Abstract
The development of Alzheimer's disease (AD) is closely connected with cholesterol metabolism. Cholesterol increases the production and deposition of amyloid-β (Aβ) peptides that result in the formation of amyloid plaques, a hallmark of the pathology. In the brain, cholesterol is synthesized in situ but cannot be degraded nor cross the blood–brain barrier. The major exportable form of brain cholesterol is 24S-hydroxycholesterol, an oxysterol generated by the neuronal cholesterol 24-hydroxylase encoded by the CYP46A1 gene. We report that the injection of adeno-associated vector (AAV) encoding CYP46A1 in the cortex and hippocampus of APP23 mice before the onset of amyloid deposits markedly reduces Aβ peptides, amyloid deposits and trimeric oligomers at 12 months of age. The Morris water maze (MWM) procedure also demonstrated improvement of spatial memory at 6 months, before the onset of amyloid deposits. AAV5-wtCYP46A1 vector injection in the cortex and hippocampus of amyloid precursor protein/presenilin 1 (APP/PS) mice after the onset of amyloid deposits also reduced markedly the number of amyloid plaques in the hippocampus, and to a less extent in the cortex, 3 months after the injection. Our data demonstrate that neuronal overexpression of CYP46A1 before or after the onset of amyloid plaques significantly reduces Aβ pathology in mouse models of AD.
Introduction
Alzheimer's disease (AD) is the most common neurodegenerative disease of the elderly but no efficient treatment is currently available. A hallmark of all forms of AD is an abnormal accumulation of amyloid-β (Aβ) peptides in specific brain regions.1 Aβ peptides are generated by sequential proteolytic processing of the amyloid precursor protein (APP) by β-site APP-cleaving enzyme 1 (BACE1) and γ-secretase.2
Further to the identification of ε4 isoform of apolipoprotein E (APOE) as the main risk factor for AD,3 there is growing evidence that cholesterol regulates the generation and clearance of APP proteolytic products.4,5 Cholesterol content in cultured cells and in transgenic mouse models of AD is paralleled by the production of Aβ peptides.6,7 Moreover, senile plaques are enriched in cholesterol8 and the postmortem analysis of AD brains revealed an abnormal retention of cholesterol in neurons.9
Drugs that modify cholesterol homeostasis are currently considered as potential therapies for AD. Cholesterol synthesis inhibitors like statins reduce amyloid burden in guinea-pig and transgenic mouse models of the disease10 but this positive effect awaits validation in prospective clinical trials.11 An inhibitor of acyl-coenzyme A: cholesterol acyltransferase was also shown to reduce amyloid pathology in APP23 mice, a mouse model of AD.12 However, these drugs not only modify cholesterol metabolism in the brain but also in peripheral tissues and plasma.
Like all mammalian cells, neurons cannot degrade cholesterol by themselves and blood–brain barrier restricts its export out of the brain.13 The major mechanism of cerebral cholesterol clearance involves the conversion of cholesterol into 24S-hydroxycholesterol by cholesterol 24-hydroxylase, an enzyme expressed only in neurons and encoded by the CYP46A1 gene.14 24S-hydroxycholesterol can freely cross the blood–brain barrier and is degraded in the liver. Following interaction with liver-X receptors (LXRs), 24S-hydroxycholesterol regulates in vitro the expression of several adenosine triphosphate–binding cassette transporters (ABCA1, ABCA2, ABCG1, ABCG4, and ABCG5) and APOE genes involved in cholesterol transport and homeostasis in the brain.15,16
We hypothesized that increasing neuronal cholesterol 24-hydroxylase activity will reduce amyloidogenesis from APP. As no small molecule is able to stimulate cholesterol 24-hydroxylase activity in vitro or in vivo, we tested whether the brain delivery of CYP46A1 gene through injection of adeno-associated vector (AAV), before or after the onset of amyloid plaques, could be a strategy to decrease the deposition of amyloid peptides. This hypothesis was evaluated: (i) before the onset of amyloid deposits in APP23 mice that develop memory deficits at 3 months and amyloid plaques at 6 months17,18 and (ii) after the onset of amyloid deposits in APP/presenilin 1 (PS) mice that develop amyloid plaques at 2½ months and are characterized by a more severe phenotype.19 Other viral vector approaches aiming at the same objective were previously chosen to deliver the neprilysin gene or small interfering RNAs targeting APP and BACE.20,21,22
We report that the overexpression of CYP46A1 gene in hippocampus and cortex of APP23 mice from the age of 3 months improves their spatial memory at 6 months and markedly reduces amyloid deposits, Aβ40 and Aβ42 peptides and trimeric Aβ oligomers at 12 months. The overexpression of CYP46A1 gene in hippocampus and cortex of APP/PS presenilin 1 mice from the age of 3 months also reduces the number of amyloid plaques at 6 months.
Further in vitro studies performed in murine neuroblastoid (N2a) cells overexpressing a mutated human APP gene indicate that CYP46A1 overexpression diminishes the cholesterol concentration and the recruitment of APP and presenilin 1 (PSEN1), a component of γ-secretase complex, in lipid rafts.
Our results suggest that the cerebral delivery of CYP46A1 gene can specifically reduce the cleavage of APP in vivo before or after the onset of amyloid deposits and indicate that this approach could have potential therapeutic value for treatment of AD.
Results
AAV-mediated expression of CYP46A1 increases 24S-hydroxycholesterol content in the brain of APP23 mice without global cholesterol change
AAVs expressing wild-type (wt) (AAV5-wtCYP46A1, n = 11 females) or mutated (AAV5-mtCYP46A1, n = 11 females) human CYP46A1 cDNA tagged with the hemaglutinin (HA) epitope were injected in hippocampus, frontal, and parietal cortices of both hemispheres of 3-month-old APP23 mice. The mutant (mt) CYP46A1 protein contains a missense mutation (A1309C) that destroys its heme structure, resulting in the complete lack of cholesterol 24-hydroxylase activity (L. Pradier, unpublished results).
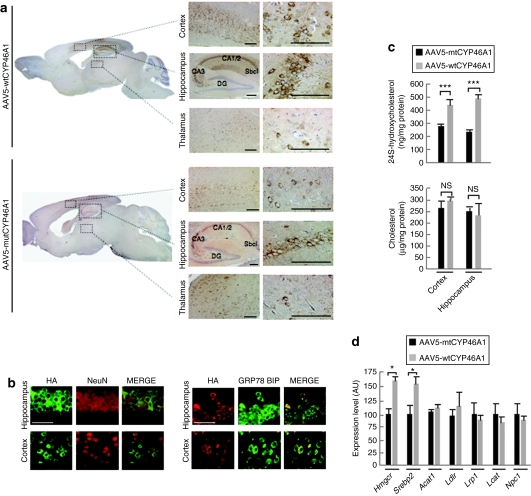
At age 6 (i.e., 4 months postinjection; n = 12) and 12 months (i.e., 10 months postinjection; n = 10), wt or mt CYP46A1 proteins showed comparable expression in neurons from the cerebral cortex and hippocampus of injected APP23 mice (Figure 1a,b). Both wt and mt CYP46A1 proteins colocalized with the GRP78 Bip marker in the endoplasmic reticulum, as did the endogenous cholesterol-24-hydroxylase23 (Figure 1b). With quantitative reverse transcriptase–PCR (RT-PCR), the level of CYP46A1 mRNA was found to be eightfold higher than the level of endogenous mouse CYP46A1 in APP23 mouse (data not shown). The overexpression of human CYP46A1 gene did not modify the expression of endogenous murine Cyp46A1 gene (data not shown). In comparison with the level observed in noninjected APP23 mice or APP23 mice injected with the AAV5-mtCYP46A1 vector, the 24S-hydroxycholesterol content increased twofold in the dissected cerebral cortex and hippocampus of mice injected with AAV5-wtCYP46A1 vector. This indicates that the recombinant wt CYP46A1 enzyme was fully active and that the mt CYP46A1 protein was inactive and did not disturb the activity of endogenous murine cholesterol 24-hydroxylase in APP23 mice. The concentrations of total cholesterol (Figure 1c) and cholesteroyl esters (data not shown) remained however unchanged in the cerebral cortex and hippocampus of 12-month-old APP23 mice injected with AAV5-wtCYP46A1 or AAV5-mtCYP46A1 vectors.
Figure 1.
