Abstract
Recent reports have linked the expression of specific microRNAs (miRNAs) with tumorigenesis and metastasis. Here, we show that microRNA (miR)-16, which is expressed at lower levels in prostate cancer cells, affects the proliferation of human prostate cancer cell lines both in vitro and in vivo. Transient transfection with synthetic miR-16 significantly reduced cell proliferation of 22Rv1, Du145, PPC-1, and PC-3M-luc cells. A prostate cancer xenograft model revealed that atelocollagen could efficiently deliver synthetic miR-16 to tumor cells on bone tissues in mice when injected into tail veins. In the therapeutic bone metastasis model, injection of miR-16 with atelocollagen via tail vein significantly inhibited the growth of prostate tumors in bone. Cell model studies indicate that miR-16 likely suppresses prostate tumor growth by regulating the expression of genes such as CDK1 and CDK2 associated with cell-cycle control and cellular proliferation. There is a trend toward lower miR-16 expression in human prostate tumors versus normal prostate tissues. Thus, this study indicates the therapeutic potential of miRNA in an animal model of cancer metastasis with systemic miRNA injection and suggest that systemic delivery of miR-16 could be used to treat patients with advanced prostate cancer.
Introduction
Advanced prostate cancer is frequently difficult to treat and causes substantial symptoms, including severe pain from metastasis to bone or other sites. Numerous experimental therapeutics are being pursued in clinical trials and offer some hope of improved treatments, but most have so far demonstrated only modest results.
Mounting evidence suggests that the altered expression of specific microRNA's (miRNA's) accuracy contributes to the development of a variety of cancers. Cancer types including prostate cancers can be classified based on their distinct miRNA expression profiles.1,2,3,4,5
MiRNAs have been implicated in prostate cancer. Volinia et al. identified >40 miRNAs with expression levels that were significantly different in prostate tumors versus normal prostate tissue.5 Furthermore, the need for additional therapies in metastasis due to hormone-refractory prostate cancer is considerable. Mattie et al. found that miRNA expression in human prostate cancer cell lines could distinguish androgen hormone–insensitive PC3 from hormone-sensitive LNCaP cells.6 LNCaP cells showed upregulation of microRNA (miR)-200c, miR-195, and several let-7 family members, whereas miR-10a, miR-27b, miR-221, miR-222, and mir-210 were lower than in PC3. The serum prostate-specific antigen is the most useful tumor marker for diagnosis and monitoring of prostate cancer. However, its low specificity in distinguishing prostate carcinoma from benign prostatic hyperplasia limits its use as an early detection biomarker. Investigators used custom designed arrays to compare the expression profiles of 319 miRNAs in prostate tumors, cancer cell lines, xenografts, and benign prostatic hyperplasia.7 MiRNAs could be used to cluster the androgen receptor status of cell lines and xenografts. Among a small set of benign prostatic hyperplasia, hormone refractory, and untreated prostate carcinomas they found 51 differentially expressed miRNAs, 37 of which were downregulated. MiRNAs in this set accurately clustered the benign prostatic hyperplasia, untreated and hormone-refractory prostate carcinomas providing evidence that miRNA expression profiles are altered by changes in disease status. More recently, Bonci et al. showed that miR-15a and miR-16-1 cluster inhibit the tumor cell proliferation and invasion via targets CCND1 (cyclinD1), WNT3A, and BCL2 in prostate cancer cell line and clinical samples.8 These miR-15a and miR-16-1 were coded on chromosome 9 13q14. In this region, loss of heterogeneity was detected in chronic lymphocytic leukemia9 and prostate cancer patients.10 These results suggest that miR-15a and/or miR-16 could be a novel target for prostate cancer therapy.
To supplement the expression studies that have been published for prostate cancer, we used a library of synthetic miRNAs to identify the small RNAs that alter the proliferation of prostate cancer cells. Among the miRNAs that were identified in a functional screen featuring 22Rv1 prostate cancer cells was miR-16, an miRNA that has been implicated in chronic lymphocytic leukemia1,11,12 and prostate cancer.8,10 Our studies of miR-16 revealed that it has the capacity to affect the proliferation of a variety of human-derived prostate cancer cells. For the evaluation of miRNA therapy for bone metastasis of prostate cancer, the mouse model of bone-metastatic prostate cancer using bioluminescence-based in vivo imaging analysis was selected. We have already established small-interfering RNA (siRNA) molecules that can be delivered to tumor cells in a bone metastatic site using an atelocollagen delivery method.13 The properties of synthetic miRNA molecules are similar to synthetic siRNA; therefore, it is speculated that synthetic miRNA can also be used for systemic treatment mediated by atelocollagen. In this article, the systemic delivery of synthetic miR-16 using atelocollagen inhibited bone-metastatic human prostate tumor growth in a mouse bone site. We further analyzed the altered expression of cancer-related genes in miR-16-transfected prostate cancer cells and verified that genes associated with cell-cycle progression were mostly affected by miR-16. These results suggest a therapeutic potency of miR-16 in bone-metastatic prostate cancer.
