Abstract
The lipid A of Rhizobium etli, a nitrogen-fixing plant endosymbiont, displays significant structural differences when compared to that of Escherichia coli. An especially striking feature of R. etli lipid A is that it lacks both the 1- and 4′phosphate groups. The 4′-phosphate moiety of the distal glucosamine unit is replaced with a galacturonic acid residue. The dephosphorylated proximal unit is present as a mixture of the glucosamine hemiacetal and an oxidized 2-aminogluconate derivative. Distinct lipid A phosphatases directed to the 1 or the 4′-positions have been identified previously in extracts of R. etli and Rhizobium leguminosarum. The corresponding structural genes, lpxE and lpxF respectively, have also been identified. Here we describe the isolation and characterization of R. etli deletion mutants in each of these phosphatase genes and the construction of a double phosphatase mutant. Mass spectrometry confirmed that the mutant strains completely lacked the wild-type lipid A species and accumulated the expected phosphate-containing derivatives. Moreover, radiochemical analysis revealed that phosphatase activity was absent in membranes prepared from the mutants. Our results indicate that LpxE and LpxF are solely responsible for selectively dephosphorylating the lipid A molecules of R. etli. All the mutant strains showed an increased sensitivity to polymyxin relative to the wild-type. However, despite the presence of altered lipid A species containing one or both phosphate groups, all the phosphatase mutants formed nitrogen-fixing nodules on Phaseolus vulgaris. Therefore, the dephosphorylation of lipid A molecules in R. etli is not required for nodulation but may instead play a role in protecting the bacteria from cationic antimicrobial peptides or other immune responses of plants.
INTRODUCTION
Gram-negative bacteria have an asymmetric outer membrane composed of an inner leaflet of glycerophospholipids and an outer leaflet of lipopolysaccharide (LPS) [1]. LPS functions as a protective barrier and consists of three domains: 1) the lipid A moiety; 2) a non-repeating core oligosaccharide; and 3) a distal O-antigen polysaccharide [1, 2]. The lipid A component of LPS is a relatively conserved structural feature of Gram-negative bacteria and serves as the membrane anchor for LPS [1]. It is required for growth in almost all Gram-negative bacteria [1] and is the “endotoxin” component of LPS associated with the complications of severe Gram-negative sepsis [3]. Lipid A is a potent stimulator of inflammation and of innate immunity in animals via the Toll-like receptor-4/MD-2 (TLR4/MD-2) complex [4].
In most enteric Gram-negative bacteria, the lipid A moiety of LPS is a hexa-acylated disaccharide of glucosamine that is phosphorylated at positions 1 and 4′ (Fig. 1A) [1]. Pharmacological studies have shown that both phosphate groups, the glucosamine disaccharide, and the proper arrangement of the fatty acyl chains are important for full activation of TLR4/MD-2 [5]. Monophosphorylated lipid A analogues, lacking the 1-phosphate group, retain some of the immune-stimulatory properties of E. coli lipid A but show greatly reduced toxicity in animals [6, 7]. Monophosphorylated lipid A preparations are therefore being developed as adjuvants in human vaccines [7].
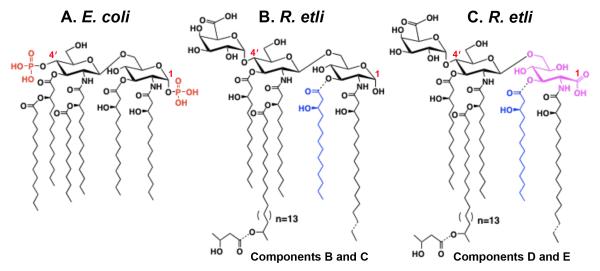
Figure 1. Structures of the major lipid A species present in E. coli and R. etli.
Panel A. The predominant lipid A moiety of E. coli LPS consists of a hexa-acylated disaccharide of glucosamine, substituted with monophosphate groups at the 1- and 4′-positions. There is very little acyl chain heterogeneity [8]. Panels B and C. The major lipid A species in R. etli and R. leguminosarum lack phosphate substituents and are more heterogeneous in with respect to fatty acyl chain length [14, 15]. Components B and C are constructed around the typical glucosamine disaccharide found in the lipid A of other Gram-negative organisms. Components D and E feature an aminogluconate unit in place of the proximal glucosamine residue, derived by LpxQ-catalyzed oxidation of B and C respectively [28, 29]. All R. etli lipid A species contain a galacturonic acid moiety at position-4′ in place of the more typical monophosphate group, and they also contain an unusual C28 secondary acyl chain that may be further derivatized at the 27-OH moiety with a β-hydroxybutyrate group [13]. Components C and E differ from B and D by the absence of a hydroxyacyl chain at position 3, which is removed by the deacylase PagL [30]. Dashed bonds highlight the most prominent micro-heterogeneity of R. etli lipid A with respect to its acyl chains and the presence of the β-hydroxybutyrate substituent. Micro-heterogeneity arising from minor branched-chain or even chain fatty acids is not indicated, but is apparent in the mass spectra shown below.
Many animal pathogens adapt strategies for modifying their lipid A structure to evade detection and early clearance by the innate immune system [8]. These lipid A modifications include dephosphorylation, changes in acylation status, or attachment of amino-sugars to neutralize the negative charges of the phosphate moieties [8]. The lipid A components of Francisella novicida and Francisella tularensis, the causative agents of tularemia in mice and humans, respectively, illustrate some of these principles. Francisella lipid A molecules lack the 4′-phosphate group and the 3′-hydroxyacyl chain [9-11], both of which are removed during the later stages of lipid A assembly [12].
Modifications to lipid A are not unique to pathogenic bacteria. For instance, some of the lipid A modifications seen in F. tularensis also occur in Rhizobium etli, a plant endosymbiont that participates in a nitrogen-fixing symbiosis with leguminous plants [8, 13]. R. etli lives intracellularly within nodules that form on roots. The lipid A of R. etli, which consists of a mixture of related components (Figs. 1B and 1C) [14, 15], lacks both the 1- and 4′-phosphate groups present in the lipid A of enteric bacteria [13]. The 4′-phosphate moiety is replaced with a galacturonosyl residue, whereas the 1-dephosphorylated proximal glucosamine residue is present either in the hemiacetal form (Fig. 1B) or as an oxidized 2-amino-2-deoxy-gluconate unit (Fig. 1C) [13-15]. R. etli lipid A also differs from that of E. coli with respect to its acylation status (Fig. 1). R. etli lipid A lacks the secondary laurate and myristate chains found in E. coli, but instead is acylated with a secondary 27-hydroxyoctacosanoate chain (27OHC28:0) at the 2′-position [13-16]. This residue may be esterified with a β-hydroxybutyrate moiety (Figs. 1B and 1C) [13-15]. Differences in fatty acyl chain length, especially at the 2 position, further contribute to R. etli lipid A micro-heterogeneity [13-15] (not shown in Fig. 1).
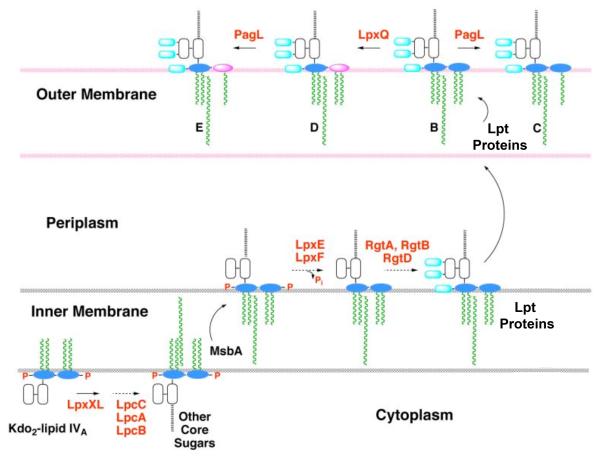
Lipid A biosynthesis in R. etli diverges from that of E. coli after the formation of the tetra-acylated intermediate Kdo2-lipid IVA, as shown schematically in Fig. 2 [8]. R. etli utilizes similar enzymes as found in E. coli to convert UDP-GlcNAc into Kdo2-lipid IVA, which is then processed differently by enzymes unique to R. etli (Fig. 2) [17]. The enzyme LpxXL incorporates the sole secondary acyl chain (27OHC28:0), which is supplied by the unique acyl carrier protein, AcpXL [18-20]. After formation of the acyloxyacyl chain and addition of other core sugars [19, 21], lipid A molecules are flipped to the outer leaflet of the inner membrane by the ABC transporter MsbA (Fig. 2) [8], where the lipid A phosphatases LpxE [22, 23] and LpxF [24, 25] dephosphorylate them. Following removal of the 4′-phosphate moiety, the enzyme RgtD is proposed to glycosylate the 4′-position with a galacturonic acid residue, using dodecaprenyl-phosphate galacturonic acid as its donor substrate [26, 27]. After removal of the 1-phosphate moiety on the outer surface of the inner membrane and transport to the outer membrane, the lipid A oxidase LpxQ oxidizes the proximal glucosamine residue to the 2-aminogluconate unit (Figs. 1C and 2) in an oxygen-dependent reaction [28, 29]. In parallel, the ester-linked hydroxyacyl chain at position 3 may be removed by the outer membrane deacylase PagL [30, 31] (Fig. 2). The functional significance of these lipid A modifications in R. etli is unknown.
Figure 2. Topography of LPS assembly and lipid A modification in R. etli.
