Abstract
AIM: To evaluate the association of the autophagy-related 16-like 1 (ATG16L1) T300A polymorphism (rs2241880) with predisposition to inflammatory bowel diseases (IBD) by means of meta-analysis.
METHODS: Publications addressing the relationship between rs2241880/T300A polymorphism of ATG16L1 and Crohn’s disease (CD) and ulcerative colitis (UC) were selected from the MEDLINE and EMBASE databases. To make direct comparisons between the data collected in these studies, the individual authors were contacted when necessary to generate a standardized set of data from these studies. From these data, odds ratio (OR) with 95% confidence interval (CI) were calculated.
RESULTS: Twenty-five studies of CD were analyzed, 14 of which involved cases of UC. The variant G allele of ATG16L1 was positively associated with CD (OR = 1.32, 95% CI: 1.26-1.39, P < 0.00001) and UC (OR = 1.06, 95% CI: 1.01-1.10, P = 0.02). For child-onset IBD, a higher G allele frequency was found for cases of CD (OR = 1.35, 95% CI: 1.16-1.57, P = 0.0001) than for cases of UC (OR = 0.98, 95% CI: 0.81-1.19, P = 0.84) relative to controls.
CONCLUSION: The ATG16L1 T300A polymorphism contributes to susceptibility to CD and UC in adults, but different in children, which implicates a role for autophagy in the pathogenesis of IBD.
Keywords: ATG16L1, Inflammatory bowel diseases, Crohn’s disease, Ulcerative colitis, Meta-analysis
INTRODUCTION
Inflammatory bowel diseases (IBD), including Crohn’s disease (CD) and ulcerative colitis (UC), are chronic inflammatory disorders of the gastrointestinal tract that have an increasing incidence and prevalence in developing countries[1]. CD can affect any part of the alimentary tract, although it most commonly involves the terminal ileum, and is characterized by transmural discontinuous lesions. UC is a continuous disease that commences in the rectum and extends for a varying distance proximally in the colon, and its continuous inflammation is limited to mucosal and submucosal layers. Currently, the etiology and pathogenesis of IBD are not completely understood. IBDs are complex diseases with a number of contributing factors such as genetic predisposition, environmental factors, intestinal microbial flora, and aberrant immune responses[2].
Advances in genetic research, especially the genome-wide association (GWA) studies, have identified a number of IBD susceptibility loci. One of these loci, the autophagy-related 16-like 1 (ATG16L1) gene, has been shown to have a role in IBD. The ATG16L1 gene is located on chromosome 2, at 2q37.1, and encodes a protein that is involved in the formation of autophagosomes during autophagy. Autophagy is a non-selective degradation system that has roles in starvation adaptation, intracellular protein and organelle clearance, development, anti-aging processes, elimination of microorganisms, cell death, tumor suppression, and antigen presentation[3]. Autophagy has also been implicated in the innate and adaptive immune responses[4].
Hampe et al[5] first identified ATG16L1 as a susceptibility gene for CD in a GWA study of 19 779 non-synonymous single nucleotide polymorphisms (SNPs) present in 735 individuals with CD and 368 controls. Further analysis showed that the marker, rs2241880, was an SNP that encodes a threonine to alanine substitution (T300A) at amino acid position 300, which was correlated with the incidence of CD in two German and one British studies of CD. The rs2241880 SNP directly correlated with the majority of risk associated with this locus. In contrast, no correlation between rs2241880 and UC was detected. Many other studies[6-29] have assessed the association of rs2241880 with predisposition to CD and UC, however, the results of these studies are inconsistent. In addition, a number of studies included child-onset IBD cases, the results of which were also confusing.
A meta-analysis was recently published that focused on the relationship between the T300A polymorphism and the incidence of CD, but not on the relationship between the T300A polymorphism and risk of UC, or the role of this polymorphism in child-onset IBD[30]. With the publication of additional studies related to the T300A polymorphism, sufficient data were available to perform a comprehensive meta-analysis of these studies in order to identify consistencies in the data to determine disease susceptibility, as well as to identify areas that need to be further addressed.
MATERIALS AND METHODS
Study selection
As of June 1, 2009, published studies in MEDLINE and EMBASE containing the following key words were included in this study: “inflammatory bowel disease”, “IBD”, “Crohn’s disease”, “CD”, “Ulcerative colitis”, “UC”, “autophagy related 16 like 1”, and “ATG16L1”. The references of the eligible publications were searched manually to identify additional relevant studies. Relevant publications were also identified using the “Related Articles” option in PubMed. Studies reported by the same authors, yet published in different journals, were checked for possible overlapping participant groups. No language restrictions were made. When pertinent data were not included, or data that were presented were unclear, the authors were directly contacted.
Inclusion criteria
To be eligible for inclusion in this meta-analysis, the following criteria were established: (1) the study must include a case-control study that addressed IBD (i.e. CD or UC) patients and unrelated controls; (2) the study must have evaluated the ATG16L1 T300A polymorphism and the risk of CD or UC; and (3) the study must have included sufficient data for extraction.
Exclusion criteria
Studies were excluded from consideration if: (1) the study was based on family data; (2) the study did not have the outcomes of comparison reported or it was not possible to determine them; or (3) the study contained a smaller sample size and overlapped others meanwhile.
Data extraction
Using a standardized form, data from published studies were extracted independently by two investigators to populate the needed information. Data collected included first author, year of publication, inclusion and exclusion criteria, study population characteristics, and sample size. Any discrepancies between the two sets of data collected were resolved.