Results
Effect of miR-16 on proliferation of human prostate cancer cell lines
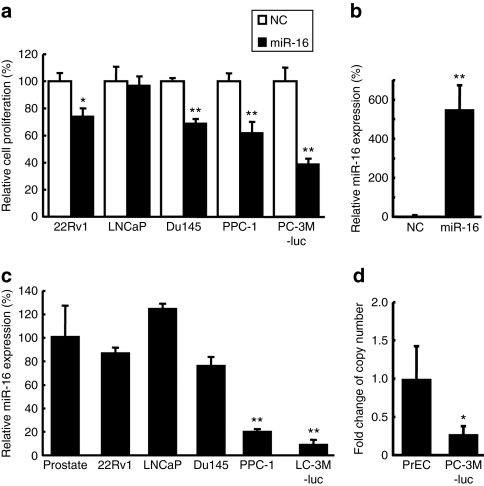
22Rv1 prostate cancer cells were transiently transfected in triplicate with individual synthetic mimics for ~200 miRNAs. Three days after transfection, the cells were monitored for proliferation and apoptotic activity. Among the most active miRNAs identified in the functional screen was miR-16, which reduced the proliferation of the prostate cancer cells by 25% and increased apoptosis by 40% (data not shown). Follow-up studies for measuring the proliferation; using the alamar blue assay with another prostate cancer cell line, PC-3M-luc, revealed that miR-16 reduces proliferation by 60% (Figure 1a) relative to the cells transfected with a negative control (NC) miRNA. Further studies of the antiproliferative effect of miR-16 on prostate cancer cells revealed that synthetic miRNA can significantly affect the expansion of cultured 22Rv1, PPC-1, and Du145 cells (Figure 1a). The only prostate cancer cell line that proved to be unaffected by the transfection of miR-16 was LNCaP (Figure 1a). The amount of miR-16 in the PC-3M-luc cells transfected with synthetic miR-16 was >500-fold higher than that in the control cells (Figure 1b). This result suggests that the induced increase of intracellular miR-16 concentrations is capable of suppressing the proliferation of the prostate cancer cells.
Figure 1.
The expression and function of miR-16 in human prostate cancer cell lines. (a) Effect of miR-16 on proliferation of human prostate cancer cell lines. Percent (%) proliferation values were normalized to values from cells treated with negative control (NC) microRNA (miRNA). Data represent the mean (n = 4) ± SD *P < 0.05, **P < 0.01 versus NC miRNA. (b) The amount of miR-16 in PC-3M-luc cells transfected with synthetic miR-16. The cellular level of miR-16 was detected by quantitative PCR. The data represent the mean (n = 3) ± SD **P < 0.01 versus NC miRNA. (c) Expression level of miR-16 in human prostate cancer cell lines. The relative expression of miR-16 for each of the cell lines was calculated by comparing the level in normal prostate tissue samples. The data represent the mean (n = 3) ± SD **P < 0.01 versus normal human prostate tissue. (d) The copy number change of the miR-16 loci on chromosome 13q14 in PC-3M-luc cells. The copy number of miR-16 genes were quantified by real-time PCR with genomic DNA. Cultured normal human prostate epithelial cells (PrEC) was used as the control for this experiment for comparison to the PC-3M-luc cells. The data represent the mean (n = 3) ± SD *P < 0.05 versus PrEC.
miR-16 expression levels in prostate cancer cell lines
Although four of the five prostate cancer cell lines exhibit significant reductions in proliferation following transfection with synthetic miR-16, it is interesting that there is a variation in the level of the effect. To address whether this might be due to variation in the levels of endogenous miR-16 in the various cell lines, we used quantitative reverse transcription (qRT)-PCR to measure the relative abundance of mature miRNA. As shown in Figure 1c, most of the cell lines expressed miR-16 at reduced levels. The extent of downregulation correlated with the phenotypic response in these cell lines: e.g., PPC-1 and PC-3M-luc cells, which showed the strongest response to miR-16, had the lowest levels of endogenous miR-16 (Figure 1a). The DNA copy numbers on chromosome 13q14, a genomic region that is frequently deleted in chronic lymphocytic leukemia and prostate cancer14 in the PC-3M-luc cells were reduced to half that of normal prostate cells (Figure 1d). However, because the DNA sequence data did not show any mutations on chromosomes coding miR-16 of other copy in the PC-3M-luc cells (data not shown), the remarkable reduction of miR-16 expression might be invoked by a combination of DNA copy number alteration and other factors to affect the expression. LNCaP cells, which showed no response to the miR-16 mimic, were the only cells that tend to have higher miR-16 expression levels than the normal prostate (Figure 1c). Additionally, the transfection of miRNAs, which are not downregulated in PC-3M-luc cells, such as miR-10a and miR-188, did not inhibit the growth of PC-3M-luc cells (data not shown). The expression and function data suggest that reduced expression of miR-16 is critical for sustained proliferation in some prostate cancer cell lines and that reintroduction of miR-16 can interfere with that phenotype.