With the exception of the UDP-diacylglucosamine hydrolase LpxH, which is replaced by LpxI (L. E. Metzler and C. R. H. Raetz, in preparation), the constitutive enzymes that generate the phosphorylated intermediate Kdo2-lipid IVA in E. coli are also present in R. etli [17]. The assembly of R. etli lipid A diverges after Kdo2-lipid IVA formation, starting with the addition of the 27-hydroxyoctacosanoate chain (27OHC28:0), catalyzed by LpxXL [20]. After completion of core glycosylation and transport to the outer surface of the inner membrane by MsbA, LpxF and LpxE remove the phosphate moieties at the 4′- and 1-positions, respectively [22, 23, 25]. Next, RgtD is thought to utilize dodecaprenyl phosphate-galacturonic acid to incorporate the 4′-galacturonic acid residue, and RgtA/RbtB similarly modify the outer Kdo unit [26, 27]. After completion of O-antigen assembly (not shown) and transport to the outer membrane by the Lpt proteins [68], the ester-linked hydroxyacyl chain at position 3 may be removed by the deacylase PagL [30, 31], and the proximal glucosamine may be converted to the aminogluconate unit by the oxidase LpxQ [28, 29]. The orientation of the LpxQ active site within the outer membrane is not established unequivocally.
A previously-reported 4′-phosphatase lpxF mutant of F. novicida is very sensitive to polymyxin and is highly attenuated in a mouse infection model [11]. In the present study we address the functional roles of the two lipid A phosphatases of R. etli encoded by lpxE and lpxF [22, 23, 25, 32] by constructing mutants with deletions in one or both of these genes. The recently-completed genome sequence of R. etli revealed only two plausible candidate genes encoding these phosphatases [33]. Deletion mutants of the individual phosphatases and a double-knockout mutant were constructed by replacing these genes with spectinomycin and/or gentamicin cassettes. The biochemical and physiological properties of these mutant strains are reported in this manuscript.
MATERIALS AND METHODS
Materials
[γ-32P] ATP was obtained from PerkinElmer Life And Analytical Sciences, Inc. (Waltham, MA). [14C]acetate (sodium salt) was obtained from GE Heathcare (Chalfont St. Giles, U.K.). Acetic acid, ammonium acetate, chloroform, formic acid, and glass-backed 0.25-mm Silica Gel-60 thin layer chromatography (TLC) plates were from EMD Chemicals, Inc. (Gibbstown, NJ). Pyridine and methanol were from Mallinckrodt Baker, Inc. (Phillipsburg, NJ). Yeast extract, agar, and tryptone were from Becton, Dickinson, and Company (Franklin Lakes, NJ). Sodium chloride was from VWR International (West Chester, PA). Bicinchoninic (BCA) protein assay reagents and Triton X-100 were from Thermo Fisher Scientific (Waltham, MA). Diethylaminoethyl (DEAE) cellulose DE52 was from Whatman, Inc. (Florham Park, NJ). GenomicPrep Cells and Tissue DNA Isolation kit was from GE Healthcare (Chalfont St. Giles, U.K.). All other chemicals were reagent grade and were purchased from either Sigma-Aldrich (St. Louis, MO) or VWR International (West Chester, PA).
Bacterial strains, growth conditions and molecular biology protocols
The bacterial strains and plasmids used and their relevant characteristics are shown in Table I. R. etli strains were grown at 29-30 °C on complex tryptone yeast extract medium (TY) supplemented with 4.5-10 mM CaCl2 [34]. Rhizobium stains were grown in media supplemented with the following concentrations of antibiotics where appropriate: neomycin (200 μg/mL), gentamicin (35 μg/mL), nalidixic acid (20 μg/mL) (streptomycin (200 μg/mL), and spectinomycin (200 μg/mL). E coli strains were cultured on LB broth at 37 °C [35]. Growth media were supplemented with the following concentrations of antibiotics where appropriate: kanamycin (50 μg/mL), gentamicin (10 μg/mL), carbenicillin (100 μg/mL), or spectinomycin (200 μg/mL). Derivatives of the suicide plasmid pK18mobsacB (Table 1) were mobilized into R. etli strains by triparental mating using the mobilizing plasmid pRK2013, as described previously [36]. Recombinant DNA techniques were performed according to standard protocols [37] using E. coli DH5α as a host strain [38]. The XL-PCR kit (Applied Biosystems) was used for amplifying DNA sequences from R. etli genomic DNA.
TABLE I.
Bacterial strains and plasmids used in this work
| Strain or Plasmid | Genotype or Description | Source or reference |
|---|---|---|
| Strains | ||
| R. etli CE3 | Spontaneous Smr derivative of wild-type strain CFN42, Nalr | [62] |
| R. etli CE3 derivatives | ||
| CS501 | ΔlpxE mutant, spectinomycin-resistant | this work |
| CS506 | ΔlpxF mutant, spectinomycin-resistant | this work |
| CS513 | ΔlpxE/ΔlpxF mutant, derived from CS506, gentamicin-resistant | this work |
| E. coli | ||
| DH5α | recA1 ϕ80 lacZΔM15 | [38] |
| Plasmids | ||
| pRK2013 | helper plasmid, kanamycin/neomycin-resistant | [63] |
| pK18mobsacB | suicide vector, kanamycin/neomycin-resistant | [64] |
| pUC18 | cloning vector, carbenicillin-resistant | [65] |
| pHY109 | spectinomycin resistance-conferring Ω interposon cloned in the EcoRI restriction sites of plasmid pCHO341 |
[66] |
| pACΩ-Gm | broad host range vector containing gentamicin resistance cassette |
[67] |
| pCCS72a | pUC18 derivative containing 1 kb of sequence upstream of lpxE as a SmaI/EcoRI fragment |
this work |
| pCCS73a | pUC18 derivative containing 1 kb of sequence upstream of lpxF as a XbaI/SmaI fragment |
this work |
| pCCS72b | pUC18 derivative containing 1 kb of sequence upstream and downstream of lpxE as XbaI/SmaI and SmaI/EcoRI fragments |
this work |
| pCCS73b | pUC18 derivative containing 1 kb of sequence upstream and downstream of lpxF as XbaI/SmaI and SmaI/EcoRI fragments |
this work |
| pCCS74a | pUC18 derivative containing spectinomycin cassette flanked by upstream and downstream regions of lpxE |
this work |
| pCCS75a | pUC18 derivative containing spectinomycin cassette flanked by upstream and downstream regions of lpxF |
this work |
| pCCS74b | suicide plasmid for construction of lpxE mutant by substitution with a spectinomycin resistance cassette |
this work |
| pCCS75b | suicide plasmid for construction of lpxF mutant by substitution with a spectinomycin resistance cassette |
this work |
| pCCS76 | suicide plasmid for construction of lpxE-/lpxF-deficient double mutant by substitution of the lpxF gene with a gentamicin cassette |
this work |
Abbreviations: Smr-Streptomycin sulfate resistant, Nalr-Nalidixic acid resistant.
Exchange of the rhizobial lpxE and lpxF genes with a spectinomycin resistance cassette
The oligonucleotide primers o1phosph_A (5′-ACG TTC TAG ATG GAT GCC GGT TTC GTC ACC GAC G-3′) and o1phosph_B (5′-ACG TCC CGG GCG CCA GCG CCT ATC AAG GGA AGC-3′) were used to amplify about 1 kb of genomic DNA upstream of the putative lpxE gene from Rhizobium etli CE3. The primers o4phosph_A (5′-ACG TTC TAG AGC ACG AGG CGG CCT TCG GCA TCG-3′) and o4phosph_B (5′-ACG TCC CGG GCA AGC AGT GCC CAC CAC GTC CAG-3′) were used to amplify about 1 kb of genomic DNA upstream of the putative lpxF gene. These primers introduced XbaI and SmaI sites (underlined) into the PCR products. After digestion with the respective enzymes, the PCR products were cloned as XbaI/SmaI fragments into pUC18, to yield plasmids pCCS72a and pCCS73a. Similarly, the primers o1phosph_C (5′-ACG TCC CGG GCG CCG GAT GGC TGG CCA CTC GC-3′) and o1phosph_D (5′-ACG TGA ATT CTC GGT GCC ACC ATG GGC CAG G-3′) were used to amplify 1 kb of genomic DNA downstream of the lpxE gene. The primers o4phosph_C (5′-ACG TCC CGG GTC TTC GCA GCG CTG CTG GCG TTA ACT G-3′), and o4phosph_D (5′-ACG TGA ATT CCT GCA TGC GTC TCC AGG AAA CCG-3′) were used to amplify about 1 kb of genomic DNA downstream of the lpxF gene. The primers introduced SmaI and EcoRI sites (underlined) into the PCR products. After digestion with the respective enzymes, the SmaI/EcoRI-digested PCR products were cloned into pCCS72a and pCCS73a to yield plasmids pCCS72b and pCCS73b. Subsequently the spectinomycin resistance cassette from the plasmid pHY109 was subcloned as a SmaI fragment between the flanking sequences in pCCS72b and pCCS73b to generate plasmids pCCS74a and pCCS75a. The whole cassettes containing the flanking regions and the spectinomycin resistance cassette were then subcloned as XbaI/EcoRI-fragments into the suicide vector pK18mobsacB (Table I) to yield plasmids pCCS74b and pCCS75b. Using pRK2013 as a helper plasmid, pCCS74b and pCCS75b were introduced by triparental mating into wild-type R. etli CE3. Transconjugants were selected on TY medium containing neomycin and spectinomycin to select for single recombinants in a first step. The single recombinants were grown under nonselective conditions in complex medium for one day before being plated on TY medium containing spectinomycin and 10% sucrose (w/v) to select for the loss of the vector backbone of pK18mobsacB from the bacterial genome. Several candidates grew after 5 days. Southern Blot analysis confirmed that the putative mutant strains were double recombinants in which the lpxE gene or lpxF genes were replaced with a spectinomycin cassette (data not shown).
Construction of a R. etli double mutant deficient in the lpxE and lpxF genes
The suicide plasmid pCCS74b (Table 1), which was used in the construction of the lpxE mutant, was digested with SmaI to liberate its spectinomycin resistance cassette. Plasmid pACΩ-Gm (Table 1) was digested with SmaI to release a gentamicin cassette, which was subsequently ligated into the SmaI-digested pCCS74b plasmid to yield plasmid pCCS76. The suicide plasmid pCCS76 was then conjugated into CS506 in order to construct the double mutant. Conjugation and subsequent selection were performed as described above for the single mutants. Double mutants were confirmed by Southern Blot analysis (data not shown), and the mutant CS513 was selected for a detailed analysis. In this strain the lpxF gene is substituted with a spectinomycin resistance cassette and the lpxE gene is substituted with a gentamicin resistance cassette.