Statistical analysis
Odds ratio (OR) and 95% confidence interval (CI) were calculated for each study using Review Manager version 4.2 software. Between-study heterogeneity was estimated using the χ2-based Q statistic. Heterogeneity was considered statistically significant when P < 0.05. I2 was also tested. If heterogeneity existed, data were analyzed using a random effects model. In the absence of heterogeneity, a fixed effects model was used.
A χ2 test was performed to examine Hardy-Weinberg equilibrium when genotype data were available. P < 0.05 was considered statistically significant. If Hardy-Weinberg disequilibrium existed, or it was impossible to evaluate this equilibrium, sensitivity analysis was performed.
RESULTS
Patients and controls
Of the 41 papers identified in a literature search of MEDLINE and EMBASE databases relevant to IBD, CD and UC, 25[5-29] were included in this meta-analysis, while 16 were excluded[31-46] (Figure 1). The reasons for exclusion are listed in Table 1. Three of the 41 papers[5,10,15] evaluated more than one study. Two of these studies contained relevant data and were combined[5,10], while the third was excluded[15]. Each of the 25 studies (Table 2) included cases of CD, with various subsets of papers considering different ethnic populations. For example, there were three studies of Asian populations[12,25,28], two of Oceanic populations[11,15], seven studies based in the United States[6,10,13,14,21,23,29], and 13 studies of European populations[5,7-9,16-20,22,24,26,27]. Of the 25 studies, 14 included both CD and UC cases and were based on data from an Asian population[25], an Oceanic population[11], two studies from the United States[10,21], and 10 European populations[5,7-9,17-20,24,26]. Seven of the included studies explored the relationship between the incidence of the T300A polymorphism of ATG16L1 and child-onset CD[6,11,13,17,20,23,24], while only two reported cases of child-onset UC[20,24]. For six of the total studies, genotype data were not available even after authors were contacted for further inquiry. Therefore, a Hardy-Weinberg equilibrium test was performed on the remaining 19 studies, and three of them showed disequilibrium[15,25,29] (P = 0.04, P = 0.00, and P = 0.03, respectively). Among the six untested studies due to unavailable data, five[5,8,10,17,26] reported Hardy-Weinberg equilibrium data in the original paper.
Figure 1.
A flowchart illustrating the selection of published studies included in this meta-analysis.
Table 1.
Reasons for study exclusion
| Studies | Reasons |
| Glas et al[31], Török et al[32] | Data overlapped those of another article[17] |
| Cooney et al[33] | Data overlapped those of another article[8] |
| Weersma et al[34], Weersma et al[35] | Data overlapped those of another article[26] |
| Franke et al[36], Franke et al[37] | Data overlapped those of another article[5] |
| Fisher et al[38], Wtccc et al[40], Parkes et al[43], Yamazaki et al[45] | Other SNPs rather than rs2241880 SNP of ATG16L1 were analyzed |
| Beckly et al[39], Barrett et al[41], Libioulle et al[42], Raelson et al[44], Kugathasan et al[46] | Data could not be extracted |
Table 2.
Studies included in the meta-analysis
| No. | First author | Year | Population |
Number of participants used in analysis |
||
| CD | UC | Controls | ||||
| 1 | Hampe et al[5]1 | 2007 | Germany, UK | 2122 | 1227 | 2056, 1032 for CD, UC respectively |
| 2 | Baldassano et al[6] | 2007 | USA | 142 | NA | 281 |
| 3 | Büning et al[7] | 2007 | Germany, Hungary, Netherlands | 614 | 296 | 707 |
| 4 | Cummings et al[8] | 2007 | UK | 645 | 676 | 1190 |
| 5 | Prescott et al[9] | 2007 | UK | 727 | 877 | 579 |
| 6 | Rioux et al[10]2 | 2007 | North-America | 1571 | 353 | 1184, 207 for CD, UC respectively |
| 7 | Roberts et al[11] | 2007 | New Zealand | 496 | 466 | 549 |
| 8 | Yamazaki et al[12] | 2007 | Japan | 481 | NA | 437 |
| 9 | Amre et al[13] | 2009 | Canada | 286 | NA | 290 |
| 10 | Baptista et al[14] | 2008 | Brazil | 180 | NA | 189 |
| 11 | Fowler et al[15]3 | 2008 | Australia | 154 | NA | 420 |
| 12 | Gaj et al[16] | 2008 | Poland | 59 | NA | 140 |
| 13 | Glas et al[17] | 2008 | Germany | 768 | 507 | 1615 |
| 14 | Lakatos et al[18] | 2008 | Hungary | 266 | 149 | 149 |
| 15 | Lappalainen et al[19] | 2008 | Finland | 240 | 459 | 190 |
| 16 | Latiano et al[20] | 2008 | Italy | 667 | 668 | 749 |
| 17 | Okazaki et al[21] | 2008 | Canada | 208 | 113 | 314 |
| 18 | Perricone et al[22] | 2008 | Italy | 163 | NA | 160 |
| 19 | Peterson et al[23] | 2008 | USA | 555 | NA | 486 |
| 20 | Van Limbergen et al[24]4 | 2008 | Scotland | 629 | 580 | 345 |
| 21 | Zhi et al[25] | 2008 | China | 40 | 40 | 50 |
| 22 | Weersma et al[26] | 2009 | Holland | 1684 | 1120 | 1350 |
| 23 | Hancock et al[27] | 2008 | UK | 586 | NA | 1156 |
| 24 | Yang et al[28] | 2009 | Korea | 377 | NA | 372 |
| 25 | Newman et al[29] | 2009 | Canada | 435 | NE | 895 |
| Total | 14 095 | 7531 | 15 849, 13 852 for CD, UC respectively | |||
1Data were extracted from the combination of panel A, panel B, panel C, and UC cohort; 2Only GWA study and replication cohort 2 study were included; 3Data from the cohort 1 study were not included in the analysis based on the use of data for controls from 251 families with IBD; 4Data of the UC group were obtained by communication with the authors of this publication. CD: Crohn’s disease; UC: Ulcerative colitis; NA: Not applicable (no such group); NE: Non-extractable or unavailable data.
