Abstract
Major depression is a serious mental illness frequently associated with devastating consequences for those affected. Suicide rates are significantly elevated, creating a sense of urgency to identify effective yet safe treatment options. A plethora of antidepressants available on the market today, designed to act on different neurotransmitter systems in the brain, provides the clinician with several treatment strategies. There is, however, very little guidance as to which antidepressant may be most successful in a certain individual. Biomarkers that can predict treatment outcome would thus be of great value, shortening the time until remission and reducing costs for the healthcare system by reducing unsuccessful treatment attempts. The proven contribution of heredity to major depression risk suggests that genetic markers may be good biomarkers for treatment outcome. The Sequenced Treatment Alternatives to Relieve Depression (STAR*D) study and a large ancillary pharmacogenetic study on 1953 STAR*D participants constitute the largest effort to date to identify genetic predictors of antidepressant treatment outcome. In this review, results of candidate gene studies carried out so far are summarized and discussed, and some future directions are proposed.
1. Introduction
Depressive disorders account for up to 80% of all psychiatric hospitalizations, and play a role in a substantial portion of hospital admissions in general.[1,2] Individuals affected by depression are more likely to experience comorbidity with other medical conditions.[3] Furthermore, depression is reported to increase the risk for suicide attempts and suicidal ideation by at least 4–6 fold.[4] Besides the personal cost for afflicted individuals, which is often devastating, the total cost to society is immense. Identification of disease causing factors, along with new and more efficient treatment options, are therefore highly relevant.
Heritable factors have consistently been shown to play an important role in susceptibility to depressive disorders (for review, see[5]), but little data exist regarding heritability of treatment response or adverse effects of medication. So far, few genetic predictors of treatment response or adverse events have been identified, and fewer still have been replicated consistently (for review, see[6]).
Over the past decades there has been an impressive increase in the availably of treatment options. A number of psychotherapies (talk-therapies, behavior therapies) and pharmacologic agents are now available to treat depression.[7,8] However, today’s treatments are not curative. Clinical observations have shown that a substantial proportion of depressed patients will fail to respond to the first-line antidepressant treatment. Often, a second choice of treatment or addition of a second agent (augmentation) is needed to achieve response, and full remission of symptoms can be difficult to achieve quickly.[7,9–11] Although most will eventually recover from the index episode, affected individuals often need a lifetime course of treatment to prevent recurrence. In addition, some reports raise the question of whether antidepressants can actually increase suicidal thinking or behavior in some patients.[12,13]
In light of all this, clinicians encounter a very difficult task when treating patients suffering from depressive disorders. Which initial treatment should be used? If this fails, what is the next best choice? Is there a need for combination therapy? Which dosage and duration of treatment should be used for achievement of full functional recovery, while minimizing adverse events?[7,10] Existing clinical data provide few answers, leading most clinicians to employ a trial-and-error strategy, albeit enhanced by their own clinical judgment. Genetic tests that help guide treatment decisions would be a great advance.
The general aim of the STAR*D study was to help the clinician with such difficult treatment decisions by elucidating which treatment options offer the greatest efficacy and tolerability. An important aim of the study was to identify alternative treatment strategies for the estimated two thirds of patients with major depression who do not achieve full remission after the initial treatment. Priority was given to identification of individual patient characteristics related to successful treatment outcomes in real-world clinical situations.
In addition, the STAR*D team joined forces with several teams of geneticists. While the approaches have varied, the goal of every team has been the identification of genetic markers of treatment outcome, including both remission and adverse events. Results from these efforts have been published in several reports[14–19] and are summarized in this review.
2. Background
2.1 Depression
According to the World Health Organization (WHO) major depression is one of the most disabling diseases worldwide.[20] It is predicted to be the second most disabling disease worldwide in the year of 2020.[21–23] The lifetime prevalence, according to these reports, is estimated to be 16–20%, the highest prevalence among the psychiatric diseases.[24,25] Women are 1.5–3.0 times more likely to be affected than men,[25,26] and MDD is more prevalent in younger people[27] and among those with other psychiatric disorders.[28]
There is a wealth of studies that have addressed the course of the disease, all varying between short and long term follow-up periods. The results give a fairly consistent picture of a chronic recurrent disorder with an average of one depressive episode every 5 years[29,30] but as many as 30% of the patients may experience a chronic course.[29,31]
Randomized clinical trials have consistently shown that most antidepressants available on the market are more effective than placebo, at least when treating more severe forms of depression in adults.[32–34] Nevertheless, a substantial minority of patients (about 20–40%, depending on initial severity) show a placebo response. Recent studies have drawn a distinction between response (usually measured as >50% improvement from baseline), where patients can still be quite impaired by their symptoms, and remission, where symptoms are essentially gone and there is no residual impairment. Remission rates are lower than response rates for both active drug and placebo, but sustained remission of moderate or severe depression is rare with placebo alone.[34–37] When treating depressive disorders, response without remission is associated with continuing disability and early relapse.[38,39]
Some reports favor particular types of antidepressants[36] whereas other reports find only marginal differences.[32] Certain forms of psychotherapy may compare favorably with pharmacological treatment, especially for less severe cases.[35] Approximately one third of patients achieve full remission when treated with antidepressants; while many of the others will continue to experience ongoing symptoms and considerable levels of disability. We clearly need new treatment strategies that maximize remission.
2.2 Suicide and Suicidal Ideation
Depression has remained the main psychiatric disorder associated with suicide, at least from the 1960’s through 1999 – 2003.[40,41] Suicide remains a leading cause of death among young people,[42] and one of the major causes of death among patients with mood disorders in all age groups.[43,44] Effective treatment of depressive disorders is the best way to prevent suicide. Paradoxically, antidepressant treatment may also provoke (or exacerbate) suicidal thoughts and behavior, especially during the early phase of treatment in young people.[12,13,45] Concern about treatment-emergent suicidal ideation led the US Food & Drug Administration to issue Black Box warnings of the risk of suicidal thoughts and behavior in adolescents and young adults treated with antidepressants.
However, doubts have persisted as to whether antidepressants are a real risk factor for suicide, even in certain subgroups of patients.[46–48] Some studies report a protection from suicidal thinking with the use of antidepressants.[49] Not all studies have taken into account the previous history of suicidal behavior[46] and many studies rely on meta-analyses with a mixed origin of the underlying data. There are important distinctions between attempts, ideation and thoughts of suicide, and actual death by suicide,[50] but many studies fail to discriminate clearly among these.
A genetic test able to predict which patients develop suicidal thinking during antidepressant treatment would be a great aid for the clinician: Those at risk could be put into special treatment regimes, and the vast majority of remaining patients would not be wrongfully advised against using the antidepressant. However, it is not clear what role, if any, genes might play in the etiology of treatment-emergent suicidal ideation. Family and twin studies certainly demonstrate that suicidal thoughts and behavior that occur outside of the context of antidepressants are both familial and heritable,[50–52] but no data exist as to the heritability of treatment-emergent suicidal thoughts or behavior, and such data would be nearly impossible to obtain.
