Abstract
The cryptomonads are a group of unicellular algae that acquired photosynthesis through the engulfment of a red algal cell, a process called secondary endosymbiosis. Here, we present the complete plastid genome sequence of the secondarily nonphotosynthetic species Cryptomonas paramecium CCAP977/2a. The ∼78 kilobase pair (Kbp) C. paramecium genome contains 82 predicted protein genes, 29 transfer RNA genes, and a single pseudogene (atpF). The C. paramecium plastid genome is approximately 50 Kbp smaller than those of the photosynthetic cryptomonads Guillardia theta and Rhodomonas salina; 71 genes present in the G. theta and/or R. salina plastid genomes are missing in C. paramecium. The pet, psa, and psb photosynthetic gene families are almost entirely absent. Interestingly, the ribosomal RNA operon, present as inverted repeats in most plastid genomes (including G. theta and R. salina), exists as a single copy in C. paramecium. The G + C content (38%) is higher in C. paramecium than in other cryptomonad plastid genomes, and C. paramecium plastid genes are characterized by significantly different codon usage patterns and increased evolutionary rates. The content and structure of the C. paramecium plastid genome provides insight into the changes associated with recent loss of photosynthesis in a predominantly photosynthetic group of algae and reveals features shared with the plastid genomes of other secondarily nonphotosynthetic eukaryotes.
Keywords: cryptomonads, plastids, genome reduction, photosynthesis, secondary endosymbiosis
Introduction
An important feature separating the known diversity of photosynthetic eukaryotes is the method by which they acquired their light harvesting apparatus: “primary” plastid-containing organisms harbor organelles thought to have evolved directly from the original cyanobacterial plastid progenitor, whereas “secondary” plastid-containing organisms acquired photosynthesis indirectly through the engulfment of a primary plastid-bearing alga (Reyes-Prieto et al. 2007; Gould et al. 2008; Archibald 2009). The loss of photosynthesis in autotrophic organisms containing both primary and secondarily derived plastids has occurred multiple times during the course of eukaryotic evolution, including instances within the heterokonts, dinoflagellates, haptophytes, and land plants (see Kim and Archibald 2009 and references therein for review). In all well-studied cases to date, the plastid itself is retained, as it is known to be the site of essential biochemical processes unrelated to photosynthesis, including fatty acid and amino acid biosynthesis (Waller and McFadden 2005; Barbrook et al. 2006; Mazumdar et al. 2006).
The cryptomonad algae are a diverse and evolutionarily significant lineage of unicellular eukaryotes known to inhabit marine, brackish, and freshwater environments (Graham and Wilcox 2000; Shalchian-Tabrizi et al. 2008). They are comprised of brown-, red-, or blue/green-pigmented photosynthetic species (called “cryptophytes”), as well as colorless secondarily nonphotosynthetic species and a single, distantly related aplastidic genus, Gonionomas (McFadden et al. 1994; Hoef-Emden et al. 2002; Hoef-Emden and Melkonian 2003; von der Heyden et al. 2004; Hoef-Emden 2008). Cryptomonads are of considerable interest to cell evolutionists by virtue of the fact that their plastids are the product of secondary endosymbiosis and, more specifically, the nucleus of the red alga that gave rise to the cryptomonad plastid persists in a vestigial form called a “nucleomorph” (Douglas et al. 1991; Maier et al. 1991; McFadden 1993; Archibald 2007). With the exception of Goniomonas, cryptomonads possess four genomes (host nuclear, mitochondrial, plastid, and nucleomorph) and are a feat of cellular integration, with two distinct cytoplasmic compartments, four membranes surrounding their plastids, and a sophisticated protein targeting apparatus used to traffic the products of nucleus-encoded, nucleomorph- and plastid-targeted proteins back to their compartment of origin (Gould et al. 2008).