Association between ATG16L1 T300A polymorphism and IBD
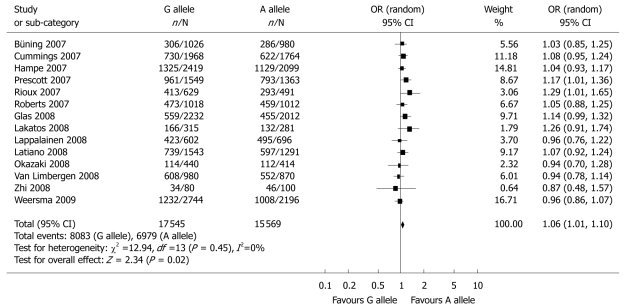
Meta-analysis of the 25 studies that fulfilled the inclusion criteria identified a significant association between the G allele of ATG16L1 and susceptibility to CD (OR = 1.32, 95% CI: 1.26-1.39, P < 0.00001) (Table 3 and Figure 2). Sensitivity analysis was performed with the omission of three studies[15,25,29] and Hardy-Weinberg disequilibrium showed similar results (data not shown). No notable change in the results of the statistic analyses was obtained when the Peterson et al[23] study was eliminated. As shown in Table 3 and Figure 3, a significant association between the G allele of ATG16L1 and the risk of UC was also detected (OR = 1.06, 95% CI: 1.01-1.10, P = 0.02). When the data of Zhi et al[25] were excluded, similar results were obtained: OR = 1.05, 95% CI: 1.01-1.10, P = 0.02.
Table 3.
Summary of the association of the rs2241880 polymorphism and IBD determined in the meta-analysis
| Comparisons | No. of studies | Effects model selection | OR (95% CI) | P-value |
| CD vs control | 25 | R | 1.32 (1.26-1.39) | < 0.00001 |
| UC vs control | 14 | F | 1.06 (1.01-1.10) | 0.02 |
| Child-onset CD vs control | 7 | R | 1.35 (1.16-1.57) | 0.0001 |
| Child-onset UC vs control | 2 | F | 0.98 (0.81-1.19) | 0.84 |
R: Random effects model; F: Fixed effects model.
Figure 2.
Meta-analysis of the association between the rs2241880 polymorphism of the ATG16L1 gene and CD for the G allele vs the A allele.
Figure 3.
Meta-analysis of the association between the rs2241880 polymorphism of the ATG16L1 gene and UC for the G allele vs the A allele.
Association between ATG16L1 T300A polymorphism and child-onset CD and UC
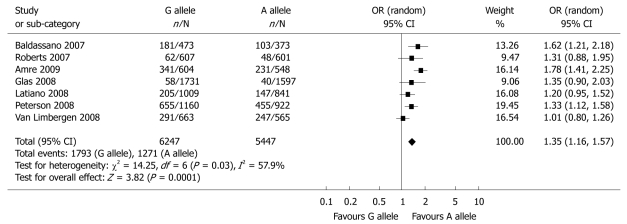
The pooled analysis for child-onset cases of CD identified the T300A polymorphism of ATG16L1 as a factor for predisposition to child-onset CD in a total of seven studies[6,11,13,17,20,23,24] (OR = 1.35, 95% CI: 1.16-1.57, P = 0.0001) (Table 3 and Figure 4). When the study of Peterson et al[23] was excluded, similar data were acquired: OR = 1.35, 95% CI: 1.11-1.64, P = 0.003. Two studies[20,24] discussed the association of the T300A polymorphism of ATG16L1 and child-onset UC, yet the results of the combined analysis indicated there was no correlation (Table 3 and Figure 5).
Figure 4.
Meta-analysis of the association between the rs2241880 polymorphism of the ATG16L1 gene and child-onset CD for the G allele vs the A allele.
Figure 5.
Meta-analysis of the association between the rs2241880 polymorphism of ATG16L1 gene and child-onset UC for the G allele vs the A allele.
DISCUSSION
Over the past few years, knowledge of the genetics of IBD has advanced tremendously since the first IBD GWA study was published in 2006[47]. Hence, several novel susceptibility loci have been identified; one of them being the ATG16L1 gene. Hampe et al[5] first identified the association of the rs2241880 SNP in ATG16L1 with cases of CD, but not with UC. Several subsequent studies have tried to verify these results, however, the results of these latter studies were inconsistent, especially regarding the relationship between ATG16L1 and UC, further perplexing our understanding of the role of ATG16L1 in IBD. Thus, we conducted a meta-analysis of the currently published reports of CD and UC to attempt to clarify the correlation between ATG16L1 and IBD. In order to obtain as much relevant data as possible, we retained some studies that presented a Hardy-Weinberg disequilibrium, however, there were no notable change in the results of our analyses whether these studies were included or omitted.
Our meta-analysis confirmed a positive association between the rs2241880 polymorphism of ATG16L1 and susceptibility to CD. Meanwhile, a modest but significant association of the rs2241880 polymorphism with predisposition to UC was also observed. Taken together, these outcomes demonstrate that the ATG16L1 variant containing the T300A substitution confers risk for both CD and UC.
