Abstract
Acquisition and quantitative analysis of high resolution images of dendritic spines are challenging tasks but are necessary for the study of animal models of neurological and psychiatric diseases. Currently available methods for automated dendritic spine detection are for the most part customized for 2D image slices, not volumetric 3D images. In this work, a fully automated method is proposed to detect and segment dendritic spines from 3D confocal microscopy images of medium-sized spiny neurons (MSNs). MSNs constitute a major neuronal population in striatum, and abnormalities in their function are associated with several neurological and psychiatric diseases. Such automated detection is critical for the development of new 3D neuronal assays which can be used for the screening of drugs and the studies of their therapeutic effects. The proposed method utilizes a generalized gradient vector flow (GGVF) with a new smoothing constraint and then detects feature points near the central regions of dendrites and spines. Then, the central regions are refined and separated based on eigen-analysis and multiple shape measurements. Finally, the spines are segmented in 3D space using the fast marching algorithm, taking the detected central regions of spines as initial points. The proposed method is compared with three popular existing methods for centerline extraction and also with manual results for dendritic spine detection in 3D space. The experimental results and comparisons show that the proposed method is able to automatically and accurately detect, segment, and quantitate dendritic spines in 3D images of MSNs.
Keywords: dendritic spine, confocal microscopy image, central region extraction, gradient vector flow, fast marching, neurological disease, psychiatric disease
1. Introduction
Dendritic spines are post-synaptic parts of glutamatergic synapses. Dendritic spines undergo activity-dependent structural remodeling, which has been proposed to be involved in learning and memory (De Roo et al., 2008; Yuste and Bonhoeffer, 2001). Spines with larger heads are dynamically more stable, express larger numbers of α-amino-3-hydroxyl-5-methyl-4-isoxazole-propionate type (AMPA-type) glutamate receptors, and contribute to stronger synaptic connections. By contrast, spines with smaller heads contribute to weaker or silent synaptic connections (Kasai et al., 2003). In addition, synaptogenesis associated with experience-dependent learning and environmental complexity is reflected in changes in the number of spines (De Roo et al., 2008; Leuner et al., 2003; Muller et al., 2002). Structural changes in dendritic spines also accompany long-term synaptic plasticity, such as long-term potentiation (LTP) and long-term depression (LTD) of excitatory synaptic transmission (Luscher et al., 2000; Yuste and Bonhoeffer, 2001). LTP-inducing stimuli have been shown to increase the proportion of large spines while LTD-inducing stimuli have been shown to decrease the proportion of large spines and cause retraction of spines (Toni et al., 1999; Zhou et al., 2004). LTP and LTD are thought to be involved in learning and memory (Bliss and Collingridge, 1993; Kandel, 2001).
The striatum is a subcortical region of the cerebrum and is the major input station of the basal ganglia system. Abnormalities in striatal function are associated with neurological and psychiatric diseases (Greengard, 2001), including Parkinsonism, schizophrenia, Attention Deficit Hyperactivity Disorder (ADHD), mental depression, and drug addiction. Compared to other brain regions, the striatum is relatively large and remarkably homogeneous. About 95% of all striatal neurons have a similar morphology and are referred to as medium-sized spiny neurons (MSNs) (Greengard et al., 1999). MSNs receive midbrain dopaminergic input, which serves to modulate excitatory glutamatergic input from the prefrontal cortex (Hyman and Malenka, 2001). The initial site of interaction between dopamine and glutamate is within the dendritic spines of MSNs. Notably, the changes of density or morphology of dendritic spines of MSNs in the striatum have been observed in several disease models and are likely associated with neuronal function and pathology (Day et al., 2006; Deutch et al., 2007; Kalivas, 2009; Kim et al., 2009; Lee et al., 2006; Robinson and Kolb, 2004). Dendritic spine morphology of MSNs is highly heterogeneous compared to that observed in other types of neurons, such as pyramidal neurons in the cerebral cortex and hippocampus (Kim et al., 2009; Shen et al., 2009). Acquisition and accurate analysis of high resolution images of dendritic spines are highly challenging but necessary tasks for better understanding of many neurological and psychiatric diseases. Modern fluorescence microscopy techniques, such as confocal scanning microscopy and 2-photon excitation laser scanning microscopy, provide powerful tools to image neurons at relatively high resolution. With these advanced modern imaging techniques, the analysis of dendritic spines, however, remains largely manual. Manually extracting spine measurements is a labor intensive process, suffers from substantial subjective bias, and often yields inaccurate spine extraction.
In order to overcome the aforementioned difficulties, researchers seek a 2D or 3D automatic way to analyze dendrites and spines. Previous works on dendritic spine detection can be roughly divided into two groups: classification-based methods (Rodriguez et al., 2008) and centerline extraction-based methods (Janoos et al., 2009; Zhang et al., 2007). Classification-based methods separate points into different groups using a certain classifier. For example, Rodriguez et al. (Rodriguez et al., 2008) proposes an automated 3D spine detection approach using point clustering. In this method, only the distances from the points to the closest point of the surface are used as the clustering criteria, which may cause spurious spines. Centerline-extraction based methods detect all the possible centerlines of certain objects in the image and treat dendritic spines as small protrusions attached to the dendrites. Koh et al. (Koh et al., 2002) adopts the thinning method to extract centerlines and applies the grassfire propagation technique to assign each dendritic point a distance to the medial axis of the dendritic structure. Since segmentation is achieved by global thresholding and very limited geometric information is considered for spine detection, this method may detect pseudo spines. Zhou et al. (Zhou et al., 2009) uses a local binary fitting model of level sets to do the spine segmentation, followed by a label-based 3D thinning strategy to do the medial axis extraction and a grassfire approach to do the spine detection. This method requires heavily designed post-processing procedures to remove pseudo medial axes from the 3D thinning method and is limited to processing relatively simple neuron structures. Zhang et al. (Zhang et al., 2007) proposes a 2D tracing algorithm that uses a curvilinear structure detector for dendrite and spine centerline analysis. This method requires several parameters to be fine-tuned and currently works only on 2D projections of images. Thus, 3D information is lost, and spines orthogonal to the projection plane are missed. Janoos et al. (Janoos et al., 2009) presents a method for dendritic skeleton structure extraction using a curve-skeletons approach based on the medial geodesic function which is defined on the reconstructed isosurfaces. Some existing commercial software tools, such as Imaris†, perform semi-automated dendrite and spine analysis and visualization, but they are limited in their scope and capability for fully automated analysis of neuron images with complex neuron structures.