Injections of AAV5-CYP46A1 vector in cerebral cortex and hippocampus of APP23 mice increase the level of 24S-hydroxycholesterol. (a) Representative expression of the wild-type (wtCYP46A1) and mutant (mtCYP46A1) forms of human CYP46A1 protein in the brain of 12-month-old APP23 mice after injection of adeno-associated vector (AAV) vector at 3 months. DG, dentate gyrus; Sbcl, subiculum; CA, Cornu Ammonis. Bar = 200 µmol/l. (b) Immunolabeling of hemaglutinin (HA)-tagged wtCYP46A1 protein in neurons (NeuN, nuclear staining, left panel) and co-localization with the endoplasmic reticulum Grp78 Bip marker (right panel). HA-tagged mtCYP46 protein has identical subcellular localization (data not shown). Bar = 200 µmol/l. (c) Cholesterol and 24S-hydroxycholesterol concentrations in the cerebral cortex and hippocampus of 12-month-old APP23 mice injected with AAV5-wtCYP46A1 or AAV5-mtCYP46A1 vectors (n = 5 mice per group). No difference in 24S-hydroxycholesterol and cholesterol content was observed between noninjected and AAV5-mtCYP46A1-injected APP23 mice. (d) Quantitative expression of murine 3-hydroxy-3-methylglutaryl-coenzyme A reductase (Hmgcr), sterol-binding protein 2 (Srebp2), acyl-coenzyme A: cholesterol acyltransferase 1 (Acat1), low-density lipoprotein–related protein 1 (Lrp1), lecithin:cholesterol acyltransferase (Lcat), and Niemann–Pick disease C1 (Npc1) genes in APP23 mice at 12 months after cerebral injections of AAV5-wtCYP46A1 or AAV5-mtCYP46A1 vectors (n = 5 mice per group). (Mann–Whitney U-test) ***P < 0.0005; NS, non significant.
We analyzed the expression of several important genes involved in cholesterol metabolism by quantitative RT-PCR. As shown in Figure 1d, the expression of 3-hydroxy-3-methylglutaryl-coenzyme A reductase (Hmgcr) and sterol-binding protein 2 (Srebp2) gene increased by 1.6-fold but no change in acyl-coenzyme A:cholesterol acyltransferase 1, low-density lipoprotein receptor (Ldlr), low-density lipoprotein–related protein 1 (Lrp1), lecithin:cholesterol acyltransferase (Lcat), and Niemann–Pick disease C1 (Npc1) gene expression was observed.
AAV5-wtCYP46A1 vector injection results in decreased amyloid deposition and Aβ peptides production in APP23 mice
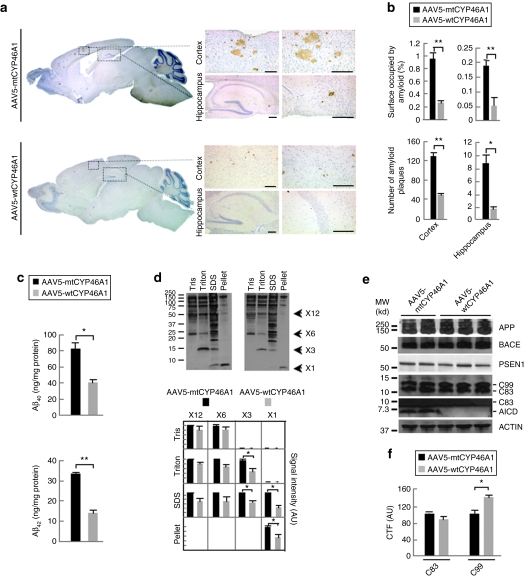
The neuronal overexpression of the CYP46A1 gene from the age of 3 months was associated with a marked reduction in the number of amyloid plaques (63 and 68%) and in the percentage of area occupied by amyloid deposits (73 and 70%) in the hippocampus and cerebral cortex of APP23 mice at 12 months of age (Figure 2a,b). Most remaining amyloid deposits were concentrated in regions where very few or no cells expressed the wtCYP46A1 protein. Hematein-eosin staining did not reveal the presence of lymphocytic infiltrates that could arise from immune reaction against the AAV or the transgene expression (data not shown). Aβ40 and Aβ42 peptides assessed by enzyme-linked immunosorbent assay decreased by 50 ± 3 and 57 ± 4%, respectively, in pooled cerebral cortex and hippocampus samples (Figure 2c). Because of their reported high neuronal toxicity,24,25 amyloid oligomers were also analyzed by western blot. The amount of hexameric and dodecameric Aβ oligomers were slightly reduced without reaching statistical significance. However, trimeric Aβ oligomers were decreased by 50% in the sodium dodecyl sulfate and Triton fractions of AAV5-wtCYP46A1-injected mice (P < 0.05, Figure 2d). As already observed in other mouse models of AD (except in 3xTg-AD mice at 20 months26), no dimeric aggregates could be detected with the 6E10 antibody in the brain homogenates of APP23 mice.27,28 These results indicate that neuronal overexpression of the CYP46A1 gene before the onset of amyloid plaques has a clear beneficial effect on the amyloidogenic processing of APP in APP23 mice at 12 months.
Figure 2.
Intracerebral delivery of CYP46A1 at 3 months markedly reduces amyloid pathology in APP23 mice at 12 months. (a) Representative immunostaining of amyloid deposits with NT2 antibody in 12-month-old APP23 mice injected with AAV5-mtCYP46A1 (upper panel) or AAV5-wtCYP46A1 (lower panel) vectors at 3 months. Bar = 200 µmol/l. (b) Stereological analyses of amyloid deposits number and surface in the cortex and hippocampus of APP23 mice injected with AAV5-mtCYP46A1 or AAV5-wtCYP46A1 vectors at 12 months (three different section levels per mouse, with three to five slices per level, n = 5 mice per group). (c) Aβ40 and Aβ42 peptide concentrations in the pooled cerebral cortex and hippocampus of 12-month-old APP23 mice injected with AAV5-wtCYP46A1 or AAV5-mtCYP46A1 vectors. Aβ peptides were quantified using enzyme-linked immunosorbent assay after a solubilization step in 5 mol/l guanidine–HCl (n = 5 mice per group). (d) Quantification at 12 months of mono-, tri-, hexa-, and dodecamers of Aβ peptides by western blotting after extraction in 10 mmol/l Tris–HCl (Tris), 10 mmol/l Tris–HCl/2% Triton (Triton) and 10 mmol/l Tris–HCl/0.5% SDS (SDS) buffers (n = 5 mice per group). (e) Representative western blot of full length APP (APPfl), BACE1, PSEN1, and CTFs (C83, C99, and AICD) in pooled cerebral cortex and hippocampus samples from 12-month-old APP23 mice injected with AAV5-wtCYP46A1 and AAV5-mtCYP46A1 vectors (n = 5 mice per group). (f) Quantification at 12 months of α- and β-secretase C-terminal fragments (C83 and C99) in pooled cerebral cortex and hippocampus samples from 12-month-old APP23 mice injected with AAV5-wtCYP46A1 and AAV5-mtCYP46A1 vectors. The amounts of C-terminal fragments are normalized to ACTIN level (n = 5 mice per group). AU, arbitrary unit. (Mann–Whitney U-test). *P < 0.05; **P < 0.005.
To determine whetherAPP processing was directly affected by the overexpression of cholesterol 24-hydroxylase, we quantified the expression level of several genes of the amyloidogenic pathway and the amounts of APP cleavage products. In AAV5-wtCYP46A1-injected mice, the expression of Adam9, -10, -17 (three enzymes that were shown to have some α-secretase activity29), Bace1 and Psen1 remained unchanged (data not shown). Western blotting experiments showed that the amount of α-secretase product C83 was unaffected whereas the β-secretase product C99 increased by 40% in AAV5-wtCYP46A1-injected mice (Figure 2e,f). APP intracellular domain (AICD) fragments generated by γ-cleavage were barely detectable (Figure 2e) in treated APP23 mice, suggesting that CYP46A1 expression induced a decreased cleavage of APP C-terminal fragments (CTFs) by γ-secretase in vivo.
Quantitative RT-PCR experiments also failed to detect any induction of LXR target genes expression such as Abca1, Abca2, Abcg1, Abcg4, Abcg5 and ApoE (data not shown), suggesting that the beneficial effect of cholesterol 24-hydroxylase overexpression on Aβ production in vivo was not due to the induction of LXR pathway.
