Abstract
Soon after birth, the neonatal intestine is confronted with a massive antigenic challenge of microbial colonization. Microbial signals are required for maturation of several physiological, anatomical, and biochemical functions of intestinal epithelial barrier (IEB) after birth. Commensal bacteria regulate intestinal innate and adaptive immunity and provide stimuli for ongoing repair and restitution of IEB. Colonization by pathogenic bacteria and/or dysmature response to microbial stimuli can result in flagrant inflammatory response as seen in necrotizing enterocolitis (NEC). Characterized by inflammation and hemorrhagic–ischemic necrosis, NEC is a devastating complication of prematurity. Although there is evidence that both prematurity and presence of bacteria, are proven contributing factors to the pathogenesis of NEC, the molecular mechanisms involved in IEB dysfunction associated with NEC have begun to emerge only recently. The metagenomic advances in the field of intestinal microecology are providing insight into the factors that are required for establishment of commensal bacteria that appear to provide protection against intestinal inflammation and NEC. Perturbations in achieving colonization by commensal bacteria such as premature birth or hospitalization in intensive care nursery can result in dysfunction of IEB and NEC. In this article, microbial modulation of functions of IEB and its relationship with barrier dysfunction and NEC are described.
Keywords: Intestine, Immune, Microbiota, Inflammation, Necrotizing enterocolitis, Microecology, Mucosa, Epithelial barrier, Prematurity, Maternal milk feedings
Introduction
It is estimated that human intestinal epithelial barrier (IEB) is habitat to 500–1,000 species of 10–100 trillion organisms [1]. There are ten times more bacterial cells than the total number of cells in human body and collective microbial genomes (microbiomes) outnumber the human genome by 100-fold [1–3]. Majority of intestinal microecology (IM) consists of bacteria. Viruses and eukaryotes (e.g., fungi) are also represented in IM, but only as a minority [1]. This article is focused on relationship of bacteria with IEB and necrotizing enterocolitis (NEC) in premature infants. The IM performs several important functions and is considered virtually an essential “organ” as it plays important role in harvesting nourishment from diet, influencing absorption and distribution of body fat, regulating mucosal development of IEB, and modulating innate and adaptive immunity [2, 4–7].
The IEB is constantly assessing luminal microecology and making adjustments to protect its frontier. Intestinal colonization by commensal bacteria prevents colonization by pathogens [4, 5]. Commensal bacteria direct immune and physiological system throughout life and are responsible for the proper education of our immune system [6, 7]. The microbiota (collective bacterial population) is responsible for the proper development of immune and inflammatory cells in the healthy gut through the “physiological” or “controlled” inflammation, and thus, confers protection against pathogens [5]. In premature infants who are hospitalized for prolonged duration in the intensive care nurseries and are exposed to numerous antibiotics, the process of normal colonization by commensal bacteria is disrupted [8, 9]. Consequently, mucosal response to abnormal IM in premature host can result in abnormal inflammatory and immune response resulting in disruption of IEB and genesis of NEC. The IEB has exhaustive task of preventing intestinal microbes and their products from translocating into internal milieu. Luminal bacterial presence and their translocations across IEB are proven essential contributors to NEC [10, 11]. Molecular mechanisms of these contributing factors through modulation of immune and inflammatory responses in premature host are now beginning to emerge. This overview (1) describes the microbial–mucosal interactions and the microbial modulation of intestinal immune responses, (2) elucidates the recent metagenomic advances in the field of intestinal microecology, and (3) presents mechanisms of microbial contribution to the genesis of NEC.
Role of IM in the development of IEB
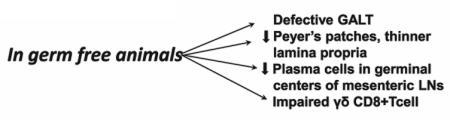
Comparisons between conventionally raised murine animals with germ-free counterparts have revealed that several key aspects of postnatal maturation of IEB are driven by IM including development of a network of vascular core in intestinal villi [12, 13]. Underdevelopment of villus-angiogenesis in germ-free mice and restoration of angiogenesis upon bacterial colonization provided evidence that microbiota play a significant role in angiogenesis. Similarly, postnatal induction of angiogenin-4, a potent bactericidal Paneth cell protein is mediated by microbiota. The expression of angiogenin-4 increases dramatically during weaning and reaches adult level [13–15]. In germ-free mice, expression of angiogenin-4 remains stunted. This microbial function was again confirmed by restoring stunted angiogenin-4 expression to normal level in germ-free animal upon introduction of commensal bacterial colonization [13–15]. Germ-free animals show extensive defects in the development of gut-associated lymphoid tissue (GALT) and antibody production. Germ-free animals also develop fewer and less cellular Peyer’s patches, a thinner and less cellular lamina propria, and fewer plasma cells in germinal centers of the mesenteric lymph nodes compared with animals raised conventionally [(Box 1); 11–15].
Box 1. Role of microbiota in the development of IEB.
Microbes contribute to angiogenesis of the villus core
Improve intestinal motility
Modulate enzyme activity
Promote intestinal restitution and repair
Promote development of intestinal Epithelial lymphocytes (IELs)
Promote high activity of angiogenin-4 (potent bactericidal protein in Paneth cells)
Contribute to development of immune system
The CD8+ lymphocytes are dominant among intestinal epithelial lymphocytes (IELs) while CD4+ cells dominate lamina propria. The IELs bearing γδ T-cell receptors are interspersed between intestinal epithelial cells on the basolateral side of epithelial tight junctions. Unlike conventional T cells, these γδ bearing IELs have the ability to secrete epithelial growth factors and to recruit inflammatory cells by producing innate cytokines and chemokines [16]. In response to mucosal injury, these cells induce a complex transcriptional program, including regulation of cytoprotective, immunomodulatory, and antibacterial factors [16]. Studies in germ-free mice revealed that commensal microbiota regulates the key components of this transcriptional program and directs γδ-bearing IELs to respond to mucosal injury by limiting bacterial penetration across injured mucosa [16]. Commensal bacteria stimulate development of enteric nervous system, therefore promote intestinal motility and modulate intestinal enzyme activity [12, 14].