Many aspects of child-onset IBD differ from adult-onset IBD, especially in regard to the type, location, and behavior of the disease, as well as sex preponderance and genetically attributable risk[48]. Considering the relatively short time of exposure to environmental factors (e.g. smoking) in children compared to adults with IBD, genetic background is hypothesized to be a more important factor in early-onset IBD. Therefore, we further analyzed the association between ATG16L1 and child-onset IBD. Our results indicated that ATG16L1 was associated with the risk of child-onset CD, but not with child-onset UC. Regarding the negative finding in early-onset UC, small sample sizes must be recognized as a contributing factor. Only two studies were included in the final analysis, therefore, this may have precluded the detection of a significant association. Additional studies are needed to determine whether an association exists in this cohort or not.
There were some limitations in the present meta-analysis. For example, because of publication limitations, some relevant studies could not be included in our analysis. Secondly, heterogeneity existed among studies of CD and child-onset CD, which had the potential to influence the results of our meta-analysis. The lack of information provided by some published studies, especially with regard to genotype data, was another limitation. Although we contacted authors of publications that did not provide a comprehensive set of data, genotype data remained unavailable for six of the total number of studies. Finally, there were only two studies that examined the relationship between ATG16L1 and child-onset UC. As a result, the small sample size available was not ideal for detecting small genetic effects.
Although the precise impact of the ATG16L1 variant on the pathogenesis of IBD remains unknown, accumulating evidence suggests that microbes play an important role in the initiation and etiopathogenesis of IBD. IBD lesions have been shown to develop preferentially in regions with the highest concentrations of bacteria[49]. In addition, enteric flora has been found to be more commonly associated with IBD patients than with control groups[50,51]. IBD animal model studies also have demonstrated that gut inflammation does not develop in a germ-free environment[52]. Autophagy is a fundamental intracellular degradation system that protects cells against various bacterial pathogens and the cytotoxic effect of bacterial toxins. Autophagy also has been identified to play a role in both innate and adaptive immune responses[53,54]. The specific role of ATG16L1 in these processes remains unclear. ATG16L1 is expressed not only in intestinal epithelial cells, but also in lymphocytes and macrophages. It interacts with two other autophagy proteins, ATG5 and ATG12, to form a complex essential for the process of autophagy. Several studies have characterized the influence of ATG16L1 deficiency. Rioux et al[10] have shown that knockdown of ATG16L1 mRNA in HeLa cells with siRNA reduced targeting of Salmonella typhimurium to autophagic vacuoles, which implicates a role for ATG16L1 in the clearance of intracellular bacteria via autophagy. Saitoh et al[55] have reported that ATG16L1 deficiency disrupts the recruitment of the ATG12-ATG5 conjugate to the isolation membrane, which results in loss of microtubule-associated protein 1 light chain 3 binding to phosphatidylethanolamine. Consequently, autophagosome formation and degradation of proteins with a long half-life are severely impaired in ATG16L1-deficient cells. ATG16L1-deficient macrophages also have been shown to produce large amounts of the inflammatory cytokines, IL-1 and IL-18, and mice that lack ATG16L1 expression in hematopoietic cells are highly predisposed to dextran sulfate sodium-induced acute colitis. Cadwell et al[56] have generated hypomorphic ATG16L1 (ATG16L1HM) mice that express ATG16L1 at 30% of its normal level. In this model, notable abnormalities have been observed in Paneth cells. These ileal epithelial cells are hypothesized to play a role in the control of intestinal microbiota by secreting granules that contained antimicrobial peptides and lysozymes. Aberrant Paneth cells exhibit deficiencies in their granule exocytosis pathway, which results in a decreased capacity to eliminate microbes. The mammalian ATG16L1 protein consists of three distinct domains: the N-terminal region mediates interactions with other autophagy proteins; a coiled-coil domain provides the capacity for ATG16L1 oligomerization and ATG5-ATG12 association; and there is a region of 7 WD repeats[57]. The T300A mutation is located within the WD repeats, which are associated with protein interactions. Therefore, the T300A mutation is hypothesized to affect protein interactions necessary for the formation of autophagosomes, which results in dysfunction of the autophagy pathway and a subsequent decrease in pathogen clearance[9]. It has been shown that the ATG16L1 coding variant is defective in mediating efficient antibacterial autophagy in cultured HeLa and Caco2 cells, and the ATG16L1*300A protein is unstable under conditions of high microbial load[58]. These observations suggest the effect of this mutation has a direct impact on autophagic effectiveness. In summary, the T300A mutation of ATG16L1 impairs specific autophagic, innate resistance mechanisms to gut commensals, which facilitate inflammation after invasion[59].
In addition to decreased ability to remove intestinal microbes directly, there may be other mechanisms that underlie the association between the ATG16L1 variant and increased risk for IBD. For example, since autophagy contributes to immune tolerance against self tissues, it seems likely that decreased autophagy may lead to a failure of immune tolerance by autoantigen presentation on major histocompatibility complex class II molecules, which causes immune inflammation[59]. Another possible mechanism involves autophagy and apoptosis. Accumulating studies have detected an acceleration in the rate of epithelial cell apoptosis and inhibition of inflammatory cell apoptosis in CD and UC[60-63]. Autophagy and apoptosis share many common triggers and cross-inhibitory interactions[64], thus, it is hypothesized that defective autophagy might alter the process of intestinal cell apoptosis, ultimately contributing to IBD pathogenesis.
In conclusion, our meta-analysis of published cases of CD and UC suggests that the ATG16L1 T300A polymorphism is associated with susceptibility to CD and UC. However, the effect of this polymorphism differs between child-onset CD and UC. These findings implicate the role of autophagy and intestinal microbes in the pathogenesis of IBD, and demonstrate the need for further studies.
