Abstract
The function of microRNAs (miRNAs) in hematopoietic stem cells (HSCs), committed progenitors, and leukemia stem cells (LSCs) is poorly understood. We show that miR-29a is highly expressed in HSC and down-regulated in hematopoietic progenitors. Ectopic expression of miR-29a in mouse HSC/progenitors results in acquisition of self-renewal capacity by myeloid progenitors, biased myeloid differentiation, and the development of a myeloproliferative disorder that progresses to acute myeloid leukemia (AML). miR-29a promotes progenitor proliferation by expediting G1 to S/G2 cell cycle transitions. miR-29a is overexpressed in human AML and, like human LSC, miR-29a-expressing myeloid progenitors serially transplant AML. Our data indicate that miR-29a regulates early hematopoiesis and suggest that miR-29a initiates AML by converting myeloid progenitors into self-renewing LSC.
Mature hematopoietic cells arise from hematopoietic stem cells (HSCs) though a hierarchically organized step-wise developmental process that proceeds through increasingly lineage-restricted progenitors that exhibit progressively decreasing proliferative capacity (Kondo et al., 2003; Bryder et al., 2006). Although both HSC and hematopoietic progenitors have the potential to develop into multiple types of hematopoietic cells, HSCs are the only cells capable of self-renewal, a property essential to their ability to maintain life-long hematopoiesis. Self-renewal is also observed in the malignant counterparts of HSC/progenitors in human acute myeloid leukemia (AML). In AML, leukemia stem cells (LSCs), or leukemia-initiating cells, are postulated to initiate a developmental hierarchy in which LSCs give rise to more differentiated non–self-renewing myeloid lineage blasts that exhibit varying degrees of maturation arrest (Lapidot et al., 1994; Bonnet and Dick, 1997). Within the leukemic blast population, only LSCs are able to self-renew. Because LSCs likely arise through the acquisition of multiple genetic and/or epigenetic changes in normal HSC/progenitors (Reya et al., 2001; Passegué et al., 2003; Weissman, 2005), understanding the mechanisms that control self-renewal and lineage commitment in normal HSC/progenitors is of fundamental importance to the prevention and treatment of hematopoietic disorders.
microRNAs (miRNAs) regulate numerous cellular processes, including proliferation, differentiation, and apoptosis (He and Hannon, 2004; Chang and Mendell, 2007), and they do so by regulating gene expression at the translational or posttranscriptional level by repressing translation from protein-encoding messenger RNAs (mRNAs) or by promoting degradation of their target mRNAs (Bartel, 2004). Given that miRNA-mediated gene regulation usually depends on imperfect match of its seed sequences of ∼6–8 nt with their target sequences within the 3′UTR of the target mRNA, one miRNA may simultaneously regulate multiple targets in the same cell (Lewis et al., 2005; Lim et al., 2005; Baek et al., 2008; Selbach et al., 2008). In the hematopoietic system, miRNAs are differentially expressed in mature cells of the hematopoietic system (Monticelli et al., 2005) and, in some cases, have been shown to regulate lineage commitment and mature effector cell function (Chen and Lodish, 2005; Garzon and Croce, 2008). However, relatively little is known about miRNA function in HSC, lineage committed progenitors, or their presumed malignant counterpart, the LSC. miRNAs are thought to play an important role in carcinogenesis, indicated by frequent deletion of chromosomal regions containing miRNAs or altered patterns of miRNA expression in various human cancers (Calin et al., 2004; Esquela-Kerscher and Slack, 2006), but direct functional demonstration of oncogenic roles of miRNAs in the hematopoietic system is limited. The miR-17∼92 polycistronic cluster has been shown to act as an oncogene by expediting formation of B cell lymphomas in the context of c-myc overexpression; however, individual miRNAs in this cluster do not exhibit oncogenic activity (He et al., 2005). Single miRNAs have been shown to exhibit activities consistent with roles as oncogenes with overexpression of miR-155 in early B cells, leading to polyclonal expansion of the pro–B cell compartment (Costinean et al., 2006) and retroviral expression of miR-155 in immature mouse hematopoietic cells, resulting in a mixed myeloproliferative/myelodysplastic disorder without progression to AML (O’Connell et al., 2008). Although these data indicate that single miRNAs may be involved in immature hematopoietic cell function and/or leukemogenesis, no evidence directly supports the contention that miRNAs regulate the function of HSC or LSC. In this paper, we show that miR-29a is highly expressed in both human and mouse HSC and down-regulated in lineage-committed progenitors. Overexpression of miR-29a in the hematopoietic system results in biased myeloid lineage development and development of a myeloproliferative disorder (MPD) that progresses to AML. Moreover, overexpression of miR-29a converts short-lived myeloid progenitors into self-renewing cells before the development of AML. Given that miR-29a is also highly expressed in the majority of human AML tested, our findings provide evidence that miRNAs may serve as robust oncogenes during leukemogenesis and that they may convert myeloid progenitors into LSC critical for the development of AML.
RESULTS
Differential expression of miR-29a in HSCs and committed hematopoietic progenitors
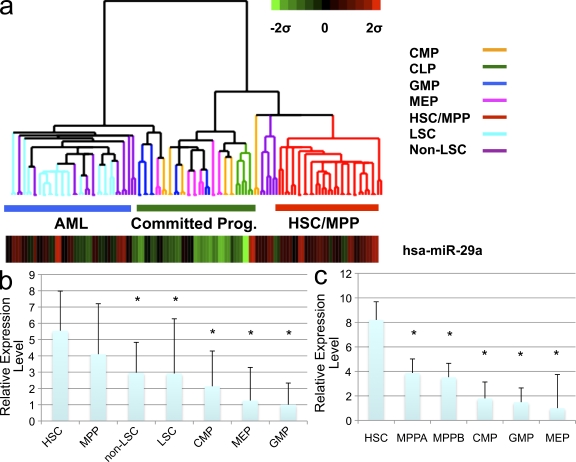
To evaluate the potential function of miRNAs in early hematopoietic development, we measured miRNA expression levels in purified human HSC (Lin−CD34+CD38−CD90+CD45RA−), multipotent progenitors (MPPs; Lin−CD34+CD38−CD90−CD45RA−), and lineage-committed progenitors, including common lymphoid progenitors (CLPs; CD34+CD38+CD10+CD19−), common myeloid progenitors (CMPs; Lin−CD34+CD38+CD123+CD45RA−), granulocyte-macrophage progenitors (GMPs; Lin−CD34+CD38+CD123+CD45RA+), and megakaryocytic-erythrocyte progenitors (MEPs; Lin−CD34+CD38−CD123loCD45RA−; Manz et al., 2002; Majeti et al., 2007). Expression of 315 mature miRNAs was measured using a sensitive multiplexed TaqMan-based real-time PCR method (Chen et al., 2005). Expression was normalized against an endogenous small RNA (sno-R2) expressed at similar levels in the tested cell populations (Fig. S1, a and b). Using unsupervised clustering and SAM analysis with a stringent cutoff (FDR < 1%), we determined that miR-29a consistently showed higher expression levels in HSC and MPP compared with more committed myeloid progenitors, with HSC/MPP showing an approximately three- to fourfold higher expression than the more mature progenitor populations (MEP and GMP; P < 0.005; Fig. 1, a and b). This pattern of expression was similar to that seen in mouse HSC (Lin−Kit+Sca+CD34−FLK2−) and committed progenitors (Fig. 1 c). Collectively, these data indicate that miR-29a exhibits an evolutionarily conserved pattern of expression, with the highest expression found in the most primitive hematopoietic cells, and suggest that the level of miR-29a expression might contribute to HSC function.
Figure 1.
miR-29a is expressed at high levels in human HSCs as well as human AML. (a) Heat map of miR-29a expression. Expression was normalized against sno-R2 and data are presented as a z-score. Lineage committed progenitors (committed prog.) include CMP, GMP, MEP, and CLP, which were sorted as described in the text. (b) Relative normalized expression levels of miR-29a in FACS-sorted normal human BM populations and human AML shows that miR-29a is expressed at highest levels in HSC. (c) Relative normalized expression levels of miR-29a expression in FACS-purified mouse HSC and lineage committed progenitors shows a dramatic decrease in miR-29a expression in MPPs (MPPA, Lin−Kit+Sca+CD34+Flk2−; MPPB, Lin−Kit+Sca+CD34+Flk2+). Asterisks signify statistically significant differential expression of indicated population compared with HSC (q < 0.05). Error bars indicate SD from at least five independent samples.