Evaluation of miRNA delivery to bone-metastatic tumors in mice
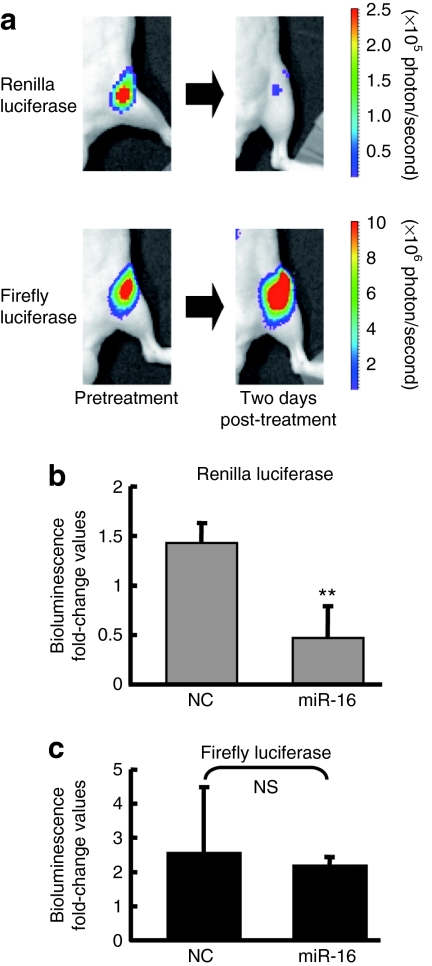
In order to assess the capacity of the synthetic miR-16 to affect prostate tumor growth in mice, we chose to use a mouse model featuring PC-3M-luc cells that have the capacity to form prostate tumors in the bones of mice.13,15,16 To evaluate that atelocollagen can efficiently deliver synthetic miRNA molecules to metastatic prostate tumors in bone, we generated a PC-3M-luc metastatic prostate cancer cell line stably expressing the renilla luciferase gene fused to the 3′UTR of Bcl2, a validated miR-16 target (Supplementary Figure S1a).17 Thus, this newly engineered cell line PC-3M-Fluc/Rluc-Bcl2 3′UTR expresses both firefly and renilla luciferase, the later of which is under control of miR-16 (Supplementary Figure S1b). As expected, transfection of cultured PC-3M-Fluc/Rluc-Bcl2 3′UTR cells with 30 nmol/l of miR-16 deceased the luminescence derived from renilla luciferase (Supplementary Figure S1c). To monitor atelocollagen-mediated delivery of miR-16 in the animal, PC-3M-luc/Rluc-Bcl2 3′UTR cells were intracardiac injected into mice and allowed the tumor cells to deposit in the bone. Nine weeks after implantation, the mice were tail-vein injected with 50 µg of miR-16 mimic that was complexed with atelocollagen. Mice injected with the miR-16/atelocollagen complex produced <50% renilla luciferase from tumors in the bone than they produced before treatment (Figure 2a,b). The signal from the firefly luciferase that represents tumor growth was unaffected by the synthetic miR-16, indicating that the inhibition observed for renilla luciferase was due to the binding of injected synthetic miR-16 to the 3′UTR of Bcl2. Synthetic miR-16 was detected in tumor tissue at >20 pg/mg tissue when injected systemically and it was observed to persist in tumors for 3 days after injection (data not shown). Thus, our dual-luciferase prostate cancer xenograft model clearly showed that atelocollagen can efficiently deliver active miRNAs into metastatic tumors in mice.
Figure 2.
Evaluation of delivery for synthetic microRNA (miRNA) molecules to tumors in bone. A dual-luciferase expressing PC-3M cells that have 3'-UTR of Bcl2 under the renilla luciferase gene, PC-3M-luc/Rluc-Bcl2 3'UTR cells, were generated. These cells were used for dual assay system, for monitoring of tumor growth by firefly luciferase, for monitoring of delivery efficacy of synthetic miR-16 by renilla luciferase. (a) Representative images of bone metastasis in the femur of mice. To examine the efficacy of synthetic miR-16 in tumor cells, PC-3M-luc/Rluc-Bcl2 3'UTR cells were injected into the heart of nude mice. Nine weeks after tumor injection, bioluminescence from renilla luciferase was detected. Intravenous injection of miR-16 complexed with atelocollagen suppressed the expression of renilla luciferase (top). In contrast, bioluminescence from firefly luciferase was not affected (bottom). (b) Normalized fold change (2 days post/pre-miR-16 administration) of bioluminescence emitted from whole body of mice. This figure is graphically shown of the results of Figure 2a by fold change of photon counts. Data represent the mean (n = 3) ± SD *P < 0.01 versus NC miRNA. NS, not significant. NC, negative control.
Inhibition of tumor growth in bone tissues in mice with systemic miR-16 treatment
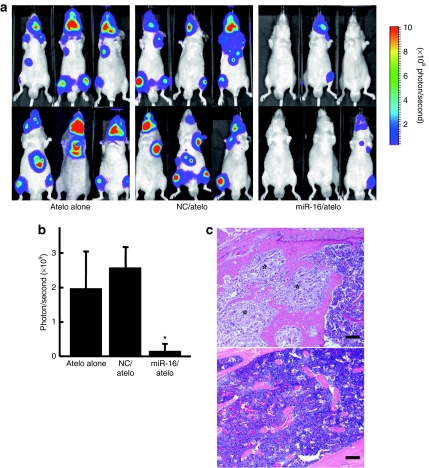
To assess the therapeutic potential of the miR-16/atelcollagen complexes, prostate tumors were initiated in the bones of mice by intracardiac injection of PC-3M-luc cells. A 50 µg of miR-16 mimic complexed with atelocollagen was administered intravenously into mice at 4, 7, and 10 days after prostate tumor initiation (Supplementary Figure S2). The development of tumor in the bone was monitored in vivo by bioluminescent imaging. At the end of the experiment on day 28, mice treated with the NC miRNA/atelocollagen complex showed the presence of tumor in the thorax, jaws, and/or legs of mice frequently (Figure 3a). In contrast, the mice injected with miR-16/atelocollagen complex exhibited no increase in luminescence during the same observation period. There are significant differences between NC and miR-16 treatment groups on day 28 (P < 0.05) (Figure 3b). Histopathological analysis also revealed that growth of PC-3M-luc cells in the bone tissues of mice was significantly inhibited by the miR-16 treatment (Figure 3c). These data suggest that atelocollagen-mediated systemic delivery of miR-16 could be a novel strategy for inhibition of prostate tumor growth in the bone tissues.
Figure 3.