COMMENTS
Background
Inflammatory bowel diseases (IBD), Crohn’s disease (CD) and ulcerative colitis (UC), are polygenic disorders. An A to G transition of the autophagy-related 16-like 1 gene (ATG16L1) has been implicated as a risk factor for IBD, but individual studies have been inconclusive or controversial.
Research frontiers
Much attention has been paid to the potential role of autophagy in the pathogenesis of IBD, since Hampe et al identified ATG16L1 as a CD susceptibility gene in 2007. Some of them have been trying to confirm the definite relationship between ATG16L1 polymorphism and IBD, and meanwhile, others attempting to uncover underlying mechanisms.
Innovations and breakthroughs
The current study demonstrated that ATG16L1 is a susceptibility gene for CD and UC in adults, but different in children.
Applications
It is seen from this study that the ATG16L1 T300A polymorphism contributes to susceptibility to CD and UC in adults, which provides an insight into the role of autophagy in the pathogenesis of IBD. Studies for further exploring the mechanism by which altered ability of autophagy predisposes to IBD are needed.
Terminology
Autophagy is a cytoplasmic homeostasis process that cleanses the interior of all eukaryotic cells. It has an important role in cell and tissue homeostasis, and has been implicated in a range of human diseases.
Peer review
This study is an interesting meta-analysis about T300A polymorphism of ATG16L1 and susceptibility to inflammatory bowel diseases. The authors carefully reviewed the available literture and oncluded that ATG16L1 T300A polymorphism contributes to susceptibility to both CD and UC in adults, but not in children, implicating a role for autophagy in the pathogenesis of IBD.
Footnotes
Peer reviewer: Dr. Marco Scarpa, PhD, Department of Surgical & Gastroenterological Sciences (Gastroenterology section), University of Padova, via Giustiniani 2, Padova, 35128, Italy
S- Editor Tian L L- Editor Kerr C E- Editor Ma WH
References
- 1.Zheng JJ, Zhu XS, Huangfu Z, Gao ZX, Guo ZR, Wang Z. Crohn’s disease in mainland China: a systematic analysis of 50 years of research. Chin J Dig Dis. 2005;6:175–181. doi: 10.1111/j.1443-9573.2005.00227.x. [DOI] [PubMed] [Google Scholar]
- 2.Baumgart DC, Carding SR. Inflammatory bowel disease: cause and immunobiology. Lancet. 2007;369:1627–1640. doi: 10.1016/S0140-6736(07)60750-8. [DOI] [PubMed] [Google Scholar]
- 3.Mizushima N. The pleiotropic role of autophagy: from protein metabolism to bactericide. Cell Death Differ. 2005;12 Suppl 2:1535–1541. doi: 10.1038/sj.cdd.4401728. [DOI] [PubMed] [Google Scholar]
- 4.Deretic V. Autophagy in innate and adaptive immunity. Trends Immunol. 2005;26:523–528. doi: 10.1016/j.it.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 5.Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, Albrecht M, Mayr G, De La Vega FM, Briggs J, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 6.Baldassano RN, Bradfield JP, Monos DS, Kim CE, Glessner JT, Casalunovo T, Frackelton EC, Otieno FG, Kanterakis S, Shaner JL, et al. Association of the T300A non-synonymous variant of the ATG16L1 gene with susceptibility to paediatric Crohn’s disease. Gut. 2007;56:1171–1173. doi: 10.1136/gut.2007.122747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Büning C, Durmus T, Molnar T, de Jong DJ. Drenth JPH., Fiedler T, Gentz E, Todorov T, Haas V, Buhner S, Sturm A. Baumgart DC., Nagy F, Lonovics J, Landt O, Kage A, Büning H, Nickel R, Büttner J, Lochs H, Schmidt HH-J, Witt H. A study in three European IBD cohorts confirms that the ATG16L1 c.898A>G (p.Thr300Ala) variant is a susceptibility factor for Crohn’s disease. J Crohn’s Colitis. 2007;1:70–76. doi: 10.1016/j.crohns.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 8.Cummings JR, Cooney R, Pathan S, Anderson CA, Barrett JC, Beckly J, Geremia A, Hancock L, Guo C, Ahmad T, et al. Confirmation of the role of ATG16L1 as a Crohn’s disease susceptibility gene. Inflamm Bowel Dis. 2007;13:941–946. doi: 10.1002/ibd.20162. [DOI] [PubMed] [Google Scholar]
- 9.Prescott NJ, Fisher SA, Franke A, Hampe J, Onnie CM, Soars D, Bagnall R, Mirza MM, Sanderson J, Forbes A, et al. A nonsynonymous SNP in ATG16L1 predisposes to ileal Crohn’s disease and is independent of CARD15 and IBD5. Gastroenterology. 2007;132:1665–1671. doi: 10.1053/j.gastro.2007.03.034. [DOI] [PubMed] [Google Scholar]
- 10.Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, Green T, Kuballa P, Barmada MM, Datta LW, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roberts RL, Gearry RB, Hollis-Moffatt JE, Miller AL, Reid J, Abkevich V, Timms KM, Gutin A, Lanchbury JS, Merriman TR, et al. IL23R R381Q and ATG16L1 T300A are strongly associated with Crohn’s disease in a study of New Zealand Caucasians with inflammatory bowel disease. Am J Gastroenterol. 2007;102:2754–2761. doi: 10.1111/j.1572-0241.2007.01525.x. [DOI] [PubMed] [Google Scholar]