2.3 The pharmacogenetic approach
Today pharmacogenetics refers to the study of individual differences in drug response due to genetic variation among individuals, whereas the term pharmacogenomics tends to emphasize the technologies used in the development of new drugs based on the knowledge of all genes in the human genome.[53] In practice, the two terms are often used interchangeably. Whether pharmacogenetics is an old discipline or was born after the completion of the Human Genome Project is still a matter of debate.[54,55]
In pharmacotherapy of psychiatric patients, clinicians have to try to help the substantial proportion of patients who do not respond sufficiently or suffer adverse effects during treatment. Of course, variable response and adverse effects may be explained by genetic variability in pharmacodynamic and pharmacokinetic pathways, but also by many other non-genetic factors, including, e.g., misclassification of disease or subtype, cultural forces, or environmental factors.[56,57]
Pharmacogenetics holds a great promise for future clinical practice to use genetic markers for prediction of treatment outcome.[56,58,59] Numerous studies have been carried out in the field, but sample sizes for most studies have so far been fairly small. Association findings in the field have recently been reviewed in detail by others.[60]
It is hoped that pharmacogenetics will lead to the development of ”personalized medicine”, with better safety and efficacy than is currently possible for many drugs.[53,61,62] But many challenges remain: How can adequate sample sizes be collected for study? What is the best way to establish robust findings? Which pharmacogenetic markers will have the greatest clinical utility? How can robust, clinically useful markers be translated most efficiently to clinical use?
2.4 The Sequenced Treatment Alternatives to Relieve Depression (STAR*D) Study
As part of a larger initiative aimed at promoting large-scale, non-industry funded clinical trials, the National Institutes of Health awarded a multi-million dollar contract to study the optimal treatment strategy for major depression in outpatients. The resulting STAR*D study included over 4000 out-patients with non-psychotic DSM-IV major depressive disorder, aged between 18 and 75, treated at 14 regional psychiatric and primary care clinics across the US over a period of 7 years[63,64]. STAR*D is by far the largest study of its kind.
Ascertainment was aimed at enrolling a representative sample of outpatients with a typical mix of race, ethnicity, and socioeconomic status. There were few exclusion criteria, and adjunctive treatment with benzodiazepines and hypnotics was allowed when needed. All participants received active treatment, since placebo randomization tends to select for an unrepresentative subset of mildly depressed patients. Many study participants were indeed quite ill: 75% experienced two or more prior episodes, or a sustained episode, of depression during the two years prior to study entry, and two-thirds reported comorbidity with another psychiatric disorder.[65]
Treatment was organized into “levels,” each with a limited number of treatment options offered for up to 12 weeks. Participants who responded well moved into follow-up; the others had the option to move on to the next level of treatment. All participants began in Level 1 with citalopram, 10–60 mg/day, as tolerated, for up to 12 weeks. Citalopram is a typical selective serotonin reuptake inhibitor (SSRI).
In order to enable future pharmacogenetic studies, STAR*D participants were asked to donate a blood sample for DNA extraction. About half agreed, and blood samples were drawn from close to 2000 participants. These samples are now available to researchers through the Rutgers Cell and DNA Repository in Piscataway, NJ (http://www.rucdr.org/).
STAR*D is now the largest pharmacogenetic study of mood disorders carried out to date. It has provided and continues to provide a tremendous opportunity to elucidate genetic determinants of treatment outcome. Some of the published pharmacogenetic findings from the first level of treatment will be described below.
3. Results from candidate gene studies
3.1 Treatment response and remission
3.1.1 HTR2A and GRIK4
One large-scale candidate gene association study of the STAR*D sample was aimed at investigating the outcome phenotypes ’remission’ and ’response.’ Response refers to improvement in depressive symptoms, while remission refers to complete or nearly-complete recovery (for details regarding the experimental design, see[17]). A split-sample design was utilized in order to address the multiple testing problem (subsequent work suggested alternative approaches to this problem[66]).
This first study included 68 genes, based on known gene function or previously reported involvement in depressive disorder or treatment outcome. Accordingly, after stringent criteria, 768 genetic markers (SNP’s) were selected. Results from the primary analysis have been published in two reports.[17,18] Two markers showed significant association that was reproduced in both split samples: One marker was located in the gene HTR2A and one was located in the gene GRIK4.
HTR2A codes for the type 2A serotonin receptor, which has been implicated in a large number of association studies in schizophrenia, depression and treatment response.[67] However, the markers that showed association in the McMahon et al study were different from, and uncorrelated with, those previously implicated.[68–70] Thus, although this study provides further evidence for involvement of the HTR2A gene in antidepressant outcome, no direct replication of results from previous studies could be established.
Interestingly, it was found that the frequency of the treatment response associated variant was lower in Black patients than in Whites. The clinical STAR*D study had shown a poorer response of Black participants to the citalopram treatment.[71,72]
This finding highlights one of the problems with using an ethnically mixed group of patients: Any polymorphism that shows a large difference in frequency between populations will show association with treatment response if such response differs greatly between these populations. In order to exclude that the observed association with citalopram treatment was an artifact induced by such stratification, McMahon et al. tested association in Whites only, which still remained highly significant. This indicates that it is unlikely that the observed association was an artifact caused by the inclusion of participants of mixed ethnical backgrounds.
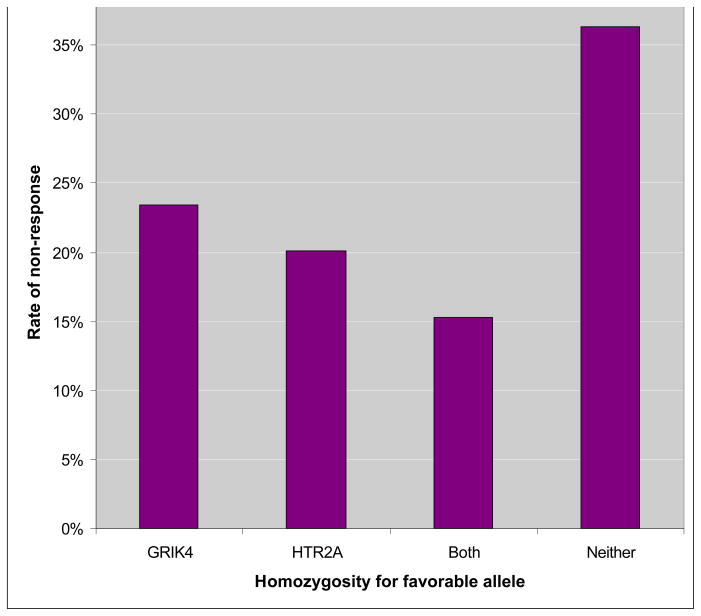
The associated marker, rs7997012, has a modest effect on treatment outcome in the STAR*D sample. Patients homozygous for the response-associated allele had an 18% lower absolute risk of non-response, compared to those homozygous for the other allele (Figure 1). Interestingly, the frequency of the response-associated allele was 42% in white participants and 6% in black participants, suggesting that this allele may contribute to the known racial differences in outcome of antidepressant treatment. The markers reside within the second intron of HTR2A, and have no clear functional consequences. This finding now awaits replication in an independent sample and functional characterization.
Figure 1.