Interactions between IM and IEB, and cellular outputs
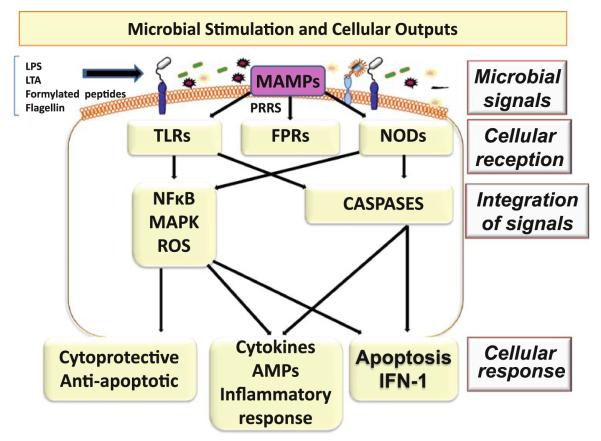
The monolayer of IEB serves as the interface between the IM and the host interior preventing microbial entry across this barrier [4, 9]. Through the process of “cross-talk” between pattern recognition receptors (PRRs) on IEB and microbial macromolecular ligands referred as microbial-associated molecular patterns (MAMPs), the IM is constantly sending signals to host interior. Distinguishing commensal bacteria from pathogens through the innate immune system, the IEB in turn is constantly making appropriate adjustments through cellular responses [12, 17] (Fig. 1). There are 11 recognized Toll-like receptors (TLRs) which are transmembrane PRRs and play a key role in microbial recognition, induction of antimicrobial genes, and control of the adaptive immune responses [17]. The intracellular PRRs are nucleotide-binding oligomerization domain-like receptors (NOD) 1 and NOD 2. Formylated peptide receptors (FPRs) are a type of transmembrane PRRs that are expressed in neutrophils where they perceive bacterial products such as formylated peptides and promote neutrophil functions such as generation of reactive oxygen species and phagocytic motility [17]. Microbial ligands (MAMPS) such as lipopolysaccharide (LPS), flagellin, lipoteichoic acid, peptidoglycans and formylated peptides generate signal to stimulate PRRS such as TLRs, FPRs, or NODs [12]. Activation of intestinal PRRs initiates regulatory pathways such as mitogen-activated protein kinase (MAPK) and nuclear factor κB (NFκB)/Rel pathways, as well as caspase-dependent pathways [17, 18]. These complex and interrelated pathways result in post-transcriptional processes and cellular responses [17, 19]. The manner in which IEB responds to microbial signal can vary depending upon how that signal is perceived. Based upon the initial perception of the triggering organism, the cellular output can be a protective response, e.g., to commensal microbiota, an inflammatory response, e.g., to a pathogen, or it can be a response that triggers apoptosis [17, 19]. Thus, the networks of transduction pathways determine if MAMPs perception warrants a cytoprotective response, an inflammatory reaction or programmed cell death [(Fig. 1); 17–19].
Fig. 1.
Microbial components such as lipopolysaccharides (LPS), lipoteichoic acid (LTA), formylated peptides, and flagellin serve as microbial-associated molecular patterns (MAMPs) and signal pattern recognition receptors (PRRs) including Toll-like receptors (TLRs), formylated peptide receptors (FPRs), or nucleotide-binding oligomerization domain-like receptors (NODs). Integration of these signals evokes cellular outputs based on the initial perception of the triggering organism. Output can be a protective response to commensal microbiota, an inflammatory response to pathogenic organism(s), or it can trigger apoptosis (reproduced with permission from Sharma et al. [12])
Mechanisms of microbial modulation of TLR and NOD/CARD mediated responses
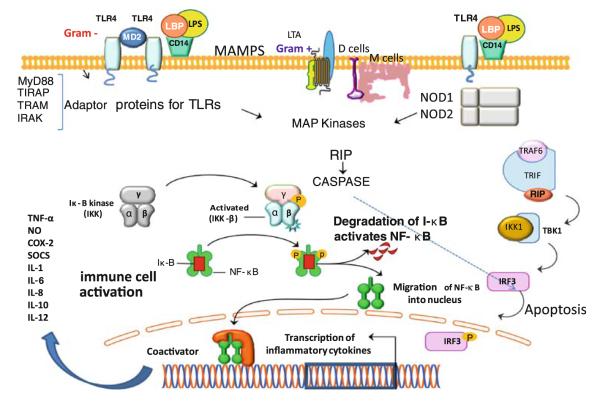
Although microbiota contributes to maintenance of health, luminal microbial overgrowth or dominance of pathogens can lead to diseases including NEC and inflammatory bowel diseases [20, 21]. In epithelial barrier, the ability of immunosensory enterocytes, M cells (microfold cells) and dendritic cells to discriminate pathogenic bacteria from commensal bacteria is mediated by the transmembrane TLRs and the intracellular NOD isoforms [(Fig. 2); 22–25]. The microbiota regulates components of the intestinal innate immune system by modulating expression of TLRs and NOD/CARD (caspase recruitment domain) mediated activation of immunosensory cells through MAMPs [17–19, 21–25]. Commensal bacteria can dampen TLR-mediated inflammatory signals and exert protective effects by attenuating proinflammatory responses, while signals from pathogens can mediate inflammatory response. One of the components of regulation of inflammatory signals is by activating IκB, the inhibitory component of nuclear factor kappa B (NFκB) activation. Manipulation of the NFκB pathways has revealed both anti-inflammatory and proinflammatory roles for this transcriptional control pathway, suggesting temporal patterns in which TLR and NFκB pathways are activated in response to distinct microbial signals [21–25]. Upon binding to their respective MAMPs, the TLRs trigger recruitment of Myd88 (myeloid differentiation primary-response gene 88). The death domain of Myd88 recruits a death domain containing protein known as IL-1R associated protein kinase (IRAK). Activation of IRAK leads to activation of NFκB, p38 MAPK (mitogen-activated protein kinase) and other regulators of gene expression (Fig. 2). These events and expression of inflammatory genes form the basis of innate immune response to IM. NODs transduce signals in the pathways of MAP kinases and NFκB and thus play a fundamental role in immune-cell activation in response to specific MAMPs [(Fig. 2); 26]. In addition to regulating the innate immune system as described above, IM also plays a critical role in regulating the adaptive immune system.
Fig. 2.