Overexpression of miR-29a in hematopoietic progenitors leads to biased myelopoiesis and an MPD
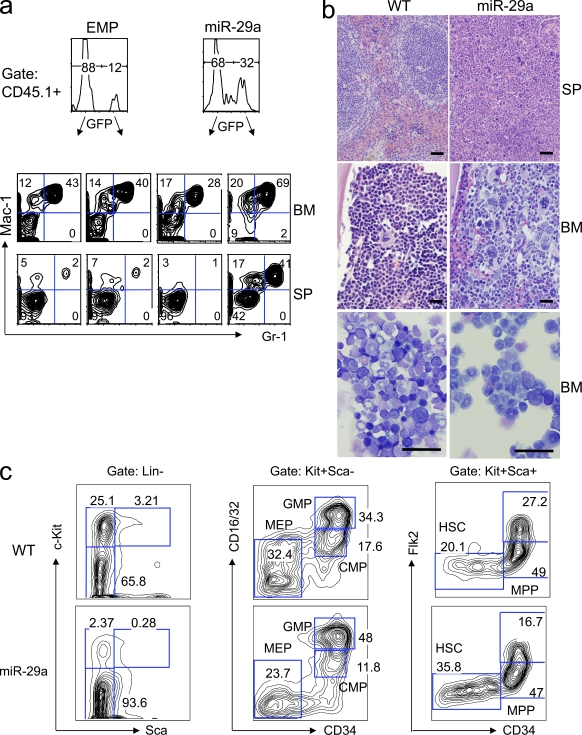
To assess miR-29a function in the hematopoietic system, we transduced mouse BM cells enriched for HSC/progenitor cells with mouse miR-29a or empty control MSCV-based GFP-expressing retrovirus (EMP). Transduced cells were transplanted into lethally irradiated recipients. Expression and appropriate processing of miR-29a was confirmed by RT-PCR and Northern blot analysis (Fig. S1 d). Flow cytometric analysis of the peripheral blood in stably engrafted primary chimeric mice showed increasing levels of donor-derived myeloid (Mac-1+) chimerism 8–12 wk after transplantation, as well as reduced percentages of donor-derived B cells (Figs. S2 and S3). This effect was specific to miR-29a, as neither EMP retrovirus nor retroviruses expressing miR-29b, miR-29c, miR-24-2, or miR-27a significantly affected engraftment levels or lineage output when transduced into mouse HSC/committed progenitors (unpublished data). Stably engrafted primary recipients (>16 wk after transplant) were necropsied to evaluate engraftment. Chimeric mice reproducibly exhibited mild splenomegaly, and flow cytometric analysis of splenocytes and BM cells consistently revealed marked statistically significant myeloid lineage bias with expansion of donor-derived granulocytes (Mac-1+ Gr-1+) and monocytes (Mac-1+Gr-1lo-int; P < 0.001; Fig. 2 a and Fig. S2 b) as well as decreased production of B lymphocytes (P < 0.001; Fig. S3 a). Histological sections of the spleen showed expanded red pulp and increased extramedullary hematopoiesis with prominent granulocytic and megakaryocytic hyperplasia. Sections and cytospin preparations of the BM showed granulocytic and megakaryocytic hyperplasia with a significant decrease in erythroid precursors (Fig. 2 b and Fig. S5 b). Collectively, these findings were consistent with an MPD.
Figure 2.
Ectopic expression of miR-29a in mouse HSC/progenitors induces myeloproliferative disease in primary chimeras. (a) Flow cytometric analysis of the BM and spleen of miR-29a–transduced or empty virus control (EMP) long-term engrafted (3–5 mo) primary chimeras reveals that miR-29a induces monocytic/granulocytic hyperplasia. (b) Primary miR-29a chimeric mice exhibit signs of myeloproliferative disease, including increased splenic extramedullary hematopoiesis, as well as granulocytic and megakaryocytic hyperplasia in both the spleen and BM. Bars, 50 µm. Cytospin preparations of the BM were Wright-Giemsa stained to reveal a predominance of maturing granulocytic precursors and near-absence of erythroid precursors in miR-29a mice relative to normal controls (bottom). Bars, 50 µm. (c) The myeloproliferative phenotype is associated with changes in the immature hematopoietic compartment, manifested by increased proportions of Lin−Kit− cells and phenotypic HSC, as well as the relative expansion of normal myeloid progenitor populations. These findings are representative of at least four independent experiments with at least 10 mice in the EMP and miR-29a–transduced groups.
miR-29a–induced biased myelopoiesis is established at myeloid progenitor stages during hematopoiesis
miR-29a–induced MPD may result from population expansion at the level of myeloid progenitors or proliferation of more mature myeloid cells. To distinguish between these possibilities, we analyzed various stages of myeloid cells in the BM by flow cytometry. Our analyses revealed that myeloid progenitors consistently exhibited changes in progenitor composition with variable increases in the frequency of GMP, MEP, CMP, and/or a novel myeloid progenitor population; however, the most frequently observed pattern was GMP expansion, which occurred in 75% of miR-29a–overexpressing mice exhibiting MPD features (Fig. 2 c and Fig. S3 d). Analysis of myeloid cells in miR-29a chimeric mice revealed statistically significant left-shifted myeloid maturation, including higher absolute numbers of Lin− cells (2.2-fold, P < 0.01) and Lin−Kit− cells (1.5-fold, P < 0.01). The immature hematopoietic compartment (Lin−Sca+Kit+, LSK) was not expanded in absolute numbers, although immunophenotypic HSC were expanded in absolute numbers (3.1-fold increase, P < 0.01; Fig. S4). There was a marked decrease in Flk2+ LSK progenitors (2.5-fold) and B cell progenitors in the BM and spleen. Consistent with the reduced B lymphoid progenitors, mature B cells were decreased in the peripheral blood, BM, and spleen (Fig. S3, b and e). The MPD phenotype was reproducibly observed in five independent sets of mice, each containing three to five mice transduced with different miR-29a retroviral preparations. Together, these data indicate that overexpression of miR-29a significantly affects hematopoietic development by altering myeloid progenitor composition and promoting myeloid differentiation.
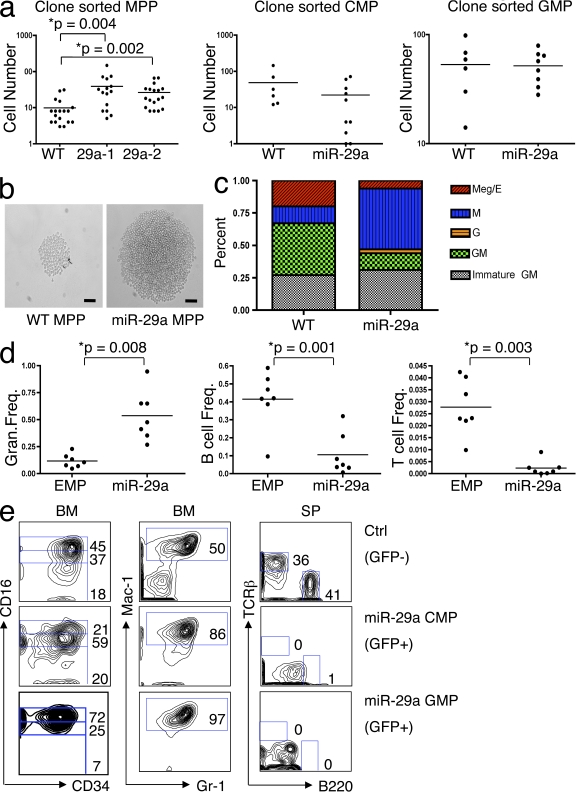
To assess the stage of hematopoiesis at which miR-29a exerts its promyeloid differentiation and proliferative effects, we sorted MPP, CMP, and GMP from miR-29a–expressing chimeric mice and assessed the ability of single sorted progenitors to proliferate and differentiate in vitro using liquid cultures. miR-29a MPP (Lin−Kit+Sca+CD34+FLK2−) exhibited an increased (approximately twofold, P < 0.005) proliferative capacity relative to WT MPP (Fig. 3, a and b). Neither miR-29a–expressing GMP nor CMP exhibited significant differences in proliferative capacity compared with their WT counterparts. Morphological evaluation of cytospin preparations from the single-cell liquid cultures revealed that miR-29a–expressing MPP exhibited a relatively decreased capacity to form megakaryocyte-containing colonies and a relative increase in the number of monocyte/macrophage colonies (Fig. 3 c). Together, these data indicate that overexpression of miR-29a acts at the level of the MPP to promote proliferation as well as to establish a granulocyte/macrophage lineage bias among myeloid progenitors.