Rates of non-response to citalopram among homozygote carriers and non-carriers of response-associated marker alleles at GRIK4 and HTR2A
The second marker significantly associated with antidepressant outcome in the STAR*D sample was located in the GRIK4 gene, coding for a kainic-acid type glutamate receptor (Paddock et al. 2007). The effect of the GRIK4 allele was also modest, but homozygote carriers of the response-associated marker alleles of both GRIK4 and HTR2A were about half as likely to experience non-response to treatment with citalopram as those who did not carry any of these alleles (Figure 1). This finding was the first direct human evidence that the glutamate system plays an important role in response to an antidepressant. Glutamate signaling and post-synaptic 5-HT signaling are known to interact via the post-synaptic complex DARRP-32 (reviewed in Svenningsson et al 2004[73]), so the additive effects of HTR2A and GRIK4 on treatment outcome may work via this pathway. GRIK4 had previously been implicated in a translocation event in a patient with schizophrenia,[74] and the marker identified in STAR*D had been associated with schizophrenia in another sample.[74] This suggests that GRIK4 may contain variants involved in several psychiatric traits. Large samples will likely be needed for replication testing.[75]
3.1.2 The serotonin transporter gene (SLC6A4)
The development of SSRIs as anti-depressant drugs has mainly been based on knowledge gathered during use of the first and second line of antidepressant medications (tricyclics and monoamine oxidase inhibitors). All of these regulate concentrations of several synaptic monoamines unselectively. The selective serotonin reuptake inhibitors were designed to block reuptake of serotonin from the synapse by the serotonin transporter. SSRIs quickly became the most widely prescribed antidepressants in Western countries, mainly due to their wide acceptability, with fewer adverse effects and greater drug safety. Since all SSRIs bind the serotonin transporter, the gene encoding this protein has become an obvious candidate for pharmacogenetic studies in depression.
The 5-HT transporter gene, SLC6A4, is modulated by a functional polymorphic promoter region, known as the linked polymorphic region or LPR, which is located upstream of the transcription start site. This polymorphism consists of a complex repeat polymorphism with many alleles, which are typically grouped into short (S) and long (L) allele sets.[76]
In vitro studies in human cell lines have shown that the LPR is associated with changes in 5HTT translation.[77] L-alleles produce higher levels of 5HTT mRNA, for review see.[78] Nakamura and collaborators have argued that a single nucleotide variant (A→G) within the LPR should to be considered when evaluating 5-HTT function.[79] Subsequent studies of these variants on the functional levels mRNA of HTTLPR also showed the importance of the single base mutation (G) in the long allele. The LG polymorphism was associated with decrease of function almost equivalent to S-allele carriers.[80]
The LPR has been reported to be associated with a large number of psychiatric and behavioural phenotypes. These include the personality trait of novelty-seeking,[81] various measures of depression and anxiety,[82] depression that follows adverse life events,[83] and lifetime risk of major depression.[84] The S-allele is often – but not always – more common in cases.
A number of pharmacogenetic studies have asked whether genetic variants in 5HTT could predict antidepressant treatment outcome. These studies are quite heterogeneous with regard to study design, sample characteristics, ethnicity, and intervention strategies (reviewed by Smiths KM et al.[85]), which complicates an overall interpretation of the results. For example, people of Asian origin differ in LPR allele frequencies.[86] In sum, previous literature suggests that the S/S genotype may be associated with risk of depression and reduced response to SSRI treatment.
5-HTT was also studied in the primary outcome candidate gene study of STAR*D. A dense set of SNP markers spanning the SLC6A4 coding region were genotyped. However, no significant association with treatment outcome was observed.[17] The LPR was investigated directly in a second study of the STAR*D sample.[14] No association was detected between treatment outcome and the L/S alleles of the LPR. There was also no significant association with treatment outcome when the LPR was treated as a tri-allelic polymorphism, based on the SNP noted above. However, a significant association of the tri-allelic polymorphism with overall side effect burden was observed. This is consistent with an earlier study that had detected a strong association between the S-allele and SSRI side effects in a geriatric sample.[87] It is possible that an association between the LPR and SSRI side effects may have influenced other, smaller pharmacogenetic studies that did not account for drug tolerability, since medication-intolerant patients would be able to tolerate only a low dose, and may be under-medicated.
Despite the rather exhaustive investigation of the serotonin transporter in the STAR*D study, there was no evidence of a role in treatment outcome. This shows that even an obvious candidate gene coding for the drug target itself and previously implicated in treatment response can render negative results in a well-powered sample. It remains to be seen whether the association between 5-HTT and side effects observed in STAR*D will prove more robust than that with treatment outcome.
3.1.3 FKBP5
Homeostatic response to stress is under strong influence of the hypothalamic-pituitary-adrenal axis (HPA-axis).[88,89] Agents such as adrenocorticotrophin (ACTH), arginine vasopressin (AVP), and corticotrophin-releasing factor (CRF) appear to have important stress-regulatory functions as well.[90] Abnormalities in HPA-axis function have been reliably detected in stress-related disorders and it has been proposed that dysregulation in this neuroendocrine axis, mainly by altered secretion of cortisol, is a major mechanism for developing depressive disorders.[91,92] This has led to the hypothesis that drugs which target the HPA-axis may be beneficial for treating depressive disorders.[93,94] Such drugs might normalize cortisol levels or modulate glucocorticoid receptor (GR) function.[93,95]
Animal studies suggest that the glucocorticoid receptor acts as a prominent regulator of the HPA axis.[96–98] Furthermore, some genetic studies have found that polymorphisms in the GR-gene may be associated with susceptibility to depression[99,100] as well as treatment response.[99]
Possible involvement of glucocorticoid receptor function in the HPA-axis regulation and depression and response to antidepressant treatment was studied by Binder and collaborators.[101] No association with depression was observed for markers in the GR-, AVP-, CRH-genes or five chaperones (BAG1, STUB1, TEBP, FKBP4 and FKBP5) of the glucocorticoid receptor, in hospitalized patients of European ancestry diagnosed major depression. However, a highly-significant association was observed between treatment response and a marker located in FKBP5, which encodes one of the chaperone molecules. The response phenotype was associated with the homozygous state of the T-allele of the functional polymorphism rs1360780. These same individuals had also experienced more lifetime episodes of depression.
Small studies did not replicate these findings in outpatients of Chinese ancestry,[102] or in an independent European sample.[103] However, these non-replications could be due to the low power of small sample sizes. The large STAR*D cohort, therefore constituted an excellent opportunity to investigate association of markers in the FKBP5-gene.
The results,[16] supported association of markers in the FKBP5-gene with disease status as well as antidepressant treatment outcome. No association with number of prior lifetime depressive episodes was detected, however. Effect sizes were clearly smaller than those reported by Binder et al. This may be reflect differences in the samples (the STAR*D enrolled outpatients with unipolar depression rather than hospitalized patients with unipolar and bipolar disorders). The ’Winner’s Curse’ also predicts that the initial study reporting an association often over-estimates effect sizes.[104,105]
Further studies are needed, but these findings appear to constitute an independently replicated association between a functional genetic polymorphism and antidepressant treatment outcome. The relative importance of this association in predicting treatment outcome prospectively remains to be determined.
3.1.4 TREK1
As a complementary approach in order to identify genetic risk factors for psychiatric disorders, Roy Perlis and collaborators[106] used findings from animal studies to generate hypotheses for genetic association testing. Four brain expressed genes were investigated, all previously involved in treatment response in a mouse model: TREK1 (KCNK2), SLC18A2 (VMAT2), S100A10 and HDAC5. When the corresponding four genomic loci where tested for association with treatment outcome based on the level 1 population, non of the markers in any of the genes provided p-values that survived correction for multiple testing. However, it should be mentioned that analysis of further levels, which is not subject of this review, yielded positive results: based on the 751 participants entering the next step of treatment, out of four markers in the TREK1-gene with nominal p-values ≤ 0.01, three markers were still significant associated with treatment response after correction with multiple testing. The TREK1 gene encodes a potassium channel and has been shown to be an important target for antidepressant drugs in mouse models.[107] Mice lacking this gene are insensitive to SSRI treatment.[108]
3.2 Treatment-Emergent Suicidal Ideation
3.2.1 GRIK2 and GRIA3
Laje et al.[15] used a candidate-gene approach in the same 68 candidate genes that were investigated in the primary outcome analysis (see above). They defined treatment-emergent suicidal ideation as the absence of suicidal thoughts at the first clinical visit followed by onset of suicidal thoughts at any later visit during the 12 weeks of Level 1. By this case definition, 120 patients were considered to display treatment-emergent suicidal ideation, although the majority of these reported only passive death wishes. The rest of the sample reported suicidal thoughts at baseline or denied suicidal thoughts throughout level 1, and was treated as “controls” (n=1742).