Lipid A extraction
Lipid A from R. etli was isolated as described previously [14, 15]; however, the conditions used for hydrolysis differed. Previous hydrolysis conditions utilized 12.5 mM sodium acetate at pH 4.5 with 1% SDS [14, 15]. In order to simplify the analysis of the lipid A species, 1% acetic acid was used for hydrolysis in the absence of SDS. Under these hydrolysis conditions, the labile glycosidic linkage between the Kdo moiety and lipid A is cleaved selectively. One liter of TY broth, supplemented with 10 mM CaCl2 and antibiotics, was inoculated with 4 mL of a fresh culture of bacteria. The flasks were shaken (225 rpm) at 30 °C overnight until the A600 reached 1.2. The cells were harvested by centrifugation at 5000 × g for 20 min at 4 °C. The cells were resuspended, washed once with 250 mL chilled phosphate-buffered saline (PBS) [39], and centrifuged once more. The washed cell pellets were frozen at −80 °C. Thawed cell pellets were resuspended in 48 mL PBS. The glycerophospholipids were extracted by adding 60 mL of chloroform and 120 mL of methanol to convert the cell suspension into a single-phase Bligh-Dyer mixture (chloroform:methanol:PBS, 1:2:0.8) [40]. The cells were extracted for 1 h and were centrifuged at 4000 × g for 20 min. The insoluble material was recovered and washed once more with 60 mL of a fresh, single-phase Bligh-Dyer mixture. The insoluble material was again recovered by centrifugation. The washed pellet was then suspended in 54 mL of 1% acetic acid and subjected to probe sonic irradiation. The suspension was boiled for 30 min and cooled to room temperature. The suspension was then converted to a two-phase Bligh-Dyer mixture (chloroform:methanol:1% acetic acid, 2:2:1.8, v/v) [40] by adding 60 mL of chloroform and 60 mL of methanol. The phases were mixed thoroughly and separated by centrifugation at 4000 × g for 20 min. The lower phase was retrieved, while the upper phase was washed once with 60 mL pre-equilibrated lower phase and centrifuged again. The second lower phase was retrieved, pooled with the first lower phase, and dried with a rotary evaporator. Remaining phospholipids were removed from the dried, crude lipid A by DEAE-cellulose column chromatography as described previously [14]. After TLC in a chloroform:methanol:water:ammonium hydroxide system (40:25:4:2, v/v), the lipids were visualized by sulfuric acid charring.
Negative ion electrospray ionization mass spectrometry (ESI/MS) of lipid A species
Lipid A species recovered from the cells were redissolved in chloroform:methanol (4:1, v/v), subjected to sonic irradiation in a bath sonicator, and subsequently diluted into chloroform:methanol (2:1). The samples were supplemented with 1% piperdine and immediately infused into the ion source at 5-10 μL/min and analyzed by ESI/MS, as described previously [10, 26, 27]. All mass spectra were acquired on a QSTAR XL quadrupole time-of-flight mass spectrometer (ABI/MDS-Sciex, Toronto, Canada). Spectra were acquired in the negative ion mode from 200 to 2400 atomic mass units. Some spectra were the accumulation of 60 scans. Data acquisition and analysis were performed using Analyst QS software version 1.1.
Preparation of membranes and substrates
All enzyme preparations were carried out at 0-4 °C. The bicinchoninic acid method [41] was used to determine protein concentrations, using bovine serum albumin as a standard. Cell-free extracts and washed membranes were prepared as described previously [20] and stored in aliquots at −80 °C. The preparation of carrier Kdo2-lipid IVA [42] and Kdo2-[4′-32P]-lipid IVA was described previously [43]. The substrates were stored as aqueous dispersions in 25 mM Tris-HCl, pH 7.8, containing 1 mM EDTA, 1mM EGTA, and 0.1% Triton X-100. Prior to use, the lipid substrates were subjected to sonic irradiation for 1 min in a bath sonicator.
In vitro assay of 4′-phosphatase activity
LpxF activity was determined as described [44]. Standard assay conditions for detecting 4′-phosphatase activity were as follows: the reaction mixture (10-20 μl) contained 50 mM MES, pH 6.5, 0.1% Triton X-100, 2 mM dithiothreitol, 2 mM EDTA, 10 mM potassium phosphate, and 5 μM Kdo2-[4′-32P]lipid IVA (1,000 cpm/nmol). Reactions were initiated with the enzyme source. The reaction mixtures were incubated at 30 °C for the indicated times. Reactions were terminated by spotting 3-μL samples onto a TLC plate, which was developed in the solvent chloroform/pyridine/88% formic acid/water (30:70:16:10, v/v). After drying and overnight exposure of the plate to a PhosphorImager screen (GE Healthcare), product formation was detected and quantified with a Storm 840 PhosphorImager equipped with ImageQuant software (GE Healthcare).
Assay of polymyxin sensitivity
The outer membrane integrity of wild-type R. etli and each of the phosphatase mutant strains was evaluated by examining sensitivity to the cationic antibacterial peptide polymyxin B sulfate (Sigma). A disk diffusion test was performed by placing sterile 6-mm filter paper disks onto a lawn of cells that had been freshly plated on TY agar using a sterile cotton swab from a culture at A600 of ~0.2. Polymyxin (1 μL of a 2, 10, or 20 mg/mL stock solution) was then applied to the disks. After 40 h of growth at 30 °C, the diameters of the zones of clearing were measured, providing an assessment of the relative polymyxin sensitivity.
In vivo labeling of R. etli strains with [14C]-acetate and quantitative analysis of lipid extracts
The lipid compositions of R. etli wild-type CE3 and the three mutant strains CS501, CS506 and CS513 were determined after labeling with [1-14C]-acetate. Cultures (1 mL) in TY medium, supplemented with 4.5 mM CaCl2, were inoculated from overnight cultures grown in the same medium. After the addition of 1 μCi [1-14C]-acetate (GE Healthcare; 60 mCi/mmol) to each culture, they were incubated for 8 h. The cells were harvested by centrifugation, washed with 500 μl of water and resuspended in 100 μl of water. The lipids were extracted according to the method of Bligh and Dyer [40]. The chloroform phase was used for lipid analysis on TLC plates, and after two-dimensional separation, the individual lipids were quantified using a Storm 820 PhosphorImager (GE Healthcare).
Plant test for symbiotic phenotype characterization
Phaseolus vulgaris cv. Negro Jamapa seeds were surface-sterilized with 1.2% sodium hypochlorite and germinated on 1% (w/v) water agar as described previously [45]. Seedlings were transferred to 250 mL flasks filled with vermiculite and nitrogen-free nutrient solution [46], and inoculated with about 105 CFU of bacteria per plant. Plants were grown in a controlled growth chamber at 28 °C with a 15 h day/ 9 h night cycle and harvested 21 d after inoculation. Nitrogenase activity of nodulated roots was determined by the acetylene reduction assay as described previously [47]. Nitrogen fixation activity per plant was normalized with respect to the nodule fresh weight per plant.
RESULTS
Candidate genes encoding the R. etli lipid A 1- and 4′-phosphatases
Using the sequence of the protein encoded by the lpxE gene from R. leguminosarum 3841 [22] and the sequence of the protein encoded by the lpxF gene from Francisella novicida [25], the genome of R. etli CFN42 [33] was searched for the corresponding candidates. The 232-amino acid protein encoded by ABC92837 is 92% identical to R. leguminosarum LpxE, and the 257-amino acid protein encoded by ABC90258 is 30% identical to F. novicida LpxF. Both genes are located on the R. etli chromosome [33], and both proteins are predicted to contain six transmembrane helices. The LpxE protein of R. etli most closely matches homologues from R. leguminosarum, Agrobacterium tumefaciens, Sinorhizobium meliloti, Mesorhizobium loti and Francisella tularensis subsp. novicida (Supporting Fig. 1), all of which are α-proteobacteria with the exception of Francisella. The putative LpxF protein from R. etli most closely matches homologues from the α-proteobacteria (Rhizobium leguminosarum, Rhodospirillum rubrum, Rhodopseudomonas palustris, Xanthobacter autotrophicus and Bradyrhizobium sp. BTAi1), the β-proteobacteria (Dechloromonas aromatica) and the γ-proteobacteria (Francisella tularensis) (Supporting Fig. 2). Both LpxE and LpxF contain variations of the lipid phosphate phosphatase sequence motif described by Stukey and Carman [48].
Lipid A from wild-type R.etli and the phosphatase knockout strains
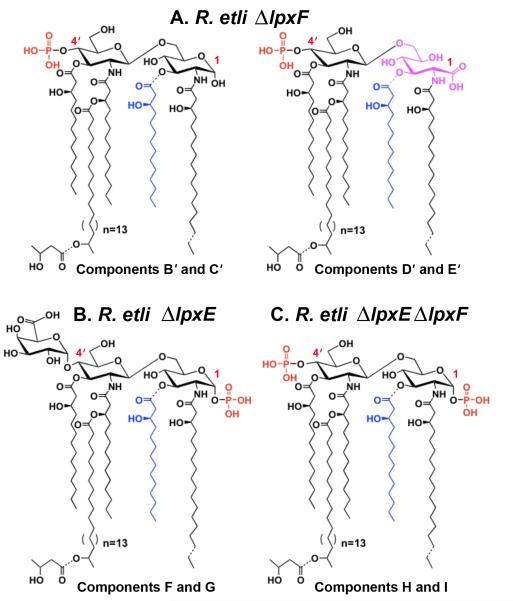
Deletion mutants of the lpxE and lpxF genes were isolated in R. etli as described in the materials and methods. The single mutants were expected to accumulate the structures shown in Fig. 3A and 3B. The double knockout of the two phosphatases was expected to accumulate lipid A species with the two phosphate groups retained at the 1 and 4′ positions, similar to the lipid A molecules found in S. meliloti (Fig. 3C) [49].