In dendritic spine detection, object centerline extraction plays an important role in analyzing the morphology of dendritic backbones and segmenting dendritic spines. Centerline extraction has been widely used in a variety of application areas. For example, researchers in computer vision utilize object skeletons to specify animation (Bloomenthal, 2002; Siddiqi et al., 2002). In visualization and computer graphics, centerlines are used as a compact representation of complex 3D models (Cornea et al., 2007; Zhou and Toga, 1999). In the medical imaging area, anatomical structure segmentation and modeling of blood vessels and nerve structures involve centerline extraction as a necessary step (Bouix et al., 2005). Centerline extraction has also been used in 3D virtual navigation to utilize symmetric features to generate meaningful paths through a scene or an object (Perchet et al., 2004; Wan et al., 2002). Conventional methods for centerline extraction can be grouped into skeletonization methods and model-based methods. Skeletonization is based on the notion that the geometric medial axis captures the object topology. Lee et al. (Lee et al., 1994) proposes a 3D topological and geometrical preserving thinning method to detect centerlines. Cornea et al.(Cornea et al., 2005) generates a repulsive force field (RFF) over a discretization of the 3D object and uses topological characteristics of the resulting vector field, such as critical points and critical curves to extract the curve-skeleton. Bouix et al.(Bouix et al., 2005) utilizes an average outward flux measure to distinguish skeletal points from non-skeletal ones and combines this measure with a topology-preserving thinning procedure. In practice, the skeletons generated by these aforementioned methods contain many spurious branches due to noise which causes irregularities on the binarized surface. In addition, many real branches are missed.
Model-based methods apply explicit geometric shape models to extract centerlines. Sato et al. (Sato et al., 1998) introduces a multi-scale Gaussian smoothing and Hessian-based technique to model the local line structure for centerline measurements. Frangi et al. (Frangi et al., 1998) proposes a generalized centerline measurement using all eigenvalues simultaneously. Manniesing et al. (Manniesing et al., 2006) improves Frangi et al.’s method by applying a non-linear anisotropic Hessian-based diffusion along the local line directions. The advantages of model-based methods include a natural ability to capture local image features, robustness to noise, and accuracy. However, most of these models on 3D image computing are limited due to increased computation complexity dealing with multi scales and difficulty in scale selection. The recently launched highly automated tool, Neuronstudio (Rodriguez et al., 2008), contains tools aimed at neuron structure tracing and reconstruction. However, it still requires user interaction while the tracing and reconstruction is not optimized for processing MSNs.
In this work, we explore the potential to improve model-based methods and apply it to the application of dendritic spine detection and segmentation in 3D high resolution images of dendritic spines. Figure 1 shows the image processing pipeline of the proposed method. First, a series of pre-processing methods are applied to the input images to remove noise and improve the image quality. Second, a gradient vector field is calculated and normalized using a Generalized Gradient Vector Flow (GGVF) framework with an enhanced smoothing strategy. Then a series of feature points are detected by tracking along the gradient vectors, and these feature points are used for further eigen-based analysis. Third, the eigen-analysis-based method is applied to the feature points to detect spines. The proposed eigen-analysis method is novel compared to previous methods, as it automates the selection of scales when calculating second-order derivatives of the image, and the scale selection is adaptive and specific for each individual point. Since the derivative calculation and eigen-analysis are applied only to those feature points, and scale selection is improved and automated, spine detection is more accurate and faster compared to previous methods. Furthermore, we propose to use a combination of various shape measurements and spine geometric features to improve the spine detection performance. Finally, a fast marching method is employed to segment the spines from the previously detected spine central regions, and spine segmentation is further improved by a series of post-processing steps, including spine head connection and pseudo spine removal.
Fig. 1.
Image Processing Pipeline for 3D dendritic spine detection and segmentation
The remainder of this paper is organized as follows: The next section is composed of our image acquisition method, the detailed spine detection algorithm and discussion on parameter selection. Section 3 reports experimental and comparison results, followed by conclusions in Section 4.
2. Materials and Methods
2.1 Image acquisition and preprocessing
Perfusion of mouse brain and preparation of brain slices labeled with the fluorescent dye, DiI, were described previously (Kim et al., 2009). Mice were anesthetized with sodium pentobarbital and perfused transcardially with 5 ml of PBS, followed by rapid perfusion with 40 ml of 4% paraformaldehyde in PBS (20 ml/min). Brains were quickly removed from the skull and post-fixed in 4% paraformaldehyde for 5 min. Brain slices (100 µm) were labeled by ballistic delivery of fluorescent dye DiI (Molecular Probes). Fluorescent dendritic images of MSNs were taken using a confocal microscope (Zeiss LSM 510) with an oil immersion lens (EC Plan-Neofluar 100×, 1.4 N.A.) and a 2.8× digital zoom. DiI was excited using a Helium/Neon 543 nm laser line. A LP 585 filter was used. Images of distal dendrites (2nd to 4th order dendrites) were examined in this study. For dendritic spines, a stack of images was acquired in the z dimension with an optical slice thickness of 0.12 μm (with oversampling). The resolution along x and y dimensions are 0.06 μm (with oversampling). A typical image volume has a size of 512×512×120 pixels. Based on the observation that a dendritic branch only covers a small part of each image (Figure 2(a)), image cropping is applied to reduce processing time so that only the neuron area is kept for further analysis (Figure 2(b)). To correct the images for the microscope’s point spread function (PSF), which causes out-of-focus objects to appear in the optical slices, a deconvolution method is adopted. Usually, the 3D microscope image h can be represented by the convolution operation between PSF p and the 3D specimen of interest f as:
| (1) |
where ε is the noise. In our experiment, the PSF is set based on the setup of the microscopy, and the de-convolution result is obtained by using Dougherty’s iterative 3D de-convolution method (Dougherty, 2005). In Dougherty’s method, a regularized Wiener filter is used as a preconditioning, and a non-negatively constrained Landweber iteration is adopted to estimate f. Figure 2(c) shows the de-convolved image, which is obtained after 50 iterations. Then, an Otsu thresholding (Otsu, 1979) procedure is employed to separate the neuron points from the background to prepare the data for further gradient vector diffusion, followed by a morphological close operation to fill small holes. In this work, the thresholding-based binarization works well since the intensity values are uniformly distributed in the image, and all foreground points are regions of interest for further analysis.
Fig. 2.