To date, six cryptomonad genomes have been sequenced. Rhodomonas salina’s plastid (Khan, Parks, et al. 2007) and mitochondrial genome (Hauth et al. 2005) are complete, and the nucleomorph and mitochondrial genomes of Hemiselmis andersenii have also been published (Lane et al. 2007; Kim et al. 2008). The model cryptomonad Guillardia theta has a sequenced nucleomorph (Douglas et al. 2001) and plastid genome (Douglas and Penny 1999), and sequencing of its nuclear genome is currently underway (http://www.jgi.doe.gov/sequencing/why/50026.html). Here, we present the complete plastid genome sequence of the nonphotosynthetic freshwater cryptomonad Cryptomonas paramecium, the first of its kind from a free-living, red algal secondary plastid-containing organism. Although multiple plastid genomes have been sequenced from parasitic land plants and apicomplexans, to date there has been just a single complete plastid genome published for a free-living, nonphotosynthetic protist, that of the euglenid Euglena longa, which possesses a green algal-derived secondary plastid (Gockel and Hachtel 2000). Comparison of the C. paramecium genome to those of the photosynthetic cryptomonads G. theta and R. salina highlights the genomic changes associated with the loss of photosynthesis in these enigmatic unicellular algae.
Materials and Methods
Cell Cultures and Organellar DNA Preparation
Cultures of C. paramecium strain 977/2a were obtained from the Culture Collection of Algae and Protozoa (CCAP) and maintained in the laboratory at room temperature in media containing 1-g sodium acetate trihydrate + 1-g “Lab Lemco” powder (Oxoid) per 1 l of ddH20. Total cellular DNA was extracted from large-scale (3–4 l) liquid cultures (∼75 l in total) as described previously (Lane et al. 2006). DNA was subjected to Hoechst dye-cesium chloride density gradient centrifugation in order to purify A + T-rich organellar DNA. Three discrete gradient fractions were isolated, purified, and rehydrated in 400 μl of Tris-EDTA buffer. Approximately 100 ng of DNA from each fraction was electrophoresed on a 0.8% agarose gel and transferred to a nylon membrane as described by Lane and Archibald (2006). Southern hybridizations were performed overnight at 45–55 °C with nucleomorph, plastid, and mitochondrial ribosomal RNA (rRNA) gene probes in order to assess the relative purity of the three fractions.
Genome Sequencing, Assembly, and Annotation
Approximately 5 μg of a sample containing plastid, mitochondrial, and nucleomorph DNAs was pyrosequenced at the US Department of Energy's Joint Genome Institute (Walnut Creek) using a Roche 454 GS-FLX standard system (454 Life Sciences). The resulting sequence data were assembled using the 454 Life Sciences Newbler Assembler (v1.1.03.24). One and one half plates were run, yielding 158,500 reads with an average read length of 206 base pairs (bp). Four contigs were identified as being of plastid origin. In order to close the gaps between these contigs, exact match polymerase chain reaction (PCR) primers were designed to each contig end and used in PCRs with previously established cryptomonad plastid genome synteny used as a guide (Douglas and Penny 1999; Khan, Parks, et al. 2007). PCR products of the expected size were purified and either directly sequenced using PCR primers or cloned using the TOPO-TA PCR IV vector, the pGEM Easy vector, or the TOPO-XL vector (Invitrogen, Promega Corp), depending on size. Sequencing reactions were performed on a Beckman-Coulter CEQ 8000 capillary DNA sequencer. The integrity of the contigs generated from 454 sequence data was verified using PCR primers designed to amplify overlapping 2–4 Kbp fragments spanning the entire genome. These and additional PCR products were sequenced as necessary to resolve all ambiguous regions, such as those with stop codons and frame shifts within open reading frames (ORFs).
Sequencher 4.5 (GeneCodes Inc) was used to combine 454 contigs with Sanger sequence data. Once a single circular mapping plastid genome sequence was obtained, genes were identified using the National Center for Biotechnology Information (NCBI) ORFinder and syntenic comparisons with plastid genomes of the photosynthetic cryptomonads R. salina CCMP1319 and G. theta CCMP327. ORFs were compared with the NCBI nonredundant database using BlastX (Altschul et al. 1997). Transfer RNA (tRNA) genes were identified with trnaScan-SE (http://lowelab.ucsc.edu/tRNAscan-SE/) using the option “search for organellar tRNAs (-O).” Small and large rRNA genes were identified by BlastN. The program Tandem Repeats Finder (http://tandem.bu.edu/trf/trf.submit.options.html) was used to identify any potential repeat regions. The circular genome map was constructed using CIRDNA (http://emboss.imb.nrc.ca). G + C content and G + C skew were analyzed using Artemis 10.0 (Rutherford et al. 2000). The C. paramecium plastid genome sequence has been deposited in GenBank under accession number GQ358203.
