Abstract
The ribosomal small subunit locus has been used for transgene expression in the rodent malaria parasites, Plasmodium berghei and P. yoelii, but this strategy utilizes single crossover integration and is thus prone to reversion by plasmid excision. Targeting of the ribosomal subunit locus may also have a negative effect on oocyst development in the mosquito. In P. berghei, the p230 paralog locus has been used for transgene expression. Here, we show that the P. yoelii S1 locus (Sporozoite expressed gene 1) (PY05712) is dispensable and can be used for stable transgene expression throughout the parasite life cycle. P. yoelii s1− parasites show no defect in blood stage replication, oocyst formation, sporozoite production, or liver stage development when compared to P. yoelii wildtype parasites. Further, we show that a fluorescent transgene can be stably expressed from this site. This demonstrates that the S1 locus can be utilized for stable expression of heterologous genes in rodent malaria parasites.
Keywords: Malaria, Plasmodium, Heterologous gene expression, Complementation locus, Reverse genetics
Malaria is caused by a protozoan parasite of the genus Plasmodium and of the four species of human malaria, Plasmodium falciparum causes the most mortality [1]. Infection starts when the bite of an infected female Anopheles mosquito deposits salivary gland sporozoites in the skin, which are then transported to the liver where they undergo liver stage development to produce tens of thousands of merozoites that proceed to infect red blood cells (reviewed in [2–4]). Gametocytes, the sexual stage of the malaria life cycle, are taken up with the blood meal during a mosquito bite [5]. Sporozoites are produced by the thousands in the maturing oocyst, which develops in the mosquito midgut, and egress into the hemolymph upon maturation. Once the sporozoites encounter the mosquito salivary gland, they traverse the epithelial cell layer and accumulate in the salivary duct, awaiting deposition into a new host [6,7].
Reverse genetics techniques have proven invaluable to understand gene function in Plasmodium and are useful for introducing deletions, mutations, and for expressing tagged proteins, and transgenes. Knocking out a gene provides an idea of which point in the parasite life cycle (if any) the gene is important. To continue to study the gene by elucidating protein localization or function, transgene expression is a valuable technique [8]. For continuous expression of a transgene or to be able to carry out experiments with stage-specific promoters, it is necessary for the targeted locus to be completely dispensable throughout the entire life cycle. Complementation and gene knock-in in rodent malaria parasite lines are often created by single homologous recombination, also known as an integration strategy, into the ribosomal RNA (rRNA) locus [9]. In P. berghei, there are four copies of the rRNA gene, two of which are transcribed during the asexual cycle (A-type) and two of which, are transcribed solely during the mosquito stage (S-type) [10]. The two S-type rRNA genes are further designated, “C” and “D”. It has been shown in P. berghei, that expression of only one of these two genes is necessary for complete mosquito stage development. When using an integration strategy, however, reversion to the wildtype can occur [9]. Additionally, it was shown that integration into the rRNA locus can have a negative effect on oocyst development [9]. An alternative strategy that produces stable parasite lines is double crossover homologous recombination [11]. The drawback of this technique in expressing transgenes is the paucity of known dispensable loci in the rodent malaria parasite. The p230 paralog locus has been successfully used for transgene expression in P. berghei [12], but to our knowledge, this locus has not been utilized for transgene expression in P. yoelii. Here, we describe a useful targeting locus for complementation and knock-ins, the S1 gene from P. yoelii (PyS1), first identified by Kaiser et al. [13]. This gene has orthologs in P. berghei and P. falciparum. Targeting PyS1 results in stable transfectants due to the ability to use the double crossover homologous recombination strategy. Here, we show that Pys1− parasites have a phenotype that is indistinguishable from WT parasites throughout the life cycle. Thus, we show that heterologous gene expression from the replaced PyS1 locus is possible throughout the parasite life cycle.
