Abstract
The haspins are divergent members of the eukaryotic protein kinase family that are conserved in many eukaryotic lineages including animals, fungi, and plants. Recently-solved crystal structures confirm that the kinase domain of human haspin has unusual structural features that stabilize a catalytically active conformation and create a distinctive substrate binding site. Haspin localizes predominantly to chromosomes and phosphorylates histone H3 at threonine-3 during mitosis, particularly at inner centromeres. This suggests that haspin directly regulates chromosome behavior by modifying histones, although it is likely that additional substrates will be identified in the future. Depletion of haspin by RNA interference in human cell lines causes premature loss of centromeric cohesin from chromosomes in mitosis and failure of metaphase chromosome alignment, leading to activation of the spindle assembly checkpoint and mitotic arrest. Haspin overexpression stabilizes chromosome arm cohesion. Haspin, therefore, appears to be required for protection of cohesion at mitotic centromeres. Saccharomyces cerevisiae homologues of haspin, Alk1 and Alk2, are also implicated in regulation of mitosis. In mammals, haspin is expressed at high levels in the testis, particularly in round spermatids, so it seems likely that haspin has an additional role in post-meiotic spermatogenesis. Haspin is currently the subject of a number of drug discovery efforts, and the future use of haspin inhibitors should provide new insight into the cellular functions of these kinases and help determine the utility of, for example, targeting haspin for cancer therapy.
Introduction
Ensuring that daughter cells each receive the correct complement of chromosomes in mitosis is simple in principle, but complicated in practice. Chromatin must be condensed and DNA decatanated to allow compaction and individualization of chromosomes, while sister chromatids are held together to allow their coordinated attachment to opposing spindle poles. A surveillance and checkpoint mechanism (the spindle assembly checkpoint or “SAC”) prevents chromatid separation until all chromosomes are correctly bioriented, and mechanical systems make sure chromosomes are segregated to opposite poles and divide the mother cell in two. A key set of mitotic kinases is critical to organize these processes, and the literature describing the activities of Cdk, Aurora, Polo, Nek, Bub, and Mps1 kinases in mitosis is extensive. Haspin is a relatively newly discovered kinase that phosphorylates histone H3 during mitosis and appears to play a role in regulating chromosome behavior during cell division. Here, I review what has been learned so far concerning this distinctive and evolutionarily conserved eukaryotic protein kinase.
Discovery and localization
Haspin mRNA was first discovered in male germ cells of mice. The gene and protein were given the names germ cell-specific gene 2 (Gsg2) and haploid cell-specific protein kinase (haspin), respectively (Tanaka et al. 1994; Tanaka et al. 1999). Northern analysis in human and mouse tissues showed that haspin is abundantly expressed in testis, with lower levels in multiple somatic tissues that have high numbers of dividing cells, including the thymus, bone marrow and spleen, and in all proliferating cell lines tested (Higgins 2001b). Endogenous haspin protein and kinase activity are also found in human tumor lines including HeLa and U2OS cells (Dai et al. 2005; Dai et al. 2009; Markaki et al. 2009). Therefore, haspin is most strongly expressed in testis, but also appears ubiquitously present in proliferating somatic cells.
Antibodies that recognize endogenous haspin in somatic cells by immunofluorescence or immunohistochemistry are not yet available, so current knowledge of haspin localization relies on the use of transfected constructs and studies of endogenous protein in spermatids. Haspin is a nuclear protein in interphase nuclei and in round spermatids (Tanaka et al. 1999; Dai et al. 2005), and is predominantly associated with chromosomes in mitosis (Dai et al. 2005). Enhanced green fluorescent protein (EGFP)-haspin can also be seen at centrosomes in mitotic cells (Dai et al. 2005). These latter observations were among the first to suggest mitotic functions for haspin.
Haspin homologues and structure
Haspin homologues have been identified in a number of eukaryotic lineages, including vertebrates, arthropods, nematodes, fungi (including microsporidia), amoebozoa, and plants. These haspin proteins all contain a divergent eukaryotic protein kinase (ePK) domain at the C-terminus and form a unique group of kinases that is not allied with other ePK families (Tanaka et al. 1999; Higgins 2001a; Higgins 2003; Kannan et al. 2007). Despite the absence of some of the highly conserved motifs found in canonical ePKs, mammalian haspin proteins clearly have serine/threonine kinase activity (Tanaka et al. 1999; Tanaka et al. 2001; Dai et al. 2005; Eswaran et al. 2009). The even more divergent budding yeast homologs Alk1 (Ygl021wp) and Alk2 (Ybl009wp) also appear to be active kinases (Nespoli et al. 2006). The N-terminal region of haspin proteins is poorly conserved, but some general features, such as a preponderance of serine and arginine/lysine residues, are shared between different species (Higgins 2003).
Two recent studies describing crystal structures of the kinase domain of human haspin confirm that it forms a bilobed structure similar to that of ePKs but with significant structural changes and a number of haspin-specific inserts (Fig. 1a, b; Eswaran et al. 2009; Villa et al. 2009). The kinase domain of haspin alone is active in vitro (Dai et al. 2005; Eswaran et al. 2009) and the structural studies confirm that it adopts an active conformation in which key residues are positioned appropriately for catalysis. Interestingly, haspin-specific structural elements including a β-hairpin insert and a number of additional α-helices surround the small lobe and appear to stabilize the active conformation. In many ePKs, an “activation segment” in the large lobe is, in the absence of phosphorylation, either disordered or locked into an inactive conformation. Phosphorylation within the segment stabilizes it in an active conformation suitable for substrate binding. The activation segment of haspin differs markedly from those in other ePKs. An additional β-strand in the segment helps stabilize the active conformation of the small lobe, and a large helical insert (αAS; Fig. 1b) replaces the “APE” motif and αEF helix found in many other kinases. No phosphorylation is observed in the activation segment in the haspin crystals, and there are no obvious candidate sites for such activating phosphorylation. The HRD motif arginine that interacts with the phosphorylated activation loop in many other kinases is instead hydrogen bonded to the main chain of the activation segment in haspin (Eswaran et al. 2009; Villa et al. 2009). This unique activation segment, together with the absence of a conserved αG helix found in other kinases (Fig. 1b), also contributes to formation of distinctive highly electronegative cleft that almost certainly serves as the binding site for the basic histone tail substrate (Fig. 1c). Other substrates of haspin identified in the future are likely to possess similar exposed basic regions.
Fig. 1.
