunnenberg E, Djerassi C, Tang SC. Letter: Oxidized cytochrome P-450. Magnetic circular dichroism evidence for thiolate ligation in the substrate-bound form Implications for the catalytic mechanism. J Am Chem Soc. 1976;98:3707–3708. doi: 10.1021/ja00428a054. [DOI] [PubMed] [Google Scholar]
- 30.Ortiz de Montellano PR, Komives EA. Branchpoint for heme alkylation and metabolite formation in the oxidation of arylacetylenes by cytochrome P-450. J Biol Chem. 1985;260:3330–3336. [PubMed] [Google Scholar]
- 31.Ortiz de Montellano PR. Mechanism-based inactivation of cytochrome P450: isolation and characterization of N-alkyl heme adducts. Methods Enzymol. 1991;206:533–540. doi: 10.1016/0076-6879(91)06123-k. [DOI] [PubMed] [Google Scholar]
- 32.Roberts ES, Hopkins NE, Alworth WL, Hollenberg PF. Mechanism-based inactivation of cytochrome P450 2B1 by 2-ethynylnaphthalene: identification of an active-site peptide. Chem Res Toxicol. 1993;6:470–479. doi: 10.1021/tx00034a013. [DOI] [PubMed] [Google Scholar]
- 33.Chan WK, Sui Z, Ortiz de Montellano PR. Determinants of protein modification versus heme alkylation: inactivation of cytochrome P450 1A1 by 1-ethynylpyrene and phenylacetylene. Chem Res Toxicol. 1993;6:38–45. doi: 10.1021/tx00031a006. [DOI] [PubMed] [Google Scholar]
- 34.Sinnokrot MO, Sherrill CD. High-accuracy quantum mechanical studies of pi-pi interactions in benzene dimers. J Phys Chem A. 2006;110:10656–10668. doi: 10.1021/jp0610416. [DOI] [PubMed] [Google Scholar]
- 35.Grimme S. Do special noncovalent pi-pi stacking interactions really exist? Angew Chem Int Ed Engl. 2008;47:3430–3434. doi: 10.1002/anie.200705157. [DOI] [PubMed] [Google Scholar]
- 36.Zimmerman SC, VanZyl CM, Hamilton GS. Rigid molecular tweezers: preorganized hosts for electron donor-acceptor complexation in organic solvents. J Am Chem Soc. 1989;111:1373–1381. [Google Scholar]
- 37.Hunter CA, Singh J, Thornton JM. Pi-pi interactions: the geometry and energetics of phenylalanine-phenylalanine interactions in proteins. J Mol Biol. 1991;218:837–846. doi: 10.1016/0022-2836(91)90271-7. [DOI] [PubMed] [Google Scholar]
- 38.Hunter CA, Sanders JKM. The nature of pi-pi interactions. J AM Chem Soc. 1990;112:5525–5534. [Google Scholar]