Inhibition of metastatic tumor growth in bone tissues by the atelocollagen-mediated miRNA treatment. Mice were injected with 2 × 106 PC-3M-luc-C6 cells into the left heart ventricle on day zero. The miR-16 and NC miRNA (50 µg) with 0.05% atelocollagen (Atelo) or Atelo alone in a 200 µl volume were injected into the tail vein on days 4, 7, and 10 after tumor injection. At the end of the experiment on day 28, the metastasis was evaluated by IVIS imaging and confirmed by subsequent necropsy. (a) All mice used in this experiment on day 28 were shown. There was an increase in luminescence in mice treated with atelocollagen alone and NC miRNA whereas the miR-16/atelocollagen-treated groups had no or low increase in luminescence during the same observation period. (b) Quantitation of bioluminescence emitted from whole body of mice on day 28. Data represent the mean (n = 6) ± SD *P <0.05 versus other groups. (c) Histopathological analysis confirmed micrometastasis in the tibia of nontreated mice (upper). Metastatic lesions are indicated by asterisk mark. In the miR-16-treated mice, any micrometastasis was not observed (lower). Bar = 100 µm.
miR-16 expression in human prostate tissues
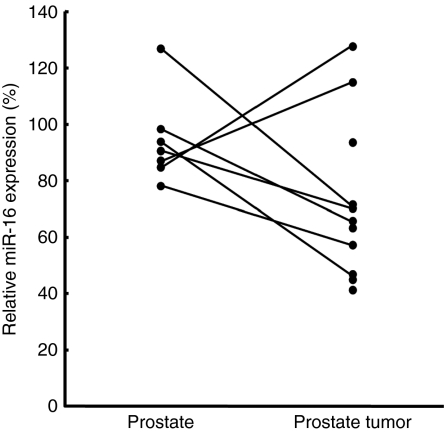
We used qRT-PCR to quantify miR-16 levels in the tumors and normal adjacent tissues of seven prostate cancer patients as well as four additional prostate tumors. The relative expression level of miR-16 in each of the samples was calculated by comparing to the average normalized miR-16 levels in prostate samples from three normal donors. The average relative expression of miR-16 in the seven prostate normal adjacent samples was 95% with a standard deviation of 16% and the eleven prostate tumors was 73% with a standard deviation of 28% (Figure 4). There is a trend toward lower miR-16 expression in prostate tumors versus normal prostate tissues, but this trend did not reach statistical significance (Figure 4).
Figure 4.
Clinical association of miR-16 expression with prostate cancer. The qRT-PCR analysis to quantify miR-16 levels in the tumors and normal adjacent tissues of seven prostate cancer patients and four additional prostate tumors was performed. The P value calculated by Student's t-test for the two sample sets was 0.08.
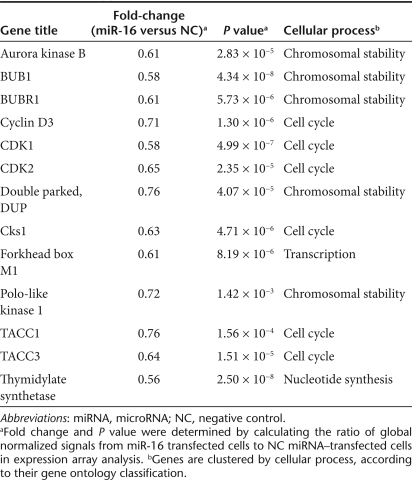
mRNA array analysis following transfection of synthetic miR-16
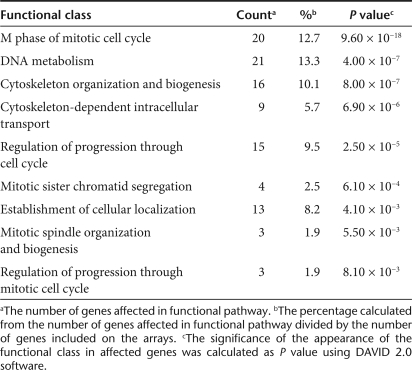
To get insight into the antioncogenic mechanism of miR-16, we transfected PC-3M-luc cells with the miR-16 mimic and analyzed the expressions of mRNA using mRNA array analysis. Exogenously, miR-16 might directly affect the mRNA levels of the target genes and indirectly affect the expression of genes that are downstream of these direct targets.18 To identify the pathways that could be affected both directly and indirectly by miR-16, total RNA was isolated from the cells 72 hours after miR-16 transfection. The mRNA array data for the miR-16-transfected samples were compared to the NC miRNA–transfected samples (Supplementary Table S1). Fold-differential and P value calculations were used to select 285 mRNAs whose expression levels were significantly altered in the miR-16-transfected samples. A selection of genes suppressed by miR-16 is listed in Table 1. Pathway analysis combining the Kyoto Encyclopedia of Genes and Genomes19,20,21 and Database for Annotation, Visualization, and Integrated Discovery22 was used to analyze the list of genes with altered expression to determine if there was a significant enrichment of genes associated with any known cellular pathways (Table 2). Overall, the statistical enrichment of pathways was moderately low, suggesting that no single pathway or network was specifically and vigorously responsive to the treatment. However, for those pathways that were considered enriched, a few strong underlying themes emerged. The gene lists were enriched for functions related to cell division and control of the cell cycle (Table 2). The functions associated with cell-cycle control were most enriched in miR-16-affected genes and these 12 genes that cover G1, S, G2, and M phase of cell cycle are mapped into the Kyoto Encyclopedia of Genes and Genomes Pathway Cell Cycle Map (Supplementary Figure S3). Thus, these data suggest that strong inhibition of prostate tumor growth in bone tissues of our animal model was due to downregulation of a key component of cell-cycle genes.
Table 1.
Genes suppressed by miR-16
Table 2.