A schematic illustration of the recognition of microbial-associated molecular patterns (MAMPs), such as lipopolysaccharides (LPS), by pattern recognition receptors (PRRs) on epithelial cells and immune-cell activation or apoptotic response. Transmembrane Toll-like receptors (TLRs) are triggered by MAMPs and stimulate PRRs. Four adaptor proteins for TLRs are involved in propagation of signals and activation of MAP kinases that can result in transcription of proinflammatory cytokines or apoptotic response through activation of NFκB (with permission, adapted from Sharma et al. [12])
Regulation of adaptive immune function by IM
The largest immune system in human body, the GALT is organized into discrete structures called Peyer’s patches which are essentially mucosal lymph nodes overlaid with M cells, the specialized epithelial immunosensory cells [9,23]. M cells are equipped with endocytic organelles that facilitate the intake of antigen particles from the intestinal lumen [27]. Microbial antigens can gain access to host interior through M cells, transcellular epithelial, or via paracellular route [21, 27, 28]. Without significant ‘cellular processing’, they can reach the underlying lymphoid Peyer’s patches which contains several different types of antigen-presenting cells such as dendritic cells and B cells [(Fig. 3); 21]. Processing of microbial antigens by dendritic cells promotes appropriate regulatory CD4 T-cell populations (Tregs). Independent of systemic adaptive immune system, this process prevents autoimmune responses. The other major route of entry involves antigen transport via a transcellular epithelial or a paracellular route. Antigens entering by this route may be taken up by antigen-presenting T cells in the lamina propria. Immune cells including antibody secreting B cells (plasma cells), T cells, and macrophages populate lamina propria [27–29].
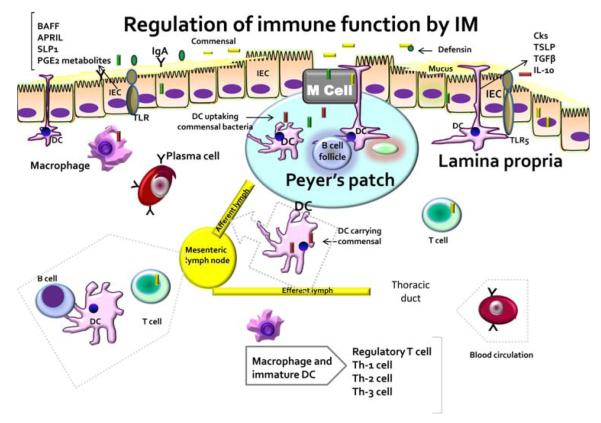
Fig. 3.
Intestinal immune-cell function can be regulated by microbiota. Microbial recognition by intestinal epithelial cell (IEC) can influence secretion of cytokines such as thymic stromal lymphopoietin (TSLP), transforming growth factor β (TGFβ) and interleukin 10 (IL-10), that can directly activate elaboration of proinflammatory cytokines by dendritic cells (DC) and macrophages in lamina propria and Peyer’s patches. Signals from commensal may influence tissue-specific functions resulting in T-cell regulation and expansion into T helper (Th)-1, Th-2, and Th-3 cells. Modulated by intestinal microbiota, other IEC-derived factors include B-cell activating factor (BAFF), APRIL (a proliferation-inducing ligand), secretory leucocyte peptidase inhibitor (SLP1), prostaglandin E2 and other metabolites. Thus, microbiota regulates functions of both antigen-presenting cells and lymphocytes in the intestinal ecosystem (reproduced with permission from Sharma et al. [4])
Signals from commensals may influence tissue-specific functions resulting in T-cell expansion and regulation of T-helper cells, Th-1, Th-2 and Th-3 cells [(Fig. 3); 21, 22, 27–29]. As a consequence of microbial interaction and stimulation, intestinal epithelial cells elaborate many cytokines including thymic stromal lymphopoietin, transforming growth factor, and IL-10. This can induce production of cytokines by dendritic cells and macrophages. Similarly, modulated by intestinal microbiota, other IEC-derived factors, including APRIL (a proliferation-inducing ligand), B-cell activating factor (BAFF), secretory leukocyte peptidase inhibitor (SLP1), prostaglandin E2, defensin, and other chemokines, cytokines, or antimicrobial peptides (AMP), directly regulate functions of both antigen-presenting cells and lymphocytes in the intestinal ecosystem. The induced T-regulator cells offer tolerance towards antigens through elaboration of cytokines (IL-10, transforming growth factor-α) that suppress effector lymphocytes [21, 22, 30, 31]. As previously described, T cells in lamina propria are primarily the CD4 cells expressing αβ T-cell receptors where as CD8 cytolytic cells expressing γδ are intraepithelial lymphocytes [16]. Transport of microbial antigen for presentation to underlying Peyer’s patches results in T-cell activation and B-cell priming, and the development of IgA+ plasma cells [30–32].
Limited in utero exposure to antigens renders a newborn with impaired adaptive immunity and neonates rely on their innate immune system that instructs the adaptive immune responses [30, 31]. Stimulation of fetal intestinal cells results in higher levels of NFκB activation and production of CXC-chemokine ligand-8 (CXCL8) and CXCL2 compared to the adult enterocytes [30–33]. After birth, the exposure of perinatal intestinal cells to LPS has demonstrated the loss of intestinal–epithelial responsiveness to LPS due to down-regulation of expression of IRAK (IL-1 receptor-associated kinase 1), an essential intermediary for epithelial TLR4 signaling [34–36]. This postnatal tolerance to endotoxin may facilitate adaptation of the newborn to subsequent microbial colonization and to support host microbe homeostasis that is required for commensal interactions [36].
Why premies are susceptible to NEC?
In the face of massive antigenic challenge after birth, immune cells distributed throughout GALT elicit hypo responsiveness to commensal bacteria while retaining their capability of responding to pathogenic challenge [33, 34]. Under the steady state, the immune system recognizes commensal bacteria and elicits tonic signals or basal signals, while pathogenic bacteria do not evoke full activation of the inflammatory immune responses [24, 33–36]. In general, the neonatal immune response is tilted towards immune tolerance (Th-1 response) [35]. Pathogenic stimulus induces production of certain chemokines and cytokines such as tumor necrosis factor-α (TNF-α), interleukins (ILs)1,6,8,10,12, cycloxygenase-2 (COX2), nitric oxide (NO) by antigen-presenting cells and neonatal monocytes that results in inflammatory response [33–36]. Thus, neonates have unique expression of TLRs, chemokines, antimicrobial proteins, and peptides. In parallel, developments of Paneth cells (a rich source of antimicrobial protein peptides) in the small intestine of neonates contribute to the clearance of pathogens and promote commensal colonization [10]. After birth there is age-dependent maturation of immune responses. Recent studies indicate that a prematurely born infant may fall short in achieving many of the immune functions that confer immune tolerance [(Box 2);11, 24]. Premature infants appear to mimic a response that is similar to fetal response characterized by hyper responsiveness to LPS [9, 24, 35, 38]. Their inability to appropriately down-regulate the response to LPS seems to be one of the mechanisms that contributes to their susceptibility to NEC [36–40].