Neuronal overexpression of CYP46A1 decreases microgliosis and astrocytosis and improves cognitive deficits in APP23 mice
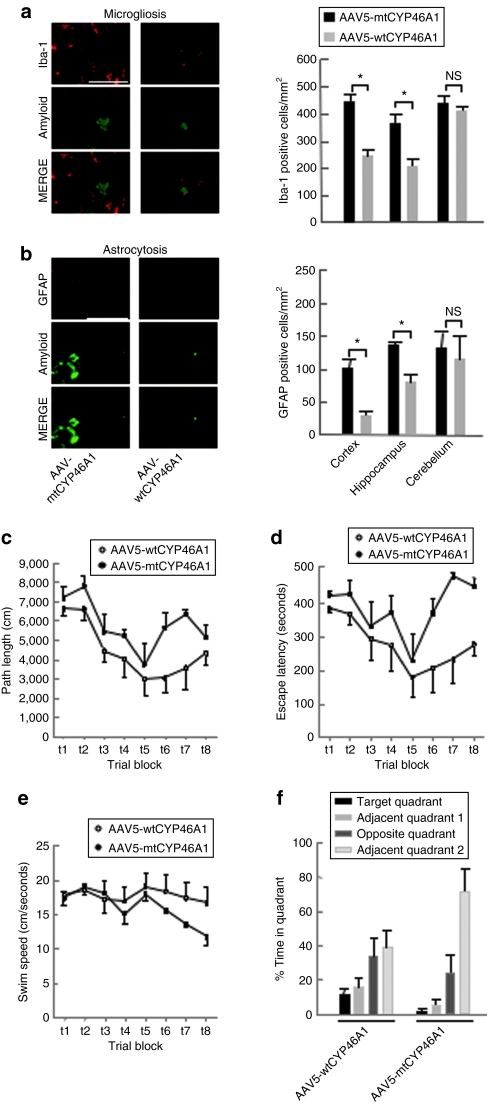
The number of microglial Iba-1 positive cells was reduced by 41 ± 7 and 46 ± 4%, respectively in the cerebral cortex and hippocampus of APP23 mice injected with AAV5-wtCYP46A1 vector (Figure 3a). Similarly, the number of glial fibrillary acidic protein -positive cells was reduced by almost 50% in APP23 mice injected with AAV5-wtCYP46A1 vector (Figure 3b). As amyloid plaques were shown to induce microglial recruitment and astrocytosis in APP23 mice,30 this decrease could be related to the lower number of amyloid deposits observed in AAV5-wtCYP46A1-injected animals. The cerebellum, a brain region that physiologically contains microglial cells31 and lacks amyloid deposits in APP23 mice, was chosen as a control brain region. As expected, no change in the recruitment of Iba-1 and glial fibrillary acidic protein-positive cells was observed in the cerebellum of treated APP23 mice (Figure 3a,b). These results indicate that neuronal overexpression of the CYP46A1 gene before the onset of amyloid plaques has a clear beneficial effect on microgliosis and astrocytosis associated with amyloid deposits in APP23 mice at 12 months.
Figure 3.
Decreased microgliosis and improvement of cognitive performances in APP23 mice treated with AAV5-wtCYP46A1 vector. (a,b) Number of Iba-1 and glial fibrillary acidic protein-positive cells in the cortex, hippocampus, and cerebellum of APP23 mice at 12 months after injections of AAV5-wtCYP46A1 or AAV5-mutCYP46A1 vectors at 3 months. The cerebellum is not affected by amyloid deposits and was thus chosen as a control brain region. Bar = 100 µmol/l. (n = 5 mice per group; three section levels per mouse were analyzed). (Mann–Whitney U-test). *P < 0.05; NS, not significant. (c) Path length, (d) escape latency, and (e) swim speed curves during the acquisition phase of the Morris water maze procedure in 6-month-old APP23 mice treated with the AAV5-wtCYP46A1 (n = 5, open circles) or AAV-mutCYP46A1 (control) vectors (n = 4, filled circles) at 3 months. Data points represent mean (± SEM) summed results of four daily trials. (f) The proportion of total time spent in each quadrant of the Morris water maze during probe trial in APP23 mice treated with the control versus the therapeutic vectors. Bars represent mean (SEM) percentage of total time in a specific quadrant.
APP23 mice develop moderate cognitive deficits at 3 months of age, whereas amyloid deposits are only detected in their brain at 6 months.18 To evaluate the effects of AAV5-wtCYP46A1 vector injection on cognitive functions, mice were tested at 6 months using the Morris water maze (MWM) procedure. All mice injected with either AAV5-wtCYP46A1 or AAV5-mtCYP46A1 vectors showed improved performances during the acquisition phase (P < 0.001) (Figure 3c,d). We observed however a significant effect of AAV5-wtCYP46A1 injection on path length parameter (P = 0.05, two-way repeated measures-analysis of variance) with no significant interaction between vector injection and trial block (P = 0.583) (Figure 3c). The absence of treatment effects on swim speed (Figure 3e) indicated that the improvement in path length was not due to a better motor performance. We observed no significant effect of vector injection on escape latency (Figure 3d). During the probe trial, mice injected with AAV5-wtCYP46A1 vector did not spend more time in the target quadrant (Figure 3f) but they crossed more frequently (1.4 ± 0.4) the previous platform position than sham-treated APP23 mice (0 ± 0, P = 0.018) indicating that their spatial memory was improved. As cognitive evaluation can only be performed on naive mice, no futher testing could be performed on the same treated mice at 12 months of age. In agreement with the beneficial effect of CYP46A1 overexpression on Aβ oligomers production, these results indicate that neuronal overexpression of the CYP46A1 gene before the onset of amyloid plaques improves cognitive functions in APP23 mice at a time amyloid deposits just begin to be detectable in their brain.
AAV5-wtCYP46A1 vector injection decreases the number of amyloid deposits in APP/PS mice
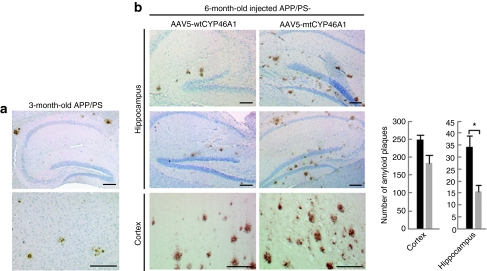
APP/PS mice develop a more severe phenotype than APP23 mice with an onset of amyloid deposits at 2½ months. To determine whether neuronal overexpression of CYP46A1 could be of benefit once amyloid pathology has become obvious, AAV5-wtCYP46A1 (n = 5 females) or AAV5-mtCYP46A1 (n = 5 females) vectors were injected in the hippocampus and cerebral cortex of both hemispheres of 3-month-old APP/PS mice. At this age, amyloid deposits are already present in the brain of these animals (Figure 4a). The number of amyloid deposits decreased by 54% (P < 0.05) in the hippocampus of APP/PS mice treated with the AAV5-wtCYP46A1 vector, and analyzed at 6 months (Figure 4b). A less marked and significant decrease (26%; P < 0.065) of amyloid deposits was observed in the cerebral cortex of treated animals.
Figure 4.
Intracerebral delivery of CYP46A1 at 3 months reduces amyloid deposits at 6 months in APP/PS mice. (a) Representative immunostaining of amyloid deposits with NT2 antibody in 3-month-old APP/PS mice (age of injection). (b) Representative immunostaining of amyloid deposits and stereological analyses of amyloid deposits in the cortex and hippocampus of 6-month-old APP/PS mice injected with AAV5-mtCYP46A1 (right panel) or AAV5-wtCYP46A1 (left panel) vectors (three different section levels per mouse, with three to five slices per level, n = 5 mice per group). Bar = 200 µmol/l. (Mann–Whitney U-test) *P < 0.05.
In vitro overexpression of CYP46A1 decreases the amount of cholesterol and PSEN1 in detergent-resistant membranes isolated from murine neuroblastoid cells expressing mutated human APP
The injection of AAV5-wtCYP46A1 vector in APP23 mice was associated with a significant decrease of Aβ peptide production without affecting the global cholesterol homeostasis or the LXR pathway. To get further insight on the mechanisms by which CYP46A1 overexpression reduces AICD fragments generated by γ-cleavage in vivo, we engineered murine neuroblastoid N2a cell lines to constitutively overexpress the human APP gene harboring the Swedish and London mutations (APPsl). We selected clone 17 (further called N2a-APP17) that was found to secrete the highest levels of Aβ40/42 peptides (Supplementary Figure S1a) and demonstrated that this clone had a decreased expression of endogenous murine CYP46A1 gene (data not shown) and a 43% reduction of 24S-hydroxycholesterol content (Supplementary Figure S1b). Identical results were observed in the two other (N2a-APP11 and -12) analyzed cells lines (data not shown). In contrast to what was observed in N2a-APP cells, no decrease in the 24S-hydroxycholesterol content could be detected in nontreated APP23 mice, as already published by Lutjohann.32
To restore the expression of cholesterol-24-hydroxylase, we engineered N2a-APP17 cells to overexpress the human CYP46A1 gene. We obtained two cell lines, called N2a-APP-CYP-A and -B that overexpressed the CYP46A1 transgene at different levels and showed an increase of 24S-hydroxycholesterol with no significant change in total cholesterol level (Supplementary Figure S1c). In these cells, a decrease of Aβ40 and Aβ42 peptides secretion was observed, which was inversely proportional to the increased 24S-hydroxycholesterol content (r2 = 0.91) (Supplementary Figure S1d). Aβ42 peptides were barely detectable in the N2a-APP-CYP-B cells that expressed the highest level of CYP46A1 gene. As previously observed in APP23 mice injected with AAV5-wtCYP46A1 vector, this effect was not due to modifications of the expression of APPsl transgene nor of murine Adam9, Adam10, Adam17, Bace1, and Psen1 involved in APP processing, nor of Hmgcr, the rate limiting enzyme of cholesterol synthesis, and nor of adenosine triphosphate–binding cassette transporter A1 (Abca1) and ApoE (data not shown).