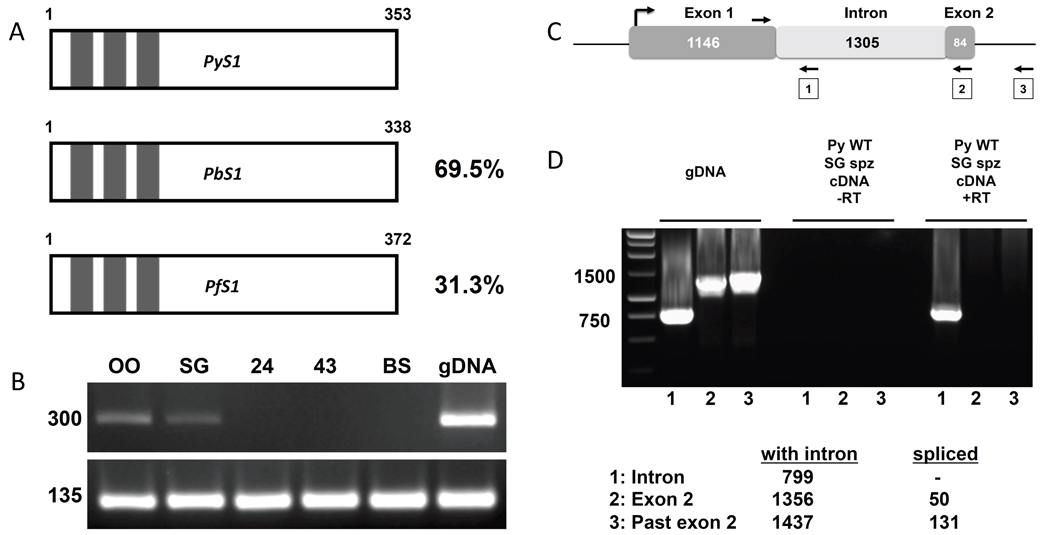
The “S genes” were identified through a subtractive hybridization assay which identified genes that were up-regulated in sporozoites (“S”) as compared to mixed blood stages [13]. It is likely that genes expressed uniquely in sporozoites could play an important role in mammalian host cell invasion and subsequent development in the hepatocyte [13,14]. P. yoelii S1 (PY05712) (PyS1), P. berghei S1 (Pb300684.00.0) (PbS1), and P. falciparum S1 (Pf10_0083) (PfS1) are listed as orthologs [15] and have relatively high amino acid sequence identity. The amino acid sequence identity between PyS1 and PbS1 is 70% and between PyS1 and PfS1 it is 31% (Figure 1A). Interestingly, all the orthologs are annotated with three C3H1 zinc finger domains [13,15]. In eukaryotes, C3H1 zinc finger domains were first identified in the tristetraprolin protein in mice [16] and have been shown to be associated with binding to the class II AU-rich elements of tumor necrosis factor alpha mRNA. This binding causes the subsequent degradation of the mRNA (reviewed in [17]). Kaiser et al. showed that PyS1 is expressed in salivary gland sporozoites but not during blood stages[13]. This has been confirmed by microarray comparing P. yoelii oocyst and salivary gland sporozoites and blood stages (GEO GSE8125) [18,19]. Here, PCR was performed with cDNA from oocyst sporozoites, salivary gland sporozoites, liver stages at 24 and 43 h post infection, and mixed blood stages from WT P. yoelii (for details, see supplemental materials and methods). These data show that PyS1 is expressed in oocyst and salivary gland sporozoites, but not in liver or blood stages (Figure 1B). The PlasmoDB annotation of PfS1 suggests that the protein is coded for by an uninterrupted open reading frame (ORF) but the PyS1 annotation has an intron separating the ORF between two exons (Figure 1C). To test whether PyS1 is coded for by two exons, a forward primer was designed with identity to the 3’ end of the presumed uninterrupted ORF of the first exon as well as three reverse primers. The first reverse primer has identity to the predicted intron; the second reverse primer has identity to the predicted second exon; and the third reverse primer has identity beyond the predicted second exon (Figure 1C). If the ORF is separated by an intron, the first primer pair should amplify a band of 799 bp from genomic DNA but should not amplify a product from cDNA as the intron would have been spliced out. If the PyS1 ORF is separated by an intron, the second pair of primers should amplify a band of 1356 bp from gDNA and 50 bp from cDNA and finally, the third pair of primers should amplify a band of 1437 bp from gDNA and 131 bp from cDNA. The PCR results for the first primer pair reveal a band of approximately 800 bp for both the gDNA and cDNA indicating that PyS1 is a single exon gene (Figure 1D). Subsequent amplification and sequencing of PyS1 cDNA from salivary gland sporozoites demonstrated the presence of an ORF of 1158 bp.