Box 2. Cellular Response to Microecology in Premature Host.
Reduced expression of IκB in enterocytes
Inability to dampen inflammatory pathways
Excessive inflammatory response even to commensals
Exaggerated response to pathogenic bacteria
Abnormal up-regulation of TLR4 in response to stress thereby increasing inflammatory signals
Increased susceptibility to apoptosis
Immaturity of biochemical and immune functions of intestinal epithelial barrier
These factors increase risk for NEC in premature infants
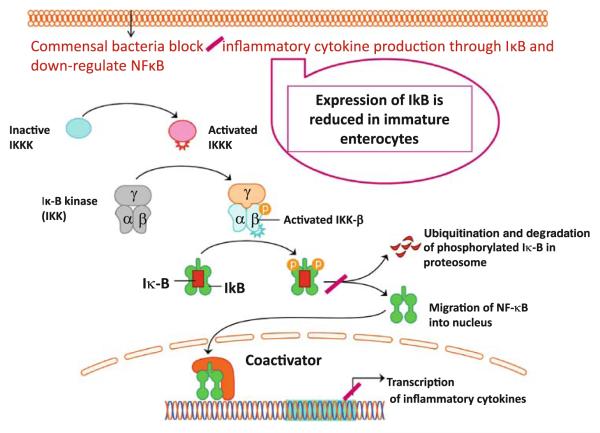
The innate mechanisms in IEB of premature infants appear to elicit exuberant inflammatory response by intestinal immunosensory cells (IECs, dendritic cells, and Paneth cells) [21, 25]. Subsequent activation of the signaling pathway causes NFκB to dissociate from inhibitor of κB (IκB) so that NFκB can translocate to the nucleus and cause transcription and translation of inter-leukin-8 (IL-8). In the presence of Toll-interacting protein, IL-1 activation is inhibited [21, 22, 24]. Toll-interacting protein inhibits autophosphorylation and kinase activity of IRAK, which prevents subsequent activation of NFκB. Expression of IκB is developmentally regulated and its expression is reduced in immature intestine [(Fig. 4); 23,25–28]. Consequently, immature enterocytes demonstrate excessive inflammatory response with both commensal and pathogenic bacteria. Moreover, other TLR-negative signal intermediates are down-regulated and expression of these intermediates is further reduced in infants who develop NEC as demonstrated by microarray analysis of intestinal samples [(Box 2); 31–35].
Fig. 4.
Commensal bacteria block activation of NFκB and thus down-regulate inflammatory cytokine production in mature host. In immature enterocyte, expression of IκB is reduced contributing to exaggerated inflammatory response
In addition, premature infants are more likely to be colonized by pathogens that may result in aberrant immune and inflammatory responses rendering them susceptible to NEC [28–30]. They also exhibit intestinal dysmotility that contributes to prolongation of the transit time of milk substrates often leading to milk stasis and this promotes colonization by pathogens. They appear to have impaired regulation of mucus secretion and less efficient function of tight junction proteins assembly [10, 40, 41]. These factors also contribute to the impaired clearance of pathogens (by mucus secretions) facilitating microbial translocation across IEB (due to leaky tight junctions), elaboration of inflammatory cytokines and NEC [10, 18, 42].
Role of commensal bacteria
In addition to the benefits of commensal bacteria as described above and as summarized in Box 3, commensal bacteria exert anti-inflammatory effect by selectively blocking transcription activation of factor NFκB in mature adult enterocytes [17]. It has been demonstrated that the commensal bacteria also strengthen tight junction protein assembly, and increase and alter characteristics of mucus secretions in a manner that inhibits microbial translocation across IEB [(Fig. 5); 12, 43, 44]. Commensal bacteria also enhance intestinal motility and provide protective nutrients against inflammation. Commensal bacteria may up-regulate the expression of intermediates that down-regulate the production of inflammatory cytokines and chemokines [12,37]. Thus, commensal bacteria serve as the driver for maturation of many innate and adaptive functions and dampen inflammatory signals (Box 3). In neonatal rat model, bifidobacteria has been found to restore tight junction proteins and protect against NEC. This protective effect is associated with reduction in inflammatory reaction in ileum, regulation of main components of mucus layer and improvement of intestinal integrity [45]. Commensal bacteria also influence immune homeostasis via direct regulation of the IL-25–IL-23–IL-17 axis. This down-regulation of IL-17 producing Th-17 cells has anti-inflammatory effect [46].
Box 3. Role of Commensal Bacteria.
Up-regulation of mucin genes
Restoration of tight junction protein assembly
Secretion of defensins
Modulation of expression of TLRS and activation of immunosensory cells through MAMPs
Regulation of NFκB signaling pathways
Dampening of TLR-mediated inflammatory signals
Serve as driver for maturation of innate and adaptive immune systems
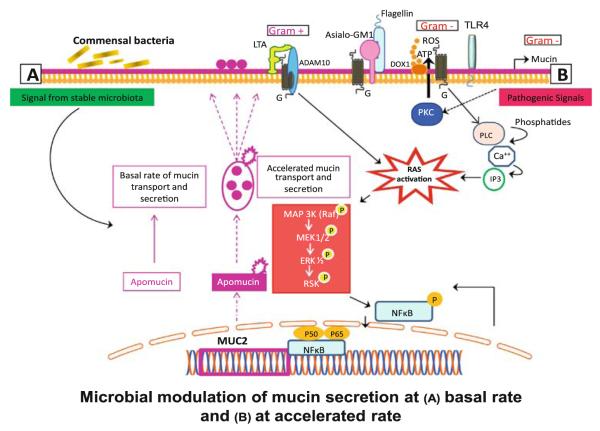
Fig. 5.