Classes of genes affected by miR-16
Discussion
The likely involvement of miR-16 in the development of prostate cancer is apparent on multiple levels. The loss of the genomic locus at 13q14 that encompasses the miR-16-1 gene has been reported to be highly associated with human prostate cancer progression.14 Dong et al. suggested loss of heterogeneity at 13q14 is associated with clinically significant high-grade and high-stage prostate cancers10 with ties to both metastasis and tumor initiation.23 Consistent with its genomic location, our qRT-PCR results showed that the miR-16 is significantly reduced in most prostate tumors and cultured prostate cancer cells relative to normal prostate tissues.
Based on our studies with cultured prostate cancer cells, the reduced expression of miR-16 is likely necessary to maintain high rates of proliferation. The relationship between miR-16 and apoptosis likely stems from the miRNA's apparent role in regulating BCL2 expression.17 Our previous data also showed that the transfection of miR-16 into 22Rv1 prostate cancer cells induced apoptosis (F. Takeshita et al., unpublished results). Although increased apoptosis is likely to be at least partially responsible for the reduced proliferation rates that we observed in miR-16-transfected PC-3M-luc cells, it appears that the small RNA also affects cell-cycle progression by regulating the expression of multiple cell-cycle genes. The transfection of prostate cancer cells with synthetic miR-16 reduced the expression of genes like Cyclin D3, CDK1, CDK2, Cks1, TAAC1, and TAAC3 that play roles in regulating cell-cycle progression. The apparent capacity of miR-16 to simultaneously regulate cell cycle and apoptosis points to the likely importance of the small RNA in maintaining normal cell function and underscores the influence that the altered expression of the miRNA likely has on tumorigenesis.
The importance of miRNAs like miR-16 as tumor suppressors is becoming increasingly clear. Myriad array and qRT-PCR studies have revealed that the expression levels of specific miRNAs are reduced in the tumors of patients with a variety of cancers.4,5 When transfected into cancer cells, many of these miRNAs affect proliferation, viability, cell cycle, or apoptosis24,25 and affect the expression of multiple known oncogenes.17,18,26,27,28 Although the growth inhibition of LNCaP cells was not induced by transfection of miR-16 in our study, Bonci et al. showed that such inhibition of LNCaP cells was induced by transduction of the miR-15a-miR-16-1 cluster by lentiviral vector.8 This discrepancy indicated that the growth inhibition of LNCaP might be induced mainly by induction of miR-15a, further careful studies are needed, considering any clinical application of miR-16.
The clinical application of these naturally occurring tumor suppressors represents a major opportunity for the future treatment of cancer patients. As with other oligonucleotide-based therapies, realizing the potential of therapeutic miRNAs will require an effective delivery technology. In a previous study, we showed that intravenous injections of EZH2 and p110α siRNA complexed with atelocollagen inhibited the tumor growth in bone tissues of the mouse model.13 These results showed that an atelocollagen-mediated systemic delivery of siRNA could reach tumor cells at metastatic sites and inhibit tumor growth in vivo. As demonstrated here, atelocollagen facilitates the accumulation of enough synthetic miRNA in the cancers cells of an existing prostate tumor to affect the expression of a target gene. Furthermore, the combination of synthetic miR-16 and atelocollagen strongly inhibited the development of human prostate tumors in the bones of mice. Interestingly, the effect of miR-16 appeared to be restricted to the prostate cancer cells, as the miR-16 treated mice showed no notable side effects. Follow-up studies featuring the treatment of larger tumors and more extensive toxicity studies will be required to demonstrate the therapeutic potential of atelocollagen-miR-16; however, these early results are extremely encouraging.
Materials and Methods
Cell culture. The human prostate cell line 22Rv1, LNCaP, DU145, and PPC-1 cells were obtained from American Type Culture Collection (Manassas, VA) and maintained in RPMI 1640 medium containing 10% fetal bovine serum. The PC-3M-luc cells continuously expressing firefly luciferase (Xenogen, Alameda, CA) were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum and 0.2 mg/ml zeocin (Invitrogen, Carlsbad, CA). For construction of 3′-UTR–renilla luciferase plasmid and reporter assays, the segment of 3′-UTR of Bcl2 gene was amplified by PCR using genomic DNA from normal human prostate epithelial cells (CT-2555; Lonza Walkersville, Walkersville, MD) as reported.17 The PCR product was inserted into the pGL4.75[HRuc/CMV] vector (Promega, Madison, WI), using XbaI site immediately downstream from the stop codon of renilla luciferase (pGL4.75[HRuc/CMV]-Bcl2 3′UTR). For reporter assays, PC-3M-luc-C6 cells were transfected with 2 µg of pGL4.75[HRuc/CMV]-Bcl2 3′UTR using LipofectAMINE 2000 (Invitrogen). Stable transfectants were selected in hygromycine (0.2 mg/ml; Invitrogen) and bioluminescence was used to screen transfected clones for renilla and firefly luciferase gene expression using dual-luciferase assay system (Promega), intensity of renilla luciferase was normalized by firefly luciferase. Clones expressing the both luciferase gene were named PC-3M-luc/Rluc-Bcl2 3′UTR. The cells were maintained in vitro at 37 °C in a humidified atmosphere of 5% CO2.