3D view of (a) the original image, with red lines indicating the cropped region, (b) cropped image, (c) deconvolved image
2.2 Feature Point Detection and Automated Scale Selection Using Gradient Vector Tracking
Our previous work (Zhang et al., 2007) has shown the effectiveness of applying model-based methods for dendritic spine detection in 2D space. This previous method models the spines as 2D lines with variational lengths and widths. Spines are detected based on 2nd order derivative calculation and eigen-analysis of the corresponding Hessian matrix. However, this method cannot be effectively extended to 3D space due to the following two reasons: (1) Computationally expensive for multi-scale selection. To effectively calculate the 2nd order derivatives, a proper scale has to be selected for the Gaussian kernel. Since the widths of the objects (including dendrites and spines) may vary greatly, multiple scales have to be used for different Gaussian kernels when dealing with different objects. The state-of-the-art method for multi-scale selection is to search within a given scale space and find the maximum responses according to a certain measurement calculation (Frangi et al., 1998). The searching and calculation is for each location in the image and the scale space usually contains a large number of scales that is enough to find the best one, both of which significantly increase the computational complexity. (2) Complex and variational spine shapes in 3D space. In 3D space, the dendritic spines are seen as different geometric shape structures: for example, as blobs, tubes, plates, and a combination of any of these three shapes. Thus, modeling a spine only as a 3D tube is not accurate enough for spine detection. In this work, we propose a novel method to automatically select scales for eigen-analysis, and the measurement calculations are only applied to a set of feature points. Furthermore, a more complex spine modeling method is explored in 3D space and a new framework is established and optimized for spine detection and segmentation of MSNs specifically.
In our method, spines are first detected, then segmented. Spine detection consists of two steps: (1) gradient vector tracking to find feature points, and (2) Hessian matrix-based eigen-analysis to detect spines. In this work, we use “central region” instead of “centerline” to indicate the central part of the spines due to various spine shapes in 3D space. We use “feature points” to indicate those points that are within or near the central region of the objects. For better computational efficiency, the derivative calculation, eigen-analysis, and spine modeling computing are only applied to the feature points. The method to detect feature points is as follows: first the gradient vector field is calculated; then a tracking from the edge point is applied along the gradient vectors. When the tracking stops under a certain stopping criterion, the current point will be selected as the feature point.
2.2.1 Gradient Vector Calculation
The conventional method to calculate the gradient vector is to convolve the image with derivatives of a Gaussian smoothing kernel (Sato et al., 1998), which requires a proper selection of the standard deviation for the Gaussian kernel. In this work, a new approach is used to avoid using the Gaussian kernel such that no standard deviation selection is required. To calculate the gradient vectors, inspired by the gradient vector field feature-preserving diffusion method (Xu and Prince, 1998), we design a new generalized gradient vector flow (GGVF) based on the distance transform information. The GGVF is defined as the gradient vector field v(x) that minimizes:
| (2) |
where x = (x, y, z) indicates the location of the point and D(x) is the Euclidean distance between the nearest edge point and point x. The weighting function g (·) is defined using the distance as follows:
| (3) |
where K is a positive constant. We will discuss how to select this value in section 2.5. By using the weighting function g (·), the central regions will have bigger smoothing factors compared to the other locations, thus leading to a smoother gradient vector field. The purpose of using the new smoothing criteria is to ensure that the gradient vector is pointing to the central regions of the local objects. Thus, the central regions of those objects can be detected from the gradient vector tracking procedure. Figure 3 compares the normalized vector fields obtained by the proposed GGVF scheme using new smoothing criteria and those obtained by conventional gradient vector flow (GVF) diffusion. It clearly illustrates the effects of the new smoothing criteria. Figure 3(b) and 3(c) show that the vectors obtained by conventional GVF are mostly pointing to the central regions of big spines or dendrites instead of small spines. This is because the conventional GVF field seeks the fastest change of intensity values, which leads the vectors pointing to the global central regions, instead of local central regions. That means the vector tracking can only detect central regions of the big spines and dendrites, but not all the spines. However, the vectors obtained by the proposed GGVF with new smoothing criteria are mostly pointing to central regions of spines (local objects), thus yielding more accurate central region detection results than those from the conventional GVF, as shown in Figure 3(b) and 3(c). After using the GGVF, all the gradient vectors will point to their central region, which is similar to the results using the proper standard deviation by Gaussian convolution. The GGVF field is further normalized as vn(x): vn (x) = v(x)/|v(x)|. The reason to normalize the GGVF field is that it helps to improve the detection of feature points, as only direction information (not magnitude information) is considered in GGVF vector tracking. Figure 4 compares the normalized gradient vector fields calculated with different methods. These methods include Gaussian kernel convolution with a small scale, with a large scale, and the GGVF method. Comparing Figure 4(a2) with Figure 4(c2) illustrates that the normalized gradient vector calculation using GGVF for spine area is equivalent to the Gaussian kernel convolution method with small scale; comparing Figure 4(b1) with Figure 4(c1) illustrates that the normalized gradient vector calculation using GGVF for dendrite area is equivalent to the Gaussian kernel convolution method with large scale. This means that the Gaussian kernel convolution method can achieve similar results on gradient vector calculation, but requires proper selection of multiple scales. The GGVF based method, however, does not require scale selection. Figure 4(a1) and Figure 4(b2) further illustrate that it is critical to select proper scales to obtain desired gradient vectors if using the Gaussian kernel convolution method, thus making the Gaussian-based method undesirable for gradient vector calculation in our application.
Fig. 3.
Comparison of gradient vector fields using the conventional GVF method and proposed GVF with strong smoothing criteria. (a) A test neuron image with gradient vector fields superimposed. (b)&(c) zoomed area 1 and 2 to better visualize gradient vector fields. Vectors in red color are obtained by the conventional GVF method, and vectors in black color are obtained by the proposed method. It is clear that by using the proposed weighting function, the vectors can be further smoothed such that they are better aligned to pointing to the central regions.
Fig. 4.
Normalized gradient vector fields using first-order derivatives of the Gaussian kernel convolved with (a) small scale, (b) large scale, and using (c) Generalized Gradient Vector Flow (GGVF) field. (a1)(a2), (b1)(b2), and (c1)(c2) are the enlarged areas of region 1 and 2 in (a), (b), and (c) respectively. Comparing (a2) with (c2) illustrates that the normalized gradient vector calculation using GGVF for spine area is equivalent to the Gaussian kernel convolution method with a small scale; by comparing (b1) with (c1) it illustrates that the normalized gradient vector calculation using GGVF for dendrite area is equivalent to the Gaussian kernel convolution method with a large scale. This shows the advantage of the proposed gradient vector calculation method: that there is no need to select multiple scales. (a1) and (b2) illustrate that proper scales have to be selected to obtain desired gradient vectors if using conventional calculation methods.
2.2.2 Feature Point Detection Using Gradient Vector Tracking
In the gradient vector field, the vectors smoothly point toward the central regions of the object (see Figure 3 and Figure 4). To detect the rough central regions (i.e., feature points), a gradient vector tracking procedure is proposed. The tracking procedure is as follows. First, the normalized vector field vn(x) = [u,v,w] is quantized into one of the 26 different directions corresponding to the 26-connected points in 3D space to indicate which one of the 26 neighborhoods x points to. The quantized discrete direction vector d(x) = [du,dv,dw] is calculated as follows:
| (4) |
and dv,dw are calculated in a similar way. By using direction quantization, searching among all the 26 neighborhoods is avoided. Then, from a starting point x0 on the surface of the foreground with a discrete vector d(x0), the next point x ' on the path is computed as:
| (5) |
At each new location x′, the angle between the two gradient vectors of x′ and its preceding x is calculated as cos−1(v(x′)·v(x)). If this angle is greater than 90°, both x′ and x will be detected as feature points and the tracking stops. Otherwise, the tracking continues until feature points are detected. Figure 5 shows an example of the feature point detection results, and it is clear that most of the detected feature points are on or near the central region of the objects. Once a tracking stops, the Euclidian distance between the starting point x0 and the detected feature point xf is calculated and stored for further scale selection.