Figure 3.
miR-29a expression alters the proliferation, differentiation, and self-renewal capacity of hematopoietic progenitors. (a) Clone-sorted miR-29a MPPs (sorted either as Lin−Kit+Sca+CD34+FLK2− or Lin−Kit+Sca+CD34+SLAM−) show a higher proliferative capacity in liquid culture compared with WT MPP, but this difference is not observed in CMP or GMP. 29a-1 and 29a-2 represent different miR-29a primary chimeric mice. (b) Photomicrographs of representative colonies formed by cloned sorted WT MPP or miR-29a MPP. Bars, 50 µm. (c) Lineage potential was assessed based on evaluation of cytospin preparations from clone-sorted MPP in vitro liquid cultures, demonstrating that miR-29a promotes monocytic differentiation and reduces megakaryocytic differentiation. The types of colonies were classified according to the types of mature myeloid cells identified, which were classified as follows: Meg/E, megakaryocyte/erythroid; G, granulocyte; M, macrophage; GM, granulocyte/macrophage; immature GM, predominance of immature myeloid precursors without evidence of megakaryocytes or erythroid precursors. (d) Statistical analysis of flow cytometric data from the peripheral blood of mice long-term engrafted (>16 wk after transplant) with sorted MPP from miR-29a MPD mice reveals a relative myeloid hyperplasia with statistically significant differences in myeloid and lymphoid output from miR-29a MPP-derived (identified as GFP+ cells) and the control recipient’s HSC/progenitors (identified as GFP− cells). Horizontal bars represent the mean. (e) Flow cytometric analysis of hematopoietic cells in sorted CMP and GMP reconstituted mice. Control cells are WT (GFP−) cells. Left, anti-CD16 and anti-CD34 staining of gated donor-derived (GFP+) Lin−c-Kit+Sca-1− BM cells; middle, anti–Mac-1 and anti–Gr-1 staining of donor-derived (GFP+) BM cells; right, anti-TCR and anti-B220 staining of donor-derived (GFP+) splenic cells. These results are representative of at least three independent transductions and greater than five mice in the EMP and miR-29a–transduced groups.
miR-29a overexpression converts non–self-renewing myeloid progenitors into self-renewing populations
In normal hematopoiesis, HSCs are the only cells capable of lympho-myeloid differentiation as well as long-term self-renewal. Because aberrant acquisition of self-renewal capacity is likely an important step in myeloid leukemogenesis (Passegué et al., 2003), we sought to determine whether miR-29a–overexpressing progenitors acquire this capacity during the early phase of MPD. We sorted MPP, CMP, and GMP from control (WT or empty retrovirus–transduced chimeric mice) or miR-29a–overexpressing primary chimeric mice before MPD development (∼1 mo after transplant) and transplanted purified progenitors (2–5 × 103) into lethally irradiated recipients together with a radioprotective dose of WT BM mononuclear cells. The BMs and spleens of transplanted mice were analyzed for donor-derived cells by flow cytometry 2–4 mo later, a time point at which normal progenitors and their progeny are no longer detectable in the peripheral blood or BM as a result of the absence of self-renewal (Christensen and Weissman, 2001). We found that although mice receiving WT MPP, CMP, and GMP possessed no detectable donor-derived cells (n = 13), 66% (8/12) of mice transplanted with the mutant MPP, 75% (3/4) of mice transplanted with the mutant CMP, and 75% (3/4) of mice transplanted with the mutant GMP contained significant numbers of donor-derived cells. The engrafted MPP cells exhibited a strong myeloid lineage bias with limited lymphoid output, which was similar to that found in the donor mice (Fig. 3 d). Although phenotypic MPPs were not detectable in the recipients that received miR-29a–overexpressing MPPs 4 mo after the transplantation, CMP and/or GMP were persistent in these mice for at least up to 6 mo (unpublished data). Mice transplanted with miR-29a–expressing CMP or GMP possessed donor-derived CMP and GMP, respectively, as well as their differentiated GFP+ progeny (Mac1+Gr-1+ cells), even 2 mo after the transplantation (Fig. 3 e). No donor-derived B or T cells were detected in the recipients, suggesting that the transplanted cells were not contaminated by HSC. Thus, these data are consistent with the conclusion that miR-29a induces aberrant self-renewal of CMP and GMP without impairing their normal ability to differentiate into monocytes or granulocytes.
Enforced miR-29a expression leads to a high incidence of AML
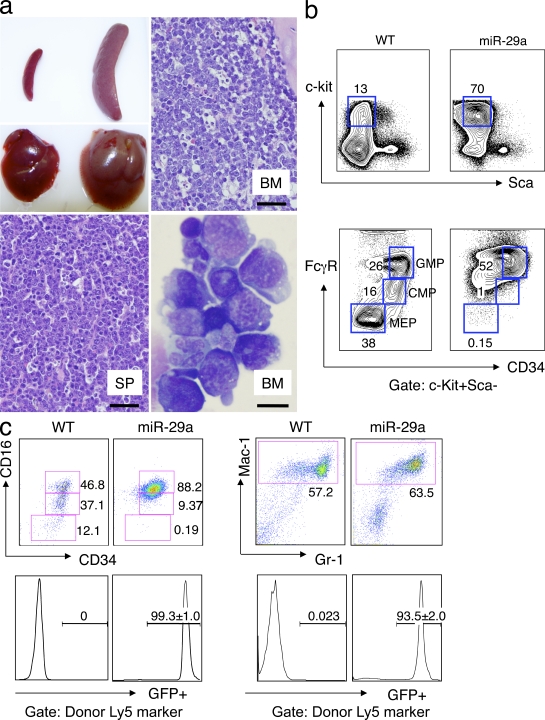
Altered miRNA expression is well documented in both solid and hematopoietic malignancies (Lu et al., 2005; Garzon et al., 2006); however, there is no functional evidence that miRNAs induce leukemias originating from immature hematopoietic cells. Abnormal acquisition of self-renewal capability by myeloid progenitors and development of MPD in miR-29a chimeric mice suggests a preleukemic state in which myeloid progenitors may function as tumor-initiating cells during progression to acute leukemia. To test this possibility, we transferred bulk splenic or BM cells from primary chimeric mice (n = 8) into three to five syngeneic recipients (secondary) mice, respectively, and then monitored the survival rates of the secondary recipients. We found that the secondary recipients began to become moribund at 4 mo after transplantation (Fig. S5 a). Among a total of 38 secondary recipients, 8 (21%) were analyzed before any sign of sickness and showed signs consistent with an MPD. The remaining 11 recipients (29%) died and 19 (50%) became severely ill and were sacrificed by 8 mo. At necropsy, the sick recipients from all eight primary chimeras exhibited profound hepatosplenomegaly, suggesting a 100% disease penetrance in the primary chimeras. Histological sections showed effacement of normal splenic and BM architecture by an immature mononuclear population, which is consistent with leukemic blasts. Morphological evaluation of cytospin preparations of total splenocytes and BM cells from leukemic mice revealed increased numbers of myeloid blasts (range 25–80%), a phenotype which is consistent with progression to AML (Fig. 4 a and Fig. S5 b). Flow cytometric analysis of splenocytes or BM cells revealed increased numbers of Lin−Kit+Sca− blast cells. These blast populations typically showed a GMP-like (GMP-L), CMP-like (CMP-L), or a previously uncharacterized myeloid progenitor phenotype within the normal Lin−Kit+Sca− progenitor compartment (Fig. 4 b). In contrast, empty retroviral vector–infected or WT spleen myeloid (Mac-1+ Gr-1+) cells contained <1% myeloid blasts (P < 0.005). Using total splenocytes or BM mononuclear cells, the leukemia was serially transplantable up to six times over a period of 18 mo (unpublished data). Southern blot analysis of retroviral integration sites using genomic DNA isolated from serially transplanted AML revealed multiple bands, indicating that the leukemia was likely oligoclonal, with no significant selection for particular clones during later passages (Fig. S6 a). Overall, these data indicate that the appearance of self-renewing miR-29a–expressing myeloid progenitor cells precedes MPD disease progression to AML, suggesting that aberrant acquisition of self-renewal by myeloid progenitors may be an early event during myeloid leukemogenesis.