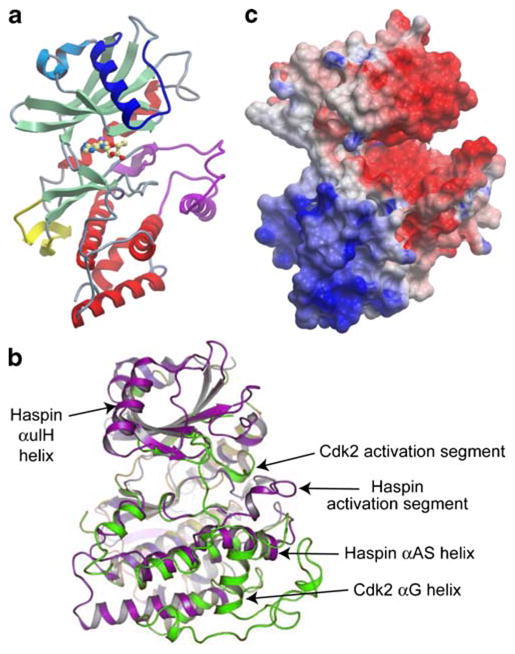
Crystal structure of the kinase domain of human haspin. a Ribbon diagram of human haspin residues 470–798. Conventional kinase domain β-strand and α-helical elements are shown in pale green and red, respectively. Unique elements include extra α-helices preceding the small lobe (light blue), an extra α-helical insert within the small lobe (αulH; dark blue), a β-hairpin near the hinge region between the two lobes (yellow), and an extended activation segment that includes the αAS helix (magenta). In this structure, the ATP-mimetic kinase inhibitor 5-iodotubercidin is bound in the active site and shown as a ball and stick representation. b Ribbon diagram superimpositions of the haspin kinase domain (magenta) and Cdk2 (green). Some of the specific features that differ between the two kinases are indicated. c Electrostatic surface potential of haspin showing the strongly electronegative putative substrate-binding groove (predominantly red region), and electropositive regions (blue)
Haspin phosphorylates histone H3T3
The only substrate of haspin identified to date (other than itself) is histone H3. Haspin immunoprecipitated from cells co-purifies with and phosphorylates histone H3, and recombinant haspin specifically phosphorylates purified and nucleosomal histone H3 in vitro (Dai et al. 2005; Patnaik et al. 2008; Eswaran et al. 2009; Villa et al. 2009). Haspin targets a single site in H3, residue T3 (H3T3ph). In cells, histone H3 is phosphorylated at a number of sites during mitosis, including T3, S10, T11, and S28 (Dai and Higgins 2005). The best characterized among these is H3S10ph, a modification carried out by the mitotic kinase Aurora B (Hsu et al. 2000; Adams et al. 2001; Giet and Glover 2001; Crosio et al. 2002). Because adjacent modifications on the densely modified tail of H3 can influence antibody recognition (Garcia et al. 2005), the distribution patterns of histone marks determined with antibodies must be interpreted with caution. Nevertheless, in mammalian cells, H3S10ph is found predominantly in pericentromeric chromatin (but not at inner centromeres) and on chromosome arms between late G2 and anaphase (Hendzel et al. 1997). H3T3ph, in contrast, appears first in late G2 or prophase at foci on chromosome arms, with a bias towards chromatin located near the nuclear periphery. During prometaphase, H3T3ph becomes concentrated at inner centromeres between the regions delineated by the centromere-specific histone CENP-A, but also remains visible on chromosome arms. H3T3ph declines during anaphase and cannot be detected in telophase cells (Polioudaki et al. 2004; Dai et al. 2005; Dai et al. 2006; Vagnarelli et al. 2008; Markaki et al. 2009). Therefore, H3T3ph and H3S10ph have distinct distributions in mitosis, but the two overlap to some extent. Indeed, H3 peptides containing both modifications have been obtained from mitotic cells (Garcia et al. 2005; Bonenfant et al. 2007). Overall, the timing of H3T3ph in mitosis, but not its location, appears similar to that of H3S10ph, suggesting distinct functions for the two modifications. Haspin RNA interference (RNAi) essentially eliminates H3T3ph in mitotic cells (Fig. 2) and, conversely, haspin overexpression causes anomalous H3T3ph generation in interphase (Dai et al. 2005; Markaki et al. 2009). Therefore, haspin appears to be the major mitotic H3T3 kinase in cultured human cells.
Fig. 2.
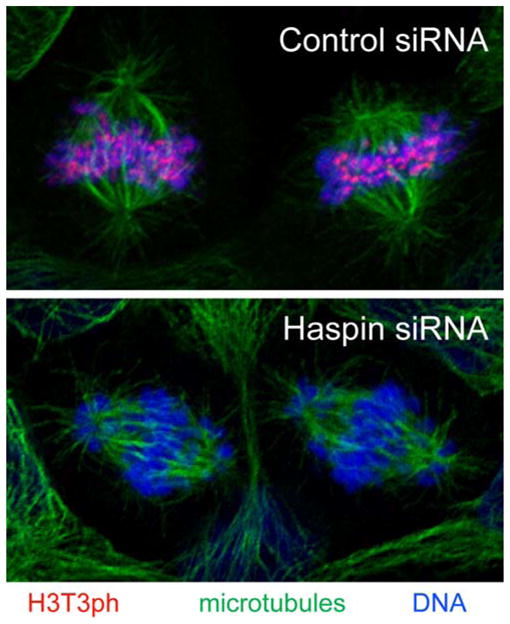
Haspin RNAi eliminates the predominantly centromeric phosphorylation of histone H3 at threonine-3 during mitosis and leads to chromosome misalignment. Human U2OS cells were transfected with control or haspin siRNAs as indicated. H3T3ph antibody staining is shown in red, microtubules (α-tubulin) in green, and DNA in blue
H3T3ph has also been examined in plants. Interestingly, the results confirm prior suggestions that the distribution of phosphorylation on H3 of mitotic chromosomes is almost reversed in plants compared to mammals. While H3S10ph is restricted to pericentromeric regions in plants, H3T3ph originates at pericentromeres in prophase and is evenly distributed along chromosome arms by prometaphase (Houben et al. 2007; Caperta et al. 2008). The reasons for these differences are unknown, but it appears that there has been considerable divergence in the roles of histone modification between plants and other organisms (Loidl 2004; Houben et al. 2007). Although plants possess haspin homologues, it remains to be determined if they are required for mitotic H3T3ph in these species. Phosphorylation of histones by recombinant budding yeast haspin homologues Alk1 and Alk2 was not observed in vitro (Nespoli et al. 2006), and as yet there are no reports of H3T3ph in fungal cells. Although these results are not definitive, they hint at the existence of alternative substrates for haspin in yeasts and other organisms.
Other H3T3 kinases
The mammalian vaccinia-related kinase-1 (VRK1) has been reported to phosphorylate both H3T3 and H3S10 in mitosis. VRK1 phosphorylates H3 in vitro, though less efficiently than the well-characterized substrate BAF, and VRK1 RNAi partially reduces H3T3ph and H3S10ph in mitosis (Kang et al. 2007). Because haspin depletion essentially eliminates H3T3ph in mitosis, an understanding of the relative contribution of VRK1 requires further study. In the green alga Chlamydomonas reinhardtii, another protein kinase, MUT9, has been shown to phosphorylate H3T3 and unidentified sites in histone H2A in vitro, and a Mut9 mutant strain is partially defective in generating H3T3ph in cells. In this case, phosphorylation in mitosis was not examined directly, but H3T3ph was found at the promoters of repressed genes, presumably in interphase. Mut9 mutant strains had decreased H3T3ph that correlated with gene de-repression, suggesting that MUT9 is necessary for gene silencing in Chlamydomonas (Casas-Mollano et al. 2008). MUT9 kinase homologs appear to be absent in animal and fungal lineages and so may represent an adaptation specific to the plant kingdom.
Haspin function in cells
Human HEK293 cells overexpressing EGFP-haspin, or a kinase-deficient deletion mutant, accumulate in G1 with 2 N DNA content (Tanaka et al. 1999). The basis for this effect is unknown but, coupled with the persistence of haspin protein in interphase, raises the little-explored prospect that haspin has functions outside mitosis. Insight into haspin function in mitosis comes primarily from RNAi studies in human cell lines. Haspin depletion leads to an increased mitotic index, with an accumulation of cells in prometaphase and a decline in the proportion of mitotic cells in anaphase, suggesting a defect in chromosome congression and a delay in exit from mitosis (Dai et al. 2005; Dai et al. 2006; Markaki et al. 2009). Haspin RNAi causes the accumulation of cells with a partial metaphase plate but with numerous non-congressed chromosomes, many of which are clustered at the spindle poles (Fig. 2; Dai et al. 2005; Dai et al. 2006). Such cells often contain Cyclin B, and the checkpoint protein Bub1 is recruited to kinetochores, indicating that haspin depletion prevents proper chromosome alignment and satisfaction of the spindle assembly checkpoint (Dai et al. 2006). Live imaging of U2OS cells expressing labeled H2B reveals that haspin depletion delays chromosome congression by about 20 min, but that metaphase-like plates do eventually form. This metaphase-like state then persists for approximately 2 h before chromosomes begin to leave the plate, eventually leading to extensive chromosome misalignment (Dai et al. 2009).