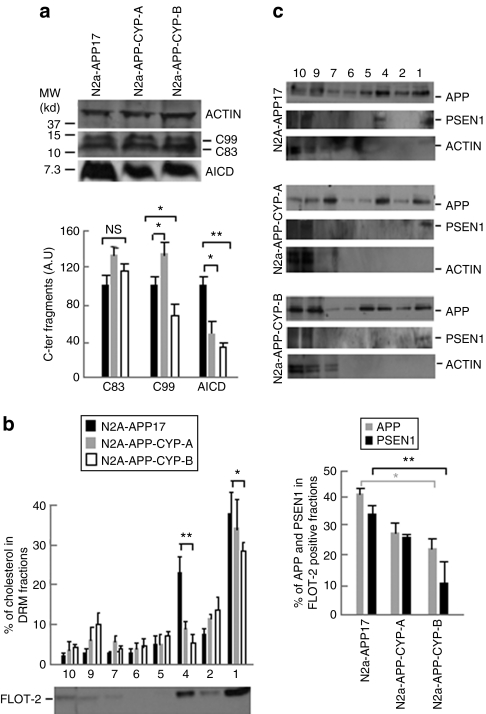
To determine whether the effect of cholesterol 24-hydroxylase overexpression in vitro was also associated with changes in γ-secretase cleavage in N2a-APP-CYP cells, we quantified the CTFs of APP by western blotting in crude cell extracts. The α-secretase cleavage product C83 content was not modified in N2a-APP-CYP-A and -B cells. The β-secretase cleavage product C99 peptide increased by 37% in N2a-APP-CYP-A cells and was reduced by 34% in N2a-APP-CYP-B cells, whereas the γ-secretase cleavage product AICD was lowered by 50–65% in both N2a-APP-CYP-A and -B cells (Figure 5a). The concomitant decrease of AICD and increase of β-CTF in N2a-APP-CYP-A cells were consistent with the hypothesis that CYP46A1 overexpression could mostly act at the level of CTF cleavage by γ-secretase, as already observed in APP23 mice injected with AAV5-wtCYP46A1 vector. However, at higher level of expression, CYP46A1 also seemed to decrease, at least in vitro, β-CTF, suggesting an additional effect on β-secretase activity in those experimental conditions.
Figure 5.
Quantification of C-terminal fragments in N2a-APP17 cells expressing the cholesterol 24-hydroxylase gene and measurement of cholesterol, APP and PSEN1 in detergent-resistant membrane (DRM). (a) Representative western blot and quantification of α, β- and γ-secretase C-terminal fragments (C83, C99, and AICD) in crude extracts from N2a-APP17, N2a-APP-CYP-A, and N2a-APP-CYP-B cells. Black bars: N2a-APP17; gray bars: N2a-APP-CYP-A; white bars: N2a-APP-CYP-B. The amounts of C-terminal fragments are normalized to ACTIN level; AU, arbitrary unit. (b) Cholesterol content in DRMs isolated after iodixanol gradient ultracentrifugation from N2a-APP17, N2a-APP-CYP-A, and N2a-APP-CYP-B cells. As expected, highest content of cholesterol is found in flotillin-2 (FLOT-2) positive fractions from N2a-APP17 cells. Black bars: N2a-APP17; gray bars: N2a-APP-CYP-A; white bars: N2a-APP-CYP-B. (c) Protein blot analysis of APP, PSEN1, and ACTIN in N2a-APP17, N2a-APP-CYP-A, and N2a-APP-CYP-B cells. The percentages of APP and PSEN1 associated with FLOT-2 positive fractions 1 and 4 are shown on the right. All experiments were done in triplicate. (analysis of variance and post hoc Student's t-test) *P < 0.05; **P < 0.005; NS, non significant.
Mounting evidence argues that lipid rafts, biochemically defined as detergent-resistant membranes (DRMs), are the principal membrane microdomains in which the amyloidogenic processing of APP occurs.33 Lipid rafts are enriched in cholesterol, and γ-secretase complex coresides with APP and β-secretase in these microdomains.34,35,36 Changes in the content of cholesterol in lipid rafts can markedly influence the production of Aβ.7,37
In N2a-APP17 cells overexpressing the CYP46A1 gene, the amount of cholesterol was decreased by 28 and 45% in the two flotillin-2 positive DRM fractions of N2a-APP-CYP-A and -B cells, respectively (Figure 5b). APP resides both in raft and nonraft domains and the percentage of APP localized in the two flotillin-2 positive DRM fractions of N2a-APP17 cells represented 40% of the total amount of APP (Figure 5c). In the same enriched cholesterol fractions from N2a-APP-CYP-A and -B cells, the amount of BACE1 was not modified (data not shown) but APP was reduced by 32 and 45% and PSEN1, a component of γ-secretase, was lowered by 25 and 67%, respectively (Figure 5c). Altogether these results suggest that the reduction of cholesterol in DRMs in N2a-APP17 cells overexpressing CYP46A1 leads to a decreased recruitment or stabilization of APP and PSEN1 in the same microdomains, resulting in a decreased cleavage of CTF by γ-secretase
Discussion
CYP46A1 encodes the cholesterol 24-hydroxylase, a master regulator of cerebral cholesterol metabolism.14,38 Our observations indicate that increasing CYP46A1 expression in the brain is a new mechanism for decreasing Aβ peptide production and amyloid deposits, the pathological hallmark of AD.
Brain tissue from AD patients shows a decreased number of neurons expressing cholesterol 24-hydroxylase, possibly because of neuronal degeneration.39 Accordingly, 24S-hydroxycholesterol concentration is lowered in the cerebrospinal fluid and peripheral circulation of AD patients at an advanced stage of the disease,40 whereas it is increased at an earlier stage.41
In our work, the cerebral delivery of human CYP46A1 gene via an AAV vector in APP23 mice before the onset of amyloid plaques induced 10 months later a twofold increase of 24S-hydroxycholesterol levels in the brain and a marked concomitant decrease of Aβ40/42 peptides and amyloid deposits in the hippocampus and cerebral cortex. Microgliosis and astrogliosis were also diminished. As the accumulation of small Aβ oligomers is often considered responsible for AD synaptic dysfunction,24,25 it may be of importance that trimeric Aβ oligomers, that were shown to fully inibit long-term potentiation,42 were reduced in the brain of APP23 mice injected with the AAV-wtCYP46A1 vector.
Cognitive performances were also improved in treated APP23 mice at 6 months of age. Despite the difficulties of MWM interpretation due to the variability of behavioral parameters,43 we documented a statistical trend for an improved acquisition learning curve based on path length values, the most reliable parameter of MWM learning, as well as an improvement in spatial memory. This effect was observed while amyloid deposits begin to be detectable in this AD mouse model.
Of note, short-term studies performed in APP/PS mice showed that the cerebral delivery of human CYP46A1 gene also decreases the number of amyloid deposits in the hippocampus when injections of AAV were performed after the onset of amyloid pathology.
In APP23 mice, CYP46A1 overexpression was neither associated with a decrease of brain cholesterol and cholesteryl esters nor with the modification of the expression of genes regulating cholesterol metabolism. Only the Hmgcr and Srebp2 genes were shown to be slightly upregulated (1.6-fold), likely contributing to maintain the steady-state level of brain cholesterol.
Previous cell-based studies demonstrated that pharmacological concentrations of 24S-hydroxycholesterol could upregulate, through the activation of the LXR pathway, the expression of APOE and of ATP-binding cassette (ABC) transporter genes that control cholesterol trafficking between neurons and astrocytes.15,16 Moreover, the use of synthetic LXR ligands was shown to induce the expression of Abca1 and ApoE both in vitro and in vivo,44,45 thus modifying the lipidation status of APOE and its capacity to degrade Aβ.46,47 In our study, even we cannot completely exclude the possibility that the beneficial effect of CYP46A1 overexpression was mediated by post-translational changes of APOE, this hypothesis seems unlikely because APOE lipidation is mostly under the control of Abca1 expression that remained unaffected in APP23 mice treated with AAV-wtCYP46A1 vector. The absence of change in ApoE, Abca1, and other Abc transporter genes in APP23 mice injected with AAV-wtCYP46A1 vector clearly indicated that the beneficial effect of CYP46A1 overexpression on amyloidogenic processing of APP was not mediated by the activation of the LXR pathway by 24S-hydroxycholesterol. These results are in agreement with recent data showing that 22R-hydroxycholesterol, another natural ligand, is a weak partial agonist of LXR receptors48 compared with synthetic molecules. However, as 24S-hydroxycholesterol concentration increased only twofold in the brain of treated APP23 mice, we cannot completely exclude that 24S-hydroxycholesterol can activate the LXR pathway in vivo, but at higher concentrations.