Mircobial–mucosal interactions regulate mucin production. Commensal bacteria induce mucin secretion at basal rate. Conversely, mucin secretion is accelerated upon activation by pathogenic organisms mediated by activation of MAP kinase system and downstream activation of NFκB with increased mucin transcription. In turn, intestinal mucin dictates the composition of bacterial community. Adhesion to specific mucin epitopes is thought to facilitate mucous colonization by commensal bacteria providing them with growth advantage when competing against pathogens (reproduced with permission from Sharma et al. [12])
Microecology in immature and mature human host
The pattern of bacterial colonization in the premature intestine is different from that of mature infants and adults [(Box 4); 41]. Microecology changes until 2 years after birth when it resembles that of adulthood [41, 47]. Adult microbiota is characterized by high population density and extensive diversity [41, 47]. Culture-based techniques demonstrated that premature infants were first colonized by enterobacteria and streptococci similar to mature infants but both organisms predominated for longer duration with delayed colonization by bifidobacteria [41]. Establishment of bifidobacteria, a commensal bacteria, is important for proper development of the immune system. Establishment of Bacteroides, Clostridium, and Lactobacillus was also delayed in premature infants [41, 47]. Thus, infants requiring intensive care acquire intestinal organisms slowly and establishment of the beneficial commensal bacteria is delayed. Culture-based techniques can only identify 20–30% of adult microbiota. Using denaturing gradient gel electrophoresis (DGGE) techniques, Schwiertz et al. [49] found that microbial profiles were very simple with very low diversity in all preterm infants that increased overtime. They also found that all preterm infants had some similarities corresponding to bands of Escherichia coli, Enterococcus sp., Clostridium and Klebsiella pneumoniae bacteria despite differences in gestational age and formula versus maternal milk feeding. Several of the hospital isolates could be detected in their fecal samples [49]. These findings indicated that initial colonization of preterm intestine is highly influenced by the hospital environment in intensive care nursery. In contrast, microbiota of maternal milk fed full-term infants who were not hospitalized were characterized by more diverse and few intra-individual similarities. These investigators also found that microbiota of full-term maternal milk fed infants was dominated by bifidobacteria and lactic acid bacteria [49]. A Japanese study reported that colonization in maternal milk fed preterm infants was characterized by high initial numbers of enterobacteria and streptococci, while bifidobacteria appeared late at 11th day of age, in contrast to full-term infants who were colonized at fourth day of age [47].
Box 4. Microecology in Immature and Mature host.
Commensal bacteria are the most prevalent organisms in the normal adult intestine
Most common commensal bacteria are: Bifidobacteria, Lactobacillus, Streptococcus thermophilus, Saccharomyces strains
Compared to pre-term, full-term neonates develop a relatively diverse (< adult) and higher number of colonies (< adult) of commensal bacteria
Premature infants have very low to nil colonization with commensal bacteria
Premature infants have low diversity of bacterial colonies that allows resident bacteria to become virulent
Premature infants have increased colonization with pathogenic bacteria
Analysis of microbiota of 16 preterm infants by Magne et al. [50] was characterized by high interindividual variations. These investigators analyzed 16S rRNA genes using PCR-TTGE (temporal temperature gradient gel electrophoresis) profiling. The main bacterial group belonged to Enterococcus, Streptococcus, and Staphylococcus genera. The preterm infants were colonized by anerobes and only 4 out of 16 by bifidobacteria. Analyzing for pattern of bifidobacterial colonization in 52 infants born at gestational age between 30 and 35 weeks, Butel et al. [51] found that with exception of gestational age, neither the birth nor mode of delivery or type of feeding influenced colonization by bifidobacteria. Another recent study evaluating microbiota of 23 neonates born at 23–32 weeks gestational age found differences in microbial diversity in infants whose mothers intended to provide maternal milk, in infants born to mother with chorioamnionitis, and in those born at <30 weeks gestation [52]. Four of these infants developed NEC and two developed systemic inflammatory response syndrome. Citrobacter-like sequences were seen only in three of four NEC cases, but the overall microbiota profiles were not significantly different from those of control infants. There was a trend of increased diversity over time and DGGE analysis revealed unique composition for each infant. Although composition changed over time, characteristic bands for each individual infant persisted over time [52]. Another group of investigators found that the intestinal bacterial colonization in all preterm infants was notable for low diversity and infants with NEC had even less diversity with an increase in abundance of Gamma-proteobacteria. Infants with NEC had received a higher mean number of previous days of exposure to antibiotics [53].
These investigations indicate that in general, preterm infants are colonized by low numbers of beneficial bacteria (e.g., bifidobacteria, lactobacilli) and high numbers of pathogenic bacteria (e.g., enterobacteria, E. coli, bacteroides, enterococci, streptococci, staphylococci, and Klebsiella species). Premature infants receiving maternal milk develop more diversity with lower number of pathogenic bacteria [54].
Molecular techniques of microbial analysis
Recent molecular techniques that are based on the 16S rRNA gene (rDNA) have become useful in efforts of cataloging human microbiome (microbial genome) [48]. The 16S rRNA gene contains highly conserved sequence domains interspersed with more variable regions. This allows comparative analysis of these variable rRNA sequences that can identify the profile of sequence patterns enabling identification of different bacterial communities. These techniques enable characterization and quantification of the microbiota while providing a classification scheme also to predict phylogenetic relationships. This technique provides comprehensive analysis of 16S rRNA content in community DNA for detection of known as well as unknown bacteria. This analysis is based on physical and chemical properties of DNA molecules. Clone libraries are set up to identify the species composition of microbiota [9, 41]. Automated ribosomal intergenic spacer analysis (ARISA) and DGGE techniques provide qualitative information of dominant microbes and information about microbial diversity. This technique is useful for diversity analyses, but does not allow direct identification of specific bacteria. Fluorescent in situ hybridization (FISH) technique can be utilized to determine abundance of particular taxa and quantitative analysis of total fecal bacteria and of targeted group of bacteria [9].