Phylogenetic Analyses
Predicted C. paramecium proteins were initially aligned with homologs (if present) from the cyanobacterium Synechocystis sp. PCC 6803 (BA000022) and the following 18 complete plastid genomes; R. salina (NC_009573), G. theta (AF041468), Emiliania huxleyi (NC_007288), Odontella sinensis (NC_001713), Phaeodactylum tricornutum (NC_008588), Thalassiosira pseudonana (EF067921), Porphyra purpurea (NC_000925), Gracilaria tenuistipitata (NC_006137), Cyanidium caldarium (NC_001840), Cyanidioschyzon merolae (NC_004799), Cyanophora paradoxa (NC_001675), Chlamydomonas reinhardtii (NC_005353), Euglena gracilis (NC_001603), Bigelowiella natans (NC_010006), Aneura mirabilis (NC_010359), E. longa (NC_002652), Epifagus virginiana (NC_001568), and Helicosporidium sp. (NC_008100). Alignments were constructed using the ClustalW (Thompson et al. 1994) option of the MEGA 4.0 sequence alignment editor (Tamura et al. 2007), and sites containing gaps were removed. Maximum likelihood phylogenetic trees were constructed for 77 individual proteins using the RaxML black box (Stamatakis et al. 2008) with the Whelan and Goldman substitution matrix and a Gamma + Invar model (four site-rate categories). Bootstrap values were calculated using the rapid bootstrap method and CAT model with 100 replicates. The same 77 alignments were used for pairwise distance calculations of proteins encoded in the C. paramecium, G. theta, and R. salina plastid genomes using the JTT matrix in MEGA 4.0. All positions containing gaps and missing data were removed from the alignments prior to distance calculations (the “complete deletion” option).
Phylogenetic analyses of various sets of concatenated plastid proteins were also performed. Homologs from the same organisms listed above were used, with the exception of E. virginiana, A. mirabilis, and E. longa. The full set contained proteins derived from the following 22 genes: atpA, atpB, atpH, rbcL, rpl2, rpl5, rpl14, rpl16, rpl20, rpoA, rpoB, rps2, rps3, rps4, rps7, rps8, rps11, rps12, rps14, rps18, rps19, and tufA. This alignment contained 5,076 amino acid positions. We also analyzed atp synthase subunit proteins as a concatenate (atpA, B, and H; 1,035 amino acids), as well as RNA polymerase subunits (rpoA and rpoB; 1,132 amino acids), small subunit ribosomal proteins (rps2, 3, 4, 7, 8, 11, 12, 14, 18, and 19; 1,329 amino acids), and large subunit ribosomal proteins (rpl2, 5, 14, 16, and 20; 728 amino acids). Phylogenetic analyses of concatenated protein alignments were performed as above except 300 rapid bootstrap replicates were performed.
Data Deposition
The C. paramecium plastid genome sequence has been deposited in GenBank under the following accession number: GQ358203.
