Abstract
Hypoxia-inducible factor-1 (HIF-1) plays crucial roles in tumor promotion by upregulating its target genes, which are involved in energy metabolism, angiogenesis, cell survival, invasion, metastasis, and drug resistance. The HIF-1α subunit, which is regulated by O2-dependent hydroxylation, ubiquitination, and degradation, has been identified as an important molecular target for cancer therapy. We have rationally designed G-rich oligodeoxynucleotides (ODNs) as inhibitors of HIF-1α for human cancer therapy. The lead compounds, JG243 and JG244, which form an intramolecular parallel G-quartet structure, selectively target HIF-1α and decreased levels of both HIF-1α and HIF-2α (IC50 < 2 µmol/l) and also inhibited the expression of HIF-1-regulated proteins [vascular endothelial growth factor (VEGF), Bcl-2, and Bcl-XL], but did not disrupt the expression of p300, Stat3, or p53. JG-ODNs induced proteasomal degradation of HIF-1α and HIF-2α that was dependent on the hydroxylase activity of prolyl-4-hydroxylase-2. JG243 and JG244 dramatically suppressed the growth of prostate, breast, and pancreatic tumor xenografts. Western blots from tumor tissues showed that JG-ODNs significantly decreased HIF-1α and HIF-2α levels and blocked the expression of VEGF. The JG-ODNs are novel anticancer agents that suppress tumor growth by inhibiting HIF-1.
Introduction
Within tumors, the availability of O2 and nutrients is limited by competition among proliferating cells, and diffusion of metabolites is inhibited by high interstitial pressure.1 Hypoxia is an important factor in the progression and therapeutic resistance of many human cancers.2 Hypoxia-inducible factor-1 (HIF-1) plays crucial roles in tumor promotion by upregulating its target genes, which are involved in energy metabolism, angiogenesis, cell survival, invasion/metastasis, and drug resistance.3 The transcription factor HIF-1 was originally shown to be induced in hypoxic cells and bound to the cis-acting hypoxia-response element located in the 3′-flanking region of the human EPO gene.4 HIF-1 is a heterodimer composed of an HIF-1α subunit and an HIF-1β subunit.5 Both HIF-1 subunits belong to the basic helix–loop–helix-containing PER-ARNT-SIM-domain family of transcription factors.6 To date, >70 putative hypoxia-inducible genes have been found to be directly regulated by HIF-1 (refs. 3,7). Under normoxic conditions the cellular half-life of HIF-1α is <5 minutes, as the protein is rapidly degraded by the ubiquitin–proteasome system, whereas under hypoxic conditions, HIF-1α is stabilized by the absence of post-translational prolyl hydroxylation at residues P402 and P564 (ref. 8). This hypoxic HIF-1α stabilization is followed by its translocation to the cell nucleus, and dimerization with HIF-1β. HIF-1 then activates transcription by binding to specific hypoxia-response elements in target genes and recruiting the coactivators p300 and CBP, which is blocked by the O2-dependent hydroxylation of asparaginyl residue N803 (ref. 8).
HIF-1α has been demonstrated to be overexpressed in many human cancers, including colon, brain, breast, gastric, lung, skin, ovarian, prostate, renal, and pancreatic carcinomas.3 Overexpression of HIF-1α, which results from intratumoral hypoxia and genetic alternations, has been associated with poor prognosis and treatment failure in a number of cancers.3 HIF-1α and HIF-2α are O2-regulated by the identical mechanism and each can form dimers with HIF-1β but have different mRNA expression patterns.9 HIF-1α is expressed ubiquitously, whereas HIF-2α expression is restricted to certain tissues. HIF-1α and HIF-2α promote angiogenesis by inducing hypoxia-induced expression of vascular endothelial growth factor (VEGF) in cancer cells.3,7,9 VEGF is essential for the proliferation and migration of vascular endothelial cells and enables the formation of new blood vessels in hypoxic tumors, leading to aggressive tumor growth. Targeting HIF-1α and HIF-2α could constitute a novel and potent cancer therapy.
G-rich sequences have been identified, cloned, and characterized in the telomeric sequences of many organisms, such as fungi, ciliates, vertebrates, and insects. The G-quartet motif was first proposed in telomeric DNA.10 G-quartets arise from the association of four G-bases into a cyclic Hoogsteen hydrogen-bonding arrangement in which each G-base makes two hydrogen bonds with its neighbor G-base. G-quartets stack on top of each other to form tetrad-helical structures. G-quartet structures have been demonstrated in vitro in telomeric sequences,11,12,13 fragile X syndrome nucleotide repeats,14 human immunodeficiency virus-1 RNA sequences,15 the immunoglobulin switch region,16 and promoter regions of several genes whose products are overexpressed in human cancer, including c-Myc, Bcl-2, VEGF, and HIF-1α.17 Depending on sequence, concentration, and base composition of the nucleic acids, G-quartet structures can be formed by an intramolecular process,12,18 by hairpin dimers,11,12 or by parallel-stranded tetramers.19,20 The stability of G-quartet structures depends on several factors including: the concentration of monovalent cations (particularly K+); the concentration of G-rich oligonucleotide (especially for dimer or tetramer formation); and the sequence of G-rich oligonucleotide, including the composition of loop sequences.21 We previously developed G-rich oligodeoxynucleotide (ODN) T40214, which forms a stable G-quartet structure and acts as an anticancer agent based on its ability to directly interact with Stat3 and block its activity.22 We also developed a novel intracellular delivery system for G-rich ODNs in order to increase drug activity in cells and in vivo.23,24 Here, we have developed novel G-rich ODNs that form an intramolecular quadruplex DNA structure with HIF-1α and HIF-2α and strongly inhibit HIF-1 activity. The two selected JG-ODNs, JG243 and JG244, significantly suppressed the growth of prostate, breast, and pancreatic tumor xenografts in nude mice.
Results
Design of G-rich ODNs as HIF-1α inhibitors
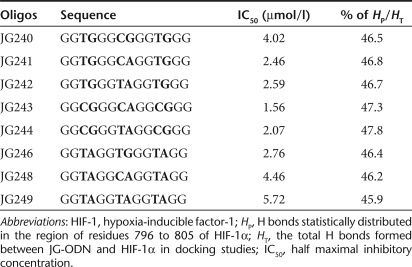
G-rich ODNs were developed as HIF-1 inhibitors based on two assumptions: (i) the nuclear magnetic resonance (NMR) structure of the C-terminal domain of HIF-1α (refs. 25,26) provides a reasonable molecular target and (ii) a computational approach can predict whether there is a stable interaction between HIF-1α and a given ODN. We designed several G-rich ODNs (JG240–JG249; Table 1) with potential to form G-quartet structures (Figure 1a) to target the HIF-1α and HIF-2α proteins. GRAMM (Global RAnge Molecular Matching; http://vakser.bioinformatics.ku.edu/main/resources_gramm1.03.php) docking was employed to model the structure of a complex formed between the two constituent molecules using their atomic coordinates. We randomly docked each JG-ODN 1,000 times onto the C-terminal domain of HIF-1α without setting any constraints and then analyzed the distribution of hydrogen (H) bonds formed between each JG-ODN and HIF-1α because H bonds play an important role in governing the interaction between proteins and DNA. The analysis showed that the hydrogen bonds formed between each JG-ODN and the C-terminal domain of HIF-1α were highly concentrated in the region of amino acids 796–805, including residue N803, which interacts with the coactivators p300 and CBP during the formation of the HIF-1 transcriptional complex (Figure 1b). The percentage of H bonds distributed in the region of residues 796–805 (HP), relative to the total H bonds formed between JG-ODN and HIF-1α (HT) for each JG-ODN is shown in Table 1.