Haspin function in chromosome cohesion
The chromosome alignment defects in haspin-depleted cells are likely due, at least in part, to premature loss of sister chromatid cohesion (Dai et al. 2006). Cohesion between replicated chromosomes is maintained by complexes consisting of the cohesins Smc1, Smc3, Rad21/Scc1, and Scc3/SA1/SA2, and perhaps also by DNA catenations (Nasmyth 2002; Peters et al. 2008). In vertebrates, cohesin removal in mitosis occurs in two steps (Losada et al. 1998; Waizenegger et al. 2000). In prophase and prometaphase, release of cohesin from chromosome arms takes place under the control of Polo-like Kinase-1 (Plk1) and Aurora B (the “prophase pathway”), while cohesion at centromeres and a few loci on the arms is maintained (Losada et al. 2002; Sumara et al. 2002; Gimenez-Abian et al. 2004). The bulk of arm cohesin is removed in a cleavage-independent fashion by a mechanism that requires SA2 phosphorylation, perhaps by Plk1 (Hauf et al. 2005). The protease separase can cleave the Rad21/Scc1 cohesin subunit (Hauf et al. 2001), but separase is kept largely inactive in early mitosis by association with securin and phosphorylation by cyclin B-Cdk1. Satisfaction of the SAC allows the anaphase promoting complex/cyclosome (APC/C) to promote degradation of securin and cyclin B and separase is activated, cleaving cohesin and triggering the separation of sister chromatids at anaphase (Nasmyth 2002; Peters et al. 2008). Topoisomerase II activity also appears to be required during mitosis to fully resolve links between sister chromatids (Downes et al. 1991; Shamu and Murray 1992; Toyoda and Yanagida 2006; Spence et al. 2007; Wang et al. 2008a). Haspin RNAi leads to loss of cohesin from chromosomes and separation of sister chromatids (Dai et al. 2006), and the chromosome alignment defects observed by live imaging of haspin-depleted cells are similar to those caused by depletion of cohesin Scc1 (Dai et al. 2009). Separation occurs in cells treated with the spindle poison nocodazole or the proteasome inhibitor MG132, suggesting that it takes place when SAC signaling is active and is independent of the APC/C pathway and mitotic exit. Conversely, overexpression of haspin prevents the normal dissociation of cohesin from chromosome arms (Dai et al. 2006). Indirect evidence indicates that haspin acts in mitosis to regulate cohesion rather than, for example, affecting cohesion establishment: H3T3ph is mitosis-specific and loss of cohesion due to haspin RNAi can be prevented by depleting the mitotic kinase Aurora B. These results suggest that haspin serves to protect centromeric cohesin from the action of the prophase pathway.
The molecular basis of the function of haspin in cohesion regulation is currently unclear. We noted that the localization pattern of H3T3ph and the residual cohesin that remains at the inner centromeres of metaphase chromosomes is similar, though not identical (Dai et al. 2006). At minimum, this shows that haspin is active in the vicinity of the protected cohesin complexes, and might indicate that H3T3ph plays a role in cohesion regulation (see below), though this is by no means the only possibility. Haspin could, for example, directly phosphorylate cohesin or have a protective function independent of kinase activity. Haspin could also collaborate with other cohesin-regulating proteins to modulate cohesion. An obvious candidate is Shugoshin-1 (Sgo1), a centromeric protein that is required for protection of cohesion at centromeres in vertebrate mitosis (Salic et al. 2004; Tang et al. 2004; Kitajima et al. 2005; McGuinness et al. 2005). In yeast and human cells, Sgo1 associates with the phosphatase PP2A (Kitajima et al. 2006; Riedel et al. 2006; Tang et al. 2006). This, coupled with observations that PP2A activity is required to maintain cohesion (Kitajima et al. 2006; Riedel et al. 2006; Tang et al. 2006), that the protective function of Sgo1 requires an intact PP2A binding site (Xu et al. 2009), and that expression of nonphosphorylatable SA2 prevents the loss of cohesion caused by Sgo1 depletion (McGuinness et al. 2005), has led to the idea that Sgo1 acts through PP2A to prevent cohesin phosphorylation and release by the prophase pathway (Kitajima et al. 2006; Riedel et al. 2006). Haspin does not appear to control Sgo1 localization because Sgo1 association with centromeres is not prevented in haspin-depleted cells, though this observation does not rule out that haspin could influence the functionality of centromeric Sgo1. Conversely, depletion of Sgo1 leads to loss of the centromeric concentration of H3T3ph and the spreading of H3T3ph throughout single chromatids (Dai et al. 2006; Eot-Houllier et al. 2008). Eventually, H3T3ph can be lost altogether after Sgo1 is depleted, although this apparently is not required for loss of cohesion (Eot-Houllier et al. 2008). These results could be interpreted to mean that haspin acts downstream of Sgo1. However, haspin and Sgo1 appear to be able to act independently of each other to protect cohesion in some experimental circumstances, such as when haspin overexpression increases arm cohesion in Sgo1-depleted cells or when Aurora B is depleted and Sgo1, but not haspin, is required for arm cohesion (Dai et al. 2006).
Interestingly, other proteins that might act independently of Sgo1 to protect centromeric cohesion have been identified. Inactivation of the acetyltransferase san in Drosophila or human cells, like haspin depletion, leads to loss of centromeric cohesin localization and cohesion in mitosis (Hou et al. 2007) and, at least in Drosophila, this is independent of separase activity (Williams et al. 2003). Loss of san does not compromise Sgo1 localization (Williams et al. 2003; Hou et al. 2007). Similarly, depletion of Prohibitin-2 (PHB2) from HeLa cells results in chromosome alignment defects and premature cohesion loss in mitotic cells, but does not prevent Sgo1 or PP2A localization to centromeres (Takata et al. 2007). PHB2 is best known as a component of inner mitochondrial membranes, but also has been reported to have nuclear functions (Osman et al. 2009). PHB2 depletion did not affect H3T3ph in mitosis, suggesting that it does not function by activating haspin kinase activity, but a number of other permutations of interaction between PHB2 (or san) and haspin can be envisioned. Clearly, further studies are required to understand the details of cohesion regulation by haspin and to determine if it is truly acting in a pathway parallel to that involving Sgo1.
Role of H3T3ph
The knowledge that haspin phosphorylates H3T3ph in mitosis raises the obvious question of the function of H3T3ph. An understanding of this is likely to require consideration of the context in which H3T3ph is found. Phosphorylation is only one of multiple types of modification that occur on histones. For example, mono, di-, and tri-methylation of lysine residues including K4, K9, and K27, dimethylation of arginine residues including R2 and R8, and acetylation of numerous lysines including K4, K9, and K14 have been reported in histone H3 alone (Kouzarides 2007; Eot-Houllier et al. 2008). This has led to a number of more or less stringent definitions of a “histone code” hypothesis stating that different combinations of histone modifications form a code that is translated by effector or “reader” proteins into distinct biological functions (Turner 1993; Strahl and Allis 2000; Fischle et al. 2003; Sims and Reinberg 2008). Therefore, the function of H3T3ph may be influenced by adjacent modifications on histone tails.