Figure 1.
P. yoelii S1 gene structure (A) A schematic representation of S1 from P. yoelii, P. berghei, and P. falciparum. Length of the predicted protein is noted in amino acids, percentages indicate amino acid identity as compared to P. yoelii, and C3H1 zinc finger domains are represented by boxes. (B) PCR analysis showing transcription during oocyst sporozoites (OO), salivary gland sporozoites (SG), and no transcription during liver stages at 24 and 43 hrs post-infection (24, 43) and mixed blood stages (BS). Amplification of Py18S was used as a positive control. (C) Schematic representation of the PyS1 locus as annotated in PlasmoDB. The arrows denote the primers used to test the intron/exon structure of PyS1. (D) PCR analysis showing that PyS1 has a single uninterrupted ORF. Lanes labeled “1” are products from the reverse primer in the putative intron, lanes labeled “2” are products from the reverse primer in the putative second exon, lanes labeled “3” are products from the reverse primer beyond the putative second exon. Expected amplicon sizes are shown in the legend.
PyS1 is uniquely expressed in sporozoites and thus PyS1 was expected to have no function during the blood stage of the life cycle. A plasmid was constructed to delete the PyS1 ORF by double crossover homologous recombination to assess the importance of PyS1 in the non-lethal P. yoelii strain, 17XNL (Figure S1A). Similar plasmids were constructed to delete S1 from the lethal strain of P. yoelii, YM, and from the non-lethal strain of P. berghei, NK65 (Figure S2A). If a gene is essential for asexual blood stage growth or replication, it is not possible to select a knockout parasite line since transfection takes place in this stage of the life cycle. PyS1 was, however, successfully deleted from the parasite genome. The Pys1− and the Pbs1− genotype of the clonal population was confirmed by PCR showing replacement at the 5’ and 3’ termini of PyS1 and lack of the PyS1 ORF (Figure S1B and S2B). To confirm the deletion of PyS1, a PCR using salivary gland sporozoite cDNA was performed. As expected, the results showed that PyS1 transcript was present in P. yoelii WT, but not in Pys1−. Primers for circumsporozoite protein were used as a positive control to amplify transcript from both WT and Pys1− (Figure S1C).