Fig. 5.
Results of the detected feature points. (a) using gradient vector tracking, the arrows indicate some of the non-central points; (b) detected spine central regions after thresholding shape measurements and removing dendrite central regions. Regions belonging to different spines are labeled in different colors. It is clear that touching spines inside the red circle in (b) are well-separated by the gradient vector tracking and non-central points are removed.
2.3 Spine Detection Using Variational Shape Modeling
After the tracking procedure, the detected feature points are on or near the central regions of the objects. However, not all the feature points belong to spines. See Figure 5 for example: some feature points belong to pseudo branches; some belong to dendrites and other objects. In this work, we use shape information to detect spines. To achieve better spine detection, a variational shape modeling concept is proposed to further detect the spine central regions from these feature points. In 3D neuron images, a spine is modeled as a tubular shape, a blob shape, a plate shape, or a combination of any of the previous three variational shapes. To detect these variational shapes, we apply an eigen-analysis-based method using the Hessian matrix and a shape measurement calculation. A Hessian matrix is calculated using the second order derivatives at point x:
| (6) |
where the second order derivatives are calculated as the image convolved with the derivatives of a Gaussian kernel Gσ(x). That is: ∂xxI(x) = I(x)*∂xxGσ(x) and * is the convolution operator. In this work, the image derivatives are calculated only at the feature points instead of over the entire image domain to reduce the computational load. The derivatives at a single point using the Gaussian kernel-based method are calculated as follows:
| (7) |
Other derivatives along different directions are calculated in a similar way. The selection of the scale σ for the Gaussian kernel is unavoidable here, as the proper selection of σ is very important to correctly calculate the derivatives and to obtain the maximum feature responses which are calculated based on the derivatives. In previous work (Zhang et al., 2007), we have proposed to use a scale searching method to find the best scale for a point x. Because it is expensive for computation and memory usage, this method is not suitable for 3D volume images. In this work, we develop another method to select a proper scale value for each feature point. The scale selection method is based on the fact that the proper scale of the Gaussian kernel is related to the sizes of the objects. We regard the cross-section of a dendrite or spine as an ellipse. The relationship between the two diameters of the ellipse (transverse diameter dt and conjugate diameter dc) and the best scale σo is obtained by examining the maximum measurements calculated using Angita’s shape measurement (Antiga, 2007) for synthetic testing images with variational ellipse-shape cross-sections. To better mimic the possible sizes of dendrites and spines in neuron images, the values for dt and dc are selected as dt = {1,2,…,30}, dc = {1,2,…,30}. These values are maintained in a table, and Figure 6 displays a surface plot of the table of the best scale corresponding to dt and dc. In practice, when a pair of real dt and dc is known, the real σo value is selected by nearest-neighbor interpolation using the four nearest neighbor grid points in the relationship table. The problem is then how to determine dt and dc for each feature point. During the gradient vector tracking procedure, the Euclidian distance between the tracking starting point x0 and the detected feature point xf is calculated. For each xf, there exist multiple such distances as multiple tracking may find the same feature point. Denoting the multiple distances as {ei},i = 1,…,L for a feature point, the pair of dt and dc for that point can be obtained by the following method:
| (8) |
Fig. 6.
Surface plot of the best scale selection according to different combinations of transverse and conjugate diameters.
Once the Hessian matrix H(xf)is calculated for each feature point xf, a special feature response, called the shape measurement, is calculated using the three eigenvalues of H(xf). The shape measurement is a value between 0 to 1 indicating what the probability is for a point belonging to a certain shape. Denoting the three eigenvalues of H(xf) as λ1,λ2λ3 in ascending order (|λ1|≤|λ2|≤3|), the shape measurement MS is calculated using Antiga’s method (Antiga, 2007):
| (9) |
where S is an integer to represent a particular shape (S = 0 for blobs, S = 1 for tubes, and S = 2 for plates). The expressions for RA,RB,RC are as follows:
| (10) |
The parameters α,β,γ are all fixed at 0.5 in our experiments. Since the scale is pre-selected and the eigen-analysis method is applied only to feature points, the accuracy and speed are significantly improved in comparison to the previous methods (Frangi et al., 1998; Sato et al., 1998).
For dendrites, only the tube measurement (M1) is considered since the dendrites can be modeled as tube-like objects; for dendritic spines, various measurements need to be considered since spines may have different geometric shapes. In this work, each voxel in the detected set of feature points is assigned a shape measurement to represent how close it is to a certain shape. Since spines have various shapes, i.e., it could be a blob, a tube, a plate, or a combination of these three shapes, we use the maximum shape measurements among all three shapes as the final shape measurement for a certain voxel. That means a spine could be partly tube and partly blob as an example. Thus, the shape measurement could be seen as a certainty value assigned to each voxel. The maximum value of all possible shape measurements is selected as follows to improve the detection of spines of various shapes:
| (11) |
where M is the shape measurement for a voxel. Since the calculation for dendrite measurement is different from that for spines, it is necessary to distinguish spine feature points from dendrite feature points. Based on the observation that dendrites usually have much bigger radii than spines, a threshold value is set for the radii (in terms of the Euclidian distances) obtained by the gradient vector tracking. Feature points with radii less than the threshold value will be treated as spine feature points; the rest will be dendrite feature points. After the measurements are obtained, the central regions of dendrites and spines can be detected by discarding those feature points with very small measurement values (ie. thresholding). Figure 5(b) shows the detected spine central regions after removing the dendrite backbones and the points with small shape measurements. Certain feature points that are not near the central regions are removed, while spines with various shapes are kept. Also, as illustrated in the red circle of Figure 5(b), the touching spines are well-separated using gradient vector-based tracking.
In summary, two separate steps, gradient vector tracking and eigen-analysis, complete the spine detection. The tracking step is to effectively find points on or near object central regions and the eigen-analysis step is to filter the points to find those belonging to spines. Both steps involve image derivative calculations which capture the image local shape and intensity features. The advantages of the proposed method are that scale selection is automated, and the calculation is only applied to the points close to the central regions of the objects, which significantly improves the computational efficiency.