Transfection with synthetic miR-16 and assay of cellular proliferation. Synthetic hsa-miR-16 (Pre-miR-hsa-miR-16; Ambion, Austin, TX) or NC miRNA (Pre-miR microRNA Precursor Molecule-Negative Control #2, cat. no. AM17111; Ambion) was delivered via lipid-based reverse transfection with 30 nmol/l final concentration of miRNA as described previously.29 As a control for inhibition of cellular proliferation, siRNA against the motor protein kinesin 11, also known as Eg5, was used. Eg5 is essential for cellular survival of most eukaryotic cells and a lack thereof leads to reduced cell proliferation and cell death.30 siEg5 was used in lipid-based transfection following the same experimental parameters that apply to miRNA. We observed 50–70% growth inhibition in all cell lines used in this study. Percent (%) proliferation values from the alamar blue assay (Invitrogen) were normalized to values from cells treated with NC miRNA.
Quantitative RT-PCR of miR-16. Human cultured cell line RNA was isolated using the ISOGEN (Wako Chemical, Tokyo, Japan). MiRNA-specific complementary DNA was generated using the TaqMan MicroRNA RT Kit (Applied Biosystems, Foster City, CA) and the miRNA-specific RT primer from the TaqMan Micro RNA Assay (Applied Biosystems). The expression of the U6 small nuclear RNA was used as an internal normalization control. miRNA levels were also measured by using the miRNA-specific probe included with TaqMan Micro RNA Assay on a Real-Time PCR System 7300 and SDS software (Applied Biosystems).
Quantitative PCR of miR-16 loci on chromosome 13q14. Genomic DNAs were extracted from PC-3M-luc and prostate epithelial cells using DNAeasy (Qiagen, Valencia, CA). Quantitative PCR for the miR-16 loci on chromosome 13q14 was performed using Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen) and primer sequences were 5′-GCA GCA CAG TTA ATA CTG GA-3′ and 5′-ATA GCT CTT ATG ATA GCA AT-3′. The house keeping gene, RNase P was also quantified as a control reference gene using Platinum Quantitative PCR SuperMix-UDG (Invitrogen) and TaqMan RNase P Detection Reagents Kit (Applied Biosystems). The reactions were incubated at 50 °C for 2 minutes, then heated to 95 °C for 2 minutes followed by 45 cycles of 15 seconds at 95 °C, and 30 seconds at 60 °C.
Evaluation of miRNA delivery to bone-metastatic tumors in mice. Animal experiments in this study were performed in compliance with the guidelines of the Institute for Laboratory Animal Research, National Cancer Center Research Institute. Seven- to ten-week-old male Balb/c athymic nude mice (CLEA Japan, Shizuoka, Japan) were anesthetized by exposure to 3% isoflurane on day zero and subsequent days. On day zero of the experiments, to generate a bone-metastatic human prostate cancer model, the anesthetized animals were injected with 2 × 106 PC-3M-luc/Rluc-Bcl2 3′UTR cells suspended in 100 µl sterile Dulbecco's phosphate-buffered saline into the left heart ventricle.13,15,16 For in vivo imaging, the mice were injected with ViviRen (5 mg/kg; Promega) by intravenous tail vein injection and imaged immediately to count the photons from animal whole bodies using the IVIS imaging system (Xenogen). After the bioluminescence from renilla luciferase disappeared, photons from firefly luciferase were counted as described previously.13
Preparation of complex with miR-16 and atelocollagen. For preparing the complexes of miRNA and atelocollagen (Koken, Tokyo, Japan), an equal volume of atelocollagen (0.1 % in phosphate-buffered saline at pH 7.4) and miRNA solution were combined and mixed by rotating for 1 hour at 4 °C. The final concentration of atelocollagen was 0.05%. Nine weeks after tumor injection, individual mice (from cohorts containing three animals) were injected with 200 µl of atelocollagen containing 50 µg of miR-16 complexed with atelocollagen, or NC miRNA/atelocollagen by intravenous tail-vein injection.
Analysis of miR-16/atelocollagen treatment for bone-metastatic prostate cancer. Mice were inoculated with PC-3M-luc cells into the left cardiac ventricle on day zero as described previously.13 The miR-16 and NC miRNA (50 µg) with 0.05% atelocollagen in a 200 µl volume were injected into the mouse tail vein on days 4, 7, and 10 postinoculation. Each experimental condition included six animals per group. At the end of the experiment on day 28, to confirm the presence of neoplastic cells, selected tissues were excised from the mice at necropsy. Tissues were fixed in 4% formaldehyde-phosphate-buffered saline(−), embedded in paraffin, cut into 5-µm sections, and stained with hematoxylin and eosin.
Clinical samples. Human prostate tissue samples derived from resected prostates from treatment-naive men with an average age of 65 (range of 52–76) diagnosed with nonmetastatic T2 or T3 prostate adenocarcinoma who gave informed consent. Gleason scores for all patients were 8 or 9. The tissues from patients were formalin-fixed, paraffin-embedded, sectioned, hematoxylin and eosin stained, and subjected to microscopic analysis. Three adjacent sections comprising 60–90% (74% average) cancerous tissue were selected as cancer samples from each patient. Three adjacent sections lacking evidence of cancer cells were selected as normal adjacent samples. RNA from the tissues were prepared using the RecoverAll Total RNA Isolation Kit (Ambion). The isolated RNA was subjected to qRT-PCR for miR-16 as described above.