Figure 3. Predicted structures of the major lipid A species in lpxF, lpxE, and lpxE/lpxF deletion mutants of R. etli.
Panel A. The proposed major lipid A species (B’, C’, D’ and E’) of the R. etli LpxF mutant should resemble that of the wild-type, except that the 4′-galacturonic acid unit would be replaced with a monophosphate group. Panel B. The proposed major lipid A species (F and G) of the R. etli LpxE mutant should be less heterogeneous than wild-type, because the proximal glucosamine unit cannot be oxidized by LpxQ. Panel C. The proposed major lipid A species (H and I) of the double mutant should retain phosphate groups at both the 1- and 4′-positions, resembling the lipid A of Sinorhizobium meliloti [49]. The exact masses predicted for these predominant lipid A molecular species are shown in Table II.
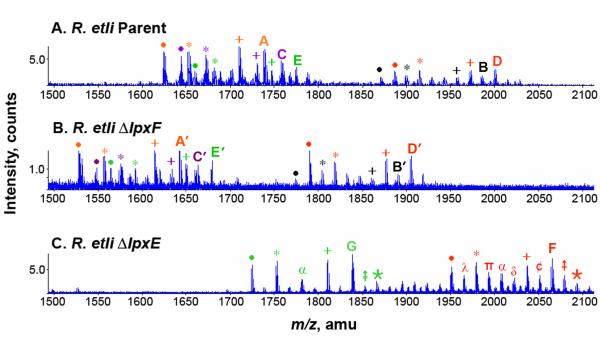
Lipid A from each of the R. etli strains was obtained by hydrolyzing delipidated cell pellets in 1% acetic acid. The lipid A components released in this manner from the wild-type migrated as multiple bands, designated A, B, C, D, and E (Fig. 4), when analyzed by TLC [14, 15]. The previously determined structures of B, C, D, and E [14, 15] are shown in Fig. 1B and 1C. Component A (Fig. 4) is an artifact arising from component D by β-elimination during the hydrolysis process (structure shown in Supporting Fig. 3 [14, 15]. The TLC patterns of the lipid A species from the lpxF and lpxE mutants were distinctly different from that of the wild-type (Fig. 4). The lpxF deletion strain CS506 contained lipid A species A’, B’, C’, D’ and E’ (Fig. 4), which migrated more slowly during TLC, consistent with the presumed retention of the 4′-phosphate group (Fig. 3A). The lipid A species from the lpxE mutant also migrated more slowly than those of the wild-type, but consisted of only two major components, designated F and G (Fig. 4). The retention of the phosphate group at the 1-position of these lipid A molecules prevents their oxidation by LpxQ (Fig. 2) [28, 29], thereby reducing their overall micro-heterogeneity (Fig 3B). The lipid A species of the lpxE/lpxF double mutant migrated even more slowly compared to the others and likewise showed less micro-heterogeneity (see below).
Figure 4. TLC analysis of lipid A species released by acetic acid hydrolysis from wild-type and phosphatase mutants.
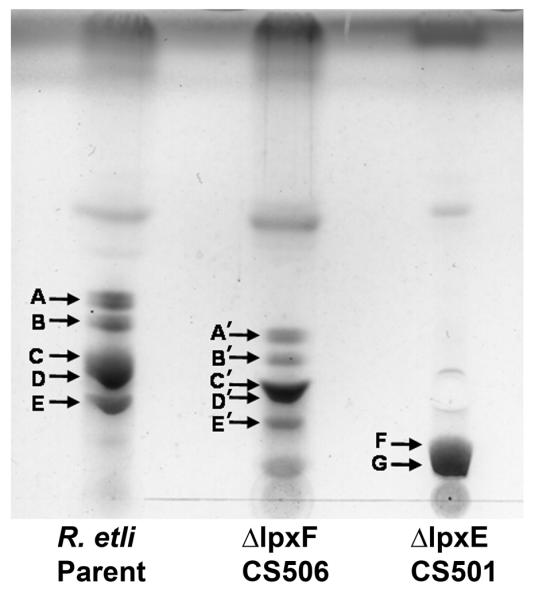
Approximately 5 μg of the lipid A molecules released from wild-type R. etli or the single phosphatase mutants by hydrolysis in 1% acetic acid were spotted onto a silica gel 60 TLC plate, which was developed in the solvent system CHCl3/MeOH/H2O/NH4OH (40:25:4:2 v/v). The lipids were detected after chromatography by spraying with 10% sulfuric acid in ethanol and charring on a hot plate. The tentative assignments of the lipid species are based on their relative migrations in this experiment, which were previously validated for the wild-type [14,15].
ESI-MS analysis of the lipid A species
The negative ion ESI mass spectra of the total lipid A species extracted from the wild-type and the single phosphatase mutants are shown in Fig. 5. The predicted masses of the major molecular species are listed in Table II. In Fig. 5A, which shows the spectrum of wild-type R. etli lipid A, there are several clusters of ions that are consistent with previous assignments [14, 15]. In the region between m/z 1850 and 2050, the most intense ion is observed at m/z 2000.49, which is interpreted as the [M-H]− ion of component D (Fig. 1C). The [M-H]− ion of component B, which is the biosynthetic precursor of D (Fig. 1B), is seen at m/z 1984.48. The [M-H]− ions of the deacylated versions of components B and D (designated C and E in Fig. 1) are seen at m/z 1758.28 and 1774.29, respectively (Fig. 5A). The intense peak at m/z 1738.26 corresponds to component A (Supporting Fig. 3), which arises from D during acid hydrolysis [14, 15]. There is significant additional micro-heterogeneity in all these lipid A species because of acyl chain length differences and/or the presence of a β-hydroxybutyryl moiety at the 27:OH position of the 2′-acyloxyacyl chain (difference of 86 amu) [13-15]. These additional differences are annotated in the legend to Fig. 5A.
Figure 5. ESI/MS analysis of lipid A species released by acetic acid hydrolysis from wild-type and single phosphatase mutants.
The lipid A released from the wild-type or the phosphatase deletion mutants by 1% acetic acid hydrolysis was analyzed by negative ion ESI/MS. Panel A. The peaks in the singly charged region corresponding to the four major components of wild-type R. etli lipid A (B, C, D, and E), shown in Figs. 1B and 1C, are labeled and color coded. Component A (Supporting Fig. 3) is an elimination artifact derived from D during hydrolysis [14, 15]. There is additional micro-heterogeneity (see below) with regard to acyl chain length or the presence of the β-hydroxybutyrate substituent for each component, in agreement with previous studies [13]. Panel B. The spectrum in the singly charged region of the lipid A from the LpxF 4′-phosphatase mutant shows the same relative pattern of peaks as the wild-type, but each component (B’, C’, D’, and E’) is shifted by 96 amu because of the replacement of the 4′-galacturonic acid moiety with a phosphate group (Fig. 3A). Panel C. The spectrum in the singly charged region of the lipid A species from the LpxE 1-phosphatase mutant (Fig. 3B) show peaks at increased m/z values because of the retention of the phosphate group at the 1-position. There are also fewer lipid A components in this mutant because oxidation of the proximal glucosamine unit by LpxQ cannot occur. The hydrolysis artifacts (A and A’), seen in wild-type R. etli and mutant CS506 respectively, are therefore not present in this strain. Additional micro-heterogeneity in fatty acid chain length (14 or 28 atomic mass units) and partial substitution with β-hydroxybutyrate (86 amu) is observed in the lipid A species of all these strains. Each major lipid A species and some of its corresponding acyl chain variants are grouped by color. Symbols indicate the following amu shifts: for Panels A and B: + = (−28) * = (−86) • = (−86 and −28); for Panel C: + = (−28) * = (−86) • = (−86 and −28)  = + 28 28)
= + 28 28) 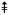 = +14 α = (−86 and + 28) ¢ = −14 π = (−86 and + 14) ∂ = (−86 and + 14 and + 28) λ = (−86 and −14).
= +14 α = (−86 and + 28) ¢ = −14 π = (−86 and + 14) ∂ = (−86 and + 14 and + 28) λ = (−86 and −14).
Table II.
Masses of lipid A components from R. etli wild-type and phosphatase knockout
| strains | ||||||
|---|---|---|---|---|---|---|
| Strain | ||||||
| R etli | Component | B | C | D | E | A |
| Molecular formula | C110H204N2O27 | C96H178N2O25 | C110H204N2O28 | C96H178N2O26 | C96H174N2O24 | |
| Exact Mass | 1985.47 | 1759.27 | 2001.46 | 1775.27 | ||
| 1739.25 | ||||||
| Observed m/z [M-H]−1 | 1984.48 | 1758.28 | 2000.49 | 1774.29 | 1738.26 | |
| CS506 | Component | B’ | C’ | D’ | E’ | A’ |
| Molecular formula | C104H197N2O24P | C90H171N2O22P | C104H197N2O25P | C90H171N2O23P | C90H167N2O21P | |
| Exact Mass | 1889.4 | 1663.21 | 1905.39 | 1679.2 | 1643.18 | |
| Observed m/z [M-H]−1 | 1888.28 | 1662.20 | 1904.40 | 1678.21 | 1642.18 | |
| CS501 | Component | F | G | |||
| Molecular formula | C110H205N2O30P | C96H179N2O28P | ||||
| Exact Mass | 2065.43 | 1839.24 | ||||
| Observed m/z [M-H]−1 | 2064.43 | 1838.24 | ||||
| CS513 | Component | H | I | |||
| Molecular formula | C104H198N2O27P2 | C90H172N2O25P2 | ||||
| Exact Mass | 1969.37 | 1743.17 | ||||
| Observed m/z [M-H]−2 | 983.64 | 870.56 | ||||
The singly-charged lipid A species from the lpxF 4′-phosphatase mutant CS506 are shown in Fig. 5B. The overall pattern of peaks in this mutant resembles that of the wild-type (Fig. 4 and Fig. 5B); however, all of the ions are shifted to m/z values that are 96 amu smaller than wild-type (Fig. 5B versus Fig. 5A). This change in mass results from the replacement of the galacturonic acid residue at the 4′-position with a phosphate moiety. No residual wild-type lipid A species are detected. The major peak at m/z 1904.40 is interpreted as the [M-H]– ion of component D’ (Fig. 3A and Fig. 4). The [M-H]– ion of component B’, the biosynthetic precursor of the D’, is observed at m/z 1888.28. The 3-O-deacylated versions of these species and the elimination product, A’, appear in the clusters of ions below m/z 1700. Additional lipid A micro-heterogeneity due to acyl chain length differences and partial substitution with the β-hydroxybutyryl moiety is annotated in the legend to Fig. 5B.