Table 1.
Effects of designed JG-ODNs on HIF-1α inhibition
Figure 1.
Analysis of isolated JG-ODNs. (a) The scheme of intramolecular G-quartet structures of JG243, JG244, and T40214. (b) Histogram of the distribution of H bonds formed between JG244 and C-terminal domain of HIF-1α. (c) Nondenaturing gel electrophoresis of T40214 and JG-ODNs (JG240–JG249). (d) NMR spectrum of JG244. In the spectrum, the chemical shifts of imino protons of all residues are located at 10.5–12.0 ppm, showing that JG244 forms a stable G-quartet molecular structure. Inset: the arrows point to the imino protons in a G-quartet formation. HIF-1 hypoxia-inducible factor-1; NMR, nuclear magnetic resonance; ODN, oligodeoxynucleotide.
Molecular structure of JG-ODNs
To gain insight into the molecular structures of JG-ODNs, we first evaluated whether the designed G-rich ODNs can form similar structures to that of T40214, which forms an intramolecular G-quadruplex structure as determined by two-dimensional NMR.27,28 The migration rate in nondenatured gels depends on the size of molecular structures of the G-rich ODNs. The similar migration rates observed for JG243, JG244, and T40214 implied that the two JG-ODNs form a G-quadruplex structure similar to that of T40214; however, other G-rich ODNs (e.g., JG249) do not seem to form a similar G-quartet structure (Figure 1c).
We also used circular dichroism and one-dimensional 1H NMR methods to characterize the structures formed by JG243 and JG244 in K+ solution. In the one-dimensional NMR spectrum, proton peaks were clearly seen at 10.5–12 ppm for JG244 (Figure 1d) and JG 243 (data not shown), characteristic of the imino protons in G-quadruplex structures. In circular dichroism, spectra of JG243 and JG244 are similar to that of T40214 (data not shown), which are characteristic of an intramolecular G-quadruplex structure (Figure 1a).
Drug delivery for JG-ODNs
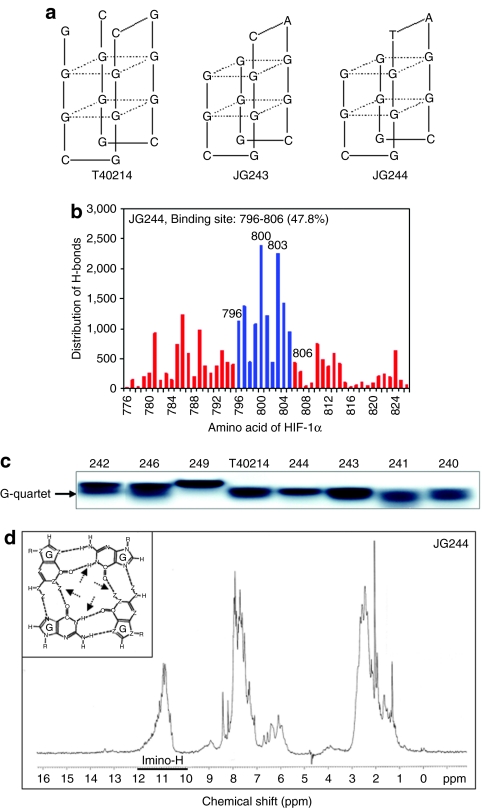
To determine whether JG-ODNs were delivered into cells and formed a G-quartet structure inside cells, we performed an assay with following steps: (i) 32P-labeled JG243 or JG244, either alone or mixed with polyethylenimine (PEI) at a molar ratio of ODN:PEI = 1:2, was added to wells containing 5 × 105 PANC1 human pancreatic cancer cells. (ii) After incubation for 3 hours, the cells were washed three times with fresh media to remove the undelivered ODNs, cultured for 24 hours, and lysed. (iii) Supernatants were fractionated by electrophoresis in a 20% nondenatured polyacrylamide gel. In Figure 2a, the migration of free T40214 (lane 1), JG243 (lane 2), and JG244 (lane 5) corresponded to a G-quartet structure. In lysates of cells exposed to JG243 or JG244 with PEI (lanes 3 and 6) two bands were observed, indicating that a portion of ODNs adhered to cell membranes (upper band) and a portion of ODNs entered into the cells and formed G-quartet structures (lower band). The analysis of the two band intensities indicated that the percentage of JG243 and JG244 that entered the cells was ~55 and 67%, respectively. Lysates of cells exposed to JG243 and JG244 without PEI (lanes 4 and 7) only showed the upper band, demonstrating that JG-ODNs alone cannot directly penetrate into cells in the absence of PEI.
Figure 2.
JG-ODN delivery and activity. (a) Nondenaturing gel electrophoresis demonstrates the intracellular delivery of JG-ODNs. Lanes 1, 2, and 5: migration of free T40214, JG243, and JG244, respectively. Lanes 3 and 6: JG243/PEI and JG244/PEI in PANC1 cell lysates. Lanes 4 and 7: JG243 and JG244 without PEI in PANC1 cell lysates. (b) JG-ODNs decreased HIF-1α levels in PANC1 cancer cells. N, nonhypoxic conditions (lane 1). Hypoxic cells were exposed to ODN concentrations from 0 to 5.0 µmol/l (lanes 2–6). GQ-ODN T40214 is a Stat3 inhibitor and ns-ODN is a control nonspecific oligonucleotide. (c) JG243 and JG244 inhibited HIF-1α and HIF-2α activations in PC3 prostate cancer cells but did not inhibit the levels of p300 and (d) p-Stat3. HIF-1 hypoxia-inducible factor-1; IL-6, interleukin-6; ns ODN, nonspecific oligodeoxynucleotide; PEI, polyethylenimine.
Levels of HIF-1α and HIF-2α are suppressed by JG-ODNs under hypoxia
We treated PANC1 cells with JG-ODNs suspended in PEI, and analyzed HIF-1α protein levels by immunoblot assay. The results (Figure 2b) demonstrated that in cells cultured at 20% O2 (lane 1), HIF-1α was barely detectable. Under hypoxic conditions (1% O2), JG-ODNs significantly decreased the level of HIF-1α in PANC1 cells compared with the level of HIF-1α in cells without JG-ODNs (lane 2). The concentration of each JG-ODN necessary for 50% inhibition of HIF-1α expression (IC50) was determined by analysis of the intensities of the HIF-1α signal (Table 1). JG243 and JG244 also suppressed HIF-1α expression in PC3 prostate cancer cells (data not shown). Based on these results, JG243 and JG244 were selected as lead compounds for further testing. Neither G-rich oligonucleotide T40214, which is a Stat3 inhibitor,24 nor a control nonspecific ODN (ns-ODN), inhibited HIF-1α expression under the same conditions. JG-ODNs inhibited HIF-1α expression at concentrations of 2.5–5.0 µmol/l.