MiR-16 functional pathway analysis. For preparation of RNA samples, PC-3M-luc cells were reverse transfected in quadruplicate by complexing miR-16 and NC miRNA and NeoFX transfection reagent (Ambion). The final concentration of miRNA was 30 nmol/l. Cells were harvested at 72 hours post-transfection. One microgram of total RNA per sample was used to prepare biotin-labeled cRNA using a MessageAmp II-based protocol (Ambion) and one round of amplification. Labeled cRNA was hybridized, washed, and scanned using Illumina's recommended protocols. Illumina BeadScan software was used to produce .idat, .xml, and .tif files for each array on a slide. Raw data were extracted using Illumina BeadStudio software, v 3.0 (Illumina, San Diego, CA). Following quality assessment, data from the replicate beads on each array were summarized into average intensity values and variances. The background subtracted data were used to compare the relative expression of mRNAs in cells transfected with miR-16, NC miRNA, and transfection agent only. analysis of variance was used to judge the significance of the variation observed between the various treatment groups. In total, 285 mRNAs exceeded the thresholds used to identify differentially expressed genes (log ratio greater than 0.5 or less than −0.5 for the average signal between miR-16 and NC miRNA or transfection agent only treatments and P values <0.001 for the 72 hour time-point).
Statistical analysis. The results are given as mean ± SD Statistical analysis was conducted using the analysis of variance with the Bonferroni correction for multiple comparisons. A P value of ≤0.05 was considered to indicate a significant difference.
SUPPLEMENTARY MATERIALFigure S1. The scheme of dual luciferase assay for monitoring of systemic miR-16 delivery. (a) A dual luciferase expressing PC-3M cells that have 3'UTR of Bcl2 under the Renilla luciferase gene, PC-3M-luc/Rluc-Bcl2 3'UTR cells, were generated. These cells were used for dual assay system, for monitoring of tumor growth by firefly luciferase, for monitoring of delivery efficacy of synthetic miR-16 by renilla luciferase. (b) When mice acquire bone metastasis derived from PC-3M-Fluc/Rluc-Bcl2 3'UTR cells, the efficacy of delivery of synthetic miR-16 into tumor cells on metastatic site can be monitored by measurement of photon counts from renilla luciferase. (c) Synthetic miR-16 was transfected into PC-3M-luc/Rluc-Bcl2 3'UTR cells. Relative repression of renilla luciferase expression was standardized to firefly luciferase. The data represent the mean (n=3) ± s.d. *P < 0.01 versus NC miRNA.Figure S2. Overview of experimental protocol for inhibition of metastatic tumor growth in bone tissues by the atelocollagen-mediated miRNA treatment. Overview of experimental protocol for inhibition of metastatic tumor growth in bone tissues by the atelocollagen-mediated miRNA Treatment. Mice were injected with 2X106 PC-3M-luc-C6 cells into the left heart ventricle on day 0. The miR-16 and NC miRNA (50 μg) with 0.05% atelocollagen (Atelo) or Atelo alone in a 200 μl volume were injected into the tail vein on days 4, 7 and 10 after tumor injection. At the end of the experiment on day 28, the metastasis was evaluated by IVIS imaging and confirmed by subsequent necropsy.Figure S3. KEGG cell cycle diagram. Genes are shown in a pathway map with genes specific to homo-sapiens shaded light green. The color red indicates down-regulated genes caused by miR-16 transfection into PC-3M-luc cells.Table S1. Data of the mRNA array for comparison of miR-16 and NC miR transfected PC-3M-luc cells.
Supplementary Material
The scheme of dual luciferase assay for monitoring of systemic miR-16 delivery. (a) A dual luciferase expressing PC-3M cells that have 3'UTR of Bcl2 under the Renilla luciferase gene, PC-3M-luc/Rluc-Bcl2 3'UTR cells, were generated. These cells were used for dual assay system, for monitoring of tumor growth by firefly luciferase, for monitoring of delivery efficacy of synthetic miR-16 by renilla luciferase. (b) When mice acquire bone metastasis derived from PC-3M-Fluc/Rluc-Bcl2 3'UTR cells, the efficacy of delivery of synthetic miR-16 into tumor cells on metastatic site can be monitored by measurement of photon counts from renilla luciferase. (c) Synthetic miR-16 was transfected into PC-3M-luc/Rluc-Bcl2 3'UTR cells. Relative repression of renilla luciferase expression was standardized to firefly luciferase. The data represent the mean (n=3) ± s.d. *P < 0.01 versus NC miRNA.
Overview of experimental protocol for inhibition of metastatic tumor growth in bone tissues by the atelocollagen-mediated miRNA treatment. Overview of experimental protocol for inhibition of metastatic tumor growth in bone tissues by the atelocollagen-mediated miRNA Treatment. Mice were injected with 2X106 PC-3M-luc-C6 cells into the left heart ventricle on day 0. The miR-16 and NC miRNA (50 μg) with 0.05% atelocollagen (Atelo) or Atelo alone in a 200 μl volume were injected into the tail vein on days 4, 7 and 10 after tumor injection. At the end of the experiment on day 28, the metastasis was evaluated by IVIS imaging and confirmed by subsequent necropsy.
KEGG cell cycle diagram. Genes are shown in a pathway map with genes specific to homo-sapiens shaded light green. The color red indicates down-regulated genes caused by miR-16 transfection into PC-3M-luc cells.
Data of the mRNA array for comparison of miR-16 and NC miR transfected PC-3M-luc cells.
Acknowledgments
We thank Ayako Inoue, Ayano Matsumoto, and Maho Kodama for their excellent technical work. We also thank Shunji Nagahara of Formulation Research Laboratories, Technology Research and Development Center, Dainippon Sumitomo Pharma Co., Ltd. for technological support and Koken Co., Ltd. for providing atelocollagen. This work was supported in part by a Grant-in-Aid for the Third-Term Comprehensive 10-Year Strategy for Cancer Control, a Grant-in-Aid for Scientific Research on Priority Areas Cancer from the Ministry of Education, Culture, Sports, Science and Technology, and the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation (NiBio), and a Takeda Science Foundation.