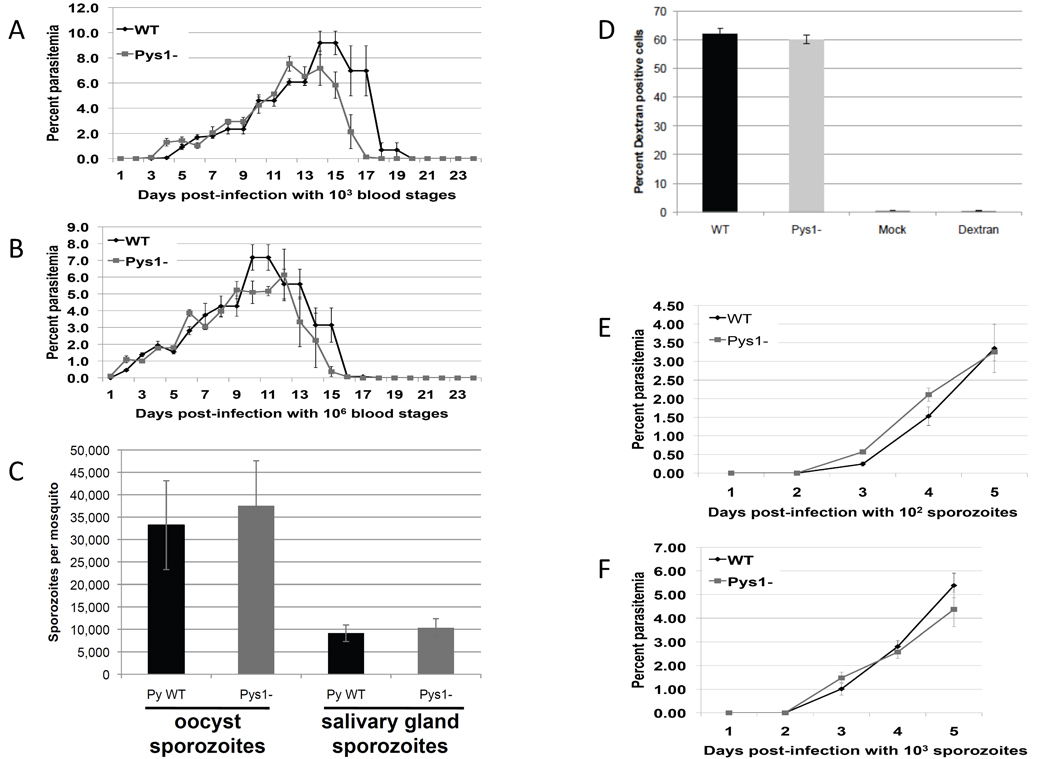
Since the “S genes” are expressed in sporozoites but not in blood stages, it is believed that they may have an important role not only in sporozoite function but also during the next step of the life cycle, i.e. invasion of and development in hepatocytes [13,14]. To assess the effect of deleting PyS1, all the life cycle stages were examined by comparing Pys1− and WT parasites. The ability to obtain the Pys1− line indicated that PyS1 is not essential during the blood stage. To determine, however, if there were any subtle changes in blood stage replication, 103 or 106 Pys1− or WT blood stages were injected IV into naïve BALB/c mice and blood stage parasitemia was measured daily (for details, see supplemental materials and methods). The rise and subsequent fall (due to clearance of the non-lethal parasites) of parasitemia was the same for both Pys1− and WT indicating there is no growth defect in Pys1− asexual stages (Figure 2A and 2B). Exflagellation by the male gametes was evaluated by microscopy of a wet mount and both Pys1− and WT male gametes exflagellated normally and in similar frequency (data not shown).
Figure 2.
Phenotypic analysis of Pys1− parasites. Either 1000 (A) or one million (B) Pys1− or WT blood stage parasites were injected into four BALB/c mice. Blood stage parasitemia was followed every day for 24 days and expressed as an average percentage. (C) Pys1− and WT infected mosquitoes were evaluated for numbers of midgut and salivary gland sporozoites at day 10 and 14, respectively. (D) P. yoelii wildtype parasites (WT), Pys1− parasites, or uninfected mosquito salivary gland material (Mock) or media supplemented with dextran only (Dextran) were incubated with hepatoma cells for one hour. Cells were then trypsinized and counted via fluorescence-activated cell sorting to determine the percentage of cells that were dextran positive. (E) 100 or (F) 1,000 PyS1− or WT sporozoites were injected IV into four BALB/c mice. Blood stage parasitemia was followed every day for five days and expressed as an average percentage.
Parasite sexual reproduction takes place in the mosquito with the subsequent development of the midgut oocyst which gives rise to sporozoites that migrate to and invade the salivary glands. To evaluate the effect of PyS1 gene deletion on sporozoite production, mosquitoes were fed an infected blood meal of Pys1− or WT mixed blood stages and midgut sporozoite numbers and salivary gland sporozoite numbers were evaluated on day 10 and 14 post infection, respectively (for details, see supplemental materials and methods). There was no significant difference in either midgut or salivary gland sporozoite production (Figure 2C). Similar results were obtained comparing Pbs1− to Pb WT (Figure S3) and PyYMs1− (data not shown). The results suggest that Pys1− parasites develop normally throughout the mosquito stage.