A recent report suggested that H3T3ph is always found within a combinatorial modification pattern with H3K4me3 and H3R8me2 (named “PMM”) in mitotic cells (Markaki et al. 2009). This assertion was based on a combination of mass spectrometry (MS) and immunochemical data that could not assign modifications with certainty. In contrast, H3T3ph-containing H3 peptides from mitotic cells observed in more direct MS/MS experiments lack H3K4me3 and H3R8me2 though in some cases they contain H3K4me1, H3K9me2, and/or H3S10ph (Garcia et al. 2005; Bonenfant et al. 2007). Further, although subject to the caveats of epitope occlusion noted earlier, immunofluorescence experiments indicate that H3K4me2, H3K4me3, and H3T3ph have distinct localizations at centromeres in mitosis (Sullivan and Karpen 2004; Dai et al. 2006). In addition, at least in vitro, haspin activity towards H3 peptides is progressively reduced as H3K4 is increasingly methylated (Eswaran et al. 2009). Casas-Mollano et al. (2008) also reported an inverse relationship between H3T3ph and H3K4me1 versus H3K4me2 and H3K4me3 at promoters in Chlamydomonas cells. Therefore, although the PMM may exist in cells (Markaki et al. 2009), it seems unlikely that H3T3ph is found only in this context. We favor the hypothesis that different (though likely partly overlapping) localizations of modifications such as H3T3ph, H3S10ph, and H3K4 methylation contribute to the creation of distinct zones of chromatin modification within centromeres and elsewhere on chromatin that have different functional attributes in mitosis. Crosstalk between modifications such as an inhibitory effect of H3K4me3 on haspin activity may play a role in establishing such regionality.
At least three (not entirely separable) molecular events that could be regulated by histone phosphorylation can be imagined: (1) phosphorylation induces a physiochemical change that influences interaction between nucleosomes or between histones and DNA. (2) Phosphorylation creates or abrogates a binding site for a histone-binding protein. (3) Phosphorylation influences the generation or functional consequences of other histone modifications. Variations of all three have been proposed for the best studied mark, H3S10ph. This modification was originally implicated in chromosome condensation in Tetrahymena (Wei et al. 1999) and it was reported to influence the interaction of H3 tails with DNA, perhaps altering nucleosome packing (Sauve et al. 1999), an example of a physiochemical mechanism. However, an H3S10 mutation does not prevent mitotic progression in budding yeast (Hsu et al. 2000) and the effects of Aurora B on condensation in multicellular organisms may be independent of H3S10ph (Adams et al. 2001; de la Barre et al. 2001; MacCallum et al. 2002; Lipp et al. 2007). An instance in which histone phosphorylation generates a protein-binding site is provided by the H3S10ph-dependent binding of 14-3-3 proteins to H3, an interaction that plays a role in regulation of gene transcription (Macdonald et al. 2005). More recently, in an example of the third mechanism mentioned above, it has been shown that H3S10ph serves as a component of a “methyl/phospho” switch to regulate the association of histone-binding proteins with chromatin. The binding of heterochromatin protein-1 (HP1) to H3 peptides is strongly enhanced by H3K9me3. The phosphorylation of H3S10 adjacent to H3K9me3, however, prevents HP1 binding and may eject it from pericentromeric heterochromatin during mitosis (Fischle et al. 2005; Hirota et al. 2005).
Similar mechanisms can be envisaged for H3T3ph. Theoretical arguments suggest that H3T3ph could contribute to changes in nucleosomal packing, though experimental approaches will be required to substantiate these ideas (Georgatos et al. 2009). There are a number of published examples where H3T3ph abrogates the interaction of proteins with histone H3. The direct binding of the inhibitor of acetyltransferases complex (INHAT) to H3 tail peptides is prevented by prior T3 phosphorylation in vitro (Schneider et al. 2004). However, H3T3ph is not unique in this, as H3S10ph and H3T11ph as well as multisite acetylation are also able to dislodge INHAT. In two other similar in vitro examples, binding of the autoimmune regulator protein AIRE and the methyltransferase-associated WDR5 protein to the tail of H3 are abrogated by either H3R2 methylation or H3T3ph (Couture et al. 2006; Koh et al. 2008; Chignola et al. 2009). In addition, the methylation of H3K4 by mixed-lineage leukemia protein is strongly reduced by H3T3ph (Southall et al. 2009). Whether H3T3 phosphorylation displaces these proteins from chromatin in cells and if this has functional significance remains to be explored.
The proximity of H3T3 to the H3R2 and H3K4 methylation sites has led to the suggestion that H3T3ph might operate in a methyl/phospho switch mechanism like that at H3K9/H3S10 (Fischle et al. 2003). Indeed, binding of chromodomain helicase DNA binding protein-1 (CHD1) to H3 methylated at K4 is abrogated by H3T3ph in vitro (Flanagan et al. 2005). Although no cell-based experiments have yet directly confirmed this idea, it is quite plausible that H3T3ph serves to displace a number of H3-binding transcription and chromatin-regulating factors from chromosomes during mitosis. This process might make way for new chromatin-binding proteins or chromatin rearrangements that are needed to accomplish chromosome segregation, without erasing epigenetic histone modifications that control gene expression. It is also possible that “clearing the deck” in this way provides a means of re-initializing cellular gene expression programs during mitosis, either to prevent the accumulation of errors over multiple cell cycles or to help establish new transcriptional profiles in differentiating cells (Egli et al. 2008). In this last scenario, H3T3ph functions during mitosis but is not directly required for mitosis itself.
In another example in which phosphorylation alters protein interaction with histones, it was recently suggested that H3T3ph plays a role in activating Aurora B at centromeres (Rosasco-Nitcher et al. 2008). In this case, prior addition of H3 peptides to inactive recombinant Aurora B/INCENP prevented activation of the kinase by TD-60 and microtubules in vitro. Such inhibition did not occur, however, if the H3 peptides carried H3T3ph, and direct binding experiments revealed that H3T3ph prevented interaction of the H3 peptides with Aurora B/INCENP. These results led to a model in which binding of H3 substrate in the active site of inactive Aurora B prevents activation of the kinase, perhaps by competing with autophosphorylation of the activation segment. Priming phosphorylation of H3T3 by haspin prevents H3 association with Aurora B and allows kinase activation to occur, particularly at centromeres where H3T3ph is concentrated. This hypothesis is currently based almost exclusively on studies of recombinant proteins in vitro, and further experiments are needed to confirm that haspin and/or H3T3ph are required for centromeric Aurora B activity in cells. It will also be of interest to determine if H3T3ph is unique among H3 modifications in its ability to facilitate Aurora B activation, and to determine if H3T3ph has a direct effect on nearby H3S10 phosphorylation by Aurora B.
Finally, though it has yet to be demonstrated, it is appealing to hypothesize the existence of proteins that bind to H3 only when phosphorylated at T3. As for H3S10ph (Hans and Dimitrov 2001), it has been speculated that H3T3ph might serve to mark chromosomes that have successfully reached metaphase (Markaki et al. 2009), though the feature(s) of mitotic progression to which H3T3 or H3S10 would be sensitive and what the reader(s) of these marks would be are unknown. Of course, a number of other roles for H3T3ph-mediated protein binding can be imagined. In particular, the centromeric accumulation of H3T3ph suggests that it might recruit proteins that function at inner centromeres. Such proteins might include cohesion factors, chromosomal passenger proteins (Ruchaud et al. 2007), or proteins that contribute to the structural or mechanical properties of centromeres.