2.4 Spine Segmentation using Fast Marching
Once spine central regions are detected, the fast marching algorithm is applied to solve spine segmentation. In this work, spine segmentation is modeled as a moving boundary problem. The moving boundary problem is that, given an initial boundary (i.e., a boundary partitioning an image into two regions) and a speed at each point on the boundary to specify the movement of the point, how to track the location of the moving boundary at a certain time. In dealing with moving boundaries, partial differential equation-based numerical methods usually build computational fluid equations to model the motion and solve them using techniques based on high-order upwind formulations. Such a numerical method has two complementary approaches, namely, the fast marching approach for certain specialized front problems and the level set approach for general front problems. The fast marching method (Sethian, 1996) is designed to deal with the situation that the boundary is always moving forward or backward. Instead of directly calculating the position of the boundary at a certain time as the conventional level set approach, the fast marching method computes the arrival time of each voxel first, and then obtains the moving boundary by extracting the locations at a certain time by interpolation. Defining a seed point as the point with the minimum arrival time, the fast marching method calculates the arrival time in an iterative way. During each iteration, only the arrival time of the neighborhood points of the current seed point is updated to speed up the evolution.
Theoretically, given an initial boundary B0 = {x,x = (x, y, z)} and a speed function Gx which indicates how fast a point x moves along its normal direction, B0 can be viewed as the zero level set of a higher dimensional function Φ x(t) at time t = 0. The level set function can be formulated as follows,
| (12) |
If Gx > 0, i.e., the boundary always moves forward, the fast marching approach can be applied to solve the level set equation (12) very efficiently. Denote Tx as the time used at which the boundary crosses the point x, then |∇Tx|Gx = 1, which means the gradient of the arriving time is inversely proportional to the speed of the boundary evolution (Sethian, 1996). Thus, the problem can be described as the evolution of a closed boundary as a function of time Tx with speed Gx in its normal direction. In our application, an initial closed boundary is the detected spine central points (inside the spine central regions); the speed function Gx is specified by the normalized gradient vectors obtained by the GGVF method (see Section 2.2); the time at which the closed boundary crosses a point can be numerically calculated by the fast marching method. Thus, the problem of spine segmentation is equivalent to a problem of finding the time that it takes each spine central point to evolve to other points in the whole volume. The input to the fast marching segmentation is the set of detected spine central regions, and the output will be the segmented spines. All points with evolution time less than infinity will be segmented as spine points. Spine segmentation using the fast marching technique can be summarized by three steps:
Step 1: Initialization
During initialization, all points are assigned with initial values of Tx and Sx, where Tx represents the time at which the boundary crosses the point x, and Sx represents the current status of point x. Here Sx ∈{PROCESSED, FREE, Jx}, where Jx is an index value representing the current index of the point x in a queue. For background points and spine central points, Sx = PROCESSED, indicating that these points will not be considered in the fast marching evolution; for all other points, Sx = FREE, indicating that these points are currently available and should be considered for fast marching evolution. If Sx has any value other than PROCESSED and FREE, then it indicates the corresponding point has been processed before and the new value of Tx should be compared with the previous one to determine whether to update Tx. For spine central points, Tx are initialized as zero since these points will be used as the starting points (called seed points) for the fast marching method; Tx are set to be INFINITE for all of the other points (called infinite points).
Step 2: Fast Marching Evolution
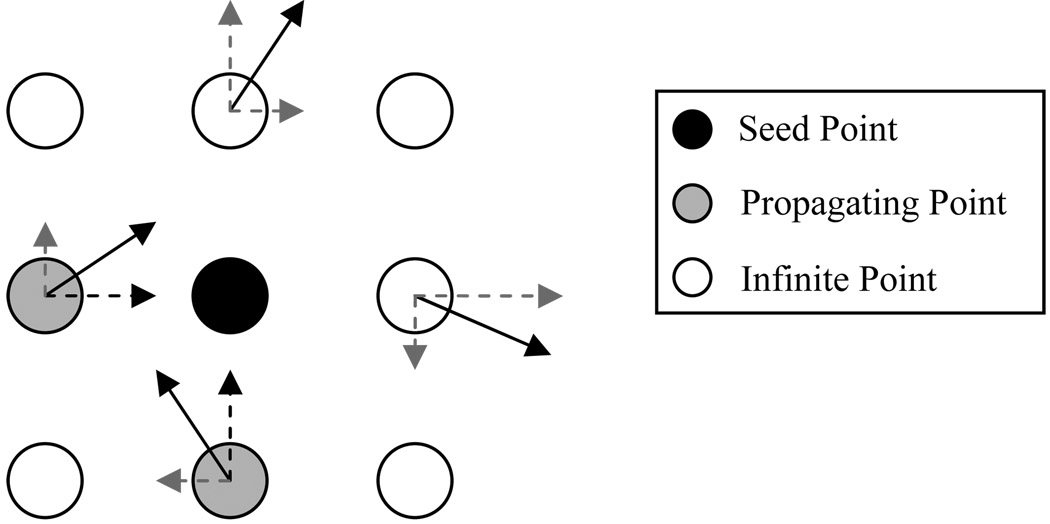
The fast marching starts at a seed point, i.e., each of the spine central points. A queue is maintained to hold all the points in the fast marching process. The marching time Tx is quantized to an integer value and used as the index in the queue. In our application, the marching is considered separately for x-, y-, and z- directions, For each x-, y-, or z- direction, there are only two possibilities for the current point to march: forward or backward. Thus, only the 6-neighborhood is considered in the 3D space: xk = {x1,2(x±1,y,z,),x3,4(x,y±1,z),x5,6(x,y,z±1)}, where (x,y,z) is the spatial location for the current starting point s. For each neighbor point xk,k =1,2,…,6, denote its normalized gradient vector flow as vn(xk) = [u(xk),v(xk),w(xk)], where u(xk),v(xk),w(xk) represents the speed values along the x-, y-, and z- directions, respectively. If its status is not processed (Sx = FREE or Jx) and any one of u(xk),v(xk),w(xk) points to the current seed point, then a new time value is calculated and compared with the previous time value for this neighbor point. Such a neighbor point is then called a propagating point. Figure 7 illustrates the fast marching propagation in the 2D case. The new time value T̃xk is calculated as follows:
| (13) |
where rx,y,z are the spacing values along the x-, y-, or z- directions, respectively. If T̃xk <T̃xk, then point xk is updated with T̃xk and added to the queue using T̃xk as the index. If point xk is in the queue already, then the position of xk is updated based on the new marching time T̃xk. All six neighbor points are processed to find all the possible propagating points along the x-, y-, and z- directions in 3D space.
Fig. 7.
Illustration for fast marching propagation in the 2D case. The black point indicates the current seed point and the solid arrows in its 4-neighborhood points represent their normalized GGVF vectors. The dashed arrows represent projected components of the corresponding GGVF vectors along the x- or y- directions. If any of the projected components points to the seed point, the neighbor point will be considered for time value calculation and comparison with the previous time value. Such a neighbor point is then called a propagating point.