Figure 4.
miR-29a myeloproliferative disease evolves to an AML that phenotypically resembles a myeloid progenitor and contains an LSC population. (a) Secondary transplant recipients become morbid ∼3–4 mo after transplantation and exhibit splenomegaly and hepatomegaly at necropsy (top left). Histological sections of the BM (top right) and spleen (bottom left) show effacement of normal architecture by sheets of myeloid blasts. Bars, 50 µm. Wright-Giemsa stain of a cytospin preparation of total splenocytes shows a marked expansion of immature blasts that exhibit myeloid and monocytic cytological features with the presence of frequent cytoplasmic granules (bottom right). Bar, 10 µm. (b) Flow cytometric phenotypic analysis of BM cells reveals a marked expansion of GMP-like (GMP-L) cells (Lin−Kit+Sca−CD34+CD16/32+) cells. Results are representative of at least three independent transductions with more than six mice in EMP and miR-29a–transduced groups. (c) Sorted GMP-L cells are highly enriched for LSC. Transplantation of as few as 20 GMP-L cells recapitulates the primary leukemia phenotypes, with increased numbers of GMP-L as well as more differentiated leukemic cells. Grafts were evaluated by gating on donor-derived (CD45.1) cells. Nontransplanted WT mice myeloid progenitor profiles are shown for comparison.
miR-29a–overexpressing myeloid progenitors are functional LSCs
Human AML is organized hierarchically, which is similar to the normal hematopoietic system, and possesses an LSC population that is capable of self-renewal and differentiation (Lapidot et al., 1994; Bonnet and Dick, 1997). As miR-29a–induced AML exhibited serial transplantability, we sought to determine whether it also contained a definable LSC population. As BMs and spleens of miR-29a–induced AML mice contained increased numbers of immunophenotypic GMP-L as well as more mature Mac-1+ cells, we sorted these two populations from leukemic mice to >99% purity and transplanted them into unirradiated recipients. After transplantation, recipient mice were monitored for engraftment. Sorted GMP-L stably engrafted recipients with AML within 2 mo after transfer, and serial dilution experiments demonstrated that as few as 20 GMP-L cells could serially transplant disease and give rise to an expanded GMP-L cell population without evidence of HSC or other normal myeloid progenitor contamination (Fig. S6 b). These GMP-L cells were not only capable of self-renewal but also gave rise to more mature monocytes/macrophages and granulocytes (Fig. 4 c). In contrast, mice that received up to 5 × 104 purified GFP+ Mac-1+/Gr-1+ or 5 × 104 c-Kit− BM cells failed to engraft AML during an 8-mo observation period (unpublished data). In situations in which leukemic blasts exhibited a CMP-L phenotype, the CMP-L blasts were similarly capable of transplanted disease. These findings indicate that miR-29a transformed myeloid progenitors are highly enriched for cells exhibiting the ability to self-renew and initiate a leukemic developmental hierarchy, which is consistent with LSC activity.
miR-29a is highly expressed in human AML
Because overexpression of miR-29a induces mouse AML, we decided to examine whether miR-29a is also overexpressed in human AML. To do so, human LSC (Lin−CD34+CD38−) and non-LSC (Lin−CD34+CD38+) blasts from 12 diagnostic AML BM patient samples were purified by FACS sorting (Fig. S6 c). miR-29a expression in the purified LSC and non-LSC blast populations was quantified by real-time PCR and compared with normal human BM HSC/committed progenitors. We found that miR-29a was expressed at high levels in all AML samples tested, with expression levels in both LSC and non-LSC blast populations similar to normal MPP. Compared with CMP, miR-29a expression levels in AML samples (both LSC and non-LSC blast populations) were significantly increased (at least threefold, q < 0.01; Fig. 1, a and b). These data indicate that a significant proportion of human AML exhibit enhanced miR-29a expression and suggest that miR-29a may also play a role in human myeloid leukemogenesis.
miR-29a regulates the G1 to S phase transition of the cell cycle
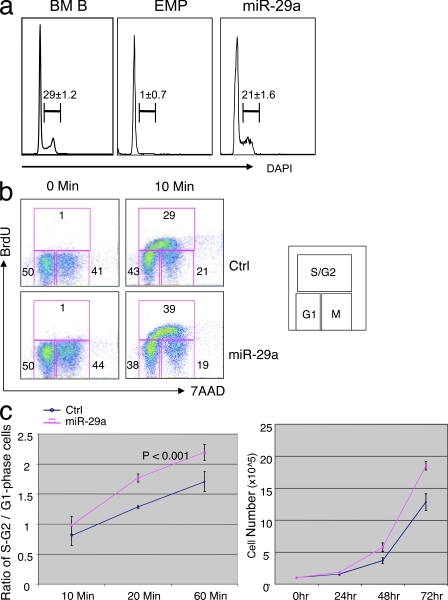
Development of myeloid leukemia in miR-29a primary chimeric mice and increased proliferation of miR-29a–overexpressing MPP suggest that miR-29a promotes cell cycling or survival of myeloid cells. To distinguish between these two possibilities, we analyzed the cell cycle status of myeloid cells in MPD mice by flow cytometry. DAPI staining of splenocytes revealed that although WT Mac-1+ Gr-1+ cells were not proliferating, ∼20% of miR-29a–expressing cells contained >2n DNA content, which is consistent with active cell cycling (Fig. 5 a). To determine whether miR-29a positively regulates cell cycle progression, and given that it is difficult to efficiently infect primary myeloid progenitors with retroviral vector, we compared the G1 to S/G2 phase transition between WT 293T and 293T cells overexpressing miR-29a by in vitro BrdU incorporation assays. We found that miR-29a–overexpressing 293T cells entered S/G2 phase more rapidly than WT 293T cells, with ∼30% more cells entering cell cycle after 60 min of BrdU labeling (Fig. 5, b and c). Consistently, the proliferation rate of miR-29a–overexpressing 293T cells was significantly enhanced when compared with WT cells (Fig. 5 c). There was no apparent effect of miR-29a overexpression on basal levels of apoptosis or apoptosis induced by serum starvation, suggesting that ectopic expression of miR-29a does not exhibit a significant impact on cell survival (unpublished data).
Figure 5.
miR-29a Overexpression expedites cell cycle progression. (a) Mac-1+Gr-1+ splenic granulocytes were evaluated for proliferation status by DAPI staining during the MPD phase of disease, revealing that miR-29a granulocytes contain a higher fraction of proliferating granulocytes. BM B cells were used as positive control for DAPI staining. These data are representative of greater than five mice in the EMP and miR-29a–transduced groups from at least two independent miR-29a retroviral transductions. (b) Analysis of cell cycle status by BrdU incorporation in 293T cells reveals increased numbers of miR-29a 293T cells in S/G2. (c) Increased numbers of miR-29a 293T cells in S/G2 phase corresponds to increased rate of proliferation as assessed by cell counts. Error bars represent SD from at least three independent experiments.
miR-29a leukemogenesis may require coordination of multiple target genes
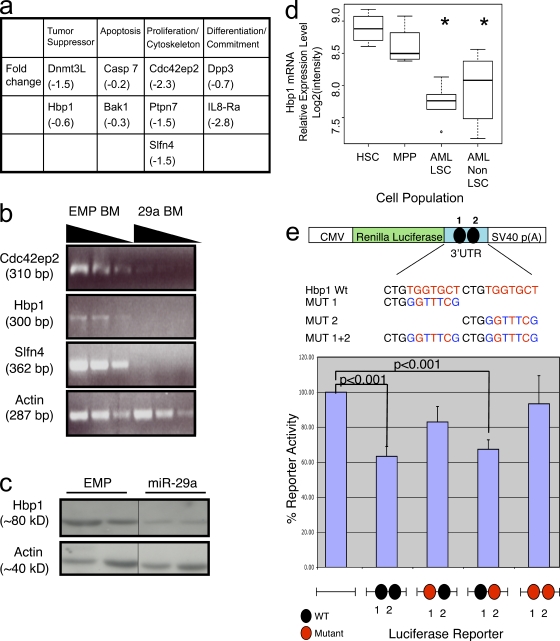
As miRNAs exert their biological effect by regulating mRNA degradation and/or protein translation, we sought to identify mRNA targets of miR-29a by comparing mRNA expression in sorted Mac-1+ Gr-1+ cells from WT mice and miR-29a overexpressing primary chimeras by mRNA gene expression array. From genes predicted to be targets of miR-29a, we identified several tumor suppressors and cell cycle regulators that were significantly decreased in miR-29a granulocytes compared with WT, including Slfn4, Dnmt3L, Cdc42ep2, and HBP1 (Fig. 6 a and Fig. S7 a). HBP1 is a known negative regulator of cell cycle progression at the G1 to S/G2 phase transition (Tevosian et al., 1997). Consistent with being physiological targets of miR-29a, decreased expression was confirmed for a subset of the genes by Western blot or semiquantitative PCR analyses, including Cdc42ep2, Slfn4, and HBP1 (Fig. 6, b and c, and Fig. S7 b). Among the predicted targets, we chose to investigate HBP1 further because of its previously described antiproliferative activity in human cell lines and association with breast cancer invasiveness (Tevosian et al., 1997; Yao et al., 2005; Paulson et al., 2007). Evaluation of Hbp-1 mRNA levels in purified human HSC/committed progenitors and AML blast subsets from primary human AML samples revealed decreased expression of Hbp-1 mRNA in human AML LSC and non-LSC relative to HSC and MPP in 89% of samples tested (8/9; Fig. 6 d). The mRNA encoding HBP1 contains two putative miR-29a binding sites within its 3′UTR, with the 5′ site representing a nonconserved site and the 3′ site representing a conserved site (Targetscan v4.1; Lewis et al., 2003, 2005; Grimson et al., 2007). To evaluate whether Hbp1 is an authentic target of miR-29a, we generated luciferase reporter fusion genes containing the renilla luciferase coding gene and either the WT or mutated 3′UTR of Hbp1. Fusion of the WT 3′UTR of Hbp1 to the luciferase reporter significantly attenuated luciferase activity, but mutation of both sites resulted in abrogation of the miR-29a–mediated inhibitory effect, indicating that at least one of the predicted miR-29a binding sites is critical for mediating miR-29a inhibition of HBP1 expression (Fig. 6 e). To better map the miR-29a binding sites, we generated mutants for each of the putative miR-29a binding sites. Of these, only the mutant containing the 5′ conserved putative miR-29a binding site reversed the inhibitory effect of miR-29a in a statistically significant manner, demonstrating its primary role in mediating miR-29a’s regulatory effect. To determine whether repression of HBP1 in hematopoietic cells is sufficient to recapitulate miR-29a’s effects in vivo, we generated a short hairpin RNA (shRNA) knockdown retroviral construct for Hbp1. This viral vector was used to transduce 5-FU–treated mouse BM cells and generate BM chimeras. Although Hbp1 shRNA was efficiently expressed in primary chimeric mice, no significant change in donor granulocyte or lymphoid chimerism was observed when compared with empty vector transduced controls (Fig. S8). These results suggest that although Hbp1 is an authentic target of miR-29a, the function of miR-29a in MPD and leukemogenesis must involve other targets that may act alone or in concert with Hbp1.