Results and Discussion
General Features of the C. paramecium Plastid Genome
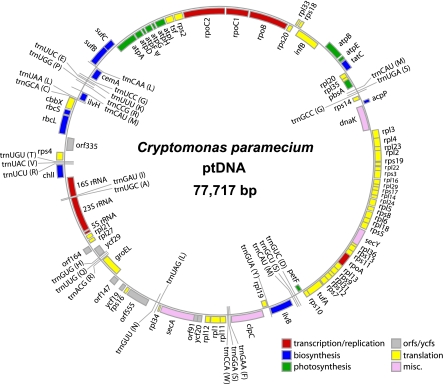
The plastid genome of the nonphotosynthetic cryptomonad C. paramecium is 77,717 bp in size (fig. 1), substantially smaller than those of its photosynthetic relatives R. salina (135,854 bp; Khan, Parks, et al. 2007) and G. theta (121,524 bp; Douglas and Penny 1999; table 1). It is nevertheless one of the least reduced nonphotosynthetic plastid genomes to be sequenced thus far. Red algal and red algal-derived plastid genomes tend to be more gene-rich than their green algal counterparts (McFadden 2001), although the smallest known plastid genomes are from the red secondary plastids of nonphotosynthetic apicomplexans such as the ∼35 Kbp “apicoplast” genome of the malaria parasite Plasmodium (Wilson et al. 1996; Waller and McFadden 2005). The C. paramecium genome presented here is not reduced in size or gene content (see below) to such an extent, being larger than that of the green algal pathogens Helicosporidium sp. (37.4 Kbp; de Koning and Keeling 2006) and Prototheca wickerhamii (54.1 Kbp; Knauf and Hachtel 2002) but comparable in size to the plastid genomes of the parasitic plant E. virginiana (70.0 Kbp; Depamphilis and Palmer 1990) and the free-living euglenoid E. longa (73.3 Kbp; Gockel and Hachtel 2000). The genome of Helicosporidium sp. has undergone a 4-fold reduction compared with its photosynthetic green algal relative Chlorella vulgaris, P. wickerhamii a 3-fold reduction (also relative to C. vulgaris), and E. virginiana and E. longa both around a 2-fold reduction compared with their respective closest photosynthetic relatives (as calculated by de Koning and Keeling 2006). The plastid genome of a relatively close photosynthetic relative of the apicomplexans has yet to be sequenced, although such an organism has recently been found (Moore et al. 2008; Obornik et al. 2008). The C. paramecium genome is only ∼1.5-fold smaller than the G. theta and R. salina genomes, consistent with the molecular phylogenetic analyses of Hoef-Emden (2005), which indicate that the heterotroph C. paramecium shares very recent common ancestry with photosynthetic species within the genus Cryptomonas (whose plastid genomes have yet to be investigated). Intriguingly, this same study concluded that colorless heterotrophs have evolved within Cryptomonas on at least two other occasions (Hoef-Emden 2005; see below).
FIG. 1.—
A circular mapping diagram of the plastid genome of Cryptomonas paramecium CCAP977/2a. The 77,717 bp genome contains a single rRNA operon, 82 predicted protein genes, and 29 tRNA genes. Genes shown on the outside of the circle are transcribed clockwise. Annotated genes are colored according to the functional categories shown in the lower right.
Table 1.
Summary of Three Cryptomonad Plastid Genome Sequences
| Size (bp) | G + C% | PGs | tRNAs | IR? | Coding% | |
| Guillardia theta | 121,524 | 32% | 147 | 30 | Yes | 87.7% |
| Rhodomonas salina | 135,854 | 34% | 146 | 31 | Yes | 80.8% |
| Cryptomonasparamecium | 77,717 | 38% | 82 | 29 | No | 87.0% |
NOTE.—The two photosynthetic cryptomonads G. theta and R. salina are included with the nonphotosynthetic C. paramecium. The size, G + C percentage, number of protein genes (PGs), tRNAs, presence of IRs, and overall coding capacity are summarized.
Including structural RNA genes, 87.0% of the C. paramecium plastid genome sequence is predicted to be coding. This is similar to the plastid genome of the photosynthetic cryptomonad G. theta, which is 87.7% coding, and slightly higher than R. salina (80.8%; table 1). The reduced genomes of nonphotosynthetic plastids range from ∼95% coding in the apicomplexans and Helicosporidium to only 58% in the angiosperm E. virginiana. The mean intergenic distance in the C. paramecium genome is 85 bp (assigning overlapping genes a value of zero). This value is an intermediate between that seen in the parasitic plants (mean intergenic space of 135 bp) and the parasitic algae and apicomplexans (ranging from 24 to 36 bp; de Koning and Keeling 2006).
The average G + C content of the C. paramecium plastid genome is 38%, whereas the R. salina and G. theta genomes are 34% and 32% GC, respectively. As one would predict, G + C richness in C. paramecium differs between protein-coding regions and the rRNA operon, with the latter being higher in G + C content (49% G + C vs. 38% G + C). A G + C skew analysis (data not shown) reveals a marked change in direction of skew just upstream of the single rRNA operon (i.e., in the region of chlI; fig. 1). Changes in G + C skew are thought to be potential origin of replication sites (Grigoriev 1998; de Koning and Keeling 2006).