Immunoblot assays were performed to determine whether JG-ODNs decreased the levels of HIF-2α, which can also dimerize with HIF-1β and activate hypoxia-induced transcription,9 or p300, which interacts with HIF-1α and HIF-2α under hypoxic conditions and is required for HIF-1-dependent transcription.26 JG243 and JG244 significantly decreased the levels of both HIF-1α and HIF-2α, but did not decrease p300 levels (Figure 2c). The drug carrier PEI facilitates delivery of JG-ODNs by endocytosis, and treatment with PEI alone had no effect on HIF-1α protein levels. JG243 and JG244 had no effect on phosphorylated Stat3 (p-Stat3) activation (Figure 2d). Taken together, these results demonstrate that JG243 and JG244 selectively target and decrease HIF-1α and HIF-2α expression in hypoxic cells.
Mechanism of JG-ODNs targeting HIF-1α and HIF-2α in hypoxic cells
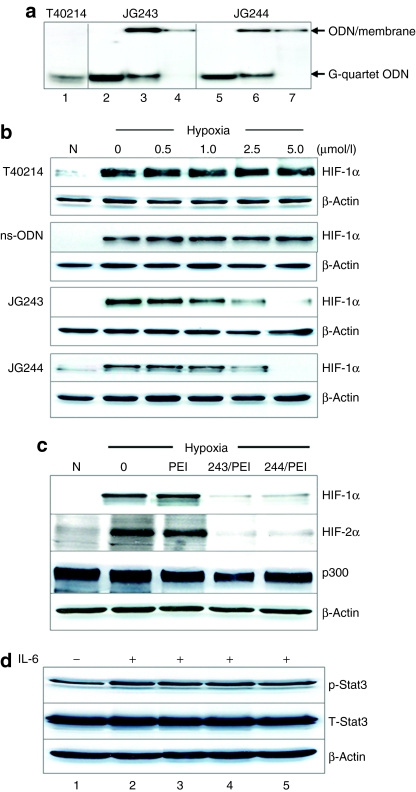
A glutathione S-transferase (GST) pull-down assay was employed to determine whether JG-ODNs forming G-quartet structure specifically interact with HIF-1α. The previous results demonstrated that both JG243 and JG244 form G-quartet structures similar to those observed with T40214 and T40214 has been demonstrated to specifically target the phospho-Stat3 dimer.29,30 Fluorescence labeled JG243, JG244, JG249, and T40214 were each dissolved in 120 mmol/l KCl, heated for 10 minutes at 90 °C, and cooled at 4 °C overnight to form G-quartet structures, and incubated with a GST-HIF-1α fusion protein containing residues 531–826 of HIF-1α, including the putative binding site for JG-ODNs (residues 796–806). The results of a glutathione pull-down assay (Figure 3a) demonstrated that compared with free JG244 and T40214 (lanes 1 and 9), JG243 and JG244 specifically interact with the fusion protein (lanes 2 and 4) but not with GST (lanes 3 and 5). T40214, JG249, and an ns-ODN that does not form a G-quartet structure did not interact with GST-HIF-1α (lanes 6–8). The binding energy between C-terminal of HIF-1α and JG244, which forms a chair-like G-quartet structure, is about −71 kcal and much lower than that between HIF-1α and T40214 (−32 kcal), which forms a symmetric G-quartet, leading to the observed stable binding interaction of HIF-1α with JG244 but not with T40214 (Figure 3b).
Figure 3.
Mechanism of JG-ODN interacting with HIF-1α and HIF-2α. (a) The results of pull-down assay A DNA gel shows the fluorescent-labeled JG244, and T40214 alone (lanes 1 and 8); GST plus JG243 or JG244 pull-down, showing no interaction between GST and JG-ODNs (lanes 3 and 5); and the pull-down of GST-HIF-1 plus JG243, JG244, JG249, ns-ODN, or T40214, respectively (lanes, 2, 4, and 6-8), showing JG243- and JG244-specific binding with HIF-1 protein but T40214, JG249, and ns-ODN do not. GST-HIF-1 contains the residues from 531 to 826 of HIF-1α. (b) Modeling of JG244 binding within C-terminal of HIF-1α. (c) HIF-1α and HIF-2α levels were determined in lysates from hypoxic untreated MDA-MB-468 breast cancer cells (control) and cells treated with JG243 or JG244 in the presence (+) or absence (−) of MG132. (d) HIF-1α and HIF-2α levels were determined in lysates from normoxia untreated MDA-MB-468 breast cancer cells (control) and cells treated with JG243 or JG244 in the presence (+) or absence (−) of CoCl2. GST, glutathione S-transferase; HIF-1, hypoxia-inducible factor-1; ns ODN, nonspecific oligodeoxynucleotide.
To determine whether JG-ODNs promote the proteasomal degradation of HIF-1α and HIF-2α in hypoxic cells, breast cancer cells (MDA-MB-468) were exposed to JG-ODNs (2.5 µmol/l) with PEI (at molar ratio of 1:2) for 3 hours, incubated in fresh medium at 1% O2 for 13 hours, and exposed to the proteasome inhibitor MG132 (10 µmol/l) for 6 hours in hypoxia before harvesting. Alternatively, after exposure to JG-ODNs for 3 hours, we added CoCl2 (100 µmol/l), which blocks prolyl hydroxylation and VHL (von Hippel–Lindau tumor-suppressor protein) binding, and then the cells were incubated 17 hours in normoxia. The results of immunoblot assays demonstrated that JG-ODNs significantly inhibited the hypoxia-induced expression of HIF-1α and HIF-2α in the absence, but not in the presence, of MG132 (Figure 3c). JG-ODNs did not inhibit the expression of HIF-1α and HIF-2α in the presence of CoCl2 under normoxia (Figure 3d). These results suggest that the treatment of hypoxic cells with JG-ODNs induces the proteasomal degradation of HIF-1α and HIF-2α, and that this effect is dependent upon hydroxylase activity.
JG-ODNs inhibit the expression of VEGF, GLUT, Bcl-2, and Bcl-XL
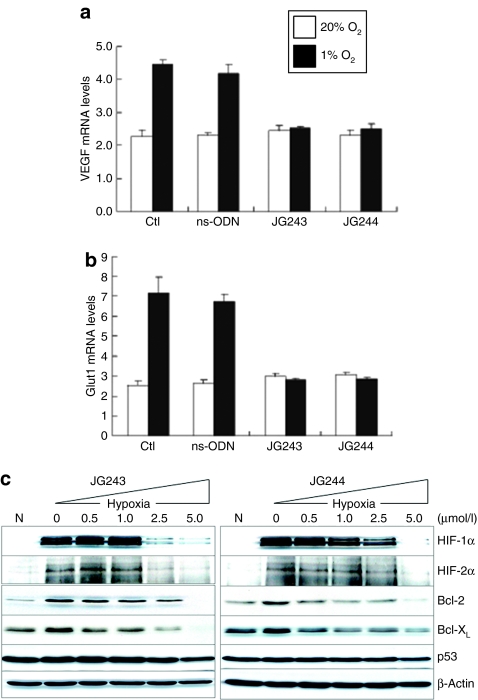
Expression of VEGF, which is a key stimulator of angiogenesis, is hypoxia-induced by HIF-1-dependent transcriptional activation.7 Quantitative real-time reverse transcriptase-PCR showed that JG243 and JG244 suppressed the twofold increase in VEGF mRNA expression in response to hypoxia (Figure 4a). The same result was observed for the levels of GLUT1 mRNA, which encodes a glucose transporter (Figure 4b). The ns-ODN did not inhibit VEGF or GLUT1 mRNA expression.