The singly-charged lipid A species obtained from the lpxE 1-phosphatase mutant CS501 are shown in Fig. 5C. As in the lpxF mutant, no peaks corresponding to residual wild-type lipid A molecules are detected. Two new major lipid A species are present in this strain (Fig. 4). The peaks at m/z 2064.43 and m/z 1838.24 are interpreted as the [M-H]– ions of components F and G respectively (Fig. 3B and Fig. 4). There are no aminogluconate-containing lipid A derivatives in this strain because the presence of the 1-phosphate group prevents oxidation by LpxQ (Fig. 3B). The peaks are therefore all shifted to correspondingly higher m/z values. Additional lipid A micro-heterogeneity arising from acyl chain differences and the β-hydroxybutyryl moiety is annotated in the legend to Fig. 5C.
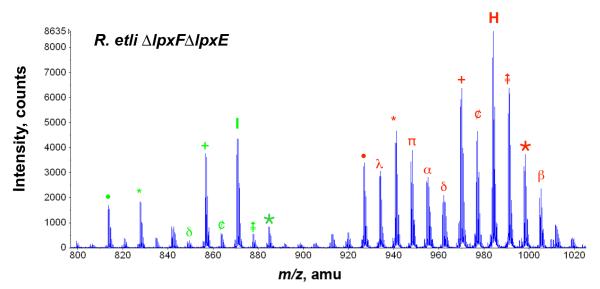
ESI-MS of the lipid A species from the double lpxE/lpxF mutant
The lipid A molecules of the lpxE/lpxF double mutant contain two phosphate moieties. Therefore, the predominant lipid A ions seen in the negative mode are doubly-charged, as shown in Fig. 6. The [M-2H]2– ions of components H and I (Fig. 3C and Table 2) are observed at m/z 983.64 and 870.56 respectively (Fig. 6). These species resemble those of the lpxE single mutant in that they lack the components harboring the aminogluconate modification in the proximal unit. Their masses are shifted to lower values compared to wild-type because of the replacement of the 4′-galacturonic acid residue with a phosphate moiety. The additional micro-heterogeneity arising from acyl chain differences and partial substitution with the β-hydroxybutyryl moiety is indicated in the legend to Fig 6.
Figure 6. ESI/MS analysis of lipid A species released by acetic acid hydrolysis from the double phosphatase mutant.
The spectrum in the negative ion mode was the accumulation of 60 scans from 200 to 2400 atomic mass units. Only the doubly-charged ion of the spectrum are shown, because the peaks of the singly charged are very small. The two major lipid A components of this mutant (H and I) lack the galacturonic modification at the 4′-position, which is replaced by a phosphate group, and do not contain the aminogluconate unit (Fig. 3C). Heterogeneity in fatty acid chain length (14 or 28 amu) and partial substitution with β-hydroxybutyrate (86 amu) is observed. Symbols indicate the following amu shifts: + = (−28) * = (−86) • = (−86 and −28) 28)  = + 28 28)
= + 28 28) 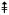 = +14 β = (+28 and +14) ¢ = −14 π = (−86 and + 14) α = (−86 and + 28) ∂ = (−86 and + 14 and + 28) λ = (−86 and −14).
= +14 β = (+28 and +14) ¢ = −14 π = (−86 and + 14) α = (−86 and + 28) ∂ = (−86 and + 14 and + 28) λ = (−86 and −14).
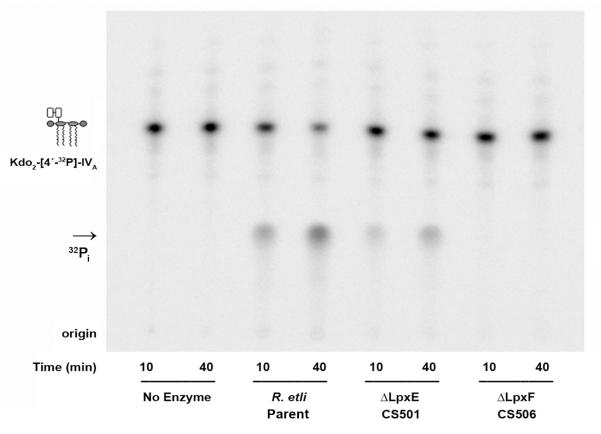
Determination of lipid A phosphatase activity in wild-type R. etli and the lpxF mutant
The in vitro LpxF activity of the wild-type and the two single mutants was measured using washed membranes as the enzyme source. In this assay system, the substrate Kdo2-[4′-32P]-lipid IVA is dephosphorylated and free inorganic phosphate is released as 32Pi. As shown in Fig. 7, the robust 4′-phosphatase activity that is characteristic of wild-type R. etli membranes [24] is absent in the lpxF mutant CS506 but is present in the lpxE mutant CS501.
Figure 7. Absence of 4′-phosphatase activity in membranes R. etli lpxF deletion mutant CS506.
Membranes of the indicated strains (0.25 mg/mL) were assayed for 4′-phosphatase activity at 30 °C with 5 μM [4′-32P]Kdo2-lipid IVA. The products were separated by TLC using the solvent chloroform:pyridine:88% formic acid:water, (30:70:16:10, v/v), followed by detection with a PhosphorImager. No enzyme; parent, R. etli CE3 wild-type; CS501, R. etli ΔlpxE; CS506, R. etli ΔlpxF.
The 1-phosphatase activity encoded by R. etlilpxE cannot be assayed in crude membranes preparations, and therefore its absence in lpxE mutant membranes could not be demonstrated (not shown). However, in R. leguminosarum and F. novicida membranes, LpxE activity is easily detected with Kdo2-[4′-32P]-lipid IVA as the substrate under otherwise similar conditions [22, 23]. It may be that R. etli extracts contain an inhibitor of LpxE activity that interferes with the assay.
Polymyxin sensitivity of R. etli strains
Polymyxin-resistant strains of E. coli and S. typhimurium survive by modifying their lipid A phosphate groups with 4-amino-4-deoxy-L-arabinose and phosphoethanolamine [8]. The reduction of the net negative charge on lipid A helps reduce the binding of polymyxin and other cationic antimicrobial peptides. R. etli removes the phosphate groups from its lipid A during biosynthesis (Fig. 2), and replaces them with galacturonic acid and aminogluconate residues (Figs. 1 and 2). The substitution of phosphate groups with these moieties would also reduce the net negative charge of lipid A. To determine if retention of one or both phosphate groups leads to polymyxin hyper-sensitivity, disk diffusion assays with polymyxin B were performed. Both the lpxE and lpxF single mutants showed increased sensitivity to polymyxin B compared to wild-type (Table III), and the double mutant showed the greatest sensitivity. Several other antibiotics (chlroamphenicol, tetracycline, tobramycin, neomycin, rifampicin, novobiocin, erythromycin, and ciprofloxacin) were also tested, but no major differences were observed.
Table III.
Increased polymyxin sensitivity of R. etli CE3 mutants lacking the lipid A phosphatases
| Zone of clearing diameter (mm) |
|||
|---|---|---|---|
| Amount of polymyxin added per disk | |||
| Strain | (2 μg) | (10 μg) | (20 μg) |
| R. etli CE3 | 8 | 10 | 12 |
| CS501 (ΔlpxE) | 9 | 12 | 14 |
| CS506 (ΔlpxF) | 10 | 14 | 15 |
| CS513 (ΔlpxE/ΔlpxF) | 15 | 17 | 18 |
The diameter of the disk is 6 mm, and the diameter of the zone of clearing shown above includes the disk diameter. The precision of the measurement of the diameter is ± 0.5 mm.
Membrane lipid composition of R. etli strains
The R. etli lipid A phosphatases LpxE and LpxF contain sequence motifs that are present in many other lipid phosphate phosphatases [48], suggesting a conserved structure and mechanism of action. However, some LpxE orthologues, such as that found in A. tumefaciens, do not show lipid A 1-phosphatase activity. Instead, the putative A. tumefaciens LpxE orthologue dephosphorylates phosphatidylglycerol phosphate [32]. The overall membrane lipid composition of the R. etli wild-type and the phosphatase knockouts was therefore examined to exclude the possibility of other alterations in lipid composition. The strains were grown in TY medium, and their membrane lipids were labeled with [14C]-acetate. The lipids were extracted, separated by TLC and quantified by PhosphorImager analysis (Table IV). No significant differences were observed between the wild type and the mutants. The major membrane lipids in each of these strains were phosphatidylcholine, phosphatidylethanolamine, phosphatidyldimethylethanolamine, phosphatidylglycerol, cardiolipin, sulfolipid and ornithine lipid, consistent with published data [50].
Table IV.