The analysis revealed two markers which, after correction for multiple testing, were significantly associated with treatment-emergent suicidal ideation. The markers resided in the ionotrophic glutamate receptor genes GRIK2 and GRIA3. Both markers reside within introns, and have no obvious functional significance. The GRIK2 marker, rs2818224, conferred a rather high odds ratio of about 8 in the homozygous state, but this genotype was uncommon (12%). The risk-allele in GRIA3, while more common, conferred a more modest OR of 1.9. This was the first study to demonstrate a significant, overall association between TESI and genetic markers, although a study published earlier that same year found suggestive evidence of association with alleles in another gene -- CREB1 – in males,[109] discussed below.
Although these findings have evoked substantial interest in the scientific community and the media,[110] a clinically useful genetic test for TESI does not seem to be imminent. Replication in an independent sample is the essential next step. Independent replication will not only verify true positive associations, but will also give a better estimation of thetrue effect size, free of the Winner’s Curse. If such a test should emerge in the future, it might offer a useful tool for clinicians who wish to identify patients in need of closer monitoring or alternative treatments.
4. Discussion
We have reviewed some of the early pharrmacogenetic findings from the STAR*D trial. Some previously well-established findings (5HTT) were not supported, such as the association between 5-HTT and treatment outcome, but several previous association findings received direct (FKBP5) or indirect (HTR2A) support. Several novel findings also emerged, which will need to be tested in independent samples. So far we have examined only a small fraction of the known common genetic variation and of the possible outcome phenotypes and adverse events. Outcomes and adverse events in treatment levels beyond level 1 remain to be studied, although sample sizes fall rapidly as patients move through the levels. Genome-wide association studies in the STAR*D cohort may provide further answers.
As seen so often in the field of complex genetics, we learn from the STAR*D pharmacogenetic approach that having a large sample is good, but having an even larger sample is better. Most of the effect sizes detected in the STAR*D genetics studies so far are small – and the only large effect (GRIK2 and TESI) was seen for an uncommon genotype. Thus very large samples will be needed in order test these findings for replication in other samples. Recent successful replication studies in type-2-diabetes[111–114] have demonstrated the value of very large sample sizes, probably an order of magnitude larger than what was available in the STAR*D cohort.
Clinical tests will likely require information from a panel of many genetic markers, the majority of which still remain to be identified. If there are strong interactions among the markers, then many specific multi-allele combinations will need to be considered. The numbers of patients who carry any particular multi-allele combination will thus be necessarily low, leading to a test with low sensitivity. On the other hand, if there is strong heterogeneity of effects, with some alleles being important in only some populations, then test specificity may prove a problem.
5. Future Directions: Moving pharmacogenetics findings from the bench to the bedside
There is a sense of urgency to use pharmacogenetic information to inform treatment decisions.[115] Patients might benefit from being matched with medications to which they are likely to respond well without serious side effects. Health care systems might benefit from more efficient treatment selection and consequent more rapid recovery of patients. Society might benefit from decreased costs related to reduced health care utilization, recouped productivity by more treatment-responsive patients, and decreased use of unnecessary expensive treatments and polypharmacy.
However, a too-rapid movement from laboratory to clinic poses significant risks. Poorly-validated tests might actually mislead physicians into prescribing less than optimal treatments.[116] Inadequate data on the range of appropriate treatment-contexts and target populations may lead to the misapplication of particular tests, reducing their value in clinical decision-making. Even valid tests, properly used, may not really affect the choice of treatments or dosages, especially when treatment alternatives are limited or other clinical factors take precedence. The recent report of The Evaluation of Genomic Applications in Practice and Prevention (EGAPP) working group is a good example: Although CYP450 polymorphisms can be reliably genotyped and are associated with blood levels of many SSRIs, CYP450 genotyping was found to have little clinical impact and no real utility in clinical decision making for typical major depressive disorder.[117]
In light of these issues, a consensus seems to be emerging that proposed pharmacogenetic tests should be required to meet some criteria before widespread clinical application is warranted. Criteria under discussion include[117]: 1) Analytic validity: is the genetic test to be used an accurate reflection of the underlying DNA sequence? 2) Clinical validity: is the genetic marker reliably (reproducibly) associated with the outcome? Is the reported sensitivity and specificity valid in the targeted clinical population? 3) Clinical utility: Will the results of the test actually affect clinical decision-making in a way that improves patient outcomes?
Analytical validity is relatively easy to establish in this era of high-throughput genomic technologies, but should not be taken for granted. Unforeseen variation within the target sequence of PCR primers, the existence of highly-homologous pseudogenes, and the potential impact of copy-number variation can all deleteriously affect the accuracy of a particular DNA assay. Clinical validity is often a major hurdle for pharmacogenetics, since genetic associations are inherently difficult to replicate and background-genetic and environmental factors can have a major impact on predictive value in different patient groups.[118] Clinical utility is in many ways the most difficult hurdle. A test with high clinical utility has a large impact on clinical decision-making, changing choice of treatment, dosage, etc. To meet this standard, a pharmacogenetic test must not only be valid, but compelling in its predictive value, and must confer unique information that is not adequately captured by the standard clinical history and physical examination.
A few pharmacogenetic tests – none in the field of psychiatry – may be close fulfilling these criteria. Such tests may blaze a path to the future. Meanwhile, caveat emptor.
Acknowledgments
Supported by the Swedish Research Council, the Swedish Foundation for Strategic Research, Karolinska Institutet foundations, the Center for Gender Medicine at Karolinska Institutet, the National Alliance for Research in Schizophrenia and Depression (F.J.M. and S.P.), and the NIMH Intramural Research Program (FJM).