Probiotics
Increasing awareness that the neonatal flora is a major contributor to their health and disease has led to different therapeutic strategies to manipulate the flora. However, evidence with respect to the characteristics of neonatal IM is very limited, let alone that of premature infants in whom NEC is more common. We are yet to enrich our existing knowledge with regards to how feeding practices (maternal milk feeding vs. formula feedings), frequent and prolonged exposure of antibiotics, delay in enteral feedings, and other morbidities related to prematurity itself influence IM. We have learnt from culture-based studies that full-term infants receiving maternal milk have abundance of bifidobacteria and lactobacilli which predominate over potentially harmful bacteria such as coliforms, enterococci, and Bacteroides that predominate formula-fed infants [41]. We have also learnt that maternal milk feedings decrease the risk for NEC in premature infants [55, 56]. Maternal milk feedings promote colonization with bifidobacteria that confers health benefits to the infant and appear to protect against intestinal diseases. Administration of such live microbial supplements (probiotics) that provide health benefits in neonatal diet has been investigated as one of the approaches to manipulate neonatal microbiome in order to reduce risk of NEC. Studies enlisted in Table 1 have demonstrated significant reduction in NEC by maneuvering IM with probiotics. These findings are reassuring. However, the safety and long-term immunological consequences of manipulating IM with live probiotic bacteria have not been established. Other unanswered questions relate to: Which species or strains that are best suited for this purpose? And whether one should administer a live or attenuated probiotic agent? Bacteremia and neonatal sepsis associated with probiotic has been reported [57].
Table 1.
Clinical trials of probiotics to prevent NEC
| Studies | Choice of probiotics | Total subjects |
Design | Results, incidence of NEC |
|---|---|---|---|---|
| Hoyos [57] |
Lactobacillus acidophilus,
Bifidobacterium infantis |
1,237 | Non-RCT, mean GA = 37 wks | 2.7 versus 6.6% (p < 0.0002) |
| Dani et al. [58] | Lactobacillus GG | 585 | DB-RCT, GA < 33 wks or BW < 1,500 g | 1.4 versus 2.7% (NS) |
| Bin-Nun et al. [59] |
Bifidobacterium infantis,
Streptococcus thermophilus, Bifidobacterium bifidus |
145 | RCT, BW < 1,500 g | 4 versus 16.4% (p = 0.03) |
| Lin et al. [60] |
Lactobacillus acidophilus,
Bifidobacterium bifidus |
434 | DB-RCT, BW < 1,500 g | 1.8 versus 6.4% (p < 0.05) |
DB double blind, RCT randomized controlled trial, GA gestational age at birth, BW birth weight, wks weeks, g gram
Studies in pediatric and adult populations are providing clues that intestinal microbiota plays a seminal role in human health and inflammatory intestinal diseases [2, 5]. Recently emerging investigations conducted in premature neonatal population are revealing that this population experiences delayed and aberrant intestinal microbial colonization [47, 49–54]. So far only handful of such studies have been conducted. More studies on microbiota of premature neonatal population are required in order to understand how it relates to their prolonged and often turbulent hospital course characterized by delayed enteral feedings and repeated exposures to antibiotics that are bound to cause perturbations in luminal environment. Abnormal microecology combined with immaturity of intestinal barrier and immune functions make them susceptible to intestinal inflammation and NEC [41]. Use of probiotics has certainly demonstrated reduction in risk for NEC but the long-term effect on immune functions in premature infants have not been ascertained conclusively [56, 61]. Future progress in understanding the microbial-mucosal modulation of IEB and mechanisms that regulate intestinal immunology of premature infants is likely to provide benefits against inflammatory intestinal diseases including NEC.
Acknowledgments
This work was supported by the National Institute of Health grant 1RO1 HD059143-01.
Abbreviations
- IEB
Intestinal epithelial barrier
- NEC
Necrotizing enterocolitis
- IM
Intestinal microecology
- GALT
Gut-associated lymphoid tissue
- IEL
Intestinal epithelial lymphocyte
- PRRs
Pattern recognition receptors
- MAMPs
Microbial-associated molecular patterns
- TLR
Toll-like receptor
- NOD
Nucleotide-binding oligomerization domain
- FPRs
Formylated peptide receptors
- LPS
Lipopolysaccharide
- MAPK
Mitogen-activated protein kinase
- NFκB
Nuclear factor kappa B
- Rel
Proteins coded by rel oncogenes
- M-cells
Microfold-cells
- CARD
Caspase recruitment domain
- Myd88
Myeloid differentiation primary-response gene 88
- T-reg
T regulatory
- Th
T helper
- APRIL
A proliferation-inducing ligand
- BAFF
B-cell activating factor
- SLP1
Secretory leucocyte peptidase inhibitor 1
- AMP
Antimicrobial peptide
- CXC
Chemokine
- IRAK
IL-1-R associated protein kinase
- TNF-α
Tumor necrosis factor-α
- NO
Nitric oxide
- IL
Interleukin
- DGGE
Denaturing gradient gel electrophoresis
- PCR-TTGE
Polymerase chain reaction-temporal temperature gradient gel electrophoresis
- rRNA
Ribosomal RNA
- ARISA
Automated ribosomal intergenic spacer analysis
- FISH
Fluorescent in situ hybridization
Contributor Information
Renu Sharma, Division of Neonatology, Department of Pediatrics, University of Florida at Jacksonville, 655 West 8th Street, Jacksonville, FL 32209, USA.
Joseph J. Tepas, III, Department of Surgery, University of Florida at Jacksonville, Jacksonville, USA.