REFERENCES
- Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, et al. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- Calin GA., and , Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66:7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattie MD, Benz CC, Bowers J, Sensinger K, Wong L, Scott GK, et al. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol Cancer. 2006;5:24. doi: 10.1186/1476-4598-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL., and , Visakorpi T. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67:6130–6135. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14:1271–1277. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- Bullrich F., and , Croce CM.2001Molecular biology of chronic lymphocytic leukemia. In: Cheson, B (ed.) Chronic Lymphoid LeukemiaDekker: New York, 9–32.
- Dong JT, Boyd JC., and , Frierson HF. Loss of heterozygosity at 13q14 and 13q21 in high grade, high stage prostate cancer. Prostate. 2001;49:166–171. doi: 10.1002/pros.1131. [DOI] [PubMed] [Google Scholar]
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA., and , Croce CM. Genomics of chronic lymphocytic leukemia microRNAs as new players with clinical significance. Semin Oncol. 2006;33:167–173. doi: 10.1053/j.seminoncol.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Takeshita F, Minakuchi Y, Nagahara S, Honma K, Sasaki H, Hirai K, et al. Efficient delivery of small interfering RNA to bone-metastatic tumors by using atelocollagen in vivo. Proc Natl Acad Sci USA. 2005;102:12177–12182. doi: 10.1073/pnas.0501753102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z, Spitz MR, Babaian RJ, Strom SS, Troncoso P., and , Kagan J. Limiting the location of a putative human prostate cancer tumor suppressor gene at chromosome 13q14.3. Oncogene. 1999;18:7576–7583. doi: 10.1038/sj.onc.1203203. [DOI] [PubMed] [Google Scholar]
- Arguello F, Furlanetto RW, Baggs RB, Graves BT, Harwell SE, Cohen HJ, et al. Incidence and distribution of experimental metastases in mutant mice with defective organ microenvironments (genotypes Sl/Sld and W/Wv) Cancer Res. 1992;52:2304–2309. [PubMed] [Google Scholar]
- Jenkins DE, Yu SF, Hornig YS, Purchio T., and , Contag PR. In vivo monitoring of tumor relapse and metastasis using bioluminescent PC-3M-luc-C6 cells in murine models of human prostate cancer. Clin Exp Metastasis. 2003;20:745–756. doi: 10.1023/b:clin.0000006817.25962.87. [DOI] [PubMed] [Google Scholar]
- Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, et al. 2008KEGG for linking genomes to life and the environment Nucleic Acids Res 36Database issue): D480–D484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., and , Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, et al. 2006From genomics to chemical genomics: new developments in KEGG Nucleic Acids Res 34Database issue): D354–D357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Lu W, Takahashi H, Furusato M, Maekawa S, Nakano M, Meng C, et al. Allelotyping analysis at chromosome 13q of high-grade prostatic intraepithelial neoplasia and clinically insignificant and significant prostate cancers. Prostate. 2006;66:405–412. doi: 10.1002/pros.20363. [DOI] [PubMed] [Google Scholar]
- Chan JA, Krichevsky AM., and , Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- Cheng AM, Byrom MW, Shelton J., and , Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB., and , Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Ovcharenko D, Jarvis R, Hunicke-Smith S, Kelnar K., and , Brown D. High-throughput RNAi screening in vitro: from cell lines to primary cells. RNA. 2005;11:985–993. doi: 10.1261/rna.7288405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil D, Garçon L, Harper M, Duménil D, Dautry F., and , Kress M. Targeting the kinesin Eg5 to monitor siRNA transfection in mammalian cells. BioTechniques. 2002;33:1244–1248. doi: 10.2144/02336st01. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The scheme of dual luciferase assay for monitoring of systemic miR-16 delivery. (a) A dual luciferase expressing PC-3M cells that have 3'UTR of Bcl2 under the Renilla luciferase gene, PC-3M-luc/Rluc-Bcl2 3'UTR cells, were generated. These cells were used for dual assay system, for monitoring of tumor growth by firefly luciferase, for monitoring of delivery efficacy of synthetic miR-16 by renilla luciferase. (b) When mice acquire bone metastasis derived from PC-3M-Fluc/Rluc-Bcl2 3'UTR cells, the efficacy of delivery of synthetic miR-16 into tumor cells on metastatic site can be monitored by measurement of photon counts from renilla luciferase. (c) Synthetic miR-16 was transfected into PC-3M-luc/Rluc-Bcl2 3'UTR cells. Relative repression of renilla luciferase expression was standardized to firefly luciferase. The data represent the mean (n=3) ± s.d. *P < 0.01 versus NC miRNA.
Overview of experimental protocol for inhibition of metastatic tumor growth in bone tissues by the atelocollagen-mediated miRNA treatment. Overview of experimental protocol for inhibition of metastatic tumor growth in bone tissues by the atelocollagen-mediated miRNA Treatment. Mice were injected with 2X106 PC-3M-luc-C6 cells into the left heart ventricle on day 0. The miR-16 and NC miRNA (50 μg) with 0.05% atelocollagen (Atelo) or Atelo alone in a 200 μl volume were injected into the tail vein on days 4, 7 and 10 after tumor injection. At the end of the experiment on day 28, the metastasis was evaluated by IVIS imaging and confirmed by subsequent necropsy.
KEGG cell cycle diagram. Genes are shown in a pathway map with genes specific to homo-sapiens shaded light green. The color red indicates down-regulated genes caused by miR-16 transfection into PC-3M-luc cells.
Data of the mRNA array for comparison of miR-16 and NC miR transfected PC-3M-luc cells.