After entering the mammalian liver, sporozoites typically traverse several hepatocytes before invading the hepatocyte within which the parasite develops [20]. The ability of Pys1− parasites to traverse hepatocytes was tested in a cell traversal assay and compared to the WT (for details, see supplemental materials and methods). Pys1− and WT sporozoites traversed hepatoma cells at similar rates, which suggests that Pys1− parasites have no defect in cell traversal (Figure 2D). In vivo analysis of liver stage development was carried out by infecting BALB/c mice with an IV injection of either 100 or 1,000 Pys1− or WT sporozoites. Blood stage parasitemia was evaluated daily for five days by Giemsa stained blood smears. Pys1− and WT parasites showed blood stage parasitemia on day 3 post-infection and parasitemia increased at a similar rate in the two subsequent days that parasitemia was assayed (Figure 2E and F). There were also no differences in the pre-patent period for Pbs1− (Table S2) and for PyYMs1− (data not shown) as compared with Pb WT and PyYM WT respectively. Equal length of the pre-patent period as well as a similar increase in blood stage parasitemia afterwards indicates that Pys1− parasites are able to complete the liver stage at the same rate as WT parasites.
The lack of discernable phenotype during the entire life cycle of Pys1− parasites as well as Pbs1− and for PyYMs1− suggests that S1 is a dispensable gene for P. yoelii and P. berghei. The identification of such a locus allows for the use of this site for heterologous gene expression.
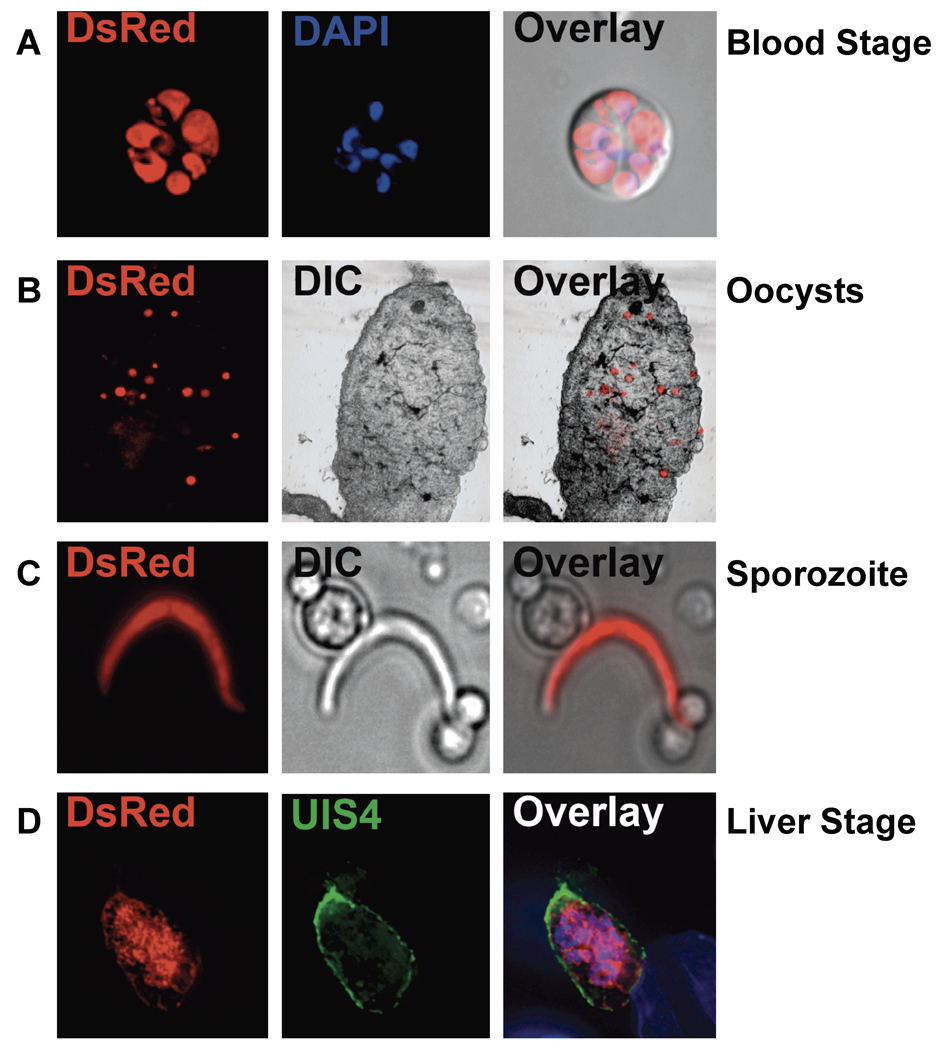
To test whether the PyS1 locus could be used for heterologous gene expression, DsRed under the control of the P. berghei elongation factor 1 α promoter and stabilized by the P. berghei dihydrofolate reductase 3’ UTR was targeted to the S1 locus (Figure S1A; for details, see supplemental materials and methods). The P. berghei elongation factor 1 α is a constitutively expressed gene and thus its promoter should result in the expression of DsRed throughout the life cycle. Fluorescent parasites were detected during the erythrocytic cycle, in mosquito midgut oocysts and salivary gland sporozoites, as well as during liver stage development (Figure 3). This clearly demonstrates that the S1 locus can be used for heterologous gene expression and as a site for complementation of knockout lines. The S1 site is not only useful for gene expression, but can be used for testing stage specificity of promoters. Finally, the S1 site can be used to express P. falciparum genes in P. yoelii or P. berghei parasites. It is quite useful to be able to express P. falciparum genes in rodent parasite lines without affecting the endogenous gene. In this way, insights into the function of P. falciparum genes can be discovered using the rodent malaria model systems [21]. The P. falciparum S1 could also be targeted for complementation or heterologous gene expression.