Effects on centrosomes
EGFP-haspin can be found at centrosomes in mitotic HeLa cells (Dai et al. 2005), and haspin RNAi leads to the emergence of multiple acentriolar centrosome-like foci during mitosis (Dai et al. 2009). These observations might lead one to suspect a role for haspin in maintaining centrosome integrity, particularly since recent work has suggested that cohesin system proteins localize to centrosomes and regulate centriole disengagement and centrosome-initiated spindle assembly (Tsou and Stearns 2006; Wang et al. 2008b; Kong et al. 2009). More detailed analyses revealed that similar centrosomal defects occur when other cohesin system proteins are depleted, and that they are most likely an indirect effect of cohesion loss at chromosomes. Indeed, treatment of haspin-depleted mitotic cells with topoisomerase II inhibitors to prevent chromosome separation was able to substantially “rescue” the centrosomal defects (Dai et al. 2009). It is possible that loss of chromosome cohesion disturbs the normal integration of chromosome-dependent and chromosome-independent pathways of spindle formation that are necessary to maintain bipolarity. It should be emphasized that these findings do not rule out functions for haspin (or other cohesin-system proteins) at centrosomes, only that such a role is not necessary to explain the appearance of multiple centrosome-like foci when cohesion factors are depleted. Further work is needed to determine if endogenous haspin is found at centrosomes and if it has any function at this location.
Regulation of haspin activity
Haspin phosphorylates histone H3 during mitosis, raising the question of how it is regulated during the cell cycle. Unlike mitotic kinases such as Aurora B and Plk1 that are degraded at the end of mitosis (Nigg 2001), human haspin is expressed at near-constant levels throughout the cell cycle (Dai et al. 2005; Wang F and Higgins JMG, unpublished data). Haspin is, however, very strongly phosphorylated during mitosis (Dai et al. 2005; Nousiainen et al. 2006), and we have identified approximately 30 mitotic phosphorylation sites in the N-terminal domain of human haspin (Ulyanova NP and Higgins JMG, unpublished data). Despite this, haspin immunoprecipitated from interphase and mitotic cells has similar kinase activity (Dai et al. 2005). Therefore, phosphorylation of haspin does not appear to significantly affect its intrinsic kinase activity. These findings are reflected in the crystallographic observations discussed earlier revealing that the kinase domain of haspin is in an active confirmation that does not require activation segment phosphorylation (Eswaran et al. 2009; Villa et al. 2009). In vitro analysis of haspin activity reveals that the N-domain alters the enzymatic properties of the kinase domain, perhaps due to a stabilizing effect on the kinase domain (Eswaran et al. 2009; Villa et al. 2009). Modification or protein binding to the N-domain therefore has the potential to modulate haspin activity. Phosphorylation of haspin on its N-domain may alter the access of haspin to its substrates, or might regulate the interaction of haspin with putative regulatory proteins that control its activity in cells.
Function of haspin in budding yeast
The functions of the two haspin homologues in budding yeast have not been extensively studied. For reasons that are unknown, the original deposition of ALK1 in the sequence databases was accompanied by an annotation suggesting it to be a “novel DNA damage response gene”. Nespoli et al (2006) later identified Alk1 in a yeast two-hybrid screen with the DNA damage checkpoint factor Ddc1 as bait. Although they found that both Alk1 and Alk2 are phosphorylated in response to DNA damaging agents, they were unable to determine a role for either protein in the DNA damage checkpoint or in DNA damage sensitivity.
The abundance of both budding yeast haspin homologues is regulated during the cell cycle. The ALK1 mRNA is a member of the “CLB2 cluster” of 33 genes that peak in expression in early mitosis (Spellman et al. 1998). Correspondingly, Alk1 protein levels peak in mitosis (Nespoli et al. 2006). The amount of ALK2 mRNA is relatively constant during the cell cycle, but the level of Alk2 protein peaks in early G2 and falls rapidly during anaphase (Sullivan et al. 2004; Nespoli et al. 2006). Degradation of Alk2 depends upon the presence of “KEN” and “Destruction” (D) boxes within the protein and appears to be carried out by the APC/C pathway. Alk1 and Alk2 are also both phosphorylated during mitosis (Nespoli et al. 2006). These properties are consistent with mitotic roles for Alk1 and Alk2. Indeed, although strains of Saccharomyces cerevisiae lacking ALK1, ALK2 or both genes are viable, overexpression of Alk2 causes a strong block in mitosis. This arrest is substantially, though not completely, eliminated when a kinase-defective form of Alk2 is used (Nespoli et al. 2006). Alk2 kinase activity, therefore, appears to have a so far undetermined role in mitosis.
Germ-line functions of haspin
Although the phosphorylation of H3T3 by haspin and mitotic defects following haspin depletion has focused attention on its roles in mitosis, it would be a mistake to discount a role for the kinase in other contexts. In particular, haspin is strongly expressed in post-meiotic round spermatids (Tanaka et al. 1999). It is tempting to think that haspin functions to modify chromatin in the special context of spermiogenesis, where histones are replaced by transition proteins and then protamines to facilitate compaction of chromosomes into the heads of developing sperm (Wykes and Krawetz 2003). This possibility is made all the more intriguing by the finding that a small percentage of histones are retained in mature sperm at loci involved in embryo development, and that modifications such as H3K4me2 and H3K4me3 are enriched on particular subsets of these histones (Hammoud et al. 2009). It will be interesting to determine if haspin and H3T3ph influence the retention, removal, or function of histones during spermiogenesis and early embryogenesis. Possible roles of haspin in meiosis, including in oocytes, as well as in interphase of somatic cells also remain to be addressed.
Conclusions
Haspin and H3T3 phosphorylation were more recently discovered and have been less investigated than other mitotic kinase families and histone modifications. Nevertheless, a number of studies in the last few years have begun to outline features and functions of this evolutionarily conserved kinase, and to raise new questions for the future. The recognition of haspin orthologs in model organisms, the description of haspin depletion and overexpression phenotypes, the production of H3T3ph-specific antibodies, and the determination of crystal structures of the haspin kinase domain will all facilitate further work. The recent identification and further development of selective chemical inhibitors of haspin (Patnaik et al. 2008; Eswaran et al. 2009) should soon allow temporally controlled kinase inhibition in a wide variety of systems that has previously been impractical. These compounds will also facilitate evaluation of haspin inhibition as a means to curb cancer cell proliferation (de Cárcer et al. 2007; Schmidt and Bastians 2007) or curtail spermatogenesis. Many other key questions remain. How direct is regulation of cohesion by haspin? What are the functions of H3T3ph? What are other substrates of haspin? Are all functions of haspin dependent on kinase activity? How is haspin activity regulated? Does haspin have functions outside mitosis? Studies aimed at answering these questions are likely to provide new insight into the mechanisms of chromosome segregation, and have the potential to illuminate other fields of research including chromatin modification and spermatogenesis.
Acknowledgments
I would like to thank Andrea Musacchio for communicating unpublished results, Stefan Knapp and Jun Dai for providing pictures for Figs. 1 and 2, and Anna Kateneva and Fangwei Wang for their comments on the manuscript. Work in the Higgins laboratory is supported by the American Cancer Society (RSG-05-134-01-GMC) and the National Institutes of Health (GM074210 and CA122608).