Reference List
- 1.Shapiro S, Skinner EA, Kessler LG, et al. Utilization of health and mental health services. Three Epidemiologic Catchment Area sites. Arch Gen Psychiatry. 1984;41:971–8. doi: 10.1001/archpsyc.1984.01790210053007. [DOI] [PubMed] [Google Scholar]
- 2.Narrow WE, Regier DA, Rae DS, et al. Use of services by persons with mental and addictive disorders. Findings from the National Institute of Mental Health Epidemiologic Catchment Area Program. Arch Gen Psychiatry. 1993;50:95–107. doi: 10.1001/archpsyc.1993.01820140017002. [DOI] [PubMed] [Google Scholar]
- 3.Gruenberg AM, Goldstein RD. Depressive Disorders. (1) 1997:990–1019. [Google Scholar]
- 4.Waldrop AE, Hanson RF, Resnick HS, et al. Risk factors for suicidal behavior among a national sample of adolescents: implications for prevention. J Trauma Stress. 2007;20:869–79. doi: 10.1002/jts.20291. [DOI] [PubMed] [Google Scholar]
- 5.Levinson DF. The genetics of depression: a review. Biol Psychiatry. 2006;60:84–92. doi: 10.1016/j.biopsych.2005.08.024. [DOI] [PubMed] [Google Scholar]
- 6.Jones DS, Perlis RH. Pharmacogenetics, race, and psychiatry: prospects and challenges. Harv Rev Psychiatry. 2006;14:92–108. doi: 10.1080/10673220600642895. [DOI] [PubMed] [Google Scholar]
- 7.Fava M. Augmentation and combination strategies in treatment-resistant depression. J Clin Psychiatry. 2001;62 (Suppl 18):4–11. [PubMed] [Google Scholar]
- 8.Kupfer DJ. The pharmacological management of depression. Dialogues Clin Neurosci. 2005;7:191–205. doi: 10.31887/DCNS.2005.7.3/dkupfer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fava M, Rush AJ. Current status of augmentation and combination treatments for major depressive disorder: a literature review and a proposal for a novel approach to improve practice. Psychother Psychosom. 2006;75:139–53. doi: 10.1159/000091771. [DOI] [PubMed] [Google Scholar]
- 10.Marangell LB. Switching antidepressants for treatment-resistant major depression. J Clin Psychiatry. 2001;62 (Suppl 18):12–7. [PubMed] [Google Scholar]
- 11.Pridmore S, Turnier-Shea Y. Medication options in the treatment of treatment-resistant depression. Aust N Z J Psychiatry. 2004;38:219–25. doi: 10.1080/j.1440-1614.2004.01335.x. [DOI] [PubMed] [Google Scholar]
- 12.King RA, Riddle MA, Chappell PB, et al. Emergence of self-destructive phenomena in children and adolescents during fluoxetine treatment. J Am Acad Child Adolesc Psychiatry. 1991;30:179–86. doi: 10.1097/00004583-199103000-00003. [DOI] [PubMed] [Google Scholar]
- 13.Teicher MH, Glod C, Cole JO. Emergence of intense suicidal preoccupation during fluoxetine treatment. Am J Psychiatry. 1990;147:207–10. doi: 10.1176/ajp.147.2.207. [DOI] [PubMed] [Google Scholar]
- 14.Hu XZ, Rush AJ, Charney D, et al. Association between a functional serotonin transporter promoter polymorphism and citalopram treatment in adult outpatients with major depression. Arch Gen Psychiatry. 2007;64:783–92. doi: 10.1001/archpsyc.64.7.783. [DOI] [PubMed] [Google Scholar]
- 15.Laje G, Paddock S, Manji H, et al. Genetic markers of suicidal ideation emerging during citalopram treatment of major depression. Am J Psychiatry. 2007;164:1530–8. doi: 10.1176/appi.ajp.2007.06122018. [DOI] [PubMed] [Google Scholar]
- 16.Lekman M, Laje G, Charney D, et al. The FKBP5-Gene in Depression and Treatment Response-an Association Study in the Sequenced Treatment Alternatives to Relieve Depression (STAR*D) Cohort. Biol Psychiatry. 2008 doi: 10.1016/j.biopsych.2007.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McMahon FJ, Buervenich S, Charney D, et al. Variation in the gene encoding the serotonin 2A receptor is associated with outcome of antidepressant treatment. Am J Hum Genet. 2006;78:804–14. doi: 10.1086/503820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paddock S, Laje G, Charney D, et al. Association of GRIK4 with outcome of antidepressant treatment in the STAR*D cohort. Am J Psychiatry. 2007;164:1181–8. doi: 10.1176/appi.ajp.2007.06111790. [DOI] [PubMed] [Google Scholar]
- 19.Perlis RH, Purcell S, Fava M, et al. Association between treatment-emergent suicidal ideation with citalopram and polymorphisms near cyclic adenosine monophosphate response element binding protein in the STAR*D study. Arch Gen Psychiatry. 2007;64:689–97. doi: 10.1001/archpsyc.64.6.689. [DOI] [PubMed] [Google Scholar]
- 20.Lopez AD. The evolution of the Global Burden of Disease framework for disease, injury and risk factor quantification: developing the evidence base for national, regional and global public health action. Global Health. 2005;1:5. doi: 10.1186/1744-8603-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopez AD, Murray CC. The global burden of disease, 1990–2020. Nat Med. 1998;4:1241–3. doi: 10.1038/3218. [DOI] [PubMed] [Google Scholar]
- 22.Mathers CD, Loncar D. Projections of global mortality and burden of disease from 2002 to 2030. PLoS Med. 2006;3:e442. doi: 10.1371/journal.pmed.0030442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murray CJ, Lopez AD. Evidence-based health policy--lessons from the Global Burden of Disease Study. Science. 1996;274:740–3. doi: 10.1126/science.274.5288.740. [DOI] [PubMed] [Google Scholar]
- 24.Kessler RC, McGonagle KA, Zhao S, et al. Lifetime and 12-month prevalence of DSM-III-R psychiatric disorders in the United States. Results from the National Comorbidity Survey. Arch Gen Psychiatry. 1994;51:8–19. doi: 10.1001/archpsyc.1994.03950010008002. [DOI] [PubMed] [Google Scholar]
- 25.Kessler RC, Berglund P, Demler O, et al. Lifetime prevalence and age-of-onset distributions of DSM-IV disorders in the National Comorbidity Survey Replication. Arch Gen Psychiatry. 2005;62:593–602. doi: 10.1001/archpsyc.62.6.593. [DOI] [PubMed] [Google Scholar]
- 26.Kessler RC, McGonagle KA, Swartz M, et al. Sex and depression in the National Comorbidity Survey. I: Lifetime prevalence, chronicity and recurrence. J Affect Disord. 1993;29:85–96. doi: 10.1016/0165-0327(93)90026-g. [DOI] [PubMed] [Google Scholar]
- 27.Wittchen HU, Knauper B, Kessler RC. Lifetime risk of depression. Br J Psychiatry Suppl. 1994:16–22. [PubMed] [Google Scholar]
- 28.Kessler RC, Berglund P, Demler O, et al. The epidemiology of major depressive disorder: results from the National Comorbidity Survey Replication (NCS-R) JAMA. 2003;289:3095–105. doi: 10.1001/jama.289.23.3095. [DOI] [PubMed] [Google Scholar]
- 29.Mueller TI, Leon AC. Recovery, chronicity, and levels of psychopathology in major depression. Psychiatr Clin North Am. 1996;19:85–102. doi: 10.1016/s0193-953x(05)70275-6. [DOI] [PubMed] [Google Scholar]