Inverted repeats (IRs) consisting of an rRNA operon (and in some cases a few additional genes) are found in most plastid genomes and may represent an ancestral feature (Stoebe and Kowallik 1999; Palmer 2003; Kim and Archibald 2009). Indeed, such repeats are present in the G. theta and R. salina genomes (Douglas and Penny 1999; Khan, Parks, et al. 2007), as well as in the genomes of other red secondary plastid-containing algae (e.g., the haptophyte E. huxleyi [Sanchez Puerta et al. 2005] and several diatoms [Oudot-Le Secq et al. 2007]). Interestingly, the C. paramecium genome lacks this arrangement, containing only a single rRNA operon in a 16S-trnI-trnA-23S-5S configuration (fig. 1 and table 1). To confirm that the apparent absence of an IR was not a genome assembly artifact, PCR amplicons were generated to verify the region around the single rRNA operon. The resulting products were identical to what would be predicted from our genome assembly (data not shown).
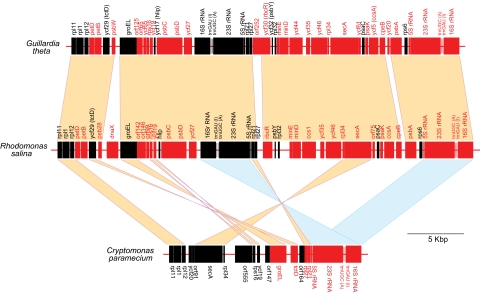
Despite their widespread distribution, rRNA operon-containing IRs are occasionally lost or rearranged (Kim and Archibald 2009). In the photosynthetic green secondary plastid-containing chlorarachniophyte alga B. natans, for example, an inversion has occurred such that within each repeat, the 16S rRNA gene is on the opposite strand as the 23S and 5S genes (Rogers et al. 2007). In the parasitic green alga Helicosporidium sp., the IR has been lost and the remaining rRNA operon split up such that the 16S gene resides on the opposite side of the genome from the 23S and 5S loci (de Koning and Keeling 2006). In the case of cryptomonads, it is interesting that although a high degree of synteny exists between the G. theta and R. salina genomes in the regions surrounding the IR, the C. paramecium genome has undergone inversions and gene losses in precisely this area (fig. 2). It thus seems reasonable to speculate that the loss of the IR in C. paramecium was associated with the intense genome reduction and compaction that accompanied the loss of photosynthesis. More plastid genome sequences, in particular from photosynthetic members of the genus Cryptomonas, will be needed to more accurately pinpoint when and how this occurred.
FIG. 2.—
Representative inversions and deletions in the plastid genome of Cryptomonas paramecium. A large region of gene synteny shared between the genomes of the photosynthetic cryptomonads Guillardia theta and Rhodomonas salina is shown aligned with the single ribosomal operon-containing region of the C. paramecium genome. Genes in black are oriented left-to-right, those in red right-to-left. Blocks of synteny are highlighted, as are inversions between C. paramecium and G. theta/R. salina. tRNA genes are not shown for simplicity, with the exception of those within the rRNA operon.
Gene Content and Synteny
Compared with its closest cryptomonad relatives, the C. paramecium plastid genome has a slightly reduced tRNA gene set. The G. theta genome has 30 tRNAs, R. salina has 31, and C. paramecium has 29. This number is still larger than the minimal set of tRNAs found in the parasitic alga Helicosporidium; C. paramecium has redundant isotypes for the amino acids glycine, serine, arginine, and leucine as well as three distinct methionine tRNAs. Just as in the cryptomonads R. salina and G. theta, as well as in Helicosporidium sp., a minimum set of tRNAs would seem to preclude the requirement for the C. paramecium plastid to import tRNAs from outside the organelle.