References
- Adams RR, Maiato H, Earnshaw WC, Carmena M. Essential roles of Drosophila inner centromere protein (INCENP) and aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. J Cell Biol. 2001;153:865–880. doi: 10.1083/jcb.153.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonenfant D, Towbin H, Coulot M, Schindler P, Mueller DR, van Oostrum J. Analysis of dynamic changes in post-translational modifications of human histones during cell cycle by mass spectrometry. Mol Cell Proteomics. 2007;6:1917–1932. doi: 10.1074/mcp.M700070-MCP200. [DOI] [PubMed] [Google Scholar]
- Caperta AD, Rosa M, Delgado M, Karimi R, Demidov D, Viegas W, Houben A. Distribution patterns of phosphorylated Thr 3 and Thr 32 of histone H3 in plant mitosis and meiosis. Cytogenet Genome Res. 2008;122:73–79. doi: 10.1159/000151319. [DOI] [PubMed] [Google Scholar]
- Casas-Mollano JA, Jeong BR, Xu J, Moriyama H, Cerutti H. The MUT9p kinase phosphorylates histone H3 threonine 3 and is necessary for heritable epigenetic silencing in Chlamydomonas. Proc Natl Acad Sci U S A. 2008;105:6486–6491. doi: 10.1073/pnas.0711310105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chignola F, Gaetani M, Rebane A, Org T, Mollica L, Zucchelli C, Spitaleri A, Mannella V, Peterson P, Musco G. The solution structure of the first PHD finger of autoimmune regulator in complex with non-modified histone H3 tail reveals the antagonistic role of H3R2 methylation. Nucleic Acids Res. 2009;37:2951–2961. doi: 10.1093/nar/gkp166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couture JF, Collazo E, Trievel RC. Molecular recognition of histone H3 by the WD40 protein WDR5. Nat Struct Mol Biol. 2006;13:698–703. doi: 10.1038/nsmb1116. [DOI] [PubMed] [Google Scholar]
- Crosio C, Fimia GM, Loury R, Kimura M, Okano Y, Zhou H, Sen S, Allis CD, Sassone-Corsi P. Mitotic phosphorylation of histone H3: spatio-temporal regulation by mammalian Aurora kinases. Mol Cell Biol. 2002;22:874–885. doi: 10.1128/MCB.22.3.874-885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Higgins JMG. Haspin: a mitotic histone kinase required for metaphase chromosome alignment. Cell Cycle. 2005;4:665–668. doi: 10.4161/cc.4.5.1683. [DOI] [PubMed] [Google Scholar]
- Dai J, Sultan S, Taylor SS, Higgins JMG. The kinase haspin is required for mitotic histone H3 Thr 3 phosphorylation and normal metaphase chromosome alignment. Genes Dev. 2005;19:472–488. doi: 10.1101/gad.1267105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Sullivan BA, Higgins JMG. Regulation of mitotic chromosome cohesion by haspin and Aurora B. Dev Cell. 2006;11:741–750. doi: 10.1016/j.devcel.2006.09.018. [DOI] [PubMed] [Google Scholar]
- Dai J, Kateneva AV, Higgins JMG. Studies of haspin-depleted cells reveal that spindle-pole integrity in mitosis requires chromosome cohesion. J Cell Sci. 2009;122:4168–4176. doi: 10.1242/jcs.054122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Cárcer G, de Castro IP, Malumbres M. Targeting cell cycle kinases for cancer therapy. Curr Med Chem. 2007;14:969–985. doi: 10.2174/092986707780362925. [DOI] [PubMed] [Google Scholar]
- de la Barre AE, Angelov D, Molla A, Dimitrov S. The N-terminus of histone H2B, but not that of histone H3 or its phosphorylation, is essential for chromosome condensation. EMBO J. 2001;20:6383–6393. doi: 10.1093/emboj/20.22.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes CS, Mullinger AM, Johnson RT. Inhibitors of DNA topoisomerase II prevent chromatid separation in mammalian cells but do not prevent exit from mitosis. Proc Natl Acad Sci U S A. 1991;88:8895–8899. doi: 10.1073/pnas.88.20.8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli D, Birkhoff G, Eggan K. Mediators of reprogramming: transcription factors and transitions through mitosis. Nat Rev Mol Cell Biol. 2008;9:505–516. doi: 10.1038/nrm2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eot-Houllier G, Fulcrand G, Watanabe Y, Magnaghi-Jaulin L, Jaulin C. Histone deacetylase 3 is required for centromeric H3K4 deacetylation and sister chromatid cohesion. Genes Dev. 2008;22:2639–2644. doi: 10.1101/gad.484108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswaran J, Patnaik D, Filippakopoulos P, Wang F, Stein RL, Murray J, Higgins JMG, Knapp S. Structure and functional characterization of the atypical human kinase haspin. Proc Natl Acad Sci USA. 2009;106:20198–20203. doi: 10.1073/pnas.0901989106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Allis CD. Binary switches and modification cassettes in histone biology and beyond. Nature. 2003;425:475–479. doi: 10.1038/nature02017. [DOI] [PubMed] [Google Scholar]
- Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H, Allis CD. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438:1116–1122. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- Flanagan JF, Mi LZ, Chruszcz M, Cymborowski M, Clines KL, Kim Y, Minor W, Rastinejad F, Khorasanizadeh S. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- Garcia BA, Barber CM, Hake SB, Ptak C, Turner FB, Busby SA, Shabanowitz J, Moran RG, Allis CD, Hunt DF. Modifications of human histone H3 variants during mitosis. Biochemistry. 2005;44:13202–13213. doi: 10.1021/bi050906n. [DOI] [PubMed] [Google Scholar]
- Georgatos SD, Markaki Y, Christogianni A, Politou AS. Chromatin remodeling during mitosis: a structure-based code? Front Biosci. 2009;14:2017–2027. doi: 10.2741/3360. [DOI] [PubMed] [Google Scholar]
- Giet R, Glover DM. Drosophila aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J Cell Biol. 2001;152:669–682. doi: 10.1083/jcb.152.4.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez-Abian JF, Sumara I, Hirota T, Hauf S, Gerlich D, de la Torre C, Ellenberg J, Peters JM. Regulation of sister chromatid cohesion between chromosome arms. Curr Biol. 2004;14:1187–1193. doi: 10.1016/j.cub.2004.06.052. [DOI] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hans F, Dimitrov S. Histone H3 phosphorylation and cell division. Oncogene. 2001;20:3021–3027. doi: 10.1038/sj.onc.1204326. [DOI] [PubMed] [Google Scholar]
- Hauf S, Waizenegger IC, Peters JM. Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science. 2001;293:1320–1323. doi: 10.1126/science.1061376. [DOI] [PubMed] [Google Scholar]
- Hauf S, Roitinger E, Koch B, Dittrich CM, Mechtler K, Peters JM. Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol. 2005;3:e69. doi: 10.1371/journal.pbio.0030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendzel MJ, Wei Y, Mancini MA, Van Hooser A, Ranalli T, Brinkley BR, Bazett-Jones DP, Allis CD. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 1997;106:348–360. doi: 10.1007/s004120050256. [DOI] [PubMed] [Google Scholar]
- Higgins JMG. Haspin-like proteins: A new family of evolutionarily conserved putative eukaryotic protein kinases. Prot Sci. 2001a;10:1677–1684. doi: 10.1110/ps.49901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JMG. The haspin gene: location in an intron of the Integrin αE gene, associated transcription of an Integrin αE-derived RNA and expression in diploid as well as haploid cells. Gene. 2001b;267:55–69. doi: 10.1016/s0378-1119(01)00387-0. [DOI] [PubMed] [Google Scholar]