- 30.Solomon DA, Keller MB, Leon AC, et al. Multiple recurrences of major depressive disorder. Am J Psychiatry. 2000;157:229–33. doi: 10.1176/appi.ajp.157.2.229. [DOI] [PubMed] [Google Scholar]
- 31.Judd LL, Akiskal HS, Maser JD, et al. A prospective 12-year study of subsyndromal and syndromal depressive symptoms in unipolar major depressive disorders. Arch Gen Psychiatry. 1998;55:694–700. doi: 10.1001/archpsyc.55.8.694. [DOI] [PubMed] [Google Scholar]
- 32.Frank E, Karp JF, Rush AJ. Efficacy of treatments for major depression. Psychopharmacol Bull. 1993;29:457–75. [PubMed] [Google Scholar]
- 33.Schulberg HC, Katon W, Simon GE, et al. Treating major depression in primary care practice: an update of the Agency for Health Care Policy and Research Practice Guidelines. Arch Gen Psychiatry. 1998;55:1121–7. doi: 10.1001/archpsyc.55.12.1121. [DOI] [PubMed] [Google Scholar]
- 34.Thase ME, Haight BR, Richard N, et al. Remission rates following antidepressant therapy with bupropion or selective serotonin reuptake inhibitors: a meta-analysis of original data from 7 randomized controlled trials. J Clin Psychiatry. 2005;66:974–81. doi: 10.4088/jcp.v66n0803. [DOI] [PubMed] [Google Scholar]
- 35.Casacalenda N, Perry JC, Looper K. Remission in major depressive disorder: a comparison of pharmacotherapy, psychotherapy, and control conditions. Am J Psychiatry. 2002;159:1354–60. doi: 10.1176/appi.ajp.159.8.1354. [DOI] [PubMed] [Google Scholar]
- 36.Thase ME, Entsuah AR, Rudolph RL. Remission rates during treatment with venlafaxine or selective serotonin reuptake inhibitors. Br J Psychiatry. 2001;178:234–41. doi: 10.1192/bjp.178.3.234. [DOI] [PubMed] [Google Scholar]
- 37.Van LL, Molenaar RP, Goekoop JG, et al. Three- to 5-year prospective follow-up of outcome in major depression. Psychol Med. 1998;28:731–5. doi: 10.1017/s0033291797006466. [DOI] [PubMed] [Google Scholar]
- 38.Judd LL, Akiskal HS, Maser JD, et al. Major depressive disorder: a prospective study of residual subthreshold depressive symptoms as predictor of rapid relapse. J Affect Disord. 1998;50:97–108. doi: 10.1016/s0165-0327(98)00138-4. [DOI] [PubMed] [Google Scholar]
- 39.Rush AJ, Kraemer HC, Sackeim HA, et al. Report by the ACNP Task Force on response and remission in major depressive disorder. Neuropsychopharmacology. 2006;31:1841–53. doi: 10.1038/sj.npp.1301131. [DOI] [PubMed] [Google Scholar]
- 40.Fergusson DM, Woodward LJ, Horwood LJ. Risk factors and life processes associated with the onset of suicidal behaviour during adolescence and early adulthood. Psychol Med. 2000;30:23–39. doi: 10.1017/s003329179900135x. [DOI] [PubMed] [Google Scholar]
- 41.Rockett IR, Wang S, Lian Y, et al. Suicide-associated comorbidity among US males and females: a multiple cause-of-death analysis. Inj Prev. 2007;13:311–5. doi: 10.1136/ip.2007.015230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anderson RN, Smith BL. Center for disease control and prevention. 2003 [Google Scholar]
- 43.Conwell Y, Duberstein PR, Cox C, et al. Relationships of age and axis I diagnoses in victims of completed suicide: a psychological autopsy study. Am J Psychiatry. 1996;153:1001–8. doi: 10.1176/ajp.153.8.1001. [DOI] [PubMed] [Google Scholar]
- 44.Henriksson MM, Aro HM, Marttunen MJ, et al. Mental disorders and comorbidity in suicide. Am J Psychiatry. 1993;150:935–40. doi: 10.1176/ajp.150.6.935. [DOI] [PubMed] [Google Scholar]
- 45.Perlis RH, Beasley CM, Jr, Wines JD, Jr, et al. Treatment-associated suicidal ideation and adverse effects in an open, multicenter trial of fluoxetine for major depressive episodes. Psychother Psychosom. 2007;76:40–6. doi: 10.1159/000096363. [DOI] [PubMed] [Google Scholar]
- 46.Brent D. Antidepressants and suicidal behavior: cause or cure? Am J Psychiatry. 2007;164:989–91. doi: 10.1176/ajp.2007.164.7.989. [DOI] [PubMed] [Google Scholar]
- 47.Hirschfeld RM. History and evolution of the monoamine hypothesis of depression. J Clin Psychiatry. 2000;61 (Suppl 6):4–6. [PubMed] [Google Scholar]
- 48.Sakinofsky I. Treating suicidality in depressive illness. Part 2: does treatment cure or cause suicidality? Can J Psychiatry. 2007;52:85S–101S. [PubMed] [Google Scholar]
- 49.Simon GE, Savarino J, Operskalski B, et al. Suicide risk during antidepressant treatment. Am J Psychiatry. 2006;163:41–7. doi: 10.1176/appi.ajp.163.1.41. [DOI] [PubMed] [Google Scholar]
- 50.Baldessarini RJ, Hennen J. Genetics of suicide: an overview. Harv Rev Psychiatry. 2004;12:1–13. doi: 10.1080/10673220490425915. [DOI] [PubMed] [Google Scholar]
- 51.Brent DA, Mann JJ. Family genetic studies, suicide, and suicidal behavior. Am J Med Genet C Semin Med Genet. 2005;133:13–24. doi: 10.1002/ajmg.c.30042. [DOI] [PubMed] [Google Scholar]
- 52.Zalsman G, Frisch A, Apter A, et al. Genetics of suicidal behavior: candidate association genetic approach. Isr J Psychiatry Relat Sci. 2002;39:252–61. [PubMed] [Google Scholar]
- 53.Johnson JA. Pharmacogenetics: potential for individualized drug therapy through genetics. Trends Genet. 2003;19:660–6. doi: 10.1016/j.tig.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 54.Meyer UA. Pharmacogenetics - five decades of therapeutic lessons from genetic diversity. Nat Rev Genet. 2004;5:669–76. doi: 10.1038/nrg1428. [DOI] [PubMed] [Google Scholar]
- 55.Nebert DW. Pharmacogenetics and pharmacogenomics: why is this relevant to the clinical geneticist? Clin Genet. 1999;56:247–58. doi: 10.1034/j.1399-0004.1999.560401.x. [DOI] [PubMed] [Google Scholar]
- 56.Malhotra AK, Murphy GM, Jr, Kennedy JL. Pharmacogenetics of psychotropic drug response. Am J Psychiatry. 2004;161:780–96. doi: 10.1176/appi.ajp.161.5.780. [DOI] [PubMed] [Google Scholar]
- 57.Ng CH, Schweitzer I, Norman T, et al. The emerging role of pharmacogenetics: implications for clinical psychiatry. Aust N Z J Psychiatry. 2004;38:483–9. doi: 10.1080/j.1440-1614.2004.01400.x. [DOI] [PubMed] [Google Scholar]
- 58.Perlis RH. Pharmacogenetic studies of antidepressant response: how far from the clinic? Psychiatr Clin North Am. 2007;30:125–38. doi: 10.1016/j.psc.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 59.Roses AD. Pharmacogenetics. Hum Mol Genet. 2001;10:2261–7. doi: 10.1093/hmg/10.20.2261. [DOI] [PubMed] [Google Scholar]
- 60.Binder EB, Holsboer F. Pharmacogenomics and antidepressant drugs. Ann Med. 2006;38:82–94. doi: 10.1080/07853890600551045. [DOI] [PubMed] [Google Scholar]
- 61.Roses AD. Pharmacogenetics and the practice of medicine. Nature. 2000;405:857–65. doi: 10.1038/35015728. [DOI] [PubMed] [Google Scholar]
- 62.Roses AD. Pharmacogenetics and future drug development and delivery. Lancet. 2000;355:1358–61. doi: 10.1016/S0140-6736(00)02126-7. [DOI] [PubMed] [Google Scholar]