Figure 6.
Hbp1 is an authentic target of miR-29a. (a) Summary of genes differentially down-regulated in miR-29a expressing granulocytes (Mac-1+Gr-1+) versus WT granulocytes. (b) Semiquantitative RT-PCR for additional predicted target genes of miR-29a identifies several additional potential targets. The sloped line indicates decreasing amounts of input RNA for the reactions. (c) Western blot analysis for HBP1 protein expression in sorted primary granulocytes from control and miR-29a–expressing mice. HBP1 expression is diminished in miR-29a cells. Black lines indicate that intervening lanes have been spliced out. (d) Hbp-1 mRNA expression is decreased in human AML LSC and non-LSC compared with HSC and MPP. mRNA gene expression data from FACS-purified normal HSC/MPP and human AML LSC (Lin−CD34+CD38−) and non-LSC (Lin−CD34+CD38+) reveal decreased Hbp-1 mRNA expression in AML blast populations (n = 9). Asterisks denote a statistically significant difference in Hbp-1 mRNA expression in marked population compared with HSC or MPP (q < 0.05). (e) Luciferase reporter assays reveal that the predicted 3′ conserved miR-29a binding site in the 3′UTR of Hbp1 mediates miR-29a’s inhibitory effect on gene expression. Error bars signify SD from five to eight independent samples
DISCUSSION
Although several miRNAs have been shown to regulate hematopoietic lineage fates or mature effector cell function, there is little information available regarding their functional roles in the most immature hematopoietic cells, HSC and committed progenitors (Chen and Lodish, 2005; Garzon and Croce, 2008; Lodish et al., 2008). Our studies demonstrate that miR-29a shows a unique pattern of expression, being highly expressed in normal HSC and primary human AML but down-regulated in more committed progenitors. Ectopic expression of miR-29a in immature mouse hematopoietic cells reveals multiple functions of miR-29a in normal hematopoietic progenitors. Not only does miR-29a promote myeloid differentiation and proliferation at the level of MPP, it also regulates self-renewal as demonstrated by the aberrant acquisition of self-renewal in CMP and GMP. Consistent with function as a bona fide oncogene, miR-29a overexpression results in the development of an MPD that progresses to AML. The miR-29 family of miRNA has three members, miR-29a, miR-29b, and miR-29c, and miR-29b has recently been implicated as a tumor suppressor in human AML (Garzon et al., 2009). However, overexpression of miR-29b or miR-29c does not recapitulate the phenotype of miR-29a overexpression, indicating that miR-29a uniquely induces an MPD and AML. Thus, miR-29a is a novel example of an miRNA that is sufficient to induce an initial step in leukemic transformation.
Although miRNAs are postulated to act as tumor suppressors and oncogenes in human cancers (Calin and Croce, 2006), there are limited data establishing such roles for miRNAs using in vivo experimental models. The first oncomir, the miR-17∼92 cluster, was shown to decrease the latency of lymphoma development in the c-myc transgenic setting (He et al., 2005), but there is little evidence that overexpression of a single miRNA is sufficient to induce cancer in a WT genetic setting. miR-155 was shown to induce oligoclonal expansion of pre–B/pro–B cells when expressed as a B cell–specific transgene (Costinean et al., 2006) and also induced an MPD using a similar experimental approach to ours (O’Connell et al., 2008); however, in neither case did miR-155–induced disease progress to acute leukemia. This may be a result of the fact that miR-155 is highly expressed in human HSC but not human AML (unpublished data). In human AML, numerous miRNA profiling studies have shown that miRNA signatures may correlate with specific AML cytogenetic subgroups and that some miRNAs may serve as prognostic markers, but a pathogenic role for miRNAs in AML has not yet been established (Garzon et al., 2008a,b; Marcucci et al., 2008). In this regard, miR-29a represents the first example of an miRNA that induces AML in vivo.
Self-renewal is an important property acquired by normal committed progenitors during leukemic transformation, and this property is required for the establishment of LSC (Passegué et al., 2003). Although the molecular origins of any given leukemia may affect the timing and/or cell population endowed with enhanced self-renewal capability during leukemic progression, this process has not been studied in detail in human AML. Interestingly, although in some cases fusion genes associated with AML have been shown to enhance self-renewal of HSC, but not progenitors (Mulloy et al., 2003), in the case of blast transformation of chronic myelogenous leukemia, aberrant acquisition of self-renewal in GMP has been postulated to be a late event in leukemia development (Passegué et al., 2003; Jamieson et al., 2004; Abrahamsson et al., 2009). In contrast, our data support the model that in miR-29a–induced leukemia, overexpression of miR-29a leads to aberrant acquisition of self-renewal capability by committed progenitors before the establishment of the MPD phase of the disease. However, further experiments will be required to completely exclude other possible explanations for the long-term engraftment of miR-29a–expressing myeloid progenitors, including that long-term engrafted miR-29a–overexpressing CMP and GMP represent the progeny of rare engrafted HSC exhibiting a strongly biased myeloid differentiation potential, and that the transplanted and/or engrafted immunophenotypic myeloid progenitors may represent abnormal HSC with altered surface marker expression. Nevertheless, because self-renewal is normally a unique feature of HSC and high levels of miR-29a expression are associated with HSC and AML, we hypothesize that miR-29a contributes to HSC and LSC self-renewal. Thus, self-renewing myeloid progenitors may exist before the establishment of a fully transformed leukemic phenotype and may serve a similar function to that postulated for HSC, namely acting as a reservoir for accumulating the multiple genetic and/or epigenetic “hits” required for leukemogenesis. Our Southern blot analysis reveals that in almost all cases, miR-29a induces monoclonal or oligoclonal AML. It is therefore conceivable that the clonal expansion may already be established by the time the MPD stage of disease is established, presumably by a few clones with the highest level of miR-29a expression. Such aberrantly self-renewing progenitors could be argued to be the more likely cell population to serve as the cell of origin for the ensuing AML because they exhibit a prolonged lifespan that may, in turn, allow the accumulation of additional genetic and/or epigenetic changes required for malignant transformation. The amount of time required to accumulate such additional genetic and/or epigenetic changes may account for the observed disease latency observed in miR-29a–induced AML. This latency may also explain why significant clonal selection, as demonstrated by Southern blot analysis for the integrated miR-29a–expressing retrovirus, was not observed during serial passage of the AML because the most significant selection pressure may have been exerted during the earlier stages of miR-29a–induced disease. Given the availability of robust xenotransplantation models for human HSC/progenitors, it would be interesting to determine whether human progenitors acquire self-renewal capacity before leukemic transformation in progressive human hematopoietic malignancies including chronic myeloproliferative diseases and myelodysplastic syndromes.