Membrane lipid composition of Rhizobium etli CE3 strains
| Component | R. etli CE3 | CS501 ΔlpxE |
CS503 ΔlpxF |
CS513 ΔlpxE/ΔlpxF |
|---|---|---|---|---|
| PE | 36.8 ± 1.4 | 40.9 ± 2.7 | 38.4 ± 0.3 | 41.9 ± 1.7 |
| PC | 34.5 ± 3.9 | 33.9 ± 5.5 | 35.8 ± 1.1 | 30.2 ± 1.2 |
| OL | 5.5 ± 1.2 | 5.1 ± 0.8 | 4.5 ± 0.4 | 4.7 ± 0.8 |
| PG | 14.5 ± 1.3 | 11.2 ± 0.9 | 13.8 ± 1.7 | 14.3 ± 2.1 |
| CL | 5.3 ± 2.4 | 5.9 ± 2.0 | 4.2 ± 0.4 | 5.9 ± 1.7 |
| SL | 1.8 ± 0.8 | 1.0 ± 0.5 | 0.8 ± 0.3 | 1.0 ± 0.6 |
| DMPE | 1.9 ± 0.6 | 2.1 ± 0.7 | 1.7 ± 0.1 | 2.1 ± 0.9 |
Symbiotic phenotypes of the R. etli phosphatase mutants
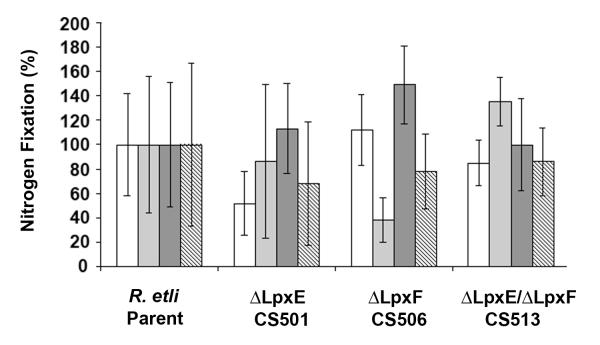
Phaseolus vulgaris roots were inoculated with R. etli, the lpxE mutant CS501, the lpxF mutant CS506, and the double mutant CS513. Eight plants were inoculated in each of four independent experiments, which are highlighted by the differential shading of the columns. One independent experiment therefore consists of 40 plants (8 control plants that were not inoculated, 8 plants inoculated with wild type CE3, 8 plants inoculated with CS501, 8 plants inoculated with CS506, and 8 plants inoculated with CS513). Three weeks after the inoculation, all four strains had induced the formation of nodules that were similar in appearance. Acetylene reduction assays were performed to determine if any of the strains were defective in nitrogen fixation. No significant differences were observed when nitrogen fixing activities per gram of nodule fresh weight were compared (Fig. 8). To exclude the possibility that the lack of a symbiotic phenotype was caused by contaminating wild-type cells, bacteria were re-isolated from the nodules and their genomic DNA probed by Southern Blotting to confirm their identity (data not shown).
Figure 8. Nitrogen fixation in nodules of wild-type and mutant R. etli.
Nitrogen fixation activity was determined using an acetylene reduction assay. Every strain was analyzed in four independent experiments, each comprised of 8 plants, as highlighted by the differential shading of the four columns. Columns show the relative nitrogen fixation activity and error bars indicate the standard deviations within each of the four independent experiments. Nitrogen fixation activity is expressed as the amount of ethylene formed per weight of fresh nodule, normalized to the nitrogen fixation activity of the wild-type CE3. Parent, R. etli CE3 wild-type; CS501, R. etli lipid A 1-phosphatase ΔlpxE mutant; CS506, R. etli lipid A 4′-phosphatase ΔlpxF mutant; CS513, R. etli mutant deficient in ΔlpxF and ΔlpxE.
DISCUSSION
Lipid A, the hydrophobic moiety of LPS, is recognized by Toll-like receptor 4 (TLR-4) in animals, triggering the innate immune response [4, 51, 52]. The constitutive lipid A biosynthetic pathway is largely conserved in Gram-negative bacteria [8], but species-specific modifications of lipid A may play a role in helping pathogens evade the innate immune response [8]. For example, Escherichia coli and S. enterica sv. typhimurium modify their lipid A with 4-aminoarabinose and phosphoethanolamine moieties in response to changes in environmental conditions found within their respective hosts, conferring resistance to cationic antimicrobial peptides [8]. These covalent modifications are controlled by the two component regulatory systems PhoP/Q and PmrA/B, which are induced by low Mg2+ and low pH, respectively [53].
Rhizobia form chronic infections on plant root hair cells during symbiosis, and like animal pathogens must survive within host-membrane-derived intracellular compartments during this process. The bacteria adapt to dramatic changes in acidity, osmolarity, and oxygen tension in these compartments. Most of the unusual covalent modifications characteristic of Rhizobium etli lipid A are observed in free living bacteria [14, 15], but it was nevertheless proposed that some of these lipid A modifications might play a role in the bacteria-plant symbiosis. Dephosphorylation of R. etli lipid A was of special interest because pharmacological studies have indicated that the presence of both phosphate groups on lipid A are important for its cytokine-inducing activities in animal systems [5]. Furthermore, a recently isolated 4′-phosphatase mutant of the animal pathogen F. novicida, was shown to display dramatically attenuated virulence in mice [11]. The Francisella novicida 4′-phosphatase mutant was viable but severely limited in its ability to kill mice compared to wild-type strains because of its hypersensitivity to cationic antimicrobial peptides [11].
We have now constructed and characterized a 1-phosphatase (lpxE) mutant, a 4′-phosphatase (lpxF) mutant, and a 1-/4′-phosphatase (lpxE/lpxF) double mutant of R. etli. The candidate phosphatase genes were found by searching the recently completed R. etli genome [33] with the LpxE sequence from R. leguminosarum [22] and the LpxF sequence from the F. novicida [25]. LpxE and LpxF both reside in the inner membrane with active sites that face the periplasm [23, 25], and both have absolute specificity in selecting the site of dephosphorylation on lipid A [23, 25, 32, 44]. LpxE and LpxF do not share significant similarity in their primary amino acid sequences [23, 25], but both contain variations of the phosphatase sequence motif described by Stukey and Carman [48].
We have demonstrated that the LpxE and LpxF mutant strains accumulate lipid A species retaining one or both phosphate moieties, as predicted by the pathway shown in Fig. 2 and Fig. 3. The retention of the phosphate groups at the 4′-position blocks the incorporation of a galacturonic acid residue (Figs. 3A and Fig. 5B), and the presence of a phosphate group at the 1-position prevents the oxidation of the proximal glucosamine residue to 2-aminogluconate (Fig. 3B and Fig. 5C). The results indicate that the two lipid A phosphatases work independently of each other, have no established order for operation, and are solely responsible for the lipid A dephosphorylaion seen in R. etli [14, 15].
The free living R. etli phosphatase mutants had no changes in growth rate, and the membrane lipid composition remained unchanged (Table 4), indicating that these phosphatases do not have any significant activity directed towards other lipid substrates. Both phosphatase mutants, however, showed an increase in sensitivity to the cationic antimicrobial drug, polymyxin, consistent with the increase in net negative charge provided by the retention of the phosphate groups. The lpxE/lpxF double mutant, which accumulates bis-phosphorylated lipid A species similar to that found in S. meliloti [49] (Fig. 3C and Fig. 6), displays the greatest sensitivity to polymyxin (Table 4).
Despite having dramatically altered lipid A structures, all the mutants were found to form normal nodules and were equally competent in nitrogen fixation when compared to the wild-type strain (Fig. 8). This implies that dephosphorylation of lipid A, as well as modification with galacturonic acid and aminogluconate residues, are not essential for establishing an effective symbiosis. However, subtle alterations in the time course of nodule formation cannot yet be excluded. These modifications may also play other roles for the bacteria in their native environment. The removal of the phosphate groups from the lipid A molecules may be important in the rhizosphere, which is much more diverse than the controlled sterile environment used in our study (Fig. 8). One likely advantage of R. etli dephosphoryating its lipid A may be resistance to cationic antimicrobial peptides, like polymyxin (Table 4). Dephosphorylation of lipid A could protect R. etli in the rhizosphere from diverse cationic antimicrobial compounds. Plants have been shown to produce defense compounds of this kind [54, 55]. The further modifications to the lipid A disaccharide backbone of lipid A molecules in R. etli following dephosphorylation could play a role in maintaining membrane stability in the absence of phosphate groups. These modifications might also function to conserve phosphorus in low-phosphate environments.
To date, the only characterized lipid A mutants of a Rhizobium species are ones that affect lipid A acylation [56, 57]. Mutants that harbor a disruption in the gene encoding the acyl carrier protein that carries the (27OHC28:0) moiety (AcpXL) have been constructed in both R. leguminosarum and S. melliloti [56, 57]. These mutants are of interest because the (27OHC28:0) moiety is found in nearly all members of the Rhizobiaceae family [16]. The acpXL mutants lacked the modification with the (27OHC28:0) moiety but were shown to contain a small amount of C18:0 at the 2′-secondary position [57]. Both mutants displayed sensitivity to detergents, acidic conditions, and changes in osmolarity, and both also displayed slow nodulation kinetics compared to wild-type cells. However, the nodules displayed a normal morphology and fixed nitrogen, but differed slightly in bacteroid development [57]. In S. meliloti, lpxXL and acpXL/lpxXL mutants were also described [58]. These mutants lacked secondary acyl chains, but were found to form nitrogen fixing nodules in a delayed manner. Hence, it was originally reported that the 28 carbon fatty acyl chain is beneficial, but not critical for symbiosis to occur. A recent report claims that a R. leguminosarum acpXL mutant supposedly regains the ability to acylate the lipid A of bacteroids with the 27-hydroxyoctacosanoic acid moiety in the nodule environment by an uncharacterized mechanism [59], the details of which require further investigation. Although improbable, it likewise remains possible that the host plant has mechanisms for removing the lipid A phosphate groups that are present in the phosphatase mutant strains in the in planta environment. An analysis of the lipid A from the mutant bacteroids will be necessary to clarify this issue.
Little is known about the response of plants to lipid A-like molecules [60]; however, plants do have systems of immunity [61]. Recognition of conserved microbial molecules like lipid A, could result in the activation of various antimicrobial effectors that ward off attacks by plant pathogens. It is not known which molecular features of lipid A are ultimately responsible for these effects. Defining how the plant perceives lipid A and other microbial molecules might aid in our understanding of how symbiotic bacteria evade these plant-associated immune responses.