The concomitant decrease of AICD and increase of β-CTF observed in APP23 mice and in N2a-APP cells overexpressing CYP46A1 suggest that the lower Aβ40/42 peptides content could be caused by a decreased cleavage of APP CTFs by γ-secretase. Cholesterol was shown to regulate the generation of Aβ peptides, particularly in lipid rafts in which APP and γ-secretase components coreside.35,36 Although the data obtained in N2a-APP cells must be interpreted with caution, our study indicates that enhanced conversion of cholesterol to 24S-hydroxycholesterol reduces cholesterol in lipid rafts. This reduction is associated with a decreased amount of APP and PSEN1, a major component of γ-secretase, in the same microdomains. Further experiments are clearly needed to clarify how the cholesterol changes in lipid rafts that are induced by CYP46A1 overexpression affect the recruitment and activity of PSEN1 and other components of γ-secretase complex in these microdomains.
Overall, these data demonstrate that selective overexpression of CYP46A1 in neurons can reduce Aβ peptides and amyloid deposits in mouse models of AD when the overexpression of CYP46A1 is induced before or after the formation of amyloid plaques. Further studies are however needed to study the long-term effect of CYP46A1 overexpression on amyloidogenic processing of APP and the effect of this therapeutic approach on amyloid pathology at different pathological stages in AD mice. This work however supports that brain gene delivery of CYP46A1 gene could be of potential therapeutic interest in Alzheimer patients.
Materials and Methods
AAV plasmid design and vectors production. Wild-type and mt CYP46A1 pcDNA plasmids were generously given by Laurent Pradier (Sanofi-Aventis, Paris, France). Three PCR-generated fragments containing the entire sequences of the 0.6-kb murine phosphoglycerate kinase (PGK) promoter, the 1.5-kb CYP46A1 cDNA and the 0.6-kb regulatory element of woodchuck hepatitis virus post-transcriptional regulatory element were cloned to generate pAAV5/PGK-hCYP46A1-woodchuck hepatitis virus post-transcriptional regulatory element plasmids. The functionality of each AAV plasmid was checked by transient transfection of 293T cells (data not shown). These plasmids were used to generate AAV5/5-PGK-HACYP46A1 (referred to as AAV5-wtCYP46A1) and AAV5/5-PGK-HACYP46A1mut (referred to as AAV5-mtCYP46A1) vectors. AAV stocks were generated by transient transfection of 293T cells and purified using CsCl ultracentrifugation gradient.49 Titers ranged from 4 to 9 × 1012 vg/ml.
Transgenic mouse lines and intracerebral injections of AAVs. The APP23 transgenic mouse line (Thy1-hAPPswe)17 was generously provided by Matthias Staufenbiel (Novartis Pharma, Basel, Switzerland). These mice overexpress the mutated human APP751 gene containing the Swedish double mutation (K670N-M671L) under the neuronal specific promoter Thy-1. APP/PS mice were kindly given by Laurent Pradier (Aventis Pharma, Paris, France). This double transgenic mouse line overexpresses both the human mutated PSEN1 (PS1M146L) and APP (APP695 containing the Swedish and London mutations) genes.19
Animals were housed in a pathogen-free animal facility. The experiments were approved by the veterinary desk of INSERM and carried out in compliance with the guide for the Care and Use of Laboratory Animals (NIH publication no.85-24) and the European Communities Council Directive (86/609/EEC). In addition, the behavioral screening protocol performed on APP23 mice was approved by the Animal Ethics Committee of the University of Antwerp.
For stereotactic intracerebral AAV injection, animals were anesthetized by intraperitoneal injection of ketamine/xylazine (0.1/0.05 mg/g body weight) and positioned on a stereotactic frame (David Kopf Instruments, Tujunga, CA). Injections of vectors were performed in the cerebral cortex (two deposits) and hippocampus (one deposit) of each hemisphere with 2 µl of viral preparation (12 × 108 vg) using a 30-gauge blunt micropipette attached to a 10-µl Hamilton syringe (Hamilton Medical, Reno, NV) at a rate of 0.2 µl/minute. Stereotactic coordinates of injection sites from bregma were (i) anteroposterior: −0.3; mediolateral: ± 2; dorsoventral: −1.5 mm; (ii) anteroposterior: −2; mediolateral: ± 1.2; dorsoventral: −1.2 mm, and anteroposterior: −2; mediolateral: ± 1.2; dorsoventral: −2 mm.
APP23 and APP/PS mice were, respectively killed at 12 and 6 months. One half-brain was used for histochemistry. The hippocampus and cerebral cortex of the other half-brain were dissected for biochemistry, gene expression and protein analysis using western blotting.
Cell lines and culture conditions. Cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 1% glutamine, and 1% penicillin and streptomycin (all from Gibco Laboratories). APPsl (containing the Swedish K670N-M671L and London V717I mutations) cDNA was obtained by PCR from APP/PS mice (Laurent Pradier, Sanofi-Aventis) and cloned under a pCMV promoter in a pIRES-PURO plasmid (Invitrogen, Eugene, OR). After transfection using the Effectene tranfection kit (Qiagen, Courtaboeuf, Fance), puromycin-resistent N2a-hAPPsl cells were selected by limit dilution method and tested by PCR. N2a-APPsl cells were then stably transfected by a pcDNA-pCMV-CYP46A1 plasmid containing a neomycin-resistant selection cassette.
Determination of cholesterol and 24S-hydroxycholesterol. The quantification of cellular cholesterol was performed using the Amplex Red cholesterol assay kit (Invitrogen). For 24S-hydroxycholesterol determination, 24-hydroxycholesterol-2H7 (Medical Isotopes, Pelham, NH) internal standard was added to brain and cell homogenates. After hydrolysis and extraction, samples were sulphatated, and 24S-hydroxycholesterol was quantified by high-performance liquid chromatography–tandem mass spectrometry (Quattro II; Micromass, Manchester, UK) using a calibration curve with electrospray ionization in the negative ion mode. Samples were loaded onto an analytical column at 0.3 ml/minute (Alltima C18, 250 × 2.1 mm, 5 µ Altech, Deerfield, IL). The mobile phase consisted of 0.1% ammonia in MilliQ (A) and acetonitril:H2O, 9:1, vol/vol (B). The following gradient was run for a total run time of 25 minutes: 0–10 minutes 85% A to 50% A, 10–20 minutes to 100% A, 20–20.1 minutes to 85% A, 20.1–25 minutes 85% A. Mass spectrometric parameters were as follows: nitrogen as nebulizing gas; argon as collision gas (2.5 × 10−3 mBar), collision energy 20 eV, capillary voltage 3 kV, source temperature 80 °C, and cone voltage 25 V. The following transitions were used to detect 24S-hydroxycholesterol:m/z 280.1→ m/z 97 and m/z 283.6→ m/z 97 for the internal standard.
Determination of Aβ levels by enzyme-linked immunosorbent assay. In vitro, secreted Aβ peptides were measured in the medium 48 hours after plating the cells. In vivo, dissected cortex and hippocampus were first homogenized in 10 mmol/l Tris–HCl buffer (pH 6.8) containing protease inhibitor cocktail (Complete; Roche, Meylan, France), adjusted to 6 mg protein/ml and 50 µl was extracted in 5 mol/l guanidine–HCl. Aβ40 and Aβ42 peptides were quantified using commercially available ELISA kits (Biosource and Innogenetics, Eugene, OR).
Western blotting. Western blot experiments were performed using a standard protocol (10% polyacrylamide gel electrophoresis–sodium dodecyl sulfate electrophoresis), except for Aβ oligomers analyses that were done using precast CRITERION 12% Bis–Tris gels (Bio-Rad, Hercules, CA) in NuPAGE MES running buffer (Invitrogen). All samples were extracted in lysis buffer containing protease inhibitor cocktail (Complete; Roche). Primary antibodies (Supplementary Table S1) were incubated for 2 hours at room temperature, followed by species-specific peroxidase-conjugated secondary antibodies. The enhanced chemiluminescence method (GE Healthcare, Saclay, France) was used for revelation. Signal quantification was done using densitometry analysis of the scanned autoradiograms with the ImageJ 1.38x National Institutes of Health software.