Figure 4.
JG-ODN suppresses the expression of HIF-1α-regulated genes. Quantitative real-time RT-PCR analysis of mRNAs encoding (a) VEGF and (b) GLUT1 in cells exposed to 20% O 2 or 1% O2. (c) JG243 and JG244 strongly suppressed the levels of HIF-1α, HIF-2α, Bcl-2, and Bcl-XL under hypoxia, but did not affect the expression of p53 or β-actin. Ctl, control; HIF-1, hypoxia-inducible factor-1; ns-ODN, nonspecific oligodeoxynucleotide.
A number of studies reported attenuated apoptosis under hypoxia of cancer cells with increased expression of the antiapoptotic proteins Bcl-2 and Bcl-XL (ref. 31) and overexpression of HIF-1α is sufficient to increase Bcl-2 and Bcl-XL levels in squamous carcinoma cells.32 The expression of mitochondrial antiapoptotic proteins in response to hypoxia may protect cells from death during cancer therapy. Expression of Bcl-2 and Bcl-XL was induced twofold in response to hypoxia (Figure 4c). JG243 and JG244 significantly inhibited the hypoxic increase of both Bcl-2 and Bcl-XL at concentrations (IC50 <2.5 µmol/l) that result in the inhibition of HIF-1α and HIF-2α. p53 expression was not disrupted by the JG-ODNs under the same conditions.
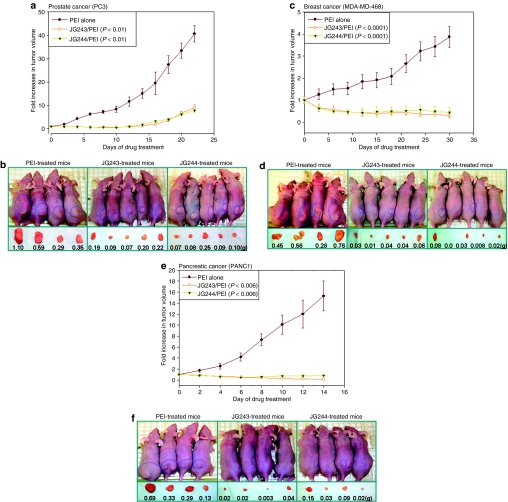
In vivo efficacy of JG-ODNs for treatment of prostate, breast, and pancreatic cancer xenografts
Hypoxia commonly develops within solid tumors and intratumoral hypoxia occurs early during tumor growth.2 In solid tumors HIF-1α is increased and treatment with JG-ODN as an inhibitor of HIF-1α may have anticancer effects. To test this hypothesis, we analyzed the in vivo efficacy of JG-ODNs in xenograft models of prostate cancer (PC3), breast cancer (MDA-MB-468), and pancreatic cancer (PNAC-1). Over 22 days of drug treatment in nude mice bearing prostate cancer xenografts, the mean volume of prostate tumors in PEI-treated mice increased 38-fold, whereas in mice treated with JG243/PEI or JG244/PEI tumor volume only increased 3.3- and 3.0-fold, respectively (Figure 5a). The mean tumor weight of PEI-treated mice was 0.58 ± 0.16 g compared to 0.15 ± 0.07 and 0.12 ± 0.03 g, in JG243/PEI- and JG244/PEI-treated mice, respectively (Figure 5b). Over 31 days of drug treatment in nude mice bearing breast cancer xenografts, the mean tumor volume in PEI-treated mice increased 4.0-fold. In contrast, the mean breast tumor volume in mice treated with JG243/PEI or JG244/PEI decreased 4.2- and 2.6-fold, respectively (Figure 5c). The mean tumor weight of PEI-treated mice was 0.45 ± 0.05 g compared to 0.04 ± 0.01 and 0.03 ± 0.03 g, for JG243/PEI- and JG244/PEI-treated mice, respectively (Figure 5d). Over 14 days of drug treatment, the mean volume of pancreatic tumor xenografts in PEI-treated mice increased 9.7-fold. The mean volume of pancreatic tumors in mice treated with JG243/PEI decreased more than fivefold and with JG244/PEI decreased 2.2-fold (Figure 5e). The mean tumor weight of PEI-treated mice was 0.36 ± 0.10 g whereas the mean tumor weights of JG243/PEI- and JG244/PEI-treated mice were 0.02 ± 0.01 and 0.07 ± 0.03 g, respectively (Figure 5f).
Figure 5.
JG-ODNs inhibit the growth of prostate and breast tumor xenografts. (a) Plot of fold increases in tumor volume of prostate tumors versus days of drug treatment. (b) Nude mice with prostate tumors (PC3) treated with PEI, JG243/PEI, and JG244/PEI, respectively. (c) Plot of fold increases in tumor volume of breast tumors versus days of drug treatment. (d) Nude mice with breast tumors (MDA-MB-468) treated with PEI, JG243/PEI, and JG244/PEI, respectively. The excised tumor and its weight (g) are shown below each mouse. (e) Plot of fold increases in tumor volume of pancreatic tumors versus days of drug treatment versus days of drug treatment. (f) Nude mice with pancreatic tumor xenografts that were treated with PEI, JG243/PEI, and JG244/PEI, respectively. ODN, oligodeoxynucleotide; PEI, polyethylenimine.
Inhibition of HIF-1α and HIF-2α by JG-ODNs in vivo
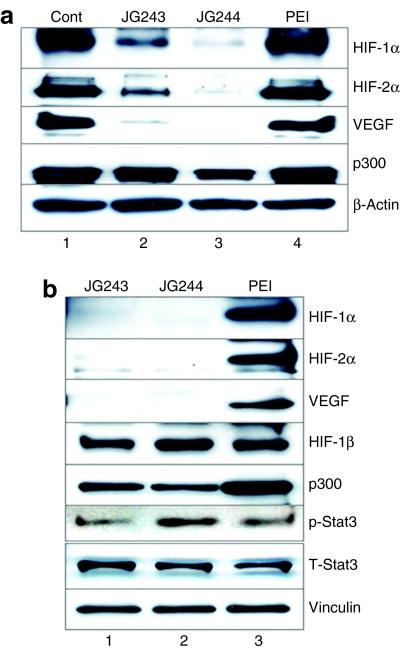
To analyze the effect of treatment with JG243 or JG244 on the intratumoral levels of HIF-1α and HIF-2α, we harvested breast and prostate tumor xenografts from untreated (as control), PEI-, JG243/PEI-, and JG244/PEI-treated mice following drug treatment. Immunoblot assays of the tumor tissues (Figure 6a,b) revealed that both JG243 and JG244 inhibited the expression of HIF-1α and HIF-2α and their downstream target VEGF, but did not inhibit the expression of p300, HIF-1β, or the activation of Stat3, consistent with the observations in cultured cancer cells that JG243 and JG244 selectively inhibited the expression of HIF-1α and HIF-2α.
Figure 6.