References
- 1.Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.J Xu, JI Gordon. Inaugural article: honor thy symbionts. Proc Natl Acad Sci USA. 2003;100:10452–10459. doi: 10.1073/pnas.1734063100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abt MC, Artis D. The intestinal microbiota in health and disease: the influence of microbial products on immune cell homeostasis. Curr Opin Gastroenterol. 2009;25:496–502. doi: 10.1097/MOG.0b013e328331b6b4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharma R, Young C, Mshvildadze M, et al. Intestinal microbiota: does it play a role in diseases of the neonate? Neoreviews. 2009;10:e166–e179. [Google Scholar]
- 5.Bäckhed F, Ley RE, Sonnenburg JL, et al. Host–bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 6.O’Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin FPJ, Sprenger N, Yap IKS, et al. Panorganismal gut microbiome–host metabolic cross talk. J Proteome Res. 2009;8:2090–2105. doi: 10.1021/pr801068x. [DOI] [PubMed] [Google Scholar]
- 8.Mariat D, Firmesse O, Levenez F, et al. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 2009;9:123. doi: 10.1186/1471-2180-9-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mshvildadze M, Neu J, Mai V. Intestinal microbiota development in the premature neonate: establishment of a lasting commensal relationship? Nutr Rev. 2008;66:658–663. doi: 10.1111/j.1753-4887.2008.00119.x. [DOI] [PubMed] [Google Scholar]
- 10.Sharma R, Tepas JJ, III, Hudak ML. Neonatal gut barrier and multiple organ failure: role of endotoxin and proinflammatory cytokines in sepsis and necrotizing enterocolitis. Pediatr Surg. 2007;42:454–461. doi: 10.1016/j.jpedsurg.2006.10.038. [DOI] [PubMed] [Google Scholar]
- 11.Lin PW, Nasr TR, Stoll BJ. Necrotizing enterocolitis: recent scientific advances in pathophysiology and prevention. Semin Perinatol. 2008;32:70–82. doi: 10.1053/j.semperi.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 12.Sharma R, Young C, Neu J. Molecular modulation of intestinal epithelial barrier: contribution of microbiota. In: Kitamura K, McCann A, Wu XR, editors. The epithelium molecular landscaping for an interactive barrier J Biomed Biotechnol. special issue. 2009. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hooper LV. Bacterial contributions to mammalian gut development. Trends Microbiol. 2004;12:129–134. doi: 10.1016/j.tim.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 14.Stappenbeck TS, Hooper LV, Gordon JI. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc Natl Acad Sci USA. 2002;99:15451–15455. doi: 10.1073/pnas.202604299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hooper LV, Stappenbeck TS, Hong CV, et al. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nat Immunol. 2003;4:269–273. doi: 10.1038/ni888. [DOI] [PubMed] [Google Scholar]
- 16.Ismail AS, Behrendt CL, Hooper LV. Reciprocal interactions between commensal bacteria and gamma delta intraepithelial lymphocytes during mucosal injury. J Immunol. 2009;182:3047–3054. doi: 10.4049/jimmunol.0802705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neish AS. Microbes in gastrointestinal health and disease. Gastroenterology. 2009;136:65–80. doi: 10.1053/j.gastro.2008.10.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conroy ME, Walker WA. Intestinal immune health. Nestle Nutr Workshop Ser Pediatr Program. 2008;62:111–121. doi: 10.1159/000146255. [DOI] [PubMed] [Google Scholar]
- 19.Akira S. Pathogen recognition by innate immunity and its signaling. Proc Jpn Acad Ser. 2009;85:143–156. doi: 10.2183/pjab.85.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Hara AM, Shanahan F. Gut microbiota: mining for therapeutic potential. Clin Gastroenterol Hepatol. 2007;5:274–284. doi: 10.1016/j.cgh.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 21.Artis D. Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat Rev Immunol. 2008;8:411–420. doi: 10.1038/nri2316. [DOI] [PubMed] [Google Scholar]
- 22.Cario E. Bacterial interactions with cells of the intestinal mucosa: Toll-like receptors and NOD2. Gut. 2005;54:1182–1193. doi: 10.1136/gut.2004.062794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duerkop BA, Vaishnava S, Hooper LV. Immune responses to the microbiota at the intestinal mucosal surface. Immunity. 2009;31:368–376. doi: 10.1016/j.immuni.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 24.Claud EC, Walker WA. Bacterial colonization, probiotics, and necrotizing enterocolitis. J Clin Gastroenterol. 2008;42(Suppl 2):S46–S52. doi: 10.1097/MCG.0b013e31815a57a8. [DOI] [PubMed] [Google Scholar]
- 25.Jilling T, Simon D, Lu J, et al. The roles of bacteria and TLR4 in rat and murine models of necrotizing enterocolitis. J Immunol. 2006;177:3273–3282. doi: 10.4049/jimmunol.177.5.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neish AS. TLRs in the gut. II Flagellin-induced inflammation and antiapoptosis. Am J Physiol Gastrointest Liver Physiol. 2007;292:G462–G466. doi: 10.1152/ajpgi.00274.2006. [DOI] [PubMed] [Google Scholar]
- 27.Macdonald TT, Monteleone G. Immunity, inflammation, and allergy in the gut. Science. 2005;307:1920–1925. doi: 10.1126/science.1106442. [DOI] [PubMed] [Google Scholar]
- 28.Macpherson AJ, Geuking MB, McCoy KD. Immune responses that adapt the intestinal mucosa to commensal intestinal bacteria. Immunology. 2005;115:153–162. doi: 10.1111/j.1365-2567.2005.02159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 30.Levy O. Innate immunity of the newborn: basic mechanisms and clinical correlates. Nat Rev Immunol. 2007;7:379–390. doi: 10.1038/nri2075. [DOI] [PubMed] [Google Scholar]
- 31.Kelly D, King T, Aminov R. Importance of microbial colonization of the gut in early life to the development of immunity. Mutat Res. 2007;622:58–69. doi: 10.1016/j.mrfmmm.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 32.Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303:1662–1665. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- 33.Coombes JL, Powrie F. Dendritic cells in intestinal immune regulation. Nat Rev Immunol. 2008;8:435–446. doi: 10.1038/nri2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abreu MT, Vora P, Faure E, et al. Decreased expression of Toll-like receptor-4 and MD-2 correlates with intestinal epithelial cell protection against dysregulated proinflammatory gene expression in response to bacterial lipopolysaccharide. J Immunol. 2001;167:1609–1616. doi: 10.4049/jimmunol.167.3.1609. [DOI] [PubMed] [Google Scholar]