Soluble Aβ oligomers analyses. Dissected cortex and hippocampus were first homogenized in a dounce homogeneizer in Tris–HCl 10 mmol/l, pH 6.8 supplemented with protease inhibitor cocktail (Complete; Roche). Protein concentration was adjusted to 6 mg/ml for all samples. A first ultracentrifugation step (100,000 g at 4 °C) was performed to collect the Tris supernatant and the pellet was dissolved into a Tris–HCl 10 mmol/l, 2% Triton buffer (pH 6.8). Another ultracentrifugation step (100,000 g, 4 °C) allowed removing the Triton supernatant and the pellet was resuspended in a Tris–HCl 10 mmol/l, 0.5% sodium dodecyl sulfate buffer (pH 6.8). After a last ultracentrifugation step (100,000 g at room temperature), the sodium dodecyl sulfate supernatant was collected and the pellet was directly dissolved in Blue Laemmli solution.
DRM isolation. Lipid rafts were isolated as previously described.36 A pellet of 10 million cells was first lysed in modified earth's solution buffer (25 mmol/l MES, 150 mmol/l NaCl, EDTA 1 mmol/l pH 6.5) containing 1% Triton X-100 and protease inhibitor cocktail (Complete; Roche). All steps were performed at 4 °C. The protein concentration was adjusted to 5 mg/ml and cell lysates were brought into 40% iodixanol (OptiPrep Dendity Gradient; Sigma, Lyon, France) diluted in appropriate buffer (0.25 mol/l sucrose, 6 mmol/l EDTA, 120 mmol/l Tricine, pH 7.6). Two layers of 30% and then 20% iodixanol buffer were overlaid at the top of the ultracentrifuge tube. After ultracentrifugation at 39,000 r.p.m. for 20 hours at 4 °C, fractions (of 1 ml) were collected and analyzed by western blotting according to standard protocol. The detection of flotillin 2 was used to identify DRM fractions.
Quantitative RT-PCR. mRNA extraction from cells or tissues was performed using the RNAble kit (Eurobio Laboratories, Les Ulis, France). Real-time quantitative RT-PCR on the ABI Prism 7700 Sequence Detection System (Perkin-Elmer Applied Biosystems, Foster City, CA) was performed as described.50
As an endogenous RNA control, we quantified transcripts of the TATA box–binding protein (TBP) gene. The amount of target transcript (Ntarget) was normalized based on the basis of the TBP content of each sample and was subsequently normalized to a basal mRNA level with the equation: Ntarget = 2 × δCt, where δCt is the Ct value of the target gene minus the Ct value of the TBP gene. Primers are listed in Supplementary Table S2.
Behavioral analysis. The MWM setting consisted of a circular pool (diameter: 150 cm, height: 30 cm) filled with opacified water (nontoxic natural paint), kept at 25 °C, and surrounded by invariable visual extramaze cues. A round acrylic glass platform (diameter: 15 cm) was placed 1 cm below the water surface at a fixed position in one of the quadrants. The acquisition phase comprised eight trial blocks of four daily trials semirandomly starting from four different positions around the border of the maze with 15-minutes intertrial intervals. In case a mouse was unable to reach the platform within 120 seconds, it was placed on the platform during 15 seconds before being returned to its home cage. Swimming trajectories were recorded using a computerized video-tracking system (EthoVision, Noldus, The Netherlands) logging path length, escape latency, and swim speed. Four days after finishing the acquisition phase, a probe trial was performed. The platform was removed from the maze, and each mouse was allowed to swim freely for 100 seconds. Spatial accuracy was expressed as the percentage of time spent in each quadrant of the MWM, and the number of crossings through the target position, i.e., the specific location of the platform during the acquisition phase.
Statistics for behavioral analysis: Two-way repeated measures-analysis of variance (repeated measures-analysis of variance), with treatment and trial block as possible sources of variation, combined with Tukey's honestly significantly different post hoc procedure, assessed the significance of differences between mean scores during the acquisition phase. Spatial acuity during the probe trial was probed using two-way analysis of variance with Tukey's honestly significantly different test. Two-tailed Student's t-test (t-test) was used to evaluate differences in the number of entries through the previous target position and path length during probe trial. All statistics were performed using Sigmastat software (SPSS, Erkrath, Germany) with the level of probability set at 95%.
Immunohistochemistry and microscopy analyses. Anesthetized animals were transcardially infused with phosphate-buffered saline. For biochemical analyses, left cortex and hippocampus were dissected, weighted, and nitrogen frozen. The right hemi-brain was postfixed in 4% paraformaldehyde in phosphate-buffered saline for 24 hours and paraffin embedded. Five-micrometer sections were sequentially (i) deparaffined in xylene, (ii) rehydrated in ethanol, (iii) permeabilized in phosphate-buffered saline 0.05% saponine, (iv) blocked in phosphate-buffered saline 0.01% saponine, 5% normal goat serum, and (v) incubated overnight with the primary antibody. For amyloid plaques labeling of amyloid plaques with the mouse monoclonal 4G8 antibody, a treatment with 80% formic acid for 30 minutes was performed. A step of antigen retrieval in citric acid (0.1 mol/l)/sodium citrate (0.1 mol/l) buffer was necessary for several antibodies. Secondary antibodies were applied 1 hour at room temperature. All the antibodies are listed in Supplementary Table S1. Images were taken with a Nikon microscope (Eclipse 800; Nikon, Champigny-sur-Marne, France) and a digital QIMAGING camera (CCD QICAM cooled plus RGB filter pixel 4.65 × 4.65 µm2; QIMAGING, Surrey, BC, Canada). Control and test slices were processed the same day and under the same condition.
Stereological analyses were performed on three section levels per mouse (the first level being located at 50 µm from the medial axis and the two next levels at 150 and 250 µm, respectively), with three to five contiguous slices per level. To determine the surface and number of amyloid deposits in anatomic region of interest, the Histolab image analyzer software (Microvision Instruments, Paris, France) was set up to automatically detect in a blind fahsion 3,3′-diaminobenzidine-labeled deposits. Plaques for which the intensity was not sufficient above the background for proper thresholding were not considered. The parameters setting remained unchanged for all analyses. Each 3,3′-diaminobenzidine positive object was considered as an amyloid plaque and the image analyzer directly measured its surface or number. For each slice, the amyloid deposits quantification was then reported to the surface of the cortex and hippocampus of the same slice. The count of glial fibrillary acidic protein and Iba-1 positive cells was performed according to the same procedure.
Statistical analyses. All statistical procedures, except for behavioral analyses, were performed using StatView 5.0 and JMP 7.0 softwares for Macintosh (SAS, Cary, NC). In vitro data were analyzed by analysis of variance followed by Student's t-test post hoc multiple comparison when appropriate. Because of small animal groups, data from in vivo experiments were analyzed using the nonparametric Mann–Whitney U-test. Error of the mean values in the text and bars on graphs stand for SEM.
SUPPLEMENTARY MATERIALFigure S1. Expression of CYP46A1 gene decreases Aβ40/42 peptide secretion in murine neuroblastoid N2a cells expressing mutated human APP (APPsl). (a) Secretion of Aβ40 and Aβ42 peptides and mRNA/protein levels of APPsl in N2a and N2a-hAPPsl cell lines (clones 11, 12 and 17). A weak signal corresponding to the non-mutated murine APP protein is detected in N2a cells. (b) Intracellular level of cholesterol and 24S-hydroxycholesterol in N2a and N2a-APP17 cells. (c) Intracellular levels of cholesterol and 24S-hydroxycholesterol in two N2a-APP17 clones expressing the human CYP46A1 gene (N2a-APP-CYP-A, and -B). (d) Decreased secretion of Aβ40/42 peptides in N2a-APP-CYP-A and -B cells. The amounts of secreted Aβ40/42 peptides are normalized to 100% in N2a-APP17 cells. All experiments were done in triplicate. (ANOVA and post-hoc Student's t-test) * P < 0.05; ** P < 0.005; *** P < 0.0005; NS: non significant.Table S1. List of primary antibodies.Table S2. List of primers used for real-time RT-PCR.
Supplementary Material
Expression of CYP46A1 gene decreases Aβ40/42 peptide secretion in murine neuroblastoid N2a cells expressing mutated human APP (APPsl). (a) Secretion of Aβ40 and Aβ42 peptides and mRNA/protein levels of APPsl in N2a and N2a-hAPPsl cell lines (clones 11, 12 and 17). A weak signal corresponding to the non-mutated murine APP protein is detected in N2a cells. (b) Intracellular level of cholesterol and 24S-hydroxycholesterol in N2a and N2a-APP17 cells. (c) Intracellular levels of cholesterol and 24S-hydroxycholesterol in two N2a-APP17 clones expressing the human CYP46A1 gene (N2a-APP-CYP-A, and -B). (d) Decreased secretion of Aβ40/42 peptides in N2a-APP-CYP-A and -B cells. The amounts of secreted Aβ40/42 peptides are normalized to 100% in N2a-APP17 cells. All experiments were done in triplicate. (ANOVA and post-hoc Student's t-test) * P < 0.05; ** P < 0.005; *** P < 0.0005; NS: non significant.