JG-ODNs specifically inhibit HIF-1α and HIF-2α expression in tumor xenografts. Lysates were prepared from (a) breast and (b) prostate tumor xenografts at the end of treatment and subjected to immunoblot assays using antibodies against the indicated proteins. Control, tumor from untreated mouse; JG243, tumor from mouse treated with JG243/PEI; JG244, tumor from mouse treated with JG244/PEI; PEI, tumor from mouse treated with PEI alone. Cont, control; HIF-1, hypoxia-inducible factor-1; ODN, oligodeoxynucleotide; PEI, polyethylenimine; VEGF, vascular endothelial growth factor.
In vivo efficacy of JG244 determined in immunocompetent mice bearing murine prostate tumors
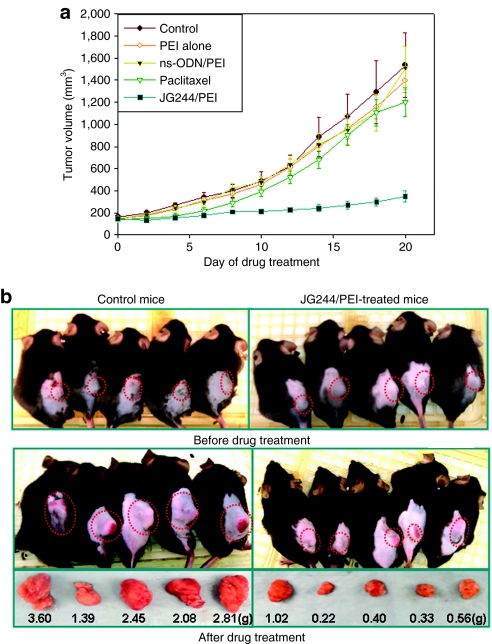
In order to determine the drug efficacy of JG244 in immunocompetent mice model, we employed normal mice (C57BL/6) bearing murine prostate tumor (TRAMP-C2). First, 200 µl of murine prostate cancer cells (TRAMP-C2) suspension (~3–5 × 106 cells) was injected subcutaneously into each mouse. When the tumor size reached ~150 mm3, the mice were randomly assigned to various groups (five mice per group): (i) untreated (control); (ii) PEI alone; (iii) ns-ODN/PEI; (iv) paclitaxel, a chemotherapeutic drug; and (v) JG244/PEI. The mice with prostate tumors were treated via intraperitoneal injection for 3 weeks. ODN and PEI were given every other day and paclitaxel was given every 4 days due to its toxicity. The results demonstrated (Figure 7 and Supplementary Table S1) that the mean tumor volumes of control, PEI-treated, ns-ODN/PEI-treated, and paclitaxel-treated mice increased from 164, 149, 145, 144 mm3 to 1,535, 1,396, 1,511, 1,201 mm3, respectively. The mean tumor volumes of JG244/PEI-treated mice only increased from 145 to 389 mm3. Also, the mean tumor weight of control, PEI-treated, ns-ODN/PEI-treated, and paclitaxel-treated mice were 2.47 ± 0.27, 1.41 ± 0.15, 1.53 ± 0.32, and 1.76 ± 0.16 g compared to 0.50 ± 0.10 g in JG244/PEI-treated mice. The results showed that JG-ODN (JG244) significantly suppressed the prostate tumor growth in immunocompetent mice as well and the treatment with JG-ODN/PEI has no sign of toxicity in the mice.
Figure 7.
JG-ODN suppresses murine prostate tumor growth. (a) Plot of tumor volume of prostate tumors versus days of drug treatment. (b) TRAMP-C2 tumor sizes and weights in the two groups of immunocompetent mice (C57BL/6) treated as control and with JG244/PEI at beginning and after 21 days, respectively. ns-ODN, nonspecific ODN; ODN, oligodeoxynucleotide; PEI, polyethylenimine.
Discussion
HIF-1 plays crucial roles in cancer progression by upregulating its target genes, which are involved in energy metabolism, angiogenesis, cell survival, invasion/metastasis, glycolysis, glucose uptake, and drug resistance. Growing evidence has demonstrated that HIF-1α is an important molecular target for cancer therapy.3 Several anticancer drugs, which were not originally developed as HIF-1 inhibitors, have been reported to inhibit HIF-1 activity,3,33 including (i) HSP90 inhibitors (e.g., radicicol, KF59333, and geldanamycin); (ii) inhibitors of topoisomerase I and topoisomerase II (e.g., Topotecan and GL331); and (iii) inhibitors of phosphatidylinositol 3-kinase (e.g., LY294002 and wortmannin) and mTOR (rapamycin). Other HIF-1 inhibitors include: YC-1, a drug that was originally developed for circulatory disorders, but which increases bleeding time and causes hypotension; PX-12 (1-methylpropyl 2-imidazolyl disulfide), which inhibits thioredoxin and thereby affects other signaling pathways; PX-478 (S-2-amino-3-[4′-N,N,-bis(2-chloroethyl)amino]), which acts by an unknown mechanism; and chetomin, which binds to the CH1 domain of p300, thereby attenuating transcription mediated by HIF-1 as well as many other transcription factors.3,33
Here, we demonstrate that the JG-ODNs, JG243 and JG244, selectively inhibit HIF-1α and HIF-2α in vitro and in vivo. The mechanism by which JG243 and JG244 inhibit HIF-1α and HIF-2α consists of three steps: (i) JG-ODNs interact with the PEI vehicle to form ODN/PEI complexes. (ii) The ODN/PEI complexes enter cells. The primary driving force for binding of ODN/PEI complexes to the cell membrane is electrostatic, whereas internalization of ODN/PEI complexes occurs mainly through endocytosis.34 (iii) JG-ODNs are released into the cytoplasm by disruption of the endosomal membrane, which is caused by interactions between cationic PEI and anionic JG-ODNs present in the membrane. After release into the cytoplasm, G-rich ODNs form an intramolecular G-quartet structure and selectively target HIF-1α and HIF-2α to promote their proteasomal degradation, leading to decreased expression of VEGF, Bcl-2, Bcl-XL, and GLUT1. Our reverse transcriptase-PCR data demonstrated that JG-ODNs suppressed hypoxia-induced but not baseline levels of VEGF and GLUT1 mRNA in cultured cells, providing further evidence that their effect was HIF-1-specific.
Previously, we developed as a potent inhibitor of phospho-Stat3 another G-quartet ODN, T40214, which significantly suppressed the growth of xenograft tumors in nude mice and has been accepted by the National Institutes of Health Rapid Access to Interventional Development program, providing additional validation of this therapeutic approach. Our results demonstrated that T40214 did not interact with HIF-1α or block the expression of HIF-1α in hypoxia, whereas JG-ODNs (JG243 and JG244) did not inhibit activation of phospho-Stat3 in cells and in tumors. Therefore, although they all form G-quartet structures, T40214 and JG-ODNs mediate their anticancer effects by specifically targeting phospho-Stat3 and HIF-1α, respectively. The distinct tertiary structures of these G-quartets provide a molecular basis for their selective interaction with the targeted proteins.