- 63.Gaynes BN, Rush AJ, Trivedi MH, et al. The STAR*D study: treating depression in the real world. Cleve Clin J Med. 2008;75:57–66. doi: 10.3949/ccjm.75.1.57. [DOI] [PubMed] [Google Scholar]
- 64.Warden D, Rush AJ, Trivedi MH, et al. The STAR*D Project results: a comprehensive review of findings. Curr Psychiatry Rep. 2007;9:449–59. doi: 10.1007/s11920-007-0061-3. [DOI] [PubMed] [Google Scholar]
- 65.Trivedi MH, Rush AJ, Wisniewski SR, et al. Evaluation of outcomes with citalopram for depression using measurement-based care in STAR*D: implications for clinical practice. Am J Psychiatry. 2006;163:28–40. doi: 10.1176/appi.ajp.163.1.28. [DOI] [PubMed] [Google Scholar]
- 66.Skol AD, Scott LJ, Abecasis GR, et al. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38:209–13. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- 67.Serretti A, Drago A, De RD. HTR2A gene variants and psychiatric disorders: a review of current literature and selection of SNPs for future studies. Curr Med Chem. 2007;14:2053–69. doi: 10.2174/092986707781368450. [DOI] [PubMed] [Google Scholar]
- 68.Cusin C, Serretti A, Zanardi R, et al. Influence of monoamine oxidase A and serotonin receptor 2A polymorphisms in SSRI antidepressant activity. Int J Neuropsychopharmacol. 2002;5:27–35. doi: 10.1017/S1461145701002711. [DOI] [PubMed] [Google Scholar]
- 69.Minov C, Baghai TC, Schule C, et al. Serotonin-2A-receptor and -transporter polymorphisms: lack of association in patients with major depression. Neurosci Lett. 2001;303:119–22. doi: 10.1016/s0304-3940(01)01704-9. [DOI] [PubMed] [Google Scholar]
- 70.Sato K, Yoshida K, Takahashi H, et al. Association between -1438G/A promoter polymorphism in the 5-HT(2A) receptor gene and fluvoxamine response in Japanese patients with major depressive disorder. Neuropsychobiology. 2002;46:136–40. doi: 10.1159/000066394. [DOI] [PubMed] [Google Scholar]
- 71.Lesser IM, Castro DB, Gaynes BN, et al. Ethnicity/race and outcome in the treatment of depression: results from STAR*D. Med Care. 2007;45:1043–51. doi: 10.1097/MLR.0b013e3181271462. [DOI] [PubMed] [Google Scholar]
- 72.Gilmer WS, Trivedi MH, Rush AJ, et al. Factors associated with chronic depressive episodes: a preliminary report from the STAR-D project. Acta Psychiatr Scand. 2005;112:425–33. doi: 10.1111/j.1600-0447.2005.00633.x. [DOI] [PubMed] [Google Scholar]
- 73.Svenningsson P, Nishi A, Fisone G, et al. DARPP-32: an integrator of neurotransmission. Annu Rev Pharmacol Toxicol. 2004;44:269–96. doi: 10.1146/annurev.pharmtox.44.101802.121415. [DOI] [PubMed] [Google Scholar]
- 74.Pickard BS, Malloy MP, Christoforou A, et al. Cytogenetic and genetic evidence supports a role for the kainate-type glutamate receptor gene, GRIK4, in schizophrenia and bipolar disorder. Mol Psychiatry. 2006;11:847–57. doi: 10.1038/sj.mp.4001867. [DOI] [PubMed] [Google Scholar]
- 75.Moonesinghe R, Khoury MJ, Liu T, et al. Required sample size and nonreplicability thresholds for heterogeneous genetic associations. Proc Natl Acad Sci U S A. 2008;105:617–22. doi: 10.1073/pnas.0705554105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ogilvie AD, Battersby S, Bubb VJ, et al. Polymorphism in serotonin transporter gene associated with susceptibility to major depression. Lancet. 1996;347:731–3. doi: 10.1016/s0140-6736(96)90079-3. [DOI] [PubMed] [Google Scholar]
- 77.Bradley SL, Dodelzon K, Sandhu HK, et al. Relationship of serotonin transporter gene polymorphisms and haplotypes to mRNA transcription. Am J Med Genet B Neuropsychiatr Genet. 2005;136:58–61. doi: 10.1002/ajmg.b.30185. [DOI] [PubMed] [Google Scholar]
- 78.Lesch KP, Gutknecht L. Pharmacogenetics of the serotonin transporter. Prog Neuropsychopharmacol Biol Psychiatry. 2005;29:1062–73. doi: 10.1016/j.pnpbp.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 79.Nakamura M, Ueno S, Sano A, et al. The human serotonin transporter gene linked polymorphism (5-HTTLPR) shows ten novel allelic variants. Mol Psychiatry. 2000;5:32–8. doi: 10.1038/sj.mp.4000698. [DOI] [PubMed] [Google Scholar]
- 80.Hu XZ, Lipsky RH, Zhu G, et al. Serotonin transporter promoter gain-of-function genotypes are linked to obsessive-compulsive disorder. Am J Hum Genet. 2006;78:815–26. doi: 10.1086/503850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Serretti A, Mandelli L, Lorenzi C, et al. Temperament and character in mood disorders: influence of DRD4, SERTPR, TPH and MAO-A polymorphisms. Neuropsychobiology. 2006;53:9–16. doi: 10.1159/000089916. [DOI] [PubMed] [Google Scholar]
- 82.Lin Z, Madras BK. Human genetics and pharmacology of neurotransmitter transporters. Handb Exp Pharmacol. 2006:327–71. doi: 10.1007/3-540-29784-7_16. [DOI] [PubMed] [Google Scholar]
- 83.Caspi A, Sugden K, Moffitt TE, et al. Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science. 2003;301:386–9. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- 84.Otte C, McCaffery J, Ali S, et al. Association of a serotonin transporter polymorphism (5-HTTLPR) with depression, perceived stress, and norepinephrine in patients with coronary disease: the Heart and Soul Study. Am J Psychiatry. 2007;164:1379–84. doi: 10.1176/appi.ajp.2007.06101617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Smits KM, Smits LJ, Schouten JS, et al. Influence of SERTPR and STin2 in the serotonin transporter gene on the effect of selective serotonin reuptake inhibitors in depression: a systematic review. Mol Psychiatry. 2004;9:433–41. doi: 10.1038/sj.mp.4001488. [DOI] [PubMed] [Google Scholar]
- 86.Serretti A, Kato M, De RD, et al. Meta-analysis of serotonin transporter gene promoter polymorphism (5-HTTLPR) association with selective serotonin reuptake inhibitor efficacy in depressed patients. Mol Psychiatry. 2007;12:247–57. doi: 10.1038/sj.mp.4001926. [DOI] [PubMed] [Google Scholar]
- 87.Murphy GM, Jr, Hollander SB, Rodrigues HE, et al. Effects of the serotonin transporter gene promoter polymorphism on mirtazapine and paroxetine efficacy and adverse events in geriatric major depression. Arch Gen Psychiatry. 2004;61:1163–9. doi: 10.1001/archpsyc.61.11.1163. [DOI] [PubMed] [Google Scholar]
- 88.Chrousos GP, Kino T. Glucocorticoid action networks and complex psychiatric and/or somatic disorders. Stress. 2007;10:213–9. doi: 10.1080/10253890701292119. [DOI] [PubMed] [Google Scholar]
- 89.Tsigos C, Chrousos GP. Hypothalamic-pituitary-adrenal axis, neuroendocrine factors and stress. J Psychosom Res. 2002;53:865–71. doi: 10.1016/s0022-3999(02)00429-4. [DOI] [PubMed] [Google Scholar]