- Higgins JMG. Structure, function and evolution of haspin and haspin-related proteins, a distinctive group of eukaryotic protein kinases. Cell Mol Life Sci. 2003;60:446–462. doi: 10.1007/s000180300038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T, Lipp JJ, Toh BH, Peters JM. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature. 2005;438:1176–1180. doi: 10.1038/nature04254. [DOI] [PubMed] [Google Scholar]
- Hou F, Chu CW, Kong X, Yokomori K, Zou H. The acetyltransferase activity of San stabilizes the mitotic cohesin at the centromeres in a shugoshin-independent manner. J Cell Biol. 2007;177:587–597. doi: 10.1083/jcb.200701043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houben A, Demidov D, Caperta AD, Karimi R, Agueci F, Vlasenko L. Phosphorylation of histone H3 in plants—a dynamic affair. Biochim Biophys Acta. 2007;1769:308–315. doi: 10.1016/j.bbaexp.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Hsu JY, Sun ZW, Li X, Reuben M, Tatchell K, Bishop DK, Grushcow JM, Brame CJ, Caldwell JA, Hunt DF, Lin R, Smith MM, Allis CD. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 2000;102:279–291. doi: 10.1016/s0092-8674(00)00034-9. [DOI] [PubMed] [Google Scholar]
- Kang TH, Park DY, Choi YH, Kim KJ, Yoon HS, Kim KT. Mitotic histone H3 phosphorylation by vaccinia-related kinase 1 in mammalian cells. Mol Cell Biol. 2007;27:8533–8546. doi: 10.1128/MCB.00018-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan N, Taylor SS, Zhai Y, Venter JC, Manning G. Structural and functional diversity of the microbial kinome. PLoS Biol. 2007;5:e17. doi: 10.1371/journal.pbio.0050017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima TS, Hauf S, Ohsugi M, Yamamoto T, Watanabe Y. Human Bub1 defines the persistent cohesion site along the mitotic chromosome by affecting Shugoshin localization. Curr Biol. 2005;15:353–359. doi: 10.1016/j.cub.2004.12.044. [DOI] [PubMed] [Google Scholar]
- Kitajima TS, Sakuno T, Ishiguro K, Iemura S, Natsume T, Kawashima SA, Watanabe Y. Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature. 2006;441:46–52. doi: 10.1038/nature04663. [DOI] [PubMed] [Google Scholar]
- Koh AS, Kuo AJ, Park SY, Cheung P, Abramson J, Bua D, Carney D, Shoelson SE, Gozani O, Kingston RE, Benoist C, Mathis D. Aire employs a histone-binding module to mediate immunological tolerance, linking chromatin regulation with organ-specific autoimmunity. Proc Natl Acad Sci U S A. 2008;105:15878–15883. doi: 10.1073/pnas.0808470105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X, Ball AR, Jr, Sonoda E, Feng J, Takeda S, Fukagawa T, Yen TJ, Yokomori K. Cohesin associates with spindle poles in a mitosis-specific manner and functions in spindle assembly in vertebrate cells. Mol Biol Cell. 2009;20:1289–1301. doi: 10.1091/mbc.E08-04-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Lipp JJ, Hirota T, Poser I, Peters JM. Aurora B controls the association of condensin I but not condensin II with mitotic chromosomes. J Cell Sci. 2007;120:1245–1255. doi: 10.1242/jcs.03425. [DOI] [PubMed] [Google Scholar]
- Loidl P. A plant dialect of the histone language. Trends Plant Sci. 2004;9:84–90. doi: 10.1016/j.tplants.2003.12.007. [DOI] [PubMed] [Google Scholar]
- Losada A, Hirano M, Hirano T. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev. 1998;12:1986–1997. doi: 10.1101/gad.12.13.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A, Hirano M, Hirano T. Cohesin release is required for sister chromatid resolution, but not for condensin-mediated compaction, at the onset of mitosis. Genes Dev. 2002;16:3004–3016. doi: 10.1101/gad.249202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCallum DE, Losada A, Kobayashi R, Hirano T. ISWI remodeling complexes in Xenopus egg extracts: identification as major chromosomal components that are regulated by INCENP-aurora B. Mol Biol Cell. 2002;13:25–39. doi: 10.1091/mbc.01-09-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald N, Welburn JP, Noble ME, Nguyen A, Yaffe MB, Clynes D, Moggs JG, Orphanides G, Thomson S, Edmunds JW, Clayton AL, Endicott JA, Mahadevan LC. Molecular basis for the recognition of phosphorylated and phosphoacetylated histone H3 by 14–3–3. Mol Cell. 2005;20:199–211. doi: 10.1016/j.molcel.2005.08.032. [DOI] [PubMed] [Google Scholar]
- Markaki Y, Christogianni A, Politou AS, Georgatos SD. Phosphorylation of histone H3 at Thr3 is part of a combinatorial pattern that marks and configures mitotic chromatin. J Cell Sci. 2009;122:2809–2819. doi: 10.1242/jcs.043810. [DOI] [PubMed] [Google Scholar]
- McGuinness BE, Hirota T, Kudo NR, Peters JM, Nasmyth K. Shugoshin prevents dissociation of cohesin from centromeres during mitosis in vertebrate cells. PLoS Biol. 2005;3:e86. doi: 10.1371/journal.pbio.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. Segregating sister genomes: the molecular biology of chromosome separation. Science. 2002;297:559–565. doi: 10.1126/science.1074757. [DOI] [PubMed] [Google Scholar]
- Nespoli A, Vercillo R, di Nola L, Diani L, Giannattasio M, Plevani P, Muzi-Falconi M. Alk1 and Alk2 are two new cell cycle-regulated haspin-like proteins in budding yeast. Cell Cycle. 2006;5:1464–1471. doi: 10.4161/cc.5.13.2914. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Mitotic kinases as regulators of cell division and its checkpoints. Nat Rev Mol Cell Biol. 2001;2:21–32. doi: 10.1038/35048096. [DOI] [PubMed] [Google Scholar]
- Nousiainen M, Sillje HH, Sauer G, Nigg EA, Korner R. Phosphoproteome analysis of the human mitotic spindle. Proc Natl Acad Sci U S A. 2006;103:5391–5396. doi: 10.1073/pnas.0507066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman C, Merkwirth C, Langer T. Prohibitins and the functional compartmentalization of mitochondrial membranes. J Cell Sci. 2009;122:3823–3830. doi: 10.1242/jcs.037655. [DOI] [PubMed] [Google Scholar]
- Patnaik D, Xian J, Glicksman MA, Cuny GD, Stein RL, Higgins JMG. Identification of small molecule inhibitors of the mitotic kinase haspin by high throughput screening using a homogeneous time-resolved fluorescence resonance energy transfer assay. J Biomol Screen. 2008;13:1025–1034. doi: 10.1177/1087057108326081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, Tedeschi A, Schmitz J. The cohesin complex and its roles in chromosome biology. Genes Dev. 2008;22:3089–3114. doi: 10.1101/gad.1724308. [DOI] [PubMed] [Google Scholar]
- Polioudaki H, Markaki Y, Kourmouli N, Dialynas G, Theodoropoulos PA, Singh PB, Georgatos SD. Mitotic phosphorylation of histone H3 at threonine 3. FEBS Lett. 2004;560:39–44. doi: 10.1016/S0014-5793(04)00060-2. [DOI] [PubMed] [Google Scholar]
- Riedel CG, Katis VL, Katou Y, Mori S, Itoh T, Helmhart W, Galova M, Petronczki M, Gregan J, Cetin B, Mudrak I, Ogris E, Mechtler K, Pelletier L, Buchholz F, Shirahige K, Nasmyth K. Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature. 2006;441:53–61. doi: 10.1038/nature04664. [DOI] [PubMed] [Google Scholar]
- Rosasco-Nitcher SE, Lan W, Khorasanizadeh S, Stukenberg PT. Centromeric Aurora-B activation requires TD-60, microtubules, and substrate priming phosphorylation. Science. 2008;319:469–472. doi: 10.1126/science.1148980. [DOI] [PubMed] [Google Scholar]