- 35.Forchielli ML, Walker WA. The role of gut-associated lymphoid tissues and mucosal defence. Br J Nutr. 2005;93(Suppl 1):S41–S48. doi: 10.1079/bjn20041356. [DOI] [PubMed] [Google Scholar]
- 36.Naik S, Kelly EJ, Meijer L, et al. Absence of Toll-like receptor 4 explains endotoxin hyporesponsiveness in human intestinal epithelium. J Pediatr Gastroenterol Nutr. 2001;32:449–453. doi: 10.1097/00005176-200104000-00011. [DOI] [PubMed] [Google Scholar]
- 37.Shibolet O, Podolsky DK. TLRs in the gut. IV. Negative regulation of Toll-like receptors, intestinal homeostasis: addition by subtraction. Am J Physiol Gastrointest Liver Physiol. 2007;292:G1469–G1473. doi: 10.1152/ajpgi.00531.2006. [DOI] [PubMed] [Google Scholar]
- 38.Neutra MR, Mantis NJ, Kraehenbuhl JP. Collaboration of epithelial cells with organized mucosal lymphoid tissues. Nat Immunol. 2001;2:1004–1009. doi: 10.1038/ni1101-1004. [DOI] [PubMed] [Google Scholar]
- 39.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neu J, Mshvildadze M, Mai V. A roadmap for understanding and preventing necrotizing enterocolitis. Curr Gastroenterol Rep. 2008;10:450–457. doi: 10.1007/s11894-008-0084-x. [DOI] [PubMed] [Google Scholar]
- 41.Cochetière MF, Rougé C, Darmaun D, et al. Intestinal microbiota in neonates and preterm infants: a review. Curr Pediatr Rev. 2007;3:21–34. [Google Scholar]
- 42.Liu Z, Li N, Neu J. Tight junctions, leaky intestines, and pediatric diseases. Acta Paediatr. 2005;94:386–393. doi: 10.1111/j.1651-2227.2005.tb01904.x. [DOI] [PubMed] [Google Scholar]
- 43.Anand RJ, Leaphart CL, Mollen KP, et al. The role of the intestinal barrier in the pathogenesis of necrotizing enterocolitis. Shock. 2007;27:124–133. doi: 10.1097/01.shk.0000239774.02904.65. [DOI] [PubMed] [Google Scholar]
- 44.Dharmani P, Srivastava V, Kissoon-Singh V, et al. Role of intestinal mucins in innate host defense mechanisms against pathogens. J Innate Immun. 2009;1:123–135. doi: 10.1159/000163037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khailova L, Dvorak K, Arganbright KM, et al. Bifidobacterium bifidum improves intestinal integrity in a rat model of necrotizing enterocolitis. Am J Physiol Gastrointest Liver Physiol. 2009 Aug 27; doi: 10.1152/ajpgi.00141.2009. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zaph C, Du Y, Saenz SA, et al. Commensal-dependent expression of IL-25 regulates the IL-23-IL-17 axis in the intestine. J Exp Med. 2008;205:2191–2198. doi: 10.1084/jem.20080720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sakata H, Yosioka H, Fujita K. Development of intestinal flora in very low birth weight infants compared to normal full-term newborns. Eur J Pediatr. 1985;144:186–190. doi: 10.1007/BF00451911. [DOI] [PubMed] [Google Scholar]
- 48.Mai V, Draganov PV. Recent advances and remaining gaps in our knowledge of associations between gut microbiota and human health. World J Gastroenterol. 2009;15:81–85. doi: 10.3748/wjg.15.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwiertz A, Gruhl B, Löbnitz M, et al. Development of the intestinal bacterial composition in hospitalized preterm infants in comparison with breast-fed, full-term infants. Pediatr Res. 2003;54:393–399. doi: 10.1203/01.PDR.0000078274.74607.7A. [DOI] [PubMed] [Google Scholar]
- 50.Magne F, Abély M, Boyer F, et al. Low species diversity and high interindividual variability in faeces of preterm infants as revealed by sequences of 16S rRNA genes and PCR-temporal temperature gradient gel electrophoresis profiles. FEMS Microbiol Ecol. 2006;57:128–138. doi: 10.1111/j.1574-6941.2006.00097.x. [DOI] [PubMed] [Google Scholar]
- 51.Butel MJ, Suau A, Campeotto F, et al. Conditions of bifidobacterial colonization in preterm infants: a prospective analysis. J Pediatr Gastroenterol Nutr. 2007;44:577–582. doi: 10.1097/MPG.0b013e3180406b20. [DOI] [PubMed] [Google Scholar]
- 52.Mshvildadze M, Neu J, Shuster J, et al. Intestinal microbial ecology in premature infants assessed with non-culture-based techniques. J Pediatr. 2009 Sep 25; doi: 10.1016/j.jpeds.2009.06.063. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Y, Hoenig JD, Malin KJ, et al. 16S rRNA gene-based analysis of fecal microbiota from preterm infants with and without necrotizing enterocolitis. ISME J. 2009;3:944–954. doi: 10.1038/ismej.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gewolb IH, Schwalbe RS, Taciak VL, et al. Stool microflora in extremely low birth weight infants. Arch Dis Child Fetal Neonatal Ed. 1999;80:F167–F173. doi: 10.1136/fn.80.3.f167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gaskins HR, Croix JA, Nakamura N, et al. Impact of the intestinal microbiota on the development of mucosal defense. Clin Infect Dis. 2008;46(Suppl 2):S80–S86. doi: 10.1086/523336. [DOI] [PubMed] [Google Scholar]
- 56.Caplan MC. Probiotic and prebiotic supplementation for the prevention of neonatal necrotizing enterocolitis. J Perinatol. 2009;29:S2–S6. doi: 10.1038/jp.2009.21. [DOI] [PubMed] [Google Scholar]
- 57.Hoyos AB. Reduced incidence of necrotizing enterocolitis associated with enteral administration of Lactobacillus acidophilus and Bifidobacterium infantis to neonates in an intensive care unit. Int J Infect Dis. 1999;3:197–202. doi: 10.1016/s1201-9712(99)90024-3. [DOI] [PubMed] [Google Scholar]
- 58.Dani C, Biadaioli R, Bertini G, et al. Probiotics feeding in prevention of urinary tract infection, bacterial sepsis and necrotizing enterocolitis in preterm infants. A prospective double-blind study. Biol Neonate. 2002;82:103–108. doi: 10.1159/000063096. [DOI] [PubMed] [Google Scholar]
- 59.Bin-Nun A, Bromiker R, Wilchanski M, et al. Oral probiotics prevent necrotizing enterocolitis in very low birth weight neonates. J Pediatr. 2005;147:192–196. doi: 10.1016/j.jpeds.2005.03.054. [DOI] [PubMed] [Google Scholar]
- 60.Lin HC, Hsu CH, Chen HL. Oral probiotics prevent necrotizing enterocolitis in very low birth weight preterm infants: a multicenter, randomized, controlled trial. Pediatrics. 2008;122:693–700. doi: 10.1542/peds.2007-3007. [DOI] [PubMed] [Google Scholar]
- 61.Neu J. Perinatal and neonatal manipulation of the intestinal microbiome: a note of caution. Nutr Rev. 2007;65:282–285. doi: 10.1301/nr.2007.jun.282-285. [DOI] [PubMed] [Google Scholar]