The points in the queue are processed using the described fast marching technique one-by-one, based on their index values. The point with the smallest marching time (i.e., index values) will be processed first. Each time a point is processed, it is removed from the queue and set Sx = PROCESSED. The processing continues until the queue is empty. Once the queue is empty, the next seed point will be processed to create a new queue and the above queue-based fast marching process is repeated.
Step 3: Spine Segmentation
Once all the seed points are processed using step 1 and step 2, each point in the 3D neuron image will have a Tx with a value from zero to INFINITE. To segment a spine is straightforward: every time a spine central region is processed, all the points with marching time Tx that are smaller than INFINITE are regarded as the spine points. Then, a unique label value is set to Tx for all of these points to indicate that they have been processed and belong to a spine. The advantages of using fast marching for spine segmentation are that the segmentation is rapid and complete and the attached spines can be separated as they have different initial boundaries and the tracking is not merged.
2.5 Parameter Selections
The proposed method requires minimal parameter selection. In GGVF diffusion, the only parameter to set is the constant K. This value is selected based on experiments. Figure 8 illustrates the diffusion fields with different K values (0.1, 0.5 and 2) on two test images. Figure 8(a) shows that if K is too small (K = 0.1), the function g would vary sharply, which causes the diffusion to be unsmooth; on the other hand, if K is set at a large value (K = 2), our proposed GGVF will have the same form as the conventional GVF method, as shown in Figure 8 (c). The experiments show that 0.5 is a good choice for K.
Fig. 8.
Two examples of GGVF diffusion fields calculated using different values of parameter K with (a) K=0.1; (b) K=0.5 and (c) K=2. If K is too small (K=0.1), the field is not smoothed; If K is set a big value (K=2), the diffused vectors are pointing to big spines or dendrites instead of local spines, similar to with conventional GVF vectors. According to the experiments, 0.5 is a good choice for K.
For central region detection, a threshold value is used to filter out non-central region points with low shape measurements. We manually select this threshold based on the observation of shape measurement from one training data and apply the same value on all the other data. Although the accuracy can be further improved by tuning this parameter for each individual image, the current method of using a fixed threshold obtained from a testing image is good enough to be used in various neuron images.
Another threshold value is used to distinguish dendrite feature points and spine feature points based on their transverse distances. Based on the observation that the dendrites are always much bigger than the spines, a value which is smaller than the dendrite radii is adopted. This value should be selected based on the width of the dendrite. If this value is set too large, some dendrite central region will be treated as spine central region; if it is too small, big spines may be missed. In our experiments, this value is determined based on the distribution of transverse distance of the spines and dendrites and applied to all the neuron images.
3. Experimental results
The proposed method is applied to fifteen dendritic images of MSNs for dendritic spine segmentation. For spine and dendrite central region extraction, the proposed method is compared with three other methods, namely, the 3D Thinning Algorithm (Lee et al., 1994), 3D Skeletonization with repulsive force field (RFF) (Cornea et al., 2005), and the flux driven centerline extraction method (Bouix et al., 2005). The parameters for these methods have been carefully selected to obtain relatively good results. As an example, for the 3D Skeletonization with RFF, the “percentage of high divergence points to return” is set to be 50%, which should return a more detailed skeleton. Figure 9 compares extracted central regions among different methods. The comparisons show that previous methods detect a lot of branches and some central regions are missing, while our proposed method is able to extract dendrite and spine central regions more accurately and completely. Table 1 compares the running time (in minutes) for the four centerline extraction methods on a PC with 2.66 GHz CPU and 4.0 Gigabytes RAM.
Fig. 9.
Comparison of central region extraction using (a)(e) 3D thinning, (b)(f) the flux driven method, (c)(g) 3D skeletonization with RFF, and (d)(h) the proposed method before backbone removal. The extracted results are shown in red.
Table 1.
Average CPU time for central region extraction using different methods.
| Methods | Thinning | Flux | Skeletonization with RFF | Proposed method |
|---|---|---|---|---|
| Processing Time(min) | 3 | 25 | 45 | 3 |
Figure 10 shows representative segmentation results using our proposed method. The segmented spines are visualized using surface rendering and labeled in color (the selection of a color for a specific spine has no meaning here). Note that some of the images are orientated vertically to save space. Figure 11 displays partial spine segmentation results, and each image is zoomed-in such that various spine shapes and their segmentations can be better inspected. Various spine shapes, including branching spines (marked with yellow arrow), plate-like spines (stubby type, marked with brown arrow), bulb-like spines (mature and thin spines, marked with pink arrow), and tube-like spines (filopodia, marked with black arrow), are represented in these images.
Fig. 10.
Spine segmentation results using our proposed method. The segmented spines are visualized using surface rendering and labeled in color (the selection of a color for a specific spine has no meaning here). Note that some of the images are orientated vertically to save space for display.
Fig. 11.
Partial spine segmentation results. Each dendrite is zoomed in such that various spine shapes and their segmentations can be better inspected. This figure illustrates various spine shapes including branching spines (yellow arrow), plate-like spines (brown arrow), bulb-like spines (pink arrow), and tube-like spines (black arrow).
Table 2 shows a detailed quantitative comparison for each test image between our proposed method and the manual segmentation results. In our experiment, one expert labels the spines on the de-convolved images manually, using the software ITK-Snap ‡ (Yushkevich et al., 2006). First, these spines are labeled using 6 kinds of labels (represented by different colors in ITK-Snap), where the touching spines are labeled with different colors. Thus, no touching spines exist on each label channel. Then, based on the region connectivity, each spine is assigned with a distinct integer value. The final manual segmentation result is the combination of the labeled images of 6 channels. In this quantitative comparison, true positive (TP) is defined as those spines that are detected accurately and completely, i.e., the overlap between the segmented results and the manual results are at least 80%; partial detected (PD) as incomplete detection (i.e., 20% < overlap < 80%); miss as the overlap less than 20%; over-segmentation (OS) if one spine is segmented as multiple ones; and under-segmentation (US) if several spines are segmented as one. Table 3 compares segmentation results in terms of the total number of true or false spines. True negative (TN) is defined as the total of PD, miss, OS, and US; false positive (FP) is defined as those detected by the proposed method but not detected by the manual method. The overall sensitivity for our method is 87.0% while the positive predictive value is 90.4%. These results show that our proposed fully-automated method is able to segment spines and dendrites more accurately and completely from neuron images with complicated spine structures.
Table 2.