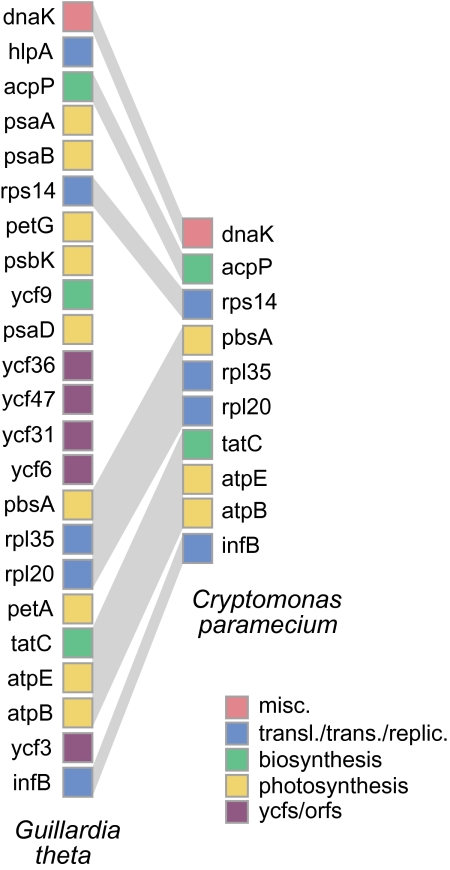
The C. paramecium plastid genome contains 82 predicted protein genes (supplementary table S1, Supplementary Material online, see below). Gene order is generally well conserved between the three cryptomonad plastid genomes, with more than 75% of the C. paramecium genome being demonstrably syntenic to the G. theta and R. salina genomes (representative regions are shown in figs. 2 and 3). This includes large tracts of complete gene order conservation, such as the highly conserved, coexpressed ribosomal protein genes and the atp gene cluster (fig. 1). Overall, there are 71 protein genes present in the G. theta and/or R. salina plastid genomes that are missing in C. paramecium. Higher level synteny is nevertheless often retained (e.g., fig. 3). Four C. paramecium ORFs share no similarity to sequences in GenBank (orf91, orf555, orf147, and orf164) and another ORF (orf335) shares similarity with other cryptomonad ORFs but no other sequences. Eight genes are missing in both the C. paramecium and G. theta plastid genomes compared with R. salina: these include dnaX, which was shown previously to be the product of lateral gene transfer and thus far appears limited to R. salina and other Rhodomonas species (Khan, Parks, et al. 2007), orf75, orf142, orf146, and ycf26, as well as a gene encoding a putative reverse transcriptase. The three light-independent protochlorophyllide reductase pseudogenes (chlB, chlN, and chlL) in R. salina are also absent. Ycf20 is shared between C. paramecium and G. theta but absent in R. salina, and there are no genes shared between the R. salina and C. paramecium genomes that are absent in G. theta.
FIG. 3.—
Gene loss and maintenance of synteny in the Cryptomonas paramecium plastid genome. A syntenic region of the genome shared between the photosynthetic cryptomonad Guillardia theta (left) and the nonphotosynthetic cryptomonad C. paramecium (right) is shown. Genes are colored according to their functional category. The diagram represents approximately 10 kbp of the C. paramecium genome (individual genes are not shown to scale).
Compared with the 26 rpl genes in G. theta, there are 25 genes for 50S ribosomal subunit proteins in the C. paramecium plastid genome. Seventeen of the eighteen 30S ribosomal protein genes present in G. theta are also found in C. paramecium. The rps6 gene, nestled next to one of the rRNA operons in both G. theta and R. salina, is conspicuously absent in the C. paramecium plastid DNA, as is the gene for rpl32 (fig. 2). As in other plastid genomes, and as noted above, many of the ribosomal protein genes in C. paramecium occur in operons, the largest being a ∼15-Kbp stretch containing 26 ribosome subunit genes and 29 consecutive genes in total (the region spanning rpl3 to rps10; fig. 1).
Many of the cell/organelle division proteins encoded in the G. theta and R. salina genomes are missing from the C. paramecium plastid genome, such as hlpA (a chromatin-associated architectural protein), dnaB (a DNA helicase), minD and minE (which prevent the creation of DNA-less “minicells” during division), and ftsH (a metalloprotease involved as a protein chaperone; Simpson and Stern 2002). Other chaperone proteins like groEL and dnaK (a member of the hsp70 family) are encoded in the C. paramecium plastid genome and presumably help with protein import and folding (Wang and Liu 1991). Whereas secG (a protein translocation gene) is absent, other components of the sec transport system are maintained (secA, secY). As well, the gene encoding the sec-independent transport protein tatC is also present, as is the proteolytic degradation pathway gene clpC. The plastid in C. paramecium thus appears to have retained its ability to import necessary proteins from the cytoplasm (e.g., proteins linked to cell division) and can mediate their degradation. Whether or not the “missing” plastid genes in C. paramecium are truly absent or have simply moved to the nuclear genome and encode proteins that are targeted posttranslationally to the organelle is unclear.