Effective delivery of JG-ODNs into target cells is a key factor for their successful utilization in clinical cancer therapy. PEI (a 25-kd polymer) has been widely used as a DNA carrier due to its high delivery efficiency.35,36 The ratio of PEI to DNA is critical for complex formation. PEI has a positively charged molecular surface that improves binding with ODNs and increases delivery efficiency. PEI also protects DNA against nuclease cleavage; however, excessive unbound PEI could interact with cell membranes and be toxic. The N/P ratio (nitrogen atoms of PEI to DNA phosphates) determines the net charge of the PEI/DNA complex, which dramatically influences the efficiency of the DNA delivery system. Previous studies suggested that when the N/P ratio is <3.0, PEI has the highest delivery efficiency and lowest toxicity in animal studies.37 Based on the structure and sequence of JG243 and JG244, the N/P ratio of JG-ODN/PEI complex was adjusted to 2.7. At this ratio, we did not find any toxicity induced by JG-ODN/PEI complexes in our experiments. Also, our in vivo results showed that after drug treatment the mean body weight of nude mice with prostate, breast, or pancreatic tumors was increased 4–6%, indicating that there is no overt toxicity of the JG-ODN/PEI complex.
In order to demonstrate the potential therapeutic efficacy of JG-ODNs, we selected prostate, breast, and pancreatic cancers for drug tests in nude mice xenografts. Our results showed that compared to tumors treated with PEI alone, JG243 and JG244 significantly suppressed, and in some instances even eliminated, these three tumor types in the xenograft model. The western blot data obtained from control tumor xenografts revealed high levels of HIF-1α and HIF-2α. JG243 and JG244 significantly reduced the levels of HIF-1α, HIF-2α, and the HIF-1-regulated protein VEGF in the tumors. Also, to investigate whether an intact immuno response can influence the drug efficacy and toxicity of JG-ODN/PEI, the immunocompetent mice (C57BL/6) bearing murine prostate tumors (TRAMP-C2) were employed to determine drug efficacy of JG244 using PEI, ns-ODN/ PEI, and paclitaxel as controls. Different from the xenograft model, TRAMP-C2 tumors grew very faster and deeper inside bodies of immunocompetent mice. The sequence of mouse HIF-1α protein is highly conserved with that of human HIF-1α protein (88%), especially in TAD (amino-terminal transactivation domain), which is 100% identical.38 Our results showed that JG244/PEI complex well controlled the tumor growth under immuno response. The mean body weight of the mice treated with JG244/PEI was increased >10%, indicating that JG244/PEI complex is well tolerated in immunocompetent mice. Together, the experimental evidence demonstrated that JG-ODN/PEI complex is a promising anticancer agent that specifically targets HIF-1α and HIF-2α for degradation, blocks HIF-1-mediated transcription, and significantly suppresses tumor growth.
Materials and Methods
Materials. All G-rich ODNs were synthesized by Midland Certified Reagent (Midland, TX) and used without further chemical modifications. PEI (~25-kd polymer) was purchased from Aldrich (St Louis, MO). The sources of antibodies are as follows: HIF-1α and HIF-1β were purchased from BD Biosciences (Franklin Lakes, NJ); HIF-2α from Novus Biologicals (Littleton, CO); VEGF from Santa Cruz Biotechnology (Santa Cruz, CA); β-actin, vinculin, and p300/CBP were from Sigma (St Louis, MO); and c-Myc, p53, Bcl-2, Bcl-XL, Stat3, and phospho-Stat3 were from Cell Signaling Technology (Danvers, MA). Human cancer cell lines PC3, MDA-MB-468, and PANC1 were purchased from American Type Culture Collection (Manassas, VA).
GRAMM docking. Three-dimensional structures of the designed JG-ODNs (JG240–JG249) were constructed by removing and replacing bases of T40214 via InsightII/Biopolymer (Accelrys, San Diego, CA). Employing the InsightII/Discover module with AMBER force field, energy minimization was performed on JG-ODNs. The intramolecular hydrogen bonds required for G-quartet formation were maintained during full geometry optimization. Conjugate-gradient energy minimization was used to optimize geometry and energy convergence was set at 0.01 kcal/mol. The GRAMM program was employed in this study.39,40 This program utilizes a geometry-based algorithm for predicting the structure of a possible complex between molecules of known structures. It can provide quantitative data related to the quality of the contact between the molecules. Three-dimensional structure of HIF-1α (PDB ID:1L8C) was obtained from the Protein Data Bank (www.rcsb.org/). Without setting any restriction, GRAMM docking of JG-ODNs to HIF-1α was carried out at low-resolution (grid step 3.3 Å, angular interval for rotation 30°) and the top 1,000 complexes obtained from the docking were analyzed to identify the primary binding site. The hydrogen bonds between JG-ODN and HIF-1α were identified in accordance with geometric standards used by the InsightII/Viewer module.
NMR. NMR experiments were performed on a Bruker DRX-600 spectrometer at 25 °C (ref. 41). Sample conditions were: 0.5 mmol/l DNA, 25 mmol/l KPO4, 100 mmol/l KCl, pH 7.0.
A pull-down assay of the GST-HIF-1 binding interaction. Binding of HIF-1α with the drugs, JG243 and JG244, were studied by GST pull-down assay as described previously42 with modifications. Escherichia coli BL21(DE3) strain was transformed with pGEX expression vector containing either GST alone or GST-HIF-1α (531–826) were induced with 0.1 mmol/l IPTG (isopropyl-β-thio galactopyranoside). Bacterial lysates were made using BugBuster Master Mix (Novagen, Madison, WI) as per manufacturer's instructions. The lysates were passed through glutathione affinity gel (Sigma) and the bound GST or GST-HIF-1α fusion protein were eluted with 10 mmol/l reduced glutathione in 50 mmol/l Tris–HCl (pH 8.6). JG243, JG244, ns-ODN, and T40214 labeled with 6-FAM (fluorophore) at 5′-end were treated to form G-quartet structures in 120 mmol/l KCl and then the 25 µg of G-quartet (JG243, JG244, ns-ODN, or T40214) were mixed with 25 µg of GST or GST-HIF-1α fusion protein in phosphate-buffered saline-Tween-20 binding buffer (Dulbecco's phosphate-buffered saline, pH 7.4, 0.1% Tween-20) incubated at dark for overnight with shaking at 4 °C, followed by further incubation of the mixtures with 70 µl of glutathione affinity gel beads for 4 hours at 4 °C. The beads were then washed with cold phosphate-buffered saline-Tween-20 buffer for five times. The bound proteins were eluted with 2× sodium dodecyl sulfate sample buffer. The above pull-down assay was carried out in duplicate and one set was resolved in 15% polyacrylamide gel, with fluorophore labeled drugs (10 µg) as such loaded in separate wells as positive controls. The gels were subsequently visualized under UV light for fluorescence. The other set of pull-down assay product was resolved in 15% sodium dodecyl sulfate–polyacrylamide gel electrophoresis followed by immunoblotting using anti GST-horseradish peroxidase antibody.
Cell culture and sample preparation. The cell lines were cultured in F-12K (for PC3) or Dulbecco's modified Eagle's medium (for PANC1 and MDA-MB-468) supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 µg/ml streptomycin and maintained in a 37 °C incubator. JG-ODNs were mixed with PEI at the ratio of 1:2. Then the JG-ODN/PEI complex was added to 6-well plates containing 6 × 105 cells/well. The concentrations of JG-ODNs added in 6-well plate were 0, 0.5, 1.0, 2.5, and 5.0 µmol/l. After incubation for 3 hours in the CO2 incubator, cells were washed three times with fresh medium and placed into a hypoxia chamber (BioSpherix, New York, NY) to continue incubation in the presence of 1% O2, 5% CO2, and 94% N2 for 16 hours before extraction. A volume of 150 µl of 1× lysis buffer (Cell Signaling Technology) was added to each well and the cells were scraped and collected within 5 minutes followed by protein extraction for western blot analysis.