List of primary antibodies.
List of primers used for real-time RT-PCR.
Acknowledgments
We thank Mathias Jucker and Ellen Kilger for their helpful comments and advices for AICD measurements, Frieda Franck for her assistance for behavioral studies, and Henk Overmars for the development of 24S-cholesterol determination. This work was supported by French Agence Nationale de la Recherche (ANR grant ANR07-neuro-009-03), the Fondation de l'Avenir (grant ET7-480), the Fondation NRJ-Institut de France, the Fund for Scientific Research-Flanders (FWO grant G.0038.05), Interuniversity Poles of Attraction (IUAP Network P6/43), Stichting voor Alzheimer Onderzoek–Fondation pour la Recherche sur la Maladie Alzheimer (SAO/FRMA), agreement between Institute Born-Bunge and University of Antwerp, the Medical Research Foundation Antwerp, the Thomas Riellaerts research fund, and Neurosearch Antwerp. D.V.D. holds a postdoctoral fellowship of the FWO. The authors declared that they have no conflict of interest.
REFERENCES
- Tanzi RE. The synaptic Abeta hypothesis of Alzheimer disease. Nat Neurosci. 2005;8:977–979. doi: 10.1038/nn0805-977. [DOI] [PubMed] [Google Scholar]
- Nunan J., and , Small DH. Regulation of APP cleavage by alpha-, beta- and gamma-secretases. FEBS Lett. 2000;483:6–10. doi: 10.1016/s0014-5793(00)02076-7. [DOI] [PubMed] [Google Scholar]
- Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, et al. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci USA. 1993;90:1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puglielli L, Tanzi RE., and , Kovacs DM. Alzheimer's disease: the cholesterol connection. Nat Neurosci. 2003;6:345–351. doi: 10.1038/nn0403-345. [DOI] [PubMed] [Google Scholar]
- Wolozin B. Cholesterol and the biology of Alzheimer's disease. Neuron. 2004;41:7–10. doi: 10.1016/s0896-6273(03)00840-7. [DOI] [PubMed] [Google Scholar]
- Levin-Allerhand JA, Lominska CE., and , Smith JD. Increased amyloid- levels in APPSWE transgenic mice treated chronically with a physiological high-fat high-cholesterol diet. J Nutr Health Aging. 2002;6:315–319. [PubMed] [Google Scholar]
- Ehehalt R, Keller P, Haass C, Thiele C., and , Simons K. Amyloidogenic processing of the Alzheimer beta-amyloid precursor protein depends on lipid rafts. J Cell Biol. 2003;160:113–123. doi: 10.1083/jcb.200207113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Paris D, Town T, Rojiani AM, Sparks DL, Delledonne A, et al. Cholesterol accumulates in senile plaques of Alzheimer disease patients and in transgenic APP(SW) mice. J Neuropathol Exp Neurol. 2001;60:778–785. doi: 10.1093/jnen/60.8.778. [DOI] [PubMed] [Google Scholar]
- Xiong H, Callaghan D, Jones A, Walker DG, Lue LF, Beach TG, et al. Cholesterol retention in Alzheimer's brain is responsible for high beta- and gamma-secretase activities and Abeta production. Neurobiol Dis. 2008;29:422–437. doi: 10.1016/j.nbd.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassbender K, Simons M, Bergmann C, Stroick M, Lutjohann D, Keller P, et al. Simvastatin strongly reduces levels of Alzheimer's disease beta -amyloid peptides Abeta 42 and Abeta 40 in vitro and in vivo. Proc Natl Acad Sci USA. 2001;98:5856–5861. doi: 10.1073/pnas.081620098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoglund K., and , Blennow K. Effect of HMG-CoA reductase inhibitors on beta-amyloid peptide levels: implications for Alzheimer's disease. CNS Drugs. 2007;21:449–462. doi: 10.2165/00023210-200721060-00002. [DOI] [PubMed] [Google Scholar]
- Hutter-Paier B, Huttunen HJ, Puglielli L, Eckman CB, Kim DY, Hofmeister A, et al. The ACAT inhibitor CP-113,818 markedly reduces amyloid pathology in a mouse model of Alzheimer's disease. Neuron. 2004;44:227–238. doi: 10.1016/j.neuron.2004.08.043. [DOI] [PubMed] [Google Scholar]
- Bjorkhem I. Crossing the barrier: oxysterols as cholesterol transporters and metabolic modulators in the brain. J Intern Med. 2006;260:493–508. doi: 10.1111/j.1365-2796.2006.01725.x. [DOI] [PubMed] [Google Scholar]
- Lund EG, Guileyardo JM., and , Russell DW. cDNA cloning of cholesterol 24-hydroxylase, a mediator of cholesterol homeostasis in the brain. Proc Natl Acad Sci USA. 1999;96:7238–7243. doi: 10.1073/pnas.96.13.7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abildayeva K, Jansen PJ, Hirsch-Reinshagen V, Bloks VW, Bakker AH, Ramaekers FC, et al. 24(S)-hydroxycholesterol participates in a liver X receptor-controlled pathway in astrocytes that regulates apolipoprotein E-mediated cholesterol efflux. J Biol Chem. 2006;281:12799–12808. doi: 10.1074/jbc.M601019200. [DOI] [PubMed] [Google Scholar]
- Koldamova R., and , Lefterov I. Role of LXR and ABCA1 in the pathogenesis of Alzheimer's disease – implications for a new therapeutic approach. Curr Alzheimer Res. 2007;4:171–178. doi: 10.2174/156720507780362227. [DOI] [PubMed] [Google Scholar]
- Sturchler-Pierrat C, Abramowski D, Duke M, Wiederhold KH, Mistl C, Rothacher S, et al. Two amyloid precursor protein transgenic mouse models with Alzheimer disease-like pathology. Proc Natl Acad Sci USA. 1997;94:13287–13292. doi: 10.1073/pnas.94.24.13287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dam D, D'Hooge R, Staufenbiel M, Van Ginneken C, Van Meir F., and , De Deyn PP. Age-dependent cognitive decline in the APP23 model precedes amyloid deposition. Eur J Neurosci. 2003;17:388–396. doi: 10.1046/j.1460-9568.2003.02444.x. [DOI] [PubMed] [Google Scholar]
- Blanchard V, Moussaoui S, Czech C, Touchet N, Bonici B, Planche M, et al. Time sequence of maturation of dystrophic neurites associated with Abeta deposits in APP/PS1 transgenic mice. Exp Neurol. 2003;184:247–263. doi: 10.1016/s0014-4886(03)00252-8. [DOI] [PubMed] [Google Scholar]
- Marr RA, Rockenstein E, Mukherjee A, Kindy MS, Hersh LB, Gage FH, et al. Neprilysin gene transfer reduces human amyloid pathology in transgenic mice. J Neurosci. 2003;23:1992–1996. doi: 10.1523/JNEUROSCI.23-06-01992.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer O, Marr RA, Rockenstein E, Crews L, Coufal NG, Gage FH, et al. Targeting BACE1 with siRNAs ameliorates Alzheimer disease neuropathology in a transgenic model. Nat Neurosci. 2005;8:1343–1349. doi: 10.1038/nn1531. [DOI] [PubMed] [Google Scholar]
- Hong CS, Goins WF, Goss JR, Burton EA., and , Glorioso JC. Herpes simplex virus RNAi and neprilysin gene transfer vectors reduce accumulation of Alzheimer's disease-related amyloid-beta peptide in vivo. Gene Ther. 2006;13:1068–1079. doi: 10.1038/sj.gt.3302719. [DOI] [PubMed] [Google Scholar]
- Ramirez DM, Andersson S., and , Russell DW. Neuronal expression and subcellular localization of cholesterol 24-hydroxylase in the mouse brain. J Comp Neurol. 2008;507:1676–1693. doi: 10.1002/cne.21605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haass C., and , Selkoe DJ. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer's amyloid beta-peptide. Nat Rev Mol Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- Lesné S, Kotilinek L., and , Ashe KH. Plaque-bearing mice with reduced levels of oligomeric amyloid-beta assemblies have intact memory function. Neuroscience. 2008;151:745–749. doi: 10.1016/j.neuroscience.2007.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oddo S, Caccamo A, Tran L, Lambert MP, Glabe CG, Klein WL, et al. Temporal profile of amyloid-beta (Abeta) oligomerization in an in vivo model of Alzheimer disease. A link between Abeta and tau pathology. J Biol Chem. 2006;281:1599–1604. doi: 10.1074/jbc.M507892200. [DOI] [PubMed] [Google Scholar]