- Ruchaud S, Carmena M, Earnshaw WC. Chromosomal passengers: conducting cell division. Nat Rev Mol Cell Biol. 2007;8:798–812. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
- Salic A, Waters JC, Mitchison TJ. Vertebrate shugoshin links sister centromere cohesion and kinetochore microtubule stability in mitosis. Cell. 2004;118:567–578. doi: 10.1016/j.cell.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Sauve DM, Anderson HJ, Ray JM, James WM, Roberge M. Phosphorylation-induced rearrangement of the histone H3 NH2-terminal domain during mitotic chromosome condensation. J Cell Biol. 1999;145:225–235. doi: 10.1083/jcb.145.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, Bastians H. Mitotic drug targets and the development of novel anti-mitotic anticancer drugs. Drug Resist Updat. 2007;10:162–181. doi: 10.1016/j.drup.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Schneider R, Bannister AJ, Weise C, Kouzarides T. Direct binding of INHAT to H3 tails disrupted by modifications. J Biol Chem. 2004;279:23859–23862. doi: 10.1074/jbc.C400151200. [DOI] [PubMed] [Google Scholar]
- Shamu CE, Murray AW. Sister chromatid separation in frog egg extracts requires DNA topoisomerase II activity during anaphase. J Cell Biol. 1992;117:921–934. doi: 10.1083/jcb.117.5.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Reinberg D. Is there a code embedded in proteins that is based on post-translational modifications? Nat Rev Mol Cell Biol. 2008;9:815–820. doi: 10.1038/nrm2502. [DOI] [PubMed] [Google Scholar]
- Southall SM, Wong PS, Odho Z, Roe SM, Wilson JR. Structural basis for the requirement of additional factors for MLL1 SET domain activity and recognition of epigenetic marks. Mol Cell. 2009;33:181–191. doi: 10.1016/j.molcel.2008.12.029. [DOI] [PubMed] [Google Scholar]
- Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence JM, Phua HH, Mills W, Carpenter AJ, Porter AC, Farr CJ. Depletion of topoisomerase II alpha leads to shortening of the metaphase interkinetochore distance and abnormal persistence of PICH-coated anaphase threads. J Cell Sci. 2007;120:3952–3964. doi: 10.1242/jcs.013730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Sullivan BA, Karpen GH. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Struct Mol Biol. 2004;11:1076–1083. doi: 10.1038/nsmb845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M, Hornig NC, Porstmann T, Uhlmann F. Studies on substrate recognition by the budding yeast separase. J Biol Chem. 2004;279:1191–1196. doi: 10.1074/jbc.M309761200. [DOI] [PubMed] [Google Scholar]
- Sumara I, Vorlaufer E, Stukenberg PT, Kelm O, Redemann N, Nigg EA, Peters JM. The dissociation of cohesin from chromosomes in prophase is regulated by Polo-like kinase. Mol Cell. 2002;9:515–525. doi: 10.1016/s1097-2765(02)00473-2. [DOI] [PubMed] [Google Scholar]
- Takata H, Matsunaga S, Morimoto A, Ma N, Kurihara D, Ono-Maniwa R, Nakagawa M, Azuma T, Uchiyama S, Fukui K. PHB2 protects sister-chromatid cohesion in mitosis. Curr Biol. 2007;17:1356–1361. doi: 10.1016/j.cub.2007.07.009. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Yoshimura Y, Nishina Y, Nozaki M, Nojima H, Nishimune Y. Isolation and characterization of cDNA clones specifically expressed in testicular germ cells. FEBS Letts. 1994;355:4–10. doi: 10.1016/0014-5793(94)01155-9. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Yoshimura Y, Nozaki M, Yomogida K, Tsuchida J, Tosaka Y, Habu T, Nakanishi T, Okada M, Nojima H, Nishimune Y. Identification and characterization of a haploid germ cell-specific nuclear protein kinase (haspin) in spermatid nuclei and its effects on somatic cells. J Biol Chem. 1999;274:17049–17057. doi: 10.1074/jbc.274.24.17049. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Iguchi N, Nakamura Y, Kohroki J, Egydio de Carvalho C, Nishimune Y. Cloning and characterization of human haspin gene encoding haploid germ cell-specific nuclear protein kinase. Mol Hum Reprod. 2001;7:211–218. doi: 10.1093/molehr/7.3.211. [DOI] [PubMed] [Google Scholar]
- Tang Z, Sun Y, Harley SE, Zou H, Yu H. Human Bub1 protects centromeric sister-chromatid cohesion through Shugoshin during mitosis. Proc Natl Acad Sci U S A. 2004;101:18012–18017. doi: 10.1073/pnas.0408600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Shu H, Qi W, Mahmood NA, Mumby MC, Yu H. PP2A is required for centromeric localization of Sgo1 and proper chromosome segregation. Dev Cell. 2006;10:575–585. doi: 10.1016/j.devcel.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Toyoda Y, Yanagida M. Coordinated requirements of human topo II and cohesin for metaphase centromere alignment under Mad2-dependent spindle checkpoint surveillance. Mol Biol Cell. 2006;17:2287–2302. doi: 10.1091/mbc.E05-11-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou MF, Stearns T. Mechanism limiting centrosome duplication to once per cell cycle. Nature. 2006;442:947–951. doi: 10.1038/nature04985. [DOI] [PubMed] [Google Scholar]
- Turner BM. Decoding the nucleosome. Cell. 1993;75:5–8. [PubMed] [Google Scholar]
- Vagnarelli P, Ribeiro SA, Earnshaw WC. Centromeres: old tales and new tools. FEBS Lett. 2008;582:1950–1959. doi: 10.1016/j.febslet.2008.04.014. [DOI] [PubMed] [Google Scholar]
- Villa F, Capasso P, Tortorici M, Forneris F, de Marco A, Mattevi A, Musacchio A. Crystal structure of the catalytic domain of haspin, an atypical kinase implicated in chromatin organization. Proc Natl Acad Sci USA. 2009;106:20204–20209. doi: 10.1073/pnas.0908485106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waizenegger IC, Hauf S, Meinke A, Peters JM. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell. 2000;103:399–410. doi: 10.1016/s0092-8674(00)00132-x. [DOI] [PubMed] [Google Scholar]
- Wang LH, Schwarzbraun T, Speicher MR, Nigg EA. Persistence of DNA threads in human anaphase cells suggests late completion of sister chromatid decatenation. Chromosoma. 2008a;117:123–135. doi: 10.1007/s00412-007-0131-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Yang Y, Duan Q, Jiang N, Huang Y, Darzynkiewicz Z, Dai W. sSgo1, a major splice variant of Sgo1, functions in centriole cohesion where it is regulated by Plk1. Dev Cell. 2008b;14:331–341. doi: 10.1016/j.devcel.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Yu L, Bowen J, Gorovsky MA, Allis CD. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell. 1999;97:99–109. doi: 10.1016/s0092-8674(00)80718-7. [DOI] [PubMed] [Google Scholar]
- Williams BC, Garrett-Engele CM, Li Z, Williams EV, Rosenman ED, Goldberg ML. Two putative acetyltransferases, san and deco, are required for establishing sister chromatid cohesion in Drosophila. Curr Biol. 2003;13:2025–2036. doi: 10.1016/j.cub.2003.11.018. [DOI] [PubMed] [Google Scholar]
- Wykes SM, Krawetz SA. The structural organization of sperm chromatin. J Biol Chem. 2003;278:29471–29477. doi: 10.1074/jbc.M304545200. [DOI] [PubMed] [Google Scholar]
- Xu Z, Cetin B, Anger M, Cho US, Helmhart W, Nasmyth K, Xu W. Structure and function of the PP2A-shugoshin interaction. Molecular Cell. 2009;35:426–441. doi: 10.1016/j.molcel.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]