Nondenatured DNA electrophoresis. The 5′-end labeling of the oligos: 400 ng of each oligo; 5 µl of reaction buffer containing 350 mmol/l Tris–HCl (pH 7.6), 50 mmol/l MgCl2, 500 mmol/l KCl, and 5 mmol/l 2-mercaptoethanol; 2 µl of γ-32P-ATP (10 µCi/µl, 3,000 Ci/mmol, from Amersham Biosciences, Piscataway, NJ) and 10 units of T4 Polynucleotide Kinase (Invitrogen, Carlsbad, CA) in a total reaction volume of 25 µl, incubated at 37 °C for 30 minutes, then added to MicroSpin G-25 Columns (Amersham Biosciences) to purify the end-labeled oligos. About 80 ng of labeled oligos were loaded in a 20% nondenatured polyacrylamide gel. The gel was run at 150 V for 4 hours at room temperature followed by autoradiography.
Western blot analysis. (i) Fifty micrograms of protein was used for each sample to load in a 10% sodium dodecyl sulfate–polyacrylamide gel. Separated proteins were transferred to Hybond-ECL nitrocellulose membranes and blocked with 5% nonfat milk in Tris-buffered saline Tween-20 for 1 hour at room temperature. Blots were then incubated with HIF-1α antibody (BD Biosciences) and β-actin (Sigma) overnight in a cold room. Blots were washed with Tris-buffered saline Tween-20 three times and subjected to horseradish peroxidase–conjugated secondary anti-mouse IgG (Cell Signaling Technology) by incubating for 1 hour at room temperature. Blots were washed with Tris-buffered saline Tween-20 and detected with the chemiluminescent kit (Amersham Biosciences). (ii) Xenografted tumors were harvested at the end of treatment, diced into small pieces, and sonicated on ice for 2 minutes. Tumor tissue (100 mg) was lysed in 300 µl of lysis buffer containing protease and phosphatase inhibitors. Tumor tissue protein (50 µg) was resolved on sodium dodecyl sulfate–polyacrylamide gel and probed by specific antibodies, as previously described. The bands were quantitated using a Personal Densitometer Scanner (version 1.30) and ImageQuant software (version 3.3) (GE Healthcare/Amersham Biosciences).
Reverse transcriptase-PCR analysis. Human GLUT1 and VEGF mRNA expression was quantified by real-time reverse transcriptase-PCR as previously described.43 RNA was isolated using Trizol (Invitrogen) followed by DNase (Ambion, Austin, TX) treatment. Primers were designed using Beacon Designer software and complementary DNA was prepared using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). cDNA samples were diluted 1:10 and real-time PCR was performed using iQ SYBR Green Supermix and the iCycler Real-time PCR Detection System (Bio-Rad). For each primer pair, annealing temperature was optimized by gradient PCR.
Animal/xenograft model. Athymic nude mice (Balb-nu/nu, 4 weeks old, weighing ~20 g) were ordered from Charles River Laboratories (Wilmington, MA). A total of 3.5 × 106 PC3, PANC1, or MDA-MB-4768 cells in 200 µl of PBS were injected subcutaneously into the right flank of each mouse. After the tumors were established (30–100 mm3), the nude mice were randomly assigned to three groups: group 1 was treated with PEI (2.5 mg/kg) alone (placebo); group 2 was treated with JG243/PEI (10 mg/kg + 2.5 mg/kg); and group 3 was treated with JG244/PEI (10 mg/kg + 2.5 mg/kg). Each group consisted of 4–5 mice. PEI and ODNs were administered every other day by intraperitoneal injection. Body weight and tumor volume were determined every other day. Tumor volume was calculated using the function (a × 0.5b2), where a equals the length and b equals the width of the tumor. The animal protocol was approved by Institutional Animal Care and Use Committee.
SUPPLEMENTARY MATERIALTable S1. In vivo effects of JG244/PEI against TRAMP-C2 murine prostate tumors in C57BL/6 mice.
Supplementary Material
In vivo effects of JG244/PEI against TRAMP-C2 murine prostate tumors in C57BL/6 mice.
Acknowledgments
We appreciate Danzhou Yong for her help to obtained NMR and CD spectra of JG-ODNs. This work was supported by NIH grant CA104035 (N.J.), The Johns Hopkins Institute for Cell Engineering (G.L.S.) and Q.Z. received support through NIH training grant T32 DK60445 (J.P.).
REFERENCES
- Fukumura D., and , Jain RK. Tumor microvasculature and microenvironment: targets for anti-angiogenesis and normalization. Microvasc Res. 2007;74:72–84. doi: 10.1016/j.mvr.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris AL. Hypoxia—a key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2:38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- Semenza GL., and , Wang GL. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol Cell Biol. 1992;12:5447–5454. doi: 10.1128/mcb.12.12.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GL., and , Semenza GL. Purification and characterization of hypoxia-inducible factor 1. J Biol Chem. 1995;270:1230–1237. doi: 10.1074/jbc.270.3.1230. [DOI] [PubMed] [Google Scholar]
- Wang GL, Jiang BH, Rue EA., and , Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci USA. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota K., and , Semenza GL. Regulation of angiogenesis by hypoxia-inducible factor 1. Crit Rev Oncol Hematol. 2006;59:15–26. doi: 10.1016/j.critrevonc.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Kaelin WG., and , Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- Gruber M., and , Simon MC. Hypoxia-inducible factors, hypoxia, and tumor angiogenesis. Curr Opin Hematol. 2006;13:169–174. doi: 10.1097/01.moh.0000219663.88409.35. [DOI] [PubMed] [Google Scholar]
- Gellert M, Lipsett MN., and , Davies DR. Helix formation by guanylic acid. Proc Natl Acad Sci USA. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundquist WI., and , Klug A. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature. 1989;342:825–829. doi: 10.1038/342825a0. [DOI] [PubMed] [Google Scholar]
- Smith FW., and , Feigon J. Quadruplex structure of Oxytricha telomeric DNA oligonucleotides. Nature. 1992;356:164–168. doi: 10.1038/356164a0. [DOI] [PubMed] [Google Scholar]
- Kang C, Zhang X, Ratliff R, Moyzis R., and , Rich A. Crystal structure of four-stranded Oxytricha telomeric DNA. Nature. 1992;356:126–131. doi: 10.1038/356126a0. [DOI] [PubMed] [Google Scholar]