Figure 3.
Fluorescence microscopy of Pys1− parasites. Expression of DsRed in Pys1− parasites was evident in (A) live blood stages whose nuclei were visualized with 4',6-diamidino-2-phenylindole (DAPI) (Blue), (B) mosquito midgut oocysts in freshly dissected midguts, (C) the salivary gland sporozoite released from dissected mosquito salivary glands by grinding, and (D) liver stages in hepatoma cells fixed and visualized with an anti-UIS4 antibody and DAPI. Fluorescent and differential interference contrast (DIC) images were captured and merged (Overlay).
Supplementary Material
Acknowledgements
This work was funded by a grant from the National Institutes of Health (R01AI053709-07A2) to S.H.I.K.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Malaria Fact Sheet no. 94 on World Wide Web URL: http://www.who.int/mediacentre/factsheets/fs094/en/index.html
- 2.Vaughan AM, Aly AS, Kappe SH. Malaria parasite pre-erythrocytic stage infection: gliding and hiding. Cell Host Microbe. 2008;4:209–218. doi: 10.1016/j.chom.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sinnis P, Coppi A. A long and winding road: the Plasmodium sporozoite's journey in the mammalian host. Parasitol Int. 2007;56:171–178. doi: 10.1016/j.parint.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mikolajczak SA, Kappe SH. A clash to conquer: the malaria parasite liver infection. Mol Microbiol. 2006;62:1499–1506. doi: 10.1111/j.1365-2958.2006.05470.x. [DOI] [PubMed] [Google Scholar]
- 5.Baton LA, Ranford-Cartwright LC. Spreading the seeds of million-murdering death: metamorphoses of malaria in the mosquito. Trends Parasitol. 2005;21:573–580. doi: 10.1016/j.pt.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 6.Matuschewski K. Getting infectious: formation and maturation of Plasmodium sporozoites in the Anopheles vector. Cell Microbiol. 2006;8:1547–1556. doi: 10.1111/j.1462-5822.2006.00778.x. [DOI] [PubMed] [Google Scholar]
- 7.Aly AS, Vaughan AM, Kappe SH. Malaria Parasite Development in the Mosquito and Infection of the Mammalian Host. Annu Rev Microbiol. 2009 doi: 10.1146/annurev.micro.091208.073403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao Y, Zhang D, Pan W. Construction of transgenic plasmodium berghei as a model for evaluation of blood-stage vaccine candidate of Plasmodium falciparum chimeric protein 2.9. PLoS ONE. 2009;4:e6894. doi: 10.1371/journal.pone.0006894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Spaendonk RM, Ramesar J, van Wigcheren A, et al. Functional equivalence of structurally distinct ribosomes in the malaria parasite, Plasmodium berghei. J Biol Chem. 2001;276:22638–22647. doi: 10.1074/jbc.M101234200. [DOI] [PubMed] [Google Scholar]