Comparison of spine detection between manual and our proposed method. TP: True Positive; PD: Partially Detected; OS: Oversegmentation; US: Undersegmentation.
| Data No. | Manual | Proposed Method | |||||
|---|---|---|---|---|---|---|---|
| Number | TP | PD | OS | US | Miss | ||
| 1 | 43 | 47 | 41 | 0 | 5 | 0 | 0 |
| 2 | 44 | 44 | 43 | 0 | 0 | 0 | 1 |
| 3 | 47 | 54 | 43 | 1 | 6 | 1 | 0 |
| 4 | 71 | 73 | 66 | 0 | 6 | 1 | 0 |
| 5 | 65 | 64 | 54 | 0 | 3 | 4 | 1 |
| 6 | 88 | 94 | 87 | 0 | 6 | 0 | 0 |
| 7 | 100 | 92 | 84 | 4 | 2 | 2 | 6 |
| 8 | 60 | 66 | 57 | 2 | 2 | 0 | 1 |
| 9 | 56 | 61 | 54 | 0 | 3 | 0 | 1 |
| 10 | 74 | 78 | 70 | 0 | 5 | 0 | 1 |
| 11 | 98 | 106 | 96 | 1 | 3 | 0 | 0 |
| 12 | 104 | 103 | 77 | 2 | 7 | 9 | 0 |
| 13 | 124 | 130 | 118 | 3 | 6 | 0 | 0 |
| 14 | 76 | 80 | 69 | 0 | 7 | 2 | 0 |
| 15 | 73 | 75 | 56 | 3 | 8 | 4 | 0 |
| Total | 1123 | 1167 | 1015 | 16 | 69 | 23 | 11 |
Table 3.
Spine number quantitative results.
| Proposed Method | |||
|---|---|---|---|
| Positive | Negative | ||
| Manual | True | 1015 | 108 |
| False | 152 | - | |
The contributions of the proposed method include a new framework of central regions detection with automated parameter selection and improved computational efficiency, a new method for detecting objects with variational shapes, and a new application of the fast marching algorithm. We believe that the proposed method can be used for other applications of object detection and segmentation because:
The method is fully automated and requires no user interaction.
The method is very computationally efficient, making it more practical to be used on various types of images and in 3D space than other methods.
The method is able to effectively detect objects with variational shapes and sizes. By extending the shape measurement using equations 9 and 10, the method can be used to detect objects with other shapes.
4. Conclusion
In this work, we propose a new method for dendritic spine detection and segmentation in 3D confocal microscopy images of MSNs. This method detects central regions for both dendrites and spines using gradient vector tracking and feature point detection. An eigen-analysis-based method is applied to each feature point to calculate a shape measurement and to remove those feature points that are not on or near the central regions of the spines. The spine central regions are then used as seed points for spine segmentation using the fast marching method. Our contributions include the proposed gradient vector tracking method for detecting dendrite and spine central regions; the automated scale selection for the Gaussian kernels for the eigen-analysis based method; and spine detection based on variational shape modeling. It is also the first successful application of the fast marching algorithm to solve spine segmentation problems. The advantages of the proposed method are that it is fully automated, requires only minimal parameter selection, and can accurately and completely detect and quantify dendritic spines of various shapes. The experimental results and comparisons show that the proposed method achieves good performance in terms of dendritic spine segmentation of MSNs in 3D space. Future work includes expanding our methods for other types of neurons or more complex assays. One possible expansion would be the design of methods to analyze dendritic spines co-stained with synaptic boutons, knoblike enlargements at the end of axons and/or with neurotransmitter receptors. Such methodological development would allow us to analyze not only the morphological aspect but also the functional state of synapses.
Acknowledgements
This research was supported by funding from the National Institutes of Health-R01 LM009161 and R01 AG028928 (Stephen Wong), and the National Institute on Drug Abuse-DA10044 (Yong Kim and Paul Greengard).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Imaris: http://www.bitplane.com/
ITK-Snap: http://www.itksnap.org/
References
- Antiga L. Generalizing vesselness with respect to dimensionality and shape. The Insight Journal 2007 July–December Issue. 2007 [Google Scholar]
- Bliss TV, Collingridge GL. A synaptic model of memory: long-term potentiation in the hippocampus. Nature. 1993;361:31–39. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- Bloomenthal J. Medial-based vertex deformation. Proceedings of the 2002 ACM SIGGRAPH/Eurographics symposium on Computer animation; ACM; San Antonio, Texas. 2002. p. 151. [Google Scholar]
- Bouix S, Siddiqi K, Tannenbaum A. Flux driven automatic centerline extraction. Medical Image Analysis. 2005;9:209–221. doi: 10.1016/j.media.2004.06.026. [DOI] [PubMed] [Google Scholar]
- Cornea N, Silver D, Min P. Curve-skeleton properties, applications, and algorithms. IEEE Transactions on Visualization and Computer Graphics. 2007;13:530–548. doi: 10.1109/TVCG.2007.1002. [DOI] [PubMed] [Google Scholar]
- Cornea ND, Silver D, Yuan X, Balasubramanian R. Computing hierarchical curve-skeletons of 3D objects. The Visual Computer. 2005;21:945–955. [Google Scholar]
- Day M, Wang Z, Ding J, An X, Ingham C, Shering A, Wokosin D, Ilijic E, Sun Z, Sampson A. Selective elimination of glutamatergic synapses on striatopallidal neurons in Parkinson disease models. Nature neuroscience. 2006;9:251–259. doi: 10.1038/nn1632. [DOI] [PubMed] [Google Scholar]
- De Roo M, Klauser P, Garcia PM, Poglia L, Muller D. Spine dynamics and synapse remodeling during LTP and memory processes. Prog Brain Res. 2008;169:199–207. doi: 10.1016/S0079-6123(07)00011-8. [DOI] [PubMed] [Google Scholar]
- Deutch AY, Colbranc RJ, Winder DJ. Striatal plasticity and medium spiny neuron dendritic remodeling in parkinsonism. Parkinsonism & Related Disorders. 2007;13:S251–S258. doi: 10.1016/S1353-8020(08)70012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty RP. Extensions of DAMAS and Benefits and Limitations of Deconvolution in Beamforming. AIAA Paper 2961. 2005 [Google Scholar]
- Frangi AF, Niessen WJ, Vincken KL, Viergever MA. Multiscale Vessel Enhancement Filtering; Proceedings of the MICCAI'98; 1998. pp. 130–137. [Google Scholar]
- Greengard P, Allen P, Nairn A. Beyond the dopamine receptor: the DARPP-32/protein phosphatase-1 cascade. Neuron. 1999;23:435–447. doi: 10.1016/s0896-6273(00)80798-9. [DOI] [PubMed] [Google Scholar]