- Fry M., and , Loeb LA. The fragile X syndrome d(CGG)n nucleotide repeats form a stable tetrahelical structure. Proc Natl Acad Sci USA. 1994;91:4950–4954. doi: 10.1073/pnas.91.11.4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundquist WI., and , Heaphy S. Evidence for interstrand quadruplex formation in the dimerization of human immunodeficiency virus 1 genomic RNA. Proc Natl Acad Sci USA. 1993;90:3393–3397. doi: 10.1073/pnas.90.8.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen D., and , Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- Dai J, Chen D, Jones RA, Hurley LH., and , Yang D. NMR solution structure of the major G-quadruplex structure formed in the human BCL2 promoter region. Nucleic Acids Res. 2006;34:5133–5144. doi: 10.1093/nar/gkl610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson JR, Raghuraman MK., and , Cech TR. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell. 1989;59:871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- Jin RZ, Breslauer KJ, Jones RA., and , Gaffney BL. Tetraplex formation of a guanine-containing nonameric DNA fragment. Science. 1990;250:543–546. doi: 10.1126/science.2237404. [DOI] [PubMed] [Google Scholar]
- Sen D., and , Gilbert W. A sodium-potassium switch in the formation of four-stranded G4-DNA. Nature. 1990;344:410–414. doi: 10.1038/344410a0. [DOI] [PubMed] [Google Scholar]
- Rhodes D., and , Giraldo R. Telomere structure and function. Curr Opin Struct Biol. 1995;5:311–322. doi: 10.1016/0959-440x(95)80092-1. [DOI] [PubMed] [Google Scholar]
- Jing N, Li Y, Xu X, Sha W, Li P, Feng L, et al. Targeting Stat3 with G-quartet oligodeoxynucleotides in human cancer cells. DNA Cell Biol. 2003;22:685–696. doi: 10.1089/104454903770946665. [DOI] [PubMed] [Google Scholar]
- Jing N, Xiong W, Guan Y, Pallansch L., and , Wang S. Potassium-dependent folding: a key to intracellular delivery of G-quartet oligonucleotides as HIV inhibitors. Biochemistry. 2002;41:5397–5403. doi: 10.1021/bi0120401. [DOI] [PubMed] [Google Scholar]
- Jing N, Li Y, Xiong W, Sha W, Jing L., and , Tweardy DJ. G-quartet oligonucleotides: a new class of signal transducer and activator of transcription 3 inhibitors that suppresses growth of prostate and breast tumors through induction of apoptosis. Cancer Res. 2004;64:6603–6609. doi: 10.1158/0008-5472.CAN-03-4041. [DOI] [PubMed] [Google Scholar]
- Dames SA, Martinez-Yamout M, De Guzman RN, Dyson HJ., and , Wright PE. Structural basis for Hif-1α/CBP recognition in the cellular hypoxic response. Proc Natl Acad Sci USA. 2002;99:5271–5276. doi: 10.1073/pnas.082121399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman SJ, Sun ZY, Poy F, Kung AL, Livingston DM, Wagner G, et al. Structural basis for recruitment of CBP/p300 by hypoxia-inducible factor-1α. Proc Natl Acad Sci USA. 2002;99:5367–5372. doi: 10.1073/pnas.082117899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing N, Gao X, Rando RF., and , Hogan ME. Potassium-induced loop conformational transition of a potent anti-HIV oligonucleotide. J Biomol Struct Dyn. 1997;15:573–585. doi: 10.1080/07391102.1997.10508967. [DOI] [PubMed] [Google Scholar]
- Jing N., and , Hogan ME. Structure-activity of tetrad-forming oligonucleotides as a potent anti-HIV therapeutic drug. J Biol Chem. 1998;273:34992–34999. doi: 10.1074/jbc.273.52.34992. [DOI] [PubMed] [Google Scholar]
- Jing N, Zhu Q, Yuan P, Li Y, Mao L., and , Tweardy DJ. Targeting signal transducer and activator of transcription 3 with G-quartet oligonucleotides: a potential novel therapy for head and neck cancer. Mol Cancer Ther. 2006;5:279–286. doi: 10.1158/1535-7163.MCT-05-0302. [DOI] [PubMed] [Google Scholar]
- Weerasinghe P, Garcia GE, Zhu Q, Yuan P, Feng L, Mao L, et al. Inhibition of Stat3 activation and tumor growth suppression of non-small cell lung cancer by G-quartet oligonucleotides. Int J Oncol. 2007;31:129–136. [PubMed] [Google Scholar]
- Shimizu S, Eguchi Y, Kamiike W, Itoh Y, Hasegawa J, Yamabe K, et al. Induction of apoptosis as well as necrosis by hypoxia and predominant prevention of apoptosis by Bcl-2 and Bcl-XL. Cancer Res. 1996;56:2161–2166. [PubMed] [Google Scholar]
- Sasabe E, Tatemoto Y, Li D, Yamamoto T., and , Osaki T. Mechanism of HIF-1α-dependent suppression of hypoxia-induced apoptosis in squamous cell carcinoma cells. Cancer Sci. 2005;96:394–402. doi: 10.1111/j.1349-7006.2005.00065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melillo G. Hypoxia-inducible factor 1 inhibitors. Meth Enzymol. 2007;435:385–402. doi: 10.1016/S0076-6879(07)35020-9. [DOI] [PubMed] [Google Scholar]
- Chesnoy S., and , Huang L. Structure and function of lipid-DNA complexes for gene delivery. Annu Rev Biophys Biomol Struct. 2000;29:27–47. doi: 10.1146/annurev.biophys.29.1.27. [DOI] [PubMed] [Google Scholar]
- Lungwitz U, Breunig M, Blunk T., and , Göpferich A. Polyethylenimine-based non-viral gene delivery systems. Eur J Pharm Biopharm. 2005;60:247–266. doi: 10.1016/j.ejpb.2004.11.011. [DOI] [PubMed] [Google Scholar]
- Neu M, Fischer D., and , Kissel T. Recent advances in rational gene transfer vector design based on poly(ethylene imine) and its derivatives. J Gene Med. 2005;7:992–1009. doi: 10.1002/jgm.773. [DOI] [PubMed] [Google Scholar]
- Boussif O, Lezoualc'h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci USA. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger RH, Rolfs A, Marti HH, Guénet JL., and , Gassmann M. Nucleotide sequence, chromosomal assignment and mRNA expression of mouse hypoxia-inducible factor-1α. Biochem Biophys Res Commun. 1996;223:54–59. doi: 10.1006/bbrc.1996.0845. [DOI] [PubMed] [Google Scholar]
- Katchalski-Katzir E, Shariv I, Eisenstein M, Friesem AA, Aflalo C., and , Vakser IA. Molecular surface recognition: determination of geometric fit between proteins and their ligands by correlation techniques. Proc Natl Acad Sci USA. 1992;89:2195–2199. doi: 10.1073/pnas.89.6.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie DW., and , Kemp GJ. Protein docking using spherical polar Fourier correlations. Proteins. 2000;39:178–194. [PubMed] [Google Scholar]
- Dai J, Carver M, Punchihewa C, Jones RA., and , Yang D. Structure of the Hybrid-2 type intramolecular human telomeric G-quadruplex in K+ solution: insights into structure polymorphism of the human telomeric sequence. Nucleic Acids Res. 2007;35:4927–4940. doi: 10.1093/nar/gkm522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek JH, Liu YV, McDonald KR, Wesley JB, Hubbi ME, Byun H, et al. Spermidine/spermine-N1-acetyltransferase 2 is an essential component of the ubiquitin ligase complex that regulates hypoxia-inducible factor 1α. J Biol Chem. 2007;282:23572–23580. doi: 10.1074/jbc.M703504200. [DOI] [PubMed] [Google Scholar]
- Zhang H, Gao P, Fukuda R, Kumar G, Krishnamachary B, Zeller KI, et al. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell. 2007;11:407–420. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
In vivo effects of JG244/PEI against TRAMP-C2 murine prostate tumors in C57BL/6 mice.