In summary, the galacturonate and aminogluconate modifications of lipid A seen in R. etli appear not to be required for nodulation. We conclude that these modifications most likely play a role in stabilizing the outer membrane of the bacteria and might also be involved in protecting the symbiont from the immune response of the host plant during pathogenic infections. We speculate that the dephosphorylation of lipid A molecules in R. etli might allow the plant to discriminate between plant pathogens and symbiotic bacteria and assist the bacteroids in evading the immune response of plants during pathogenic infections. Future work will address the roles that the lipid A components from symbiotic bacteria and plant pathogens play in eliciting or suppressing plant defenses.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Dr. Ziqiang Guan for assistance with mass spectrometry, and Dr. David Six for isolating the Kdo2-lipid IVA and reading the manuscript. This research was supported by NIH grant GM-51796 to C.R.H.R. and DGAPAUNAM (IN217907) to C.S. Dr. Z. Guan and the mass spectrometry facility at the Duke University Medical Center are supported by the LIPID MAPS Large Scale Collaborative Grant GM-069338.
ABBREVIATIONS
The abbreviations used are:
- ACP
acyl carrier protein
- BCA
bicinchoninic acid
- CL
cardiolipin
- DEAE-cellulose
diethylaminoethyl cellulose
- DMPE
dimethyl PE
- EDTA
ethylenediaminetetraacetic acid
- ESI
electrospray ionization
- Kdo
3-deoxy-D-manno-oct-2-ulosonic acid
- LPS
lipopolysaccharide
- MES
2-(N-morpholino)-ethanesulfonic acid
- MS
mass spectrometry
- OL
ornithine-containing lipid
- PBS
phosphate-buffered saline
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PG
phosphatidylglycerol
- SL
sulfolipid
- TLC
thin layer chromatography
- TLR-4
Toll-like receptor-4
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Raetz CRH, Whitfield C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Russell JA. Management of sepsis. N. Engl. J. Med. 2006;355:1699–1713. doi: 10.1056/NEJMra043632. [DOI] [PubMed] [Google Scholar]
- [4].Park BS, Song DH, Kim HM, Choi BS, Lee H, Lee JO. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 2009;458:1191–1195. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- [5].Rietschel ET, Kirikae T, Schade FU, Mamat U, Schmidt G, Loppnow H, Ulmer AJ, Zähringer U, Seydel U, Di Padova F, Schreier M, Brade H. Bacterial endotoxin: molecular relationships of structure to activity and function. FASEB Journal. 1994;8:217–225. doi: 10.1096/fasebj.8.2.8119492. [DOI] [PubMed] [Google Scholar]
- [6].Qureshi N, Takayama K, Ribi E. Purification and structural determination of non-toxic lipid A obtained from the lipopolysaccharide of Salmonella typhimurium. J. Biol. Chem. 1982;257:11808–11815. [PubMed] [Google Scholar]
- [7].Baldridge JR, McGowan P, Evans JT, Cluff C, Mossman S, Johnson D, Persing D. Taking a Toll on human disease: Toll-like receptor 4 agonists as vaccine adjuvants and monotherapeutic agents. Expert. Opin. Biol. Ther. 2004;4:1129–1138. doi: 10.1517/14712598.4.7.1129. [DOI] [PubMed] [Google Scholar]
- [8].Raetz CRH, Reynolds CM, Trent MS, Bishop RE. Lipid A modification systems in gram-negative bacteria. Annu. Rev. Biochem. 2007;76:295–329. doi: 10.1146/annurev.biochem.76.010307.145803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Vinogradov E, Perry MB, Conlan JW. Structural analysis of Francisella tularensis lipopolysaccharide. Eur. J. Biochem. 2002;269:6112–6118. doi: 10.1046/j.1432-1033.2002.03321.x. [DOI] [PubMed] [Google Scholar]
- [10].Wang X, Ribeiro AA, Guan Z, McGrath S, Cotter R, Raetz CRH. Structure and biosynthesis of free lipid A molecules that replace lipopolysaccharide in Francisella tularensis subsp. novicida. Biochemistry. 2006;45:14427–14440. doi: 10.1021/bi061767s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wang X, Ribeiro AA, Guan Z, Abraham SN, Raetz CRH. Attenuated virulence of a Francisella mutant lacking the lipid A 4′-phosphatase. Proc. Natl. Acad. Sci. U S A. 2007;104:4136–4141. doi: 10.1073/pnas.0611606104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Raetz CRH, Guan ZQ, Ingram BO, Six DA, Song F, Wang XY, Zhao JS. Discovery of new biosynthetic pathways: the lipid A story. Journal of Lipid Research. 2009;50:S103–S108. doi: 10.1194/jlr.R800060-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bhat UR, Forsberg LS, Carlson RW. The structure of the lipid A component of Rhizobium leguminosarum bv. phaseoli lipopolysaccharide. A unique, non-phosphorylated lipid A containing 2-amino-2-deoxy-gluconate, galacturonate, and glucosamine. J. Biol. Chem. 1994;269:14402–14410. [PubMed] [Google Scholar]
- [14].Que NLS, Lin S, Cotter RJ, Raetz CRH. Purification and mass spectrometry of six lipid A species from the bacterial endosymbiont Rhizobium etli; demonstration of a conserved distal and a variable proximal portion. J. Biol. Chem. 2000;275:28006–28016. doi: 10.1074/jbc.M004008200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Que NLS, Ribeiro AA, Raetz CRH. Two-dimensional NMR spectroscopy and structures of six lipid A species from Rhizobium etli CE3; detection of an acyloxyacyl residue in each component and origin of the aminogluconate moiety. J. Biol. Chem. 2000;275:28017–28027. doi: 10.1074/jbc.M004009200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Bhat UR, Carlson RW, Busch M, Mayer H. Distribution and phylogenetic significance of 27-hydroxy-octacosanoic acid in lipopolysaccharides from bacteria belonging to the alpha-2 subgroup of of Proteobacteria. Int. J. Syst. Bacteriol. 1991;41:213–217. doi: 10.1099/00207713-41-2-213. [DOI] [PubMed] [Google Scholar]
- [17].Price NPJ, Kelly TM, Raetz CRH, Carlson RW. Biosynthesis of a structurally novel lipid A in Rhizobium leguminosarum: identification and characterization of six metabolic steps leading from UDP-GlcNAc to (Kdo)2-lipid IVA. J. Bacteriol. 1994;176:4646–4655. doi: 10.1128/jb.176.15.4646-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Brozek KA, Carlson RW, Raetz CRH. A special acyl carrier protein for transferring long hydroxylated fatty acids to lipid A in Rhizobium. J. Biol. Chem. 1996;271:32126–32136. doi: 10.1074/jbc.271.50.32126. [DOI] [PubMed] [Google Scholar]
- [19].Brozek KA, Kadrmas JL, Raetz CRH. Lipopolysaccharide biosynthesis in Rhizobium leguminosarum. Novel enzymes that process precursors containing 3-deoxy-D-manno-octulosonic acid. J. Biol. Chem. 1996;271:32112–32118. [PubMed] [Google Scholar]
- [20].Basu SS, Karbarz MJ, Raetz CRH. Expression cloning and characterization of the C28 acyltransferase of lipid A biosynthesis in Rhizobium leguminosarum. J. Biol. Chem. 2002;277:28959–28971. doi: 10.1074/jbc.M204525200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kadrmas JL, Allaway D, Studholme RE, Sullivan JT, Ronson CW, Poole PS, Raetz CRH. Cloning and overexpression of glycosyltransferases that generate the lipopolysaccharide core of Rhizobium leguminosarum. J. Biol. Chem. 1998;273:26432–26440. doi: 10.1074/jbc.273.41.26432. [DOI] [PubMed] [Google Scholar]
- [22].Karbarz MJ, Kalb SR, Cotter RJ, Raetz CRH. Expression cloning and biochemical characterization of a Rhizobium leguminosarum lipid A 1-phosphatase. J. Biol. Chem. 2003;278:39269–39279. doi: 10.1074/jbc.M305830200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wang X, Karbarz MJ, McGrath SC, Cotter RJ, Raetz CRH. MsbA transporter-dependent lipid A 1-dephosphorylation on the periplasmic surface of the inner membrane: topography of Francisella novicida LpxE expressed in Escherichia coli. J. Biol. Chem. 2004;279:49470–49478. doi: 10.1074/jbc.M409078200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Price NJP, Jeyaretnam B, Carlson RW, Kadrmas JL, Raetz CRH, Brozek KA. Lipid A biosynthesis in Rhizobium leguminosarum: role of a novel Kdo activated 4′-phosphatase. Proc. Natl. Acad. Sci. U. S. A. 1995;92:7352–7356. doi: 10.1073/pnas.92.16.7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wang X, McGrath SC, Cotter RJ, Raetz CRH. Expression cloning and periplasmic orientation of the Francisella novicida lipid A 4′-phosphatase LpxF. J. Biol. Chem. 2006;281:9321–9330. doi: 10.1074/jbc.M600435200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kanjilal-Kolar S, Basu SS, Kanipes MI, Guan Z, Garrett TA, Raetz CRH. Expression cloning of three Rhizobium leguminosarum lipopolysaccharide core galacturonosyltransferases. J. Biol. Chem. 2006;281:12865–12878. doi: 10.1074/jbc.M513864200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kanjilal-Kolar S, Raetz CRH. Dodecaprenyl phosphate-galacturonic acid as a donor substrate for lipopolysaccharide core glycosylation in Rhizobium leguminosarum. J. Biol. Chem. 2006;281:12879–12887. doi: 10.1074/jbc.M513865200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Que-Gewirth NLS, Karbarz MJ, Kalb SR, Cotter RJ, Raetz CRH. Origin of the 2-amino-2-deoxy-gluconate unit in Rhizobium leguminosarum lipid A. Expression cloning of the outer membrane oxidase LpxQ. J. Biol. Chem. 2003;278:12120–12129. doi: 10.1074/jbc.M300379200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Que-Gewirth NLS, Lin S, Cotter RJ, Raetz CRH. An outer membrane enzyme that