- 12.Yamazaki K, Onouchi Y, Takazoe M, Kubo M, Nakamura Y, Hata A. Association analysis of genetic variants in IL23R, ATG16L1 and 5p13.1 loci with Crohn’s disease in Japanese patients. J Hum Genet. 2007;52:575–583. doi: 10.1007/s10038-007-0156-z. [DOI] [PubMed] [Google Scholar]
- 13.Amre DK, Mack DR, Morgan K, Krupoves A, Costea I, Lambrette P, Grimard G, Dong J, Feguery H, Bucionis V, et al. Autophagy gene ATG16L1 but not IRGM is associated with Crohn’s disease in Canadian children. Inflamm Bowel Dis. 2009;15:501–507. doi: 10.1002/ibd.20785. [DOI] [PubMed] [Google Scholar]
- 14.Baptista ML, Amarante H, Picheth G, Sdepanian VL, Peterson N, Babasukumar U, Lima HC, Kugathasan S. CARD15 and IL23R influences Crohn’s disease susceptibility but not disease phenotype in a Brazilian population. Inflamm Bowel Dis. 2008;14:674–679. doi: 10.1002/ibd.20372. [DOI] [PubMed] [Google Scholar]
- 15.Fowler EV, Doecke J, Simms LA, Zhao ZZ, Webb PM, Hayward NK, Whiteman DC, Florin TH, Montgomery GW, Cavanaugh JA, et al. ATG16L1 T300A shows strong associations with disease subgroups in a large Australian IBD population: further support for significant disease heterogeneity. Am J Gastroenterol. 2008;103:2519–2526. doi: 10.1111/j.1572-0241.2008.02023.x. [DOI] [PubMed] [Google Scholar]
- 16.Gaj P, Habior A, Mikula M, Ostrowski J. Lack of evidence for association of primary sclerosing cholangitis and primary biliary cirrhosis with risk alleles for Crohn’s disease in Polish patients. BMC Med Genet. 2008;9:81. doi: 10.1186/1471-2350-9-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glas J, Konrad A, Schmechel S, Dambacher J, Seiderer J, Schroff F, Wetzke M, Roeske D, Török HP, Tonenchi L, et al. The ATG16L1 gene variants rs2241879 and rs2241880 (T300A) are strongly associated with susceptibility to Crohn’s disease in the German population. Am J Gastroenterol. 2008;103:682–691. doi: 10.1111/j.1572-0241.2007.01694.x. [DOI] [PubMed] [Google Scholar]
- 18.Lakatos PL, Szamosi T, Szilvasi A, Molnar E, Lakatos L, Kovacs A, Molnar T, Altorjay I, Papp M, Tulassay Z, et al. ATG16L1 and IL23 receptor (IL23R) genes are associated with disease susceptibility in Hungarian CD patients. Dig Liver Dis. 2008;40:867–873. doi: 10.1016/j.dld.2008.03.022. [DOI] [PubMed] [Google Scholar]
- 19.Lappalainen M, Halme L, Turunen U, Saavalainen P, Einarsdottir E, Färkkilä M, Kontula K, Paavola-Sakki P. Association of IL23R, TNFRSF1A, and HLA-DRB1*0103 allele variants with inflammatory bowel disease phenotypes in the Finnish population. Inflamm Bowel Dis. 2008;14:1118–1124. doi: 10.1002/ibd.20431. [DOI] [PubMed] [Google Scholar]
- 20.Latiano A, Palmieri O, Valvano MR, D’Incà R, Cucchiara S, Riegler G, Staiano AM, Ardizzone S, Accomando S, de Angelis GL, et al. Replication of interleukin 23 receptor and autophagy-related 16-like 1 association in adult- and pediatric-onset inflammatory bowel disease in Italy. World J Gastroenterol. 2008;14:4643–4651. doi: 10.3748/wjg.14.4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okazaki T, Wang MH, Rawsthorne P, Sargent M, Datta LW, Shugart YY, Bernstein CN, Brant SR. Contributions of IBD5, IL23R, ATG16L1, and NOD2 to Crohn‘s disease risk in a population-based case-control study: evidence of gene-gene interactions. Inflamm Bowel Dis. 2008;14:1528–1541. doi: 10.1002/ibd.20512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perricone C, Borgiani P, Romano S, Ciccacci C, Fusco G, Novelli G, Biancone L, Calabrese E, Pallone F. ATG16L1 Ala197Thr is not associated with susceptibility to Crohn’s disease or with phenotype in an Italian population. Gastroenterology. 2008;134:368–370. doi: 10.1053/j.gastro.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 23.Peterson N, Guthery S, Denson L, Lee J, Saeed S, Prahalad S, Biank V, Ehlert R, Tomer G, Grand R, et al. Genetic variants in the autophagy pathway contribute to paediatric Crohn’s disease. Gut. 2008;57:1336–1337; author reply 1337. doi: 10.1136/gut.2008.152207. [DOI] [PubMed] [Google Scholar]
- 24.Van Limbergen J, Russell RK, Nimmo ER, Drummond HE, Smith L, Anderson NH, Davies G, Gillett PM, McGrogan P, Weaver LT, et al. Autophagy gene ATG16L1 influences susceptibility and disease location but not childhood-onset in Crohn’s disease in Northern Europe. Inflamm Bowel Dis. 2008;14:338–346. doi: 10.1002/ibd.20340. [DOI] [PubMed] [Google Scholar]
- 25.Zhi J, Zhi FC, Chen ZY, Yao GP, Guan J, Lin Y, Zhang YC. [Correlation of the autophagosome gene ATG16L1 polymorphism and inflammatory bowel disease] Nanfang Yikedaxue Xuebao. 2008;28:649–651. [PubMed] [Google Scholar]