At present, the mRNA targets required for miR-29a’s biological activities remain unclear. Self-renewal of AML stem cells and leukemic transformation has been previously linked to the lipid phosphatase PTEN (Yilmaz et al., 2006), which is predicted by some target prediction algorithms as a target of miR-29a (Lewis et al., 2005; Grimson et al., 2007); however, PTEN protein and mRNA were expressed at equivalent levels in WT and miR-29a–overexpressing Mac-1+ Gr-1+ granulocytes (unpublished data). Our gene array expression studies revealed decreased expression of HBP1, Slfn4, Dnmt3L, and Cdc42ep2 in miR-29a mutant cells compared with WT controls, and HBP1 appeared to be one of the authentic targets of miR-29a. HBP1 is a high mobility group protein that interacts with and is transcriptionally regulated by Rb, functionally inhibits G1 to G2/S phase cell cycling in leukemia cell lines, and is associated with invasive breast cancer (Tevosian et al., 1997; Shih et al., 1998; Yao et al., 2005; Paulson et al., 2007). Although shRNA knock-down of HBP1 in mouse progenitors did not recapitulate the myeloid proliferation or AML induced by miR-29 overexpression, a coordinating role of HBP1 with other factors such as Slfn4, Dnmt3L, and Cdc42ep2 in leukemic development cannot be excluded. Overexpression of miR-29a enhances proliferation of MPP and 293T cells but does not significantly alter MPP self-renewal, yet overexpression of miR-29a in CMP or GMP is sufficient to induce self-renewal but not proliferation, suggesting that miR-29a exerts stage-specific and cell context–specific effects. Although both these activities are associated with leukemogenesis, at present it is not clear whether both these events are required for leukemogenesis. Given Hbp1’s role as an inhibitor of cell proliferation, it is possible that miR-29a inhibition of Hbp1 may drive increased MPP proliferation but may not be responsible for other events such as biased myelopoiesis and self-renewal of CMP and GMP. Nevertheless, a potential role for HBP1 in AML leukemogenesis is supported by decreased Hbp-1 mRNA transcript levels in purified human AML LSC and non-LSC compared with HSC and MPP. Given the dramatic phenotype induced by miR-29a overexpression, it is likely that miR-29a targets multiple mRNAs in addition to HBP1 to induce the described phenotypes. Consistent with this possibility, recent experiments from Baek et al. (2008) and Selbach et al. (2008) demonstrate that most miRNAs target multiple proteins’ expression, each with only moderate level of inhibition.
Collectively, our data indicate that miR-29a plays an important role in the earliest stages of hematopoietic development and that abnormal expression of a single miRNA is sufficient to alter hematopoietic progenitor lineage specification, proliferation, and self-renewal. These activities have been hypothesized to be required for the fully transformed leukemic phenotype in human AML (Weissman, 2005). As high levels of miR-29a expression are also present in human AML, it will be of great interest to determine whether miR-29a may serve as a potential target for AML therapy.
MATERIALS AND METHODS
Human BM and peripheral blood samples.
Normal BM samples from healthy donors <35 yr old were purchased from All Cells, Inc. BM mononuclear cells were prepared by Ficoll density centrifugation and enriched for CD34+ cells by magnetic bead selection before staining according to the manufacturer’s protocol (CD34+ Microbead kit; Miltenyi Biotec). Primary human AML samples were collected from the Stanford Hospital Clinical Laboratory under a Stanford University Panel on Medical Human Subjects Institutional Review Board–approved protocol (#11177).
Retroviral preparation and transduction.
The MDH-PGK-GFP2.0 plasmid was a gift from C.-Z. Chen (Stanford University School of Medicine, Stanford, CA). miR-29a was PCR-amplified from mouse genomic DNA using the following primers: 5′-CCGCTCGAGTTGGTTTGGCCCTTTATC-3′ and 5′-CGGAATTCCCACCCTGCTTACCTCTG-3′. The resulting fragment was cloned into the XhoI and EcoRI sites in the 3′LTR of MDH-PGK-GFP, and the insert was confirmed by sequencing. Retroviral supernatant was generated by standard procedures after calcium phosphate transfection of MDH-PGK-GFP2.0-miR-29a and the pCLeco viral packaging construct into 293T cells.
To enrich for hematopoietic stem/progenitor cells, donor mice were injected i.p. with 5 mg 5-fluorouracil 5 d before BM harvest. BM cells were collected by flushing long bones with PBS/1% FBS and red blood cells were lysed with ACK lysis buffer (Loundon). BM cells were infected with retrovirus as per previously described protocol (Chen et al., 2004). Infected cells were resuspended in PBS then injected i.v. into lethally irradiated (9.5 Gy) recipient mice.
miRNA analysis
Total RNA was prepared from sorted human populations using mirVana RNA prep kits (Ambion) according to the manufacturer’s protocol. 100 µg of total RNA was preamplified for 14–18 rounds using a set of 315 unique probes designed by Applied Biosystems (Chen et al., 2004), and the resulting reaction product was divided equally into a 384-well plate prespotted with individual miRNA TaqMan probes. PCR was performed for 40 cycles, for 1 min at 95°C, and for 30 s at 60°C.
Mature miR-29a expression was measured using the mirVana qRT-PCR miRNA Detection kit (Ambion). In brief, total RNA was isolated using Trizol (Invitrogen). Complementary DNA (cDNA) was synthesized using U6- or miR-29a–specific RT primer sets, followed by application with their respective PCR primer sets using SYBR Green (Roche). ROX Reference dye was used for normalization of fluorescent reporter signal (Invitrogen). Expression levels were normalized to endogenous expression of U6 small nuclear RNA.
Northern blots were prepared using 20 µg of total RNA. A γ-32P-ATP end-labeled DNA antisense oligonucleotide probe to miR-29a was used for detection. Standard hybridization conditions were used and washes were performed at room temperature. U6 expression was used as a loading control.
Mice/transplantations.
C57BL/6 and C57BL/6 SJL mice were purchased from Taconic or provided by Y.R. Zou (The Feinstein Institute for Medical Research, Manhasset, NY), respectively. B6/Ka and B6 Ly5.2 mice were bred in I.L. Weissman’s mouse colony. Cells for transplant were injected i.v. into the retro-orbital sinuses of recipient mice under isoflurane or avertin anesthesia. Recipient mice were sublethally irradiated (4.7 Gy) or lethally irradiated (9.5 Gy) using a cesium radiation source and were maintained on antibiotics (Baytril; Bayer) at least 4 wk after transplantation. All mice used in this study were maintained under specific pathogen-free conditions according to institutional guidelines and animal study proposals approved by the Columbia University Institutional Animal Care and Use Committees.
Mouse tissues.
BM cells were prepared by crushing long bones and pelvis with a mortar and pestle in staining media (PBS/2% fetal calf serum). Splenocyte cell suspensions were prepared by mechanical dissociation. Red cell lysis was performed with ACK lysis buffer before flow cytometric analysis. For histology, samples were fixed in 4% paraformaldehyde or 10% neutral-buffered formalin overnight before standard processing for paraffin-embedded tissues. 10-µm sections were stained with hematoxylin-eosin. Cytospin preparations were prepared by spinning cells in a Shandon cytocentrifuge (Thermo Fisher Scientific) at 500 rpm for 5 min. Slides were stained with a modified Wright-Giemsa stain as per standard protocols.
Cell staining and flow cytometry.
Human BM cells were stained for HSC/progenitor populations as previously described (Manz et al., 2002; Majeti et al., 2007). After staining, cells were analyzed and sorted using a FACSAria (BD), with purity routinely >90%. Primary human AML LSC (Lin−CD34+CD38−) and non-LSC (Lin−CD34+CD38+) blasts were stained using a more limited lineage antibody cocktail (CD3, CD4, CD8, CD19, CD20, CD14, and CD11b).
Mouse stem and progenitor cell stains included the following monoclonal antibodies: lineage cocktail, Mac-1, Gr-1, CD3, CD4, CD8, B220, and Ter119 conjugated to Cy5-PE (eBioscience); c-Kit PE-Cy7 or APC-Cy7 (eBioscience); Sca-1 Alexa Fluor 680 or Pacific Blue (e13-161-7); CD34 FITC or biotin (eBioscience); CD16/32 (FcγRII/III) APC (BD); and CD135 (Flk-2) PE (eBioscience). Staining was performed as previously described (Akashi et al., 2000; Yang et al., 2005). Cells were then stained with streptavidin-conjugated PECy7 (eBioscience) or quantum dot 605 (Millipore). FACS data were analyzed using FlowJo software (Tree Star, Inc.).