generates the 2-aminogluconate moiety of Rhizobium leguminosarum lipid A. J. Biol. Chem. 2003;278:12109–12119. doi: 10.1074/jbc.M300378200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Basu SS, White KA, Que NL, Raetz CRH. A deacylase in Rhizobium leguminosarum membranes that cleaves the 3-O-linked beta-hydroxymyristoyl moiety of lipid A precursors. J. Biol. Chem. 1999;274:11150–11158. doi: 10.1074/jbc.274.16.11150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Rutten L, Geurtsen J, Lambert W, Smolenaers JJ, Bonvin AM, Haan A. de, van der Ley P, Egmond MR, Gros P, Tommassen J. Crystal structure and catalytic mechanism of the LPS 3-O-deacylase PagL from Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U S A. 2006;103:7071–7076. doi: 10.1073/pnas.0509392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Karbarz MJ, Six DA, Raetz CRH. Purification and characterization of the lipid A 1-phosphatase LpxE of Rhizobium leguminosarum. J. Biol. Chem. 2009;284:414–425. doi: 10.1074/jbc.M808390200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Gonzalez V, Santamaria RI, Bustos P, Hernandez-Gonzalez I, Medrano-Soto A, Moreno-Hagelsieb G, Janga SC, Ramirez MA, Jimenez-Jacinto V, Collado-Vides J, Davila G. The partitioned Rhizobium etli genome: genetic and metabolic redundancy in seven interacting replicons. Proc. Natl. Acad. Sci. U S A. 2006;103:3834–3839. doi: 10.1073/pnas.0508502103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Johnston AW, Beringer JE. Identification of the rhizobium strains in pea root nodules using genetic markers. J. Gen. Microbiol. 1975;87:343–350. doi: 10.1099/00221287-87-2-343. [DOI] [PubMed] [Google Scholar]
- [35].Miller JR. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1972. [Google Scholar]
- [36].Ruvkun GB, Ausubel FM. A general method for site-directed mutagenesis in prokaryotes. Nature. 1981;289:85–88. doi: 10.1038/289085a0. [DOI] [PubMed] [Google Scholar]
- [37].Sambrook JG, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd ed Cold Spring Harbor; Cold Spring Harbor, NY: 2001. [Google Scholar]
- [38].Hanahan D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- [39].Dulbecco R, Vogt M. Plaque formation and isolation of pure lines with poliomyelitis viruses. J. Exp. Med. 1954;99:167–182. doi: 10.1084/jem.99.2.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Bligh EG, Dyer JJ. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- [41].Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- [42].Six DA, Carty SM, Guan Z, Raetz CRH. Purification and mutagenesis of LpxL, the Lauroyltransferase of Escherichia coli lipid A biosynthesis. Biochemistry. 2008;47:8623–8637. doi: 10.1021/bi800873n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Reynolds CM, Kalb SR, Cotter RJ, Raetz CRH. A phosphoethanolamine transferase specific for the outer 3-deoxy-D-manno-octulosonic acid residue of Escherichia coli lipopolysaccharide. Identification of the eptB gene and Ca2+ hypersensitivity of an eptB deletion mutant. J. Biol. Chem. 2005;280:21202–21211. doi: 10.1074/jbc.M500964200. [DOI] [PubMed] [Google Scholar]
- [44].Basu SS, York JD, Raetz CRH. A phosphotransferase that generates phosphatidylinositol 4-phosphate (PtdIns-4-P) from phosphatidylinositol and lipid A in Rhizobium leguminosarum. A membrane-bound enzyme linking lipid A and PtdIns-4-P biosynthesis. J. Biol. Chem. 1999;274:11139–11149. doi: 10.1074/jbc.274.16.11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Vinuesa P, Reuhs BL, Breton C, Werner D. Identification of a plasmid-borne locus in Rhizobium etli KIM5s involved in lipopolysaccharide O-chain biosynthesis and nodulation of Phaseolus vulgaris. J. Bacteriol. 1999;181:5606–5614. doi: 10.1128/jb.181.18.5606-5614.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Fahraeus G. The infection of clover root hairs by nodule bacteria studied by a simple glass slide technique. J. Gen. Microbiol. 1957;16:374–381. doi: 10.1099/00221287-16-2-374. [DOI] [PubMed] [Google Scholar]
- [47].Martinez E, Pardo MA, Palacios R, Cevallos MA. Reiteration of nitrogen fixation gene sequences and specificity of Rhizobium in nodulation and nitrogen fixation in Phaseolus vulgaris. J. Gen. Microbiol. 1985;131:1779–1786. [Google Scholar]
- [48].Stukey J, Carman GM. Identification of a novel phosphatase sequence motif. Protein Sci. 1997;6:469–472. doi: 10.1002/pro.5560060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Gudlavalleti SK, Forsberg LS. Structural characterization of the lipid A component of Sinorhizobium sp. NGR234 rough and smooth form lipopolysaccharide. Demonstration that the distal amide-linked acyloxyacyl residue containing the long chain fatty acid is conserved in rhizobium and Sinorhizobium sp. J. Biol. Chem. 2003;278:3957–3968. doi: 10.1074/jbc.M210491200. [DOI] [PubMed] [Google Scholar]
- [50].Geiger O, Rohrs V, Weissenmayer B, Finan TM, Thomas-Oates JE. The regulator gene phoB mediates phosphate stress-controlled synthesis of the membrane lipid diacylglyceryl-N,N,N-trimethylhomoserine in Rhizobium (Sinorhizobium) meliloti. Mol. Microbiol. 1999;32:63–73. doi: 10.1046/j.1365-2958.1999.01325.x. [DOI] [PubMed] [Google Scholar]
- [51].Beutler B, Jiang Z, Georgel P, Crozat K, Croker B, Rutschmann S, Du X, Hoebe K. Genetic analysis of host resistance: Toll-like receptor signaling and immunity at large. Annu. Rev. Immunol. 2006;24:353–389. doi: 10.1146/annurev.immunol.24.021605.090552. [DOI] [PubMed] [Google Scholar]
- [52].Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- [53].Gibbons HS, Kalb SR, Cotter RJ, Raetz CRH. Role of Mg++ and pH in the modification of Salmonella lipid A following endocytosis by macrophage tumor cells. Mol. Microbiol. 2005;55:425–440. doi: 10.1111/j.1365-2958.2004.04409.x. [DOI] [PubMed] [Google Scholar]
- [54].Campbell GR, Reuhs BL, Walker GC. Chronic intracellular infection of alfalfa nodules by Sinorhizobium meliloti requires correct lipopolysaccharide core. Proc. Natl. Acad. Sci. U S A. 2002;99:3938–3943. doi: 10.1073/pnas.062425699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Epand RM, Vogel HJ. Diversity of antimicrobial peptides and their mechanisms of action. Biochim. Biophys. Acta. 1999;1462:11–28. doi: 10.1016/s0005-2736(99)00198-4. [DOI] [PubMed] [Google Scholar]
- [56].Sharypova LA, Niehaus K, Scheidle H, Holst O, Becker A. Sinorhizobium meliloti acpXL mutant lacks the C28 hydroxylated fatty acid moiety of lipid A and does not express a slow migrating form of lipopolysaccharide. J. Biol. Chem. 2003;278:12946–12954. doi: 10.1074/jbc.M209389200. [DOI] [PubMed] [Google Scholar]
- [57].Vedam V, Kannenberg EL, Haynes JG, Sherrier DJ, Datta A, Carlson RW. A Rhizobium leguminosarum AcpXL mutant produces lipopolysaccharide lacking 27-hydroxyoctacosanoic acid. J. Bacteriol. 2003;185:1841–1850. doi: 10.1128/JB.185.6.1841-1850.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Ferguson GP, Datta A, Carlson RW, Walker GC. Importance of unusually modified lipid A in Sinorhizobium stress resistance and legume symbiosis. Mol. Microbiol. 2005;56:68–80. doi: 10.1111/j.1365-2958.2005.04536.x. [DOI] [PubMed] [Google Scholar]
- [59].Vedam V, Kannenberg E, Datta A, Brown D, Haynes-Gann JG, Sherrier DJ, Carlson RW. The pea nodule environment restores the ability of a Rhizobium leguminosarum lipopolysaccharide acpXL mutant to add 27-hydroxyoctacosanoic acid to its lipid A. J. Bacteriol. 2006;188:2126–2133. doi: 10.1128/JB.188.6.2126-2133.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Zeidler D, Zahringer U, Gerber I, Dubery I, Hartung T, Bors W, Hutzler P, Durner J. Innate immunity in Arabidopsis thaliana: lipopolysaccharides activate nitric oxide synthase (NOS) and induce defense genes. Proc. Natl. Acad. Sc. U S A. 2004;101:15811–15816. doi: 10.1073/pnas.0404536101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- [62].Noel KD, Sanchez A, Fernandez L, Leemans J, Cevallos MA. Rhizobium phaseoli symbiotic mutants with transposon Tn5 insertions. J. Bacteriol. 1984;158:148–155. doi: 10.1128/jb.158.1.148-155.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Figurski DH, Helinski DR. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U S A. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Schafer A, Tauch A, Jager W, Kalinowski J, Thierbach G, Puhler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- [65].Yanish-Perron C, Viera L, Messing J. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- [66].Osteras M, Boncompagni E, Vincent N, Poggi MC, Le Rudulier D. Presence of a gene encoding choline sulfatase in Sinorhizobium meliloti bet operon: choline-O-sulfate is metabolized into glycine betaine. Proc. Natl. Acad. Sci. U S A. 1998;95:11394–11399. doi: 10.1073/pnas.95.19.11394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Schweizer HD. Small broad-host-range gentamycin resistance gene cassettes for site-specific insertion and deletion mutagenesis. Biotechniques. 1993;15:831–834. [PubMed] [Google Scholar]
- [68].Ruiz N, Kahne D, Silhavy TJ. Transport of lipopolysaccharide across the cell envelope: the long road of discovery. Nat. Rev. Microbiol. 2009;7:677–683. doi: 10.1038/nrmicro2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.