- 10.Li J, Gutell RR, Damberger SH, et al. Regulation and trafficking of three distinct 18 S ribosomal RNAs during development of the malaria parasite. J Mol Biol. 1997;269:203–213. doi: 10.1006/jmbi.1997.1038. [DOI] [PubMed] [Google Scholar]
- 11.Menard R, Janse C. Gene targeting in malaria parasites. Methods. 1997;13:148–157. doi: 10.1006/meth.1997.0507. [DOI] [PubMed] [Google Scholar]
- 12.Janse CJ, Franke-Fayard B, Mair GR, et al. High efficiency transfection of Plasmodium berghei facilitates novel selection procedures. Mol Biochem Parasitol. 2006;145:60–70. doi: 10.1016/j.molbiopara.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 13.Kaiser K, Matuschewski K, Camargo N, et al. Differential transcriptome profiling identifies Plasmodium genes encoding pre-erythrocytic stage-specific proteins. Mol Microbiol. 2004;51:1221–1232. doi: 10.1046/j.1365-2958.2003.03909.x. [DOI] [PubMed] [Google Scholar]
- 14.Matuschewski K, Ross J, Brown SM, et al. Infectivity-associated changes in the transcriptional repertoire of the malaria parasite sporozoite stage. J Biol Chem. 2002;277:41948–41953. doi: 10.1074/jbc.M207315200. [DOI] [PubMed] [Google Scholar]
- 15.PlasmoDB: Plasmodium Genome Resource on World Wide Web URL: http://plasmodb.org/plasmo/showRecord.do?name=GeneRecordClasses.GeneRecordClass&source_id=PY05712&project_id=PlasmoDB
- 16.Lai WS, Stumpo DJ, Blackshear PJ. Rapid insulin-stimulated accumulation of an mRNA encoding a proline-rich protein. J Biol Chem. 1990;265:16556–16563. [PubMed] [Google Scholar]
- 17.Blackshear PJ. Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem Soc Trans. 2002;30:945–952. doi: 10.1042/bst0300945. [DOI] [PubMed] [Google Scholar]
- 18.Mikolajczak SA, Silva-Rivera H, Peng X, et al. Distinct malaria parasite sporozoites reveal transcriptional changes that cause differential tissue infection competence in the mosquito vector and mammalian host. Mol Cell Biol. 2008;28:6196–6207. doi: 10.1128/MCB.00553-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tarun AS, Peng X, Dumpit RF, et al. A combined transcriptome and proteome survey of malaria parasite liver stages. Proc Natl Acad Sci U S A. 2008;105:305–310. doi: 10.1073/pnas.0710780104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mota MM, Pradel G, Vanderberg JP, et al. Migration of Plasmodium sporozoites through cells before infection. Science. 2001;291:141–144. doi: 10.1126/science.291.5501.141. [DOI] [PubMed] [Google Scholar]
- 21.Tewari R, Spaccapelo R, Bistoni F, et al. Function of region I and II adhesive motifs of Plasmodium falciparum circumsporozoite protein in sporozoite motility and infectivity. J Biol Chem. 2002;277:47613–47618. doi: 10.1074/jbc.M208453200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.