Cell cycle assays.
For BrdU incorporation assays, 10 µM BrdU was added to equal numbers of WT 293T cells and 293T cells stably expressing miR-29a cells (in duplicate for each time point). Cells were collected at indicated time points and processed according to the manufacturer’s protocol using APC-anti-BrdU and 7-AAD (BD).
Microarray analysis and validation.
Spleen myeloid cells (Mac-1+ Gr-1+) from empty vector control or mir-29a chimeric mice were sorted using a FACSAria. Total RNA was extracted using RNeasy total RNA kit (QIAGEN) and cRNA was produced from the RNA for array analysis using the Illumina TotalPrep RNA Amplification kit according to the manufacturer’s protocol (Ambion). The labeled cRNA was hybridized using the Illumina array at the Rockefeller University Genomics Resource Center.
For semiquantitative RT-PCR, cDNA was made using random primers and the Superscript reverse transcription kit (Invitrogen) according to the manufacturer’s protocol. β-actin was used for normalization, and the following PCR primers were used for detection: Hdac6, 5′-CATCAGAGAGCAACTGATCC-3′ and 5′-ATGCCATTCCGAATCTCAGC-3′; Bak1, 5′-GCTACGTTTTTTACCTCCAC-3′ and 5′-CATCTGGCGATGTAATGATG-3′; Lrrk1, 5′-GTGAGTCTTTGGAGGTCCTT-3′ and 5′-TGATGGTGGCTTCACTGTGT-3′; Slfn4, 5′-TCCACAGACATCCATAGAGC-3′ and 5′-TTCCATCTCTGGTGAAGCTG-3′; Cdc42ep2, 5′-TCTGCTCAAAAACGCCATCT-3′ and 5′-GTGATCCATAATCTGGAGGA-3′; Hbp1, 5′-CACCATTTGGCACTGCTTTC-3′ and 5′-CCGAATGACACACTCTCTTC-3′; and actin, 5′-GCTACAGCTTCACCACCACAG-3′ and 5′-GGTCTTTACGGATGTCAACGTC-3′.
Western blots were performed as per standard protocols, and protein was revealed by chemiluminescence as per the manufacturer’s protocol (ECL; Amersham). Antibodies included Hbp1 (1:800; Santa Cruz Biotechnology, Inc.).
pGL3 firefly luciferase reporter constructs were generated by cloning the 3′UTR of respective genes downstream of the luciferase open reading frame. Constructs (0.05 µg each) were cotransfected into 293T with 0.01 µg of a Renilla luciferase control vector by calcium phosphate transfection. Luciferase activity was measured 36 h after transfection and normalized against Renilla activity according to the manufacturer’s protocol (Dual-Luciferase Reporter Assay System; Promega). Constructs were generated using the following primers: Hbp1 3UTR, 5′-CTAGTCTAGATGCTTGTGTTTGTAAGTCTG-3′ and 5′-CTAGTCTAGAGGGGCAATATGTTTAACAAG-3′; Bak1 3UTR, 5′-GCTCTAGAGCATTGCACAGTTTATTTCCA-3′ and 5′-GCTCTAGACTGGCTGGACTAAACCTCT-3′; Dpp3 3UTR, 5′-GGACTAGTGAAGATCTGTGTGGTCTCTC-3′ and 5′-GGACTAGTGATGGTGGTCGTCATATTTATT-3′; E2F7 3UTR, 5′-GCTCTAGATGCTTCGGTGGGTGGGAT-3′ and 5′-GCTCTAGATAACATACACGTCTTACTAAATA-3′; and Ptpn7 3UTR, 5′-GGACTAGTCCCTCCACCAGCTCATGG-3′ and 5′-GGACTAGTGCATCCAAGATGGTTATTTATTTA-3′.
Online supplemental material.
Fig. S1 shows miRNA expression normalization and expression vector strategies. Fig. S2 summarizes granulocyte chimerism in miR-29a primary chimeras. Fig. S3 shows lineage and progenitor composition of the peripheral blood and BM of miR-29a primary or secondary transplants. Fig. S4 summarizes BM frequencies of major hematopoietic cell populations in primary chimeras. Fig. S5 summarizes the transplant data and illustrates the histological changes in miR-29a–expressing MPD mice. Fig. S6 shows Southern blot analysis of serially passaged leukemias, summarizes limiting dilution studies to determine LSC frequency, and summarizes the AML samples included in this study. Fig. S7 summarizes mRNA array expression studies and shows semiquantitative PCR validation of differentially expressed mRNAs. Fig. S8 shows the lack of significant hematopoietic changes in HBP1 shRNA chimeric mice. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20090831/DC1.
Supplementary Material
Acknowledgments
Support was provided by an Institutional Fund from Columbia University and a Fund from Irene Diamond Foundation to H. Gu, National Institutes of Health immunology training grant to Y.-C. Han (AI07525), a physician-scientist career development award from National Institutes of Health/NCI to C.Y. Park (KO8 CA1295470), and a National Institutes of Health/NCI grant to I.L. Weissman (R01 CA86017).
We thank Dr. Jinping Zhang for the miR-29a retroviral construct, Dr. C.-Z. Chen for pHDM1 retroviral vector, Drs. S. Goff and K. Calame for discussion, and Drs. R. Dalla-Favera and K. Rajewsky for reading the manuscript.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- AML
- acute myeloid leukemia
- cDNA
- complementary DNA
- CLP
- common lymphoid progenitor
- CMP
- common myeloid progenitor
- GMP
- granulocyte-macrophage progenitor
- HSC
- hematopoietic stem cell
- LSC
- leukemia stem cell
- MEP
- megakaryocytic-erythrocyte progenitor
- MPD
- myeloproliferative disorder
- MPP
- multipotent progenitor
- mRNA
- messenger RNA
- miRNA
- microRNA
- shRNA
- short hairpin RNA
References
- Abrahamsson A.E., Geron I., Gotlib J., Dao K.H., Barroga C.F., Newton I.G., Giles F.J., Durocher J., Creusot R.S., Karimi M., et al. 2009. Glycogen synthase kinase 3beta missplicing contributes to leukemia stem cell generation. Proc. Natl. Acad. Sci. USA. 106:3925–3929 10.1073/pnas.0900189106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi K., Traver D., Miyamoto T., Weissman I.L. 2000. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 404:193–197 10.1038/35004599 [DOI] [PubMed] [Google Scholar]
- Baek D., Villén J., Shin C., Camargo F.D., Gygi S.P., Bartel D.P. 2008. The impact of microRNAs on protein output. Nature. 455:64–71 10.1038/nature07242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 116:281–297 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- Bonnet D., Dick J.E. 1997. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 3:730–737 10.1038/nm0797-730 [DOI] [PubMed] [Google Scholar]
- Bryder D., Rossi D.J., Weissman I.L. 2006. Hematopoietic stem cells: the paradigmatic tissue-specific stem cell. Am. J. Pathol. 169:338–346 10.2353/ajpath.2006.060312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin G.A., Croce C.M. 2006. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 6:857–866 10.1038/nrc1997 [DOI] [PubMed] [Google Scholar]
- Calin G.A., Sevignani C., Dumitru C.D., Hyslop T., Noch E., Yendamuri S., Shimizu M., Rattan S., Bullrich F., Negrini M., Croce C.M. 2004. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. USA. 101:2999–3004 10.1073/pnas.0307323101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T.C., Mendell J.T. 2007. microRNAs in vertebrate physiology and human disease. Annu. Rev. Genomics Hum. Genet. 8:215–239 10.1146/annurev.genom.8.080706.092351 [DOI] [PubMed] [Google Scholar]
- Chen C.Z., Lodish H.F. 2005. MicroRNAs as regulators of mammalian hematopoiesis. Semin. Immunol. 17:155–165 10.1016/j.smim.2005.01.001 [DOI] [PubMed] [Google Scholar]
- Chen C.Z., Li L., Lodish H.F., Bartel D.P. 2004. MicroRNAs modulate hematopoietic lineage differentiation. Science. 303:83–86 10.1126/science.1091903 [DOI] [PubMed] [Google Scholar]
- Chen C., Ridzon D.A., Broomer A.J., Zhou Z., Lee D.H., Nguyen J.T., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R., et al. 2005. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 33:e179 10.1093/nar/gni178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen J.L., Weissman I.L. 2001. Flk-2 is a marker in hematopoietic stem cell differentiation: a simple method to isolate long-term stem cells. Proc. Natl. Acad. Sci. USA. 98:14541–14546 10.1073/pnas.261562798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costinean S., Zanesi N., Pekarsky Y., Tili E., Volinia S., Heerema N., Croce C.M. 2006. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc. Natl. Acad. Sci. USA. 103:7024–7029 10.1073/pnas.0602266103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquela-Kerscher A., Slack F.J. 2006. Oncomirs - microRNAs with a role in cancer. Nat. Rev. Cancer. 6:259–269 10.1038/nrc1840 [DOI] [PubMed] [Google Scholar]