- Hyman S, Malenka R. Addiction and the brain: the neurobiology of compulsion and its persistence. Nature Reviews Neuroscience. 2001;2:695–703. doi: 10.1038/35094560. [DOI] [PubMed] [Google Scholar]
- Janoos F, Mosaliganti K, Xu X, Machiraju R, Huang K, Wong STC. Robust 3D reconstruction and identification of dendritic spines from optical microscopy imaging. Medical Image Analysis. 2009;13:167–179. doi: 10.1016/j.media.2008.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalivas P. The glutamate homeostasis hypothesis of addiction. Nature Reviews Neuroscience. 2009;10:561–572. doi: 10.1038/nrn2515. [DOI] [PubMed] [Google Scholar]
- Kandel ER. The molecular biology of memory storage: a dialogue between genes and synapses. Science. 2001;294:1030–1038. doi: 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- Kasai H, Matsuzaki M, Noguchi J, Yasumatsu N, Nakahara H. Structure-stability-function relationships of dendritic spines. Trends Neurosci. 2003;26:360–368. doi: 10.1016/S0166-2236(03)00162-0. [DOI] [PubMed] [Google Scholar]
- Kim Y, Teylan M, Baron M, Sands A, Nairn A, Greengard P. Methylphenidate-induced dendritic spine formation and DeltaFosB expression in nucleus accumbens. Proceedings of the National Academy of Sciences. 2009:2915–2920. doi: 10.1073/pnas.0813179106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh IYY, Lindquist WB, Zito K, Nimchinsky EA, Svoboda K. An image analysis algorithm for the fine structure of neuronal dendrites. Neural Comput. 2002;14:1283–1310. doi: 10.1162/089976602753712945. [DOI] [PubMed] [Google Scholar]
- Lee K, Kim Y, Kim A, Helmin K, Nairn A, Greengard P. Cocaine-induced dendritic spine formation in D1 and D2 dopamine receptor-containing medium spiny neurons in nucleus accumbens. Proceedings of the National Academy of Sciences. 2006;103:3399–3404. doi: 10.1073/pnas.0511244103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TC, Kashyap RL, Chu CN. Building skeleton models via 3-D medial surface/axis thinning algorithms. Computer Vision, Graphics, and Image Processing. 1994;56:462–478. [Google Scholar]
- Leuner B, Falduto J, Shors TJ. Associative memory formation increases the observation of dendritic spines in the hippocampus. J Neurosci. 2003;23:659–665. doi: 10.1523/JNEUROSCI.23-02-00659.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscher C, Nicoll RA, Malenka RC, Muller D. Synaptic plasticity and dynamic modulation of the postsynaptic membrane. Nat Neurosci. 2000;3:545–550. doi: 10.1038/75714. [DOI] [PubMed] [Google Scholar]
- Manniesing R, Viergever M, Niessen W. Vessel enhancing diffusion A scale space representation of vessel structures. Medical Image Analysis. 2006;10:815–825. doi: 10.1016/j.media.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Muller D, Nikonenko I, Jourdain P, Alberi S. LTP, memory and structural plasticity. Curr Mol Med. 2002;2:605–611. doi: 10.2174/1566524023362041. [DOI] [PubMed] [Google Scholar]
- Otsu N. A threshold selection method from gray-level histograms. Systems, Man and Cybernetics, IEEE Transactions on. 1979;9:62–66. [Google Scholar]
- Perchet D, Fetita C, Prêteux F. SPIE: Image Processing. CA, USA: Algorithms and Systems III San Jose; 2004. Advanced navigation tools for virtual bronchoscopy. [Google Scholar]
- Robinson T, Kolb B. Structural plasticity associated with exposure to drugs of abuse. Neuropharmacology. 2004;47:33–46. doi: 10.1016/j.neuropharm.2004.06.025. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Ehlenberger DB, Dickstein DL, Hof PR, Wearne SL. Automated Three-Dimensional Detection and Shape Classification of Dendritic Spines from Fluorescence Microscopy Images. PLoS ONE. 2008;3:e1997. doi: 10.1371/journal.pone.0001997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Nakajima S, Shiraga N, Atsumi H, Yoshida S, Koller T, Gerig G, Kikinis R. 3D multi-scale line filter for segmentation and visualization of curvilinear structures in medical images. Medical Image Analysis. 1998;2:143–168. doi: 10.1016/s1361-8415(98)80009-1. [DOI] [PubMed] [Google Scholar]
- Sethian J. A fast marching level set method for monotonically advancing fronts. Proceedings of the National Academy of Sciences. 1996;93:1591–1595. doi: 10.1073/pnas.93.4.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Toda S, Moussawi K, Bouknight A, Zahm D, Kalivas P. Altered dendritic spine plasticity in cocaine-withdrawn rats. Journal of Neuroscience. 2009;29:2876–2884. doi: 10.1523/JNEUROSCI.5638-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqi K, Bouix S, Tannenbaum A, Zucker S. Hamilton-jacobi skeletons. International Journal of Computer Vision. 2002;48:215–231. [Google Scholar]
- Toni N, Buchs PA, Nikonenko I, Bron CR, Muller D. LTP promotes formation of multiple spine synapses between a single axon terminal and a dendrite. Nature. 1999;402:421–425. doi: 10.1038/46574. [DOI] [PubMed] [Google Scholar]
- Wan M, Liang Z, Ke Q, Hong L, Bitter I, Kaufman A. Automatic centerline extraction for virtual colonoscopy. IEEE Trans. Med. Imaging. 2002;21:1450–1460. doi: 10.1109/TMI.2002.806409. [DOI] [PubMed] [Google Scholar]
- Xu C, Prince J. Generalized gradient vector flow external forces for active contours. Signal Processing. 1998;71:131–139. [Google Scholar]
- Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. NeuroImage. 2006;31:1116–1128. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
- Yuste R, Bonhoeffer T. Morphological changes in dendritic spines associated with long-term synaptic plasticity. Annu Rev Neurosci. 2001;24:1071–1089. doi: 10.1146/annurev.neuro.24.1.1071. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhou X, Witt RM, Sabatini BL, Adjeroh D, Wong STC. Dendritic spine detection using curvilinear structure detector and LDA classifier. Neuroimage. 2007;36:346–360. doi: 10.1016/j.neuroimage.2007.02.044. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Homma KJ, Poo MM. Shrinkage of dendritic spines associated with long-term depression of hippocampal synapses. Neuron. 2004;44:749–757. doi: 10.1016/j.neuron.2004.11.011. [DOI] [PubMed] [Google Scholar]
- Zhou W, Li H, Zhou X. 3D neuron dendritic spine detection and dendrite reconstruction. International Journal of Computer Aided Engineering and Technology. 2009;1:516–531. [Google Scholar]
- Zhou Y, Toga A. Efficient skeletonization of volumetric objects. IEEE Transactions on Visualization and Computer Graphics. 1999;5:196–209. doi: 10.1109/2945.795212. [DOI] [PMC free article] [PubMed] [Google Scholar]