- 26.Weersma RK, Stokkers PC, van Bodegraven AA, van Hogezand RA, Verspaget HW, de Jong DJ, van der Woude CJ, Oldenburg B, Linskens RK, Festen EA, et al. Molecular prediction of disease risk and severity in a large Dutch Crohn’s disease cohort. Gut. 2009;58:388–395. doi: 10.1136/gut.2007.144865. [DOI] [PubMed] [Google Scholar]
- 27.Hancock L, Beckly J, Geremia A, Cooney R, Cummings F, Pathan S, Guo C, Warren BF, Mortensen N, Ahmad T, et al. Clinical and molecular characteristics of isolated colonic Crohn’s disease. Inflamm Bowel Dis. 2008;14:1667–1677. doi: 10.1002/ibd.20517. [DOI] [PubMed] [Google Scholar]
- 28.Yang SK, Park M, Lim J, Park SH, Ye BD, Lee I, Song K. Contribution of IL23R but not ATG16L1 to Crohn’s disease susceptibility in Koreans. Inflamm Bowel Dis. 2009;15:1385–1390. doi: 10.1002/ibd.20921. [DOI] [PubMed] [Google Scholar]
- 29.Newman WG, Zhang Q, Liu X, Amos CI, Siminovitch KA. Genetic variants in IL-23R and ATG16L1 independently predispose to increased susceptibility to Crohn’s disease in a Canadian population. J Clin Gastroenterol. 2009;43:444–447. doi: 10.1097/MCG.0b013e318168bdf0. [DOI] [PubMed] [Google Scholar]
- 30.Zhang HF, Qiu LX, Chen Y, Zhu WL, Mao C, Zhu LG, Zheng MH, Wang Y, Lei L, Shi J. ATG16L1 T300A polymorphism and Crohn’s disease susceptibility: evidence from 13,022 cases and 17,532 controls. Hum Genet. 2009;125:627–631. doi: 10.1007/s00439-009-0660-7. [DOI] [PubMed] [Google Scholar]
- 31.Glas J, Seiderer J, Pasciuto G, Tillack C, Diegelmann J, Pfennig S, Konrad A, Schmechel S, Wetzke M, Török HP, et al. rs224136 on chromosome 10q21.1 and variants in PHOX2B, NCF4, and FAM92B are not major genetic risk factors for susceptibility to Crohn’s disease in the German population. Am J Gastroenterol. 2009;104:665–672. doi: 10.1038/ajg.2008.65. [DOI] [PubMed] [Google Scholar]
- 32.Török HP, Glas J, Endres I, Tonenchi L, Teshome MY, Wetzke M, Klein W, Lohse P, Ochsenkühn T, Folwaczny M, et al. Epistasis between Toll-like receptor-9 polymorphisms and variants in NOD2 and IL23R modulates susceptibility to Crohn’s disease. Am J Gastroenterol. 2009;104:1723–1733. doi: 10.1038/ajg.2009.184. [DOI] [PubMed] [Google Scholar]
- 33.Cooney R, Cummings JR, Pathan S, Beckly J, Geremia A, Hancock L, Guo C, Morris A, Jewell DP. Association between genetic variants in myosin IXB and Crohn’s disease. Inflamm Bowel Dis. 2009;15:1014–1021. doi: 10.1002/ibd.20885. [DOI] [PubMed] [Google Scholar]
- 34.Weersma RK, Zhernakova A, Nolte IM, Lefebvre C, Rioux JD, Mulder F, van Dullemen HM, Kleibeuker JH, Wijmenga C, Dijkstra G. ATG16L1 and IL23R are associated with inflammatory bowel diseases but not with celiac disease in the Netherlands. Am J Gastroenterol. 2008;103:621–627. doi: 10.1111/j.1572-0241.2007.01660.x. [DOI] [PubMed] [Google Scholar]
- 35.Weersma RK, Stokkers PC, Cleynen I, Wolfkamp SC, Henckaerts L, Schreiber S, Dijkstra G, Franke A, Nolte IM, Rutgeerts P, et al. Confirmation of multiple Crohn’s disease susceptibility loci in a large Dutch-Belgian cohort. Am J Gastroenterol. 2009;104:630–638. doi: 10.1038/ajg.2008.112. [DOI] [PubMed] [Google Scholar]
- 36.Franke A, Hampe J, Rosenstiel P, Becker C, Wagner F, Häsler R, Little RD, Huse K, Ruether A, Balschun T, et al. Systematic association mapping identifies NELL1 as a novel IBD disease gene. PLoS One. 2007;2:e691. doi: 10.1371/journal.pone.0000691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Franke A, Balschun T, Karlsen TH, Hedderich J, May S, Lu T, Schuldt D, Nikolaus S, Rosenstiel P, Krawczak M, et al. Replication of signals from recent studies of Crohn’s disease identifies previously unknown disease loci for ulcerative colitis. Nat Genet. 2008;40:713–715. doi: 10.1038/ng.148. [DOI] [PubMed] [Google Scholar]
- 38.Fisher SA, Tremelling M, Anderson CA, Gwilliam R, Bumpstead S, Prescott NJ, Nimmo ER, Massey D, Berzuini C, Johnson C, et al. Genetic determinants of ulcerative colitis include the ECM1 locus and five loci implicated in Crohn’s disease. Nat Genet. 2008;40:710–712. doi: 10.1038/ng.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beckly JB, Hancock L, Geremia A, Cummings JR, Morris A, Cooney R, Pathan S, Guo C, Jewell DP. Two-stage candidate gene study of chromosome 3p demonstrates an association between nonsynonymous variants in the MST1R gene and Crohn’s disease. Inflamm Bowel Dis. 2008;14:500–507. doi: 10.1002/ibd.20365. [DOI] [PubMed] [Google Scholar]