- Garzon R., Croce C.M. 2008. MicroRNAs in normal and malignant hematopoiesis. Curr. Opin. Hematol. 15:352–358 10.1097/MOH.0b013e328303e15d [DOI] [PubMed] [Google Scholar]
- Garzon R., Fabbri M., Cimmino A., Calin G.A., Croce C.M. 2006. MicroRNA expression and function in cancer. Trends Mol. Med. 12:580–587 10.1016/j.molmed.2006.10.006 [DOI] [PubMed] [Google Scholar]
- Garzon R., Garofalo M., Martelli M.P., Briesewitz R., Wang L., Fernandez-Cymering C., Volinia S., Liu C.G., Schnittger S., Haferlach T., et al. 2008a. Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proc. Natl. Acad. Sci. USA. 105:3945–3950 10.1073/pnas.0800135105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R., Volinia S., Liu C.G., Fernandez-Cymering C., Palumbo T., Pichiorri F., Fabbri M., Coombes K., Alder H., Nakamura T., et al. 2008b. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood. 111:3183–3189 10.1182/blood-2007-07-098749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R., Liu S., Fabbri M., Liu Z., Heaphy C.E., Callegari E., Schwind S., Pang J., Yu J., Muthusamy N., et al. 2009. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 113:6411–6418 10.1182/blood-2008-07-170589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A., Farh K.K., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P. 2007. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 27:91–105 10.1016/j.molcel.2007.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Hannon G.J. 2004. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5:522–531 10.1038/nrg1379 [DOI] [PubMed] [Google Scholar]
- He L., Thomson J.M., Hemann M.T., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S.W., Hannon G.J., Hammond S.M. 2005. A microRNA polycistron as a potential human oncogene. Nature. 435:828–833 10.1038/nature03552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson C.H., Ailles L.E., Dylla S.J., Muijtjens M., Jones C., Zehnder J.L., Gotlib J., Li K., Manz M.G., Keating A., et al. 2004. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N. Engl. J. Med. 351:657–667 10.1056/NEJMoa040258 [DOI] [PubMed] [Google Scholar]
- Kondo M., Wagers A.J., Manz M.G., Prohaska S.S., Scherer D.C., Beilhack G.F., Shizuru J.A., Weissman I.L. 2003. Biology of hematopoietic stem cells and progenitors: implications for clinical application. Annu. Rev. Immunol. 21:759–806 10.1146/annurev.immunol.21.120601.141007 [DOI] [PubMed] [Google Scholar]
- Lapidot T., Sirard C., Vormoor J., Murdoch B., Hoang T., Caceres-Cortes J., Minden M., Paterson B., Caligiuri M.A., Dick J.E. 1994. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 367:645–648 10.1038/367645a0 [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B. 2003. Prediction of mammalian microRNA targets. Cell. 115:787–798 10.1016/S0092-8674(03)01018-3 [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., Bartel D.P. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 120:15–20 10.1016/j.cell.2004.12.035 [DOI] [PubMed] [Google Scholar]
- Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M. 2005. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 433:769–773 10.1038/nature03315 [DOI] [PubMed] [Google Scholar]
- Lodish H.F., Zhou B., Liu G., Chen C.Z. 2008. Micromanagement of the immune system by microRNAs. Nat. Rev. Immunol. 8:120–130 10.1038/nri2252 [DOI] [PubMed] [Google Scholar]
- Lu J., Getz G., Miska E.A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A., et al. 2005. MicroRNA expression profiles classify human cancers. Nature. 435:834–838 10.1038/nature03702 [DOI] [PubMed] [Google Scholar]
- Majeti R., Park C.Y., Weissman I.L. 2007. Identification of a hierarchy of multipotent hematopoietic progenitors in human cord blood. Cell Stem Cell. 1:635–645 10.1016/j.stem.2007.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manz M.G., Miyamoto T., Akashi K., Weissman I.L. 2002. Prospective isolation of human clonogenic common myeloid progenitors. Proc. Natl. Acad. Sci. USA. 99:11872–11877 10.1073/pnas.172384399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcucci G., Radmacher M.D., Maharry K., Mrózek K., Ruppert A.S., Paschka P., Vukosavljevic T., Whitman S.P., Baldus C.D., Langer C., et al. 2008. MicroRNA expression in cytogenetically normal acute myeloid leukemia. N. Engl. J. Med. 358:1919–1928 10.1056/NEJMoa074256 [DOI] [PubMed] [Google Scholar]
- Monticelli S., Ansel K.M., Xiao C., Socci N.D., Krichevsky A.M., Thai T.H., Rajewsky N., Marks D.S., Sander C., Rajewsky K., et al. 2005. MicroRNA profiling of the murine hematopoietic system. Genome Biol. 6:R71 10.1186/gb-2005-6-8-r71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulloy J.C., Cammenga J., Berguido F.J., Wu K., Zhou P., Comenzo R.L., Jhanwar S., Moore M.A., Nimer S.D. 2003. Maintaining the self-renewal and differentiation potential of human CD34+ hematopoietic cells using a single genetic element. Blood. 102:4369–4376 10.1182/blood-2003-05-1762 [DOI] [PubMed] [Google Scholar]
- O’Connell R.M., Rao D.S., Chaudhuri A.A., Boldin M.P., Taganov K.D., Nicoll J., Paquette R.L., Baltimore D. 2008. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J. Exp. Med. 205:585–594 10.1084/jem.20072108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passegué E., Jamieson C.H., Ailles L.E., Weissman I.L. 2003. Normal and leukemic hematopoiesis: are leukemias a stem cell disorder or a reacquisition of stem cell characteristics? Proc. Natl. Acad. Sci. USA. 100:11842–11849 10.1073/pnas.2034201100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson K.E., Rieger-Christ K., McDevitt M.A., Kuperwasser C., Kim J., Unanue V.E., Zhang X., Hu M., Ruthazer R., Berasi S.P., et al. 2007. Alterations of the HBP1 transcriptional repressor are associated with invasive breast cancer. Cancer Res. 67:6136–6145 10.1158/0008-5472.CAN-07-0567 [DOI] [PubMed] [Google Scholar]
- Reya T., Morrison S.J., Clarke M.F., Weissman I.L. 2001. Stem cells, cancer, and cancer stem cells. Nature. 414:105–111 10.1038/35102167 [DOI] [PubMed] [Google Scholar]
- Selbach M., Schwanhäusser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. 2008. Widespread changes in protein synthesis induced by microRNAs. Nature. 455:58–63 10.1038/nature07228 [DOI] [PubMed] [Google Scholar]
- Shih H.H., Tevosian S.G., Yee A.S. 1998. Regulation of differentiation by HBP1, a target of the retinoblastoma protein. Mol. Cell. Biol. 18:4732–4743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tevosian S.G., Shih H.H., Mendelson K.G., Sheppard K.A., Paulson K.E., Yee A.S. 1997. HBP1: a HMG box transcriptional repressor that is targeted by the retinoblastoma family. Genes Dev. 11:383–396 10.1101/gad.11.3.383 [DOI] [PubMed] [Google Scholar]
- Weissman I. 2005. Stem cell research: paths to cancer therapies and regenerative medicine. JAMA. 294:1359–1366 10.1001/jama.294.11.1359 [DOI] [PubMed] [Google Scholar]
- Yang L., Bryder D., Adolfsson J., Nygren J., Månsson R., Sigvardsson M., Jacobsen S.E. 2005. Identification of Lin(-)Sca1(+)kit(+)CD34(+)Flt3- short-term hematopoietic stem cells capable of rapidly reconstituting and rescuing myeloablated transplant recipients. Blood. 105:2717–2723 10.1182/blood-2004-06-2159 [DOI] [PubMed] [Google Scholar]
- Yao C.J., Works K., Romagnoli P.A., Austin G.E. 2005. Effects of overexpression of HBP1 upon growth and differentiation of leukemic myeloid cells. Leukemia. 19:1958–1968 10.1038/sj.leu.2403918 [DOI] [PubMed] [Google Scholar]
- Yilmaz O.H., Valdez R., Theisen B.K., Guo W., Ferguson D.O., Wu H., Morrison S.J. 2006. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 441:475–482 10.1038/nature04703 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.