- Lesné S, Koh MT, Kotilinek L, Kayed R, Glabe CG, Yang A, et al. A specific amyloid-beta protein assembly in the brain impairs memory. Nature. 2006;440:352–357. doi: 10.1038/nature04533. [DOI] [PubMed] [Google Scholar]
- McLaurin J, Kierstead ME, Brown ME, Hawkes CA, Lambermon MH, Phinney AL, et al. Cyclohexanehexol inhibitors of Abeta aggregation prevent and reverse Alzheimer phenotype in a mouse model. Nat Med. 2006;12:801–808. doi: 10.1038/nm1423. [DOI] [PubMed] [Google Scholar]
- Allinson TM, Parkin ET, Turner AJ., and , Hooper NM. ADAMs family members as amyloid precursor protein alpha-secretases. J Neurosci Res. 2003;74:342–352. doi: 10.1002/jnr.10737. [DOI] [PubMed] [Google Scholar]
- Bornemann KD, Wiederhold KH, Pauli C, Ermini F, Stalder M, Schnell L, et al. Abeta-induced inflammatory processes in microglia cells of APP23 transgenic mice. Am J Pathol. 2001;158:63–73. doi: 10.1016/s0002-9440(10)63945-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson LJ, Perry VH, Dri P., and , Gordon S. Heterogeneity in the distribution and morphology of microglia in the normal adult mouse brain. Neuroscience. 1990;39:151–170. doi: 10.1016/0306-4522(90)90229-w. [DOI] [PubMed] [Google Scholar]
- Lütjohann D, Brzezinka A, Barth E, Abramowski D, Staufenbiel M, von Bergmann K, et al. Profile of cholesterol-related sterols in aged amyloid precursor protein transgenic mouse brain. J Lipid Res. 2002;43:1078–1085. doi: 10.1194/jlr.m200071-jlr200. [DOI] [PubMed] [Google Scholar]
- Cordy JM, Hooper NM., and , Turner AJ. The involvement of lipid rafts in Alzheimer's disease. Mol Membr Biol. 2006;23:111–122. doi: 10.1080/09687860500496417. [DOI] [PubMed] [Google Scholar]
- Li YM, Xu M, Lai MT, Huang Q, Castro JL, DiMuzio-Mower J, et al. Photoactivated gamma-secretase inhibitors directed to the active site covalently label presenilin 1. Nature. 2000;405:689–694. doi: 10.1038/35015085. [DOI] [PubMed] [Google Scholar]
- Wahrle S, Das P, Nyborg AC, McLendon C, Shoji M, Kawarabayashi T, et al. Cholesterol-dependent gamma-secretase activity in buoyant cholesterol-rich membrane microdomains. Neurobiol Dis. 2002;9:11–23. doi: 10.1006/nbdi.2001.0470. [DOI] [PubMed] [Google Scholar]
- Vetrivel KS, Cheng H, Lin W, Sakurai T, Li T, Nukina N, et al. Association of gamma-secretase with lipid rafts in post-Golgi and endosome membranes. J Biol Chem. 2004;279:44945–44954. doi: 10.1074/jbc.M407986200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won JS, Im YB, Khan M, Contreras M, Singh AK., and , Singh I. Lovastatin inhibits amyloid precursor protein (APP) beta-cleavage through reduction of APP distribution in Lubrol WX extractable low density lipid rafts. J Neurochem. 2008;105:1536–1549. doi: 10.1111/j.1471-4159.2008.05283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björkhem I, Heverin M, Leoni V, Meaney S., and , Diczfalusy U. Oxysterols and Alzheimer's disease. Acta Neurol Scand Suppl. 2006;185:43–49. doi: 10.1111/j.1600-0404.2006.00684.x. [DOI] [PubMed] [Google Scholar]
- Brown J 3rd, Theisler C, Silberman S, Magnuson D, Gottardi-Littell N, Lee JM, et al. Differential expression of cholesterol hydroxylases in Alzheimer's disease. J Biol Chem. 2004;279:34674–34681. doi: 10.1074/jbc.M402324200. [DOI] [PubMed] [Google Scholar]
- Papassotiropoulos A, Lütjohann D, Bagli M, Locatelli S, Jessen F, Buschfort R, et al. 24S-hydroxycholesterol in cerebrospinal fluid is elevated in early stages of dementia. J Psychiatr Res. 2002;36:27–32. doi: 10.1016/s0022-3956(01)00050-4. [DOI] [PubMed] [Google Scholar]
- Kolsch H, Lutjohann D, von Bergmann K., and , Heun R. The role of 24S-hydroxycholesterol in Alzheimer's disease. J Nutr Health Aging. 2003;7:37–41. [PubMed] [Google Scholar]
- Townsend M, Shankar GM, Mehta T, Walsh DM., and , Selkoe DJ.2006Effects of secreted oligomers of amyloid beta-protein on hippocampal synaptic plasticity: a potent role for trimers J Physiol (Lond) 572Pt 2): 477–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi DT., and , Chen KS. Behavioral phenotypes of amyloid-based genetically modified mouse models of Alzheimer's disease. Genes Brain Behav. 2005;4:173–196. doi: 10.1111/j.1601-183X.2005.00124.x. [DOI] [PubMed] [Google Scholar]
- Eckert GP, Vardanian L, Rebeck GW., and , Burns MP. Regulation of central nervous system cholesterol homeostasis by the liver X receptor agonist TO-901317. Neurosci Lett. 2007;423:47–52. doi: 10.1016/j.neulet.2007.05.063. [DOI] [PubMed] [Google Scholar]
- Beaven SW., and , Tontonoz P. Nuclear receptors in lipid metabolism: targeting the heart of dyslipidemia. Annu Rev Med. 2006;57:313–329. doi: 10.1146/annurev.med.57.121304.131428. [DOI] [PubMed] [Google Scholar]
- Jiang Q, Lee CY, Mandrekar S, Wilkinson B, Cramer P, Zelcer N, et al. ApoE promotes the proteolytic degradation of Abeta. Neuron. 2008;58:681–693. doi: 10.1016/j.neuron.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzman DM. Role of apoe/Abeta interactions in the pathogenesis of Alzheimer's disease and cerebral amyloid angiopathy. J Mol Neurosci. 2001;17:147–155. doi: 10.1385/JMN:17:2:147. [DOI] [PubMed] [Google Scholar]
- Albers M, Blume B, Schlueter T, Wright MB, Kober I, Kremoser C, et al. A novel principle for partial agonism of liver X receptor ligands. Competitive recruitment of activators and repressors. J Biol Chem. 2006;281:4920–4930. doi: 10.1074/jbc.M510101200. [DOI] [PubMed] [Google Scholar]
- Sevin C, Benraiss A, Van Dam D, Bonnin D, Nagels G, Verot L, et al. Intracerebral adeno-associated virus-mediated gene transfer in rapidly progressive forms of metachromatic leukodystrophy. Hum Mol Genet. 2006;15:53–64. doi: 10.1093/hmg/ddi425. [DOI] [PubMed] [Google Scholar]
- Bièche I, Lerebours F, Tozlu S, Espie M, Marty M., and , Lidereau R. Molecular profiling of inflammatory breast cancer: identification of a poor-prognosis gene expression signature. Clin Cancer Res. 2004;10:6789–6795. doi: 10.1158/1078-0432.CCR-04-0306. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression of CYP46A1 gene decreases Aβ40/42 peptide secretion in murine neuroblastoid N2a cells expressing mutated human APP (APPsl). (a) Secretion of Aβ40 and Aβ42 peptides and mRNA/protein levels of APPsl in N2a and N2a-hAPPsl cell lines (clones 11, 12 and 17). A weak signal corresponding to the non-mutated murine APP protein is detected in N2a cells. (b) Intracellular level of cholesterol and 24S-hydroxycholesterol in N2a and N2a-APP17 cells. (c) Intracellular levels of cholesterol and 24S-hydroxycholesterol in two N2a-APP17 clones expressing the human CYP46A1 gene (N2a-APP-CYP-A, and -B). (d) Decreased secretion of Aβ40/42 peptides in N2a-APP-CYP-A and -B cells. The amounts of secreted Aβ40/42 peptides are normalized to 100% in N2a-APP17 cells. All experiments were done in triplicate. (ANOVA and post-hoc Student's t-test) * P < 0.05; ** P < 0.005; *** P < 0.0005; NS: non significant.
List of primary antibodies.
List of primers used for real-time RT-PCR.