- 40.Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmada MM, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Libioulle C, Louis E, Hansoul S, Sandor C, Farnir F, Franchimont D, Vermeire S, Dewit O, de Vos M, Dixon A, et al. Novel Crohn disease locus identified by genome-wide association maps to a gene desert on 5p13.1 and modulates expression of PTGER4. PLoS Genet. 2007;3:e58. doi: 10.1371/journal.pgen.0030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parkes M, Barrett JC, Prescott NJ, Tremelling M, Anderson CA, Fisher SA, Roberts RG, Nimmo ER, Cummings FR, Soars D, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn’s disease susceptibility. Nat Genet. 2007;39:830–832. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raelson JV, Little RD, Ruether A, Fournier H, Paquin B, Van Eerdewegh P, Bradley WE, Croteau P, Nguyen-Huu Q, Segal J, et al. Genome-wide association study for Crohn’s disease in the Quebec Founder Population identifies multiple validated disease loci. Proc Natl Acad Sci USA. 2007;104:14747–14752. doi: 10.1073/pnas.0706645104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamazaki K, Takahashi A, Takazoe M, Kubo M, Onouchi Y, Fujino A, Kamatani N, Nakamura Y, Hata A. Positive association of genetic variants in the upstream region of NKX2-3 with Crohn’s disease in Japanese patients. Gut. 2009;58:228–232. doi: 10.1136/gut.2007.140764. [DOI] [PubMed] [Google Scholar]
- 46.Kugathasan S, Baldassano RN, Bradfield JP, Sleiman PM, Imielinski M, Guthery SL, Cucchiara S, Kim CE, Frackelton EC, Annaiah K, et al. Loci on 20q13 and 21q22 are associated with pediatric-onset inflammatory bowel disease. Nat Genet. 2008;40:1211–1215. doi: 10.1038/ng.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, Abraham C, Regueiro M, Griffiths A, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Biank V, Broeckel U, Kugathasan S. Pediatric inflammatory bowel disease: clinical and molecular genetics. Inflamm Bowel Dis. 2007;13:1430–1438. doi: 10.1002/ibd.20213. [DOI] [PubMed] [Google Scholar]
- 49.Kugathasan S, Fiocchi C. Progress in basic inflammatory bowel disease research. Semin Pediatr Surg. 2007;16:146–153. doi: 10.1053/j.sempedsurg.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 50.Barnich N, Carvalho FA, Glasser AL, Darcha C, Jantscheff P, Allez M, Peeters H, Bommelaer G, Desreumaux P, Colombel JF, et al. CEACAM6 acts as a receptor for adherent-invasive E. coli, supporting ileal mucosa colonization in Crohn disease. J Clin Invest. 2007;117:1566–1574. doi: 10.1172/JCI30504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Darfeuille-Michaud A, Neut C, Barnich N, Lederman E, Di Martino P, Desreumaux P, Gambiez L, Joly B, Cortot A, Colombel JF. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn’s disease. Gastroenterology. 1998;115:1405–1413. doi: 10.1016/s0016-5085(98)70019-8. [DOI] [PubMed] [Google Scholar]
- 52.Strober W, Fuss IJ, Blumberg RS. The immunology of mucosal models of inflammation. Annu Rev Immunol. 2002;20:495–549. doi: 10.1146/annurev.immunol.20.100301.064816. [DOI] [PubMed] [Google Scholar]
- 53.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 54.Levine B, Deretic V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat Rev Immunol. 2007;7:767–777. doi: 10.1038/nri2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 56.Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xavier RJ, Huett A, Rioux JD. Autophagy as an important process in gut homeostasis and Crohn’s disease pathogenesis. Gut. 2008;57:717–720. doi: 10.1136/gut.2007.134254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kuballa P, Huett A, Rioux JD, Daly MJ, Xavier RJ. Impaired autophagy of an intracellular pathogen induced by a Crohn’s disease associated ATG16L1 variant. PLoS One. 2008;3:e3391. doi: 10.1371/journal.pone.0003391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Münz C. Enhancing immunity through autophagy. Annu Rev Immunol. 2009;27:423–449. doi: 10.1146/annurev.immunol.021908.132537. [DOI] [PubMed] [Google Scholar]
- 60.Zeissig S, Bojarski C, Buergel N, Mankertz J, Zeitz M, Fromm M, Schulzke JD. Downregulation of epithelial apoptosis and barrier repair in active Crohn‘s disease by tumour necrosis factor alpha antibody treatment. Gut. 2004;53:1295–1302. doi: 10.1136/gut.2003.036632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boirivant M, Marini M, Di Felice G, Pronio AM, Montesani C, Tersigni R, Strober W. Lamina propria T cells in Crohn’s disease and other gastrointestinal inflammation show defective CD2 pathway-induced apoptosis. Gut. 1999;116:557–565. doi: 10.1016/s0016-5085(99)70177-0. [DOI] [PubMed] [Google Scholar]
- 62.Yukawa M, Iizuka M, Horie Y, Yoneyama K, Shirasaka T, Itou H, Komatsu M, Fukushima T, Watanabe S. Systemic and local evidence of increased Fas-mediated apoptosis in ulcerative colitis. Int J Colorectal Dis. 2002;17:70–76. doi: 10.1007/s003840100340. [DOI] [PubMed] [Google Scholar]
- 63.Karamanolis DG, Kyrlagkitsis I, Konstantinou K, Papatheodoridis GV, Karameris A, Mallas E, Ladas SD, Raptis S. The Bcl-2/Bax system and apoptosis in ulcerative colitis. Hepatogastroenterology. 2007;54:1085–1088. [PubMed] [Google Scholar]
- 64.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]