- 90.Swaab DF, Bao AM, Lucassen PJ. The stress system in the human brain in depression and neurodegeneration. Ageing Res Rev. 2005;4:141–94. doi: 10.1016/j.arr.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 91.Mizoguchi K, Ishige A, Aburada M, et al. Chronic stress attenuates glucocorticoid negative feedback: involvement of the prefrontal cortex and hippocampus. Neuroscience. 2003;119:887–97. doi: 10.1016/s0306-4522(03)00105-2. [DOI] [PubMed] [Google Scholar]
- 92.Pariante CM, Miller AH. Glucocorticoid receptors in major depression: relevance to pathophysiology and treatment. Biol Psychiatry. 2001;49:391–404. doi: 10.1016/s0006-3223(00)01088-x. [DOI] [PubMed] [Google Scholar]
- 93.Pariante CM, Makoff A, Lovestone S, et al. Antidepressants enhance glucocorticoid receptor function in vitro by modulating the membrane steroid transporters. Br J Pharmacol. 2001;134:1335–43. doi: 10.1038/sj.bjp.0704368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Thomson F, Craighead M. Innovative Approaches for the Treatment of Depression: Targeting the HPA Axis. Neurochem Res. 2007 doi: 10.1007/s11064-007-9518-3. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 95.DeRijk R, de Kloet ER. Corticosteroid receptor genetic polymorphisms and stress responsivity. Endocrine. 2005;28:263–70. doi: 10.1385/ENDO:28:3:263. [DOI] [PubMed] [Google Scholar]
- 96.Barden N, Stec IS, Montkowski A, et al. Endocrine profile and neuroendocrine challenge tests in transgenic mice expressing antisense RNA against the glucocorticoid receptor. Neuroendocrinology. 1997;66:212–20. doi: 10.1159/000127240. [DOI] [PubMed] [Google Scholar]
- 97.Boyle MP, Brewer JA, Funatsu M, et al. Acquired deficit of forebrain glucocorticoid receptor produces depression-like changes in adrenal axis regulation and behavior. Proc Natl Acad Sci U S A. 2005;102:473–8. doi: 10.1073/pnas.0406458102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ridder S, Chourbaji S, Hellweg R, et al. Mice with genetically altered glucocorticoid receptor expression show altered sensitivity for stress-induced depressive reactions. J Neurosci. 2005;25:6243–50. doi: 10.1523/JNEUROSCI.0736-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.van Rossum EF, Binder EB, Majer M, et al. Polymorphisms of the glucocorticoid receptor gene and major depression. Biol Psychiatry. 2006;59:681–8. doi: 10.1016/j.biopsych.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 100.van WD, Van Den EF, Del-Favero J, et al. Glucocorticoid receptor gene-based SNP analysis in patients with recurrent major depression. Neuropsychopharmacology. 2006;31:620–7. doi: 10.1038/sj.npp.1300898. [DOI] [PubMed] [Google Scholar]
- 101.Binder EB, Salyakina D, Lichtner P, et al. Polymorphisms in FKBP5 are associated with increased recurrence of depressive episodes and rapid response to antidepressant treatment. Nat Genet. 2004;36:1319–25. doi: 10.1038/ng1479. [DOI] [PubMed] [Google Scholar]
- 102.Tsai SJ, Hong CJ, Chen TJ, et al. Lack of supporting evidence for a genetic association of the FKBP5 polymorphism and response to antidepressant treatment. Am J Med Genet B Neuropsychiatr Genet. 2007;144:1097–8. doi: 10.1002/ajmg.b.30246. [DOI] [PubMed] [Google Scholar]
- 103.Papiol S, Arias B, Gasto C, et al. Genetic variability at HPA axis in major depression and clinical response to antidepressant treatment. J Affect Disord. 2007;104:83–90. doi: 10.1016/j.jad.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 104.Bazerman MH. The Journal of Conflict Resolution. The Journal of Conflict Resolution. 1983;27:618–34. [Google Scholar]
- 105.Goring HH, Terwilliger JD, Blangero J. Large upward bias in estimation of locus-specific effects from genomewide scans. Am J Hum Genet. 2001;69:1357–69. doi: 10.1086/324471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Perlis RH, Moorjani P, Fagerness J, et al. Pharmacogenetic Analysis of Genes Implicated in Rodent Models of Antidepressant Response: Association of TREK1 and Treatment Resistance in the STAR(*)D Study. Neuropsychopharmacology. 2008 Feb 20; doi: 10.1038/npp.2008.6. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Peter D, Finn JP, Klisak I, et al. Chromosomal localization of the human vesicular amine transporter genes. Genomics. 1993;18:720–3. doi: 10.1016/s0888-7543(05)80383-0. [DOI] [PubMed] [Google Scholar]
- 108.Heurteaux C, Lucas G, Guy N, et al. Deletion of the background potassium channel TREK-1 results in a depression-resistant phenotype. Nat Neurosci. 2006;9:1134–41. doi: 10.1038/nn1749. [DOI] [PubMed] [Google Scholar]
- 109.Perlis RH, Purcell S, Fagerness J, et al. Clinical and genetic dissection of anger expression and CREB1 polymorphisms in major depressive disorder. Biol Psychiatry. 2007;62:536–40. doi: 10.1016/j.biopsych.2006.10.034. [DOI] [PubMed] [Google Scholar]
- 110.Carey B. Genes Tied to Bad Reactions to Antidepressant Drug. New York Times. 2007 [Google Scholar]
- 111.Gloyn AL, Weedon MN, Owen KR, et al. Large-scale association studies of variants in genes encoding the pancreatic beta-cell KATP channel subunits Kir6.2 (KCNJ11) and SUR1 (ABCC8) confirm that the KCNJ11 E23K variant is associated with type 2 diabetes. Diabetes. 2003;52:568–72. doi: 10.2337/diabetes.52.2.568. [DOI] [PubMed] [Google Scholar]
- 112.Grant SF, Thorleifsson G, Reynisdottir I, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006;38:320–3. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 113.Sladek R, Rocheleau G, Rung J, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–5. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 114.Zeggini E, Weedon MN, Lindgren CM, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–41. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Laje G, McMahon FJ. The pharmacogenetics of major depression: past, present, and future. Biol Psychiatry. 2007;62:1205–7. doi: 10.1016/j.biopsych.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 116.Janssens AC, Gwinn M, Bradley LA, et al. A critical appraisal of the scientific basis of commercial genomic profiles used to assess health risks and personalize health interventions. Am J Hum Genet. 2008;82:593–9. doi: 10.1016/j.ajhg.2007.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Working Group. Recommendations from the EGAPP Working Group: testing for cytochrome P450 polymorphisms in adults with nonpsychotic depression treated with selective serotonin reuptake inhibitors. Genet Med. 2007;9:819–25. doi: 10.1097/gim.0b013e31815bf9a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ioannidis JP, Boffetta P, Little J, et al. Assessment of cumulative evidence on genetic associations: interim guidelines. Int J Epidemiol. 2008;37:120–32. doi: 10.1093/ije/dym159. [DOI] [PubMed] [Google Scholar]