The C. paramecium plastid genome possesses a nearly full complement of the atp synthase subunit genes found in the photosynthetic cryptomonads examined thus far (supplementary table S1, Supplementary Material online). These 6 genes are present in the heterotrophic green alga P. wickerhamii but are absent in the nonphotosynthetic euglenid E. longa, the parasitic alga Helicosporidium sp., the parasitic plant Epifagus virgianiana, and the apicomplexan Plasmodium falciparum. These genes have varying degrees of sequence conservation between the three cryptomonads, with atpF and atpG being particularly highly divergent. Indeed, we have designated atpF as a pseudogene: It lacks an obvious start codon and is truncated at its 5′ and 3′ ends relative to the G. theta and R. salina genes.
Photosynthetic Genes
Not surprisingly, the bulk of the gene loss in the C. paramecium genome has occurred in the category of photosynthesis (supplementary table S1, Supplementary Material online). The gene encoding the β subunit of phycoerythrin (cpeB), which is part of the phycobiliprotein complex in cryptomonads, is present in R. salina and G. theta (Douglas and Penny 1999; Khan, Parks, et al. 2007) but missing in C. paramecium as well as in the genomes of the photosynthetic heterokonts and haptophytes (Sanchez Puerta et al. 2005; Oudot-Le Secq et al. 2007). The photosynthetic regulator and electron transfer gene ftrB is absent in the plastid genome of C. paramecium, as is the hlip gene (also known as ycf17). As in E. longa and other dramatically reduced plastid genomes, an rbcL gene encoding the large subunit of ribulose 1,5-bisphosphate carboxylase/oxygenase is present, presumably functioning in a nonphotosynthetic capacity (Wickett et al. 2008), as well as the small subunit rbcS gene.
The psb and psa gene families encode protein subunits of photosystem II (PSII) and photosystem I, respectively. In the two photosynthetic cryptomonad plastid genomes sequenced thus far, there are a total of 18 psb genes (Douglas and Penny 1999; Khan, Parks, et al. 2007), all of which are absent in C. paramecium. In the parasitic liverwort A. mirabilis, five of the psb genes are pseudogenes (Wickett et al. 2008), and in the substantially reduced genome of the parasitic plant E. virginiana, which is thought to have lost photosynthesis earlier than A. mirabilis, all PSII genes are gone except for psbA and psbB pseudogenes (Depamphilis and Palmer 1990). It is hypothesized that the psb family of genes is the first to disappear from a nonphotosynthetic plastid (an exception being the ndh genes, if they are present [Wickett et al. 2008]). The E. longa plastid genome is also completely devoid of the psb gene family, which is in contrast to its closest photosynthetic relative E. gracilis, which has 11 psb genes (Hallick et al. 1993). In total, the loss of 18 psb genes in C. paramecium accounts for approximately 7.5 Kbp of missing plastid DNA. Similarly, although there are 11 psa genes in R. salina and G. theta (both photosynthetic cryptomonads), none are present in C. paramecium. The E. longa plastid genome has lost all 5 psa genes compared with its photosynthetic relative E. gracilis. The parasitic plant E. virginiana has also lost all five genes, and whereas four intact psa genes remain in A. mirabilis, there are also two pseudogenes (Wickett et al. 2008).
The last set of photosynthesis-related genes that have nearly entirely disappeared from C. paramecium is the pet family. In photosynthetic organisms, pet proteins create a complex required for oxygenic photosynthesis, in particular the noncyclic electron flow mediated by the cytochrome b6f complex. Eight pet genes are present in the G. theta and R. salina plastid genomes but in secondarily nonphotosynthetic organisms (e.g., E. virginiana, E. longa, and A. mirabilis), the pet genes are all either missing or have become pseudogenes (supplementary table S1, Supplementary Material online). Similarly, all the pet genes have been lost in C. paramecium, with the curious exception of petF. The petF coding region appears to be intact and the predicted protein is surprisingly well conserved, sharing 76% amino acid sequence identity with the R. salina and G. theta homologs, and >60% identity with homologs in diatoms, haptophytes and even cyanobacteria. The function(s) of a stand-alone petF gene in C. paramecium is not obvious. In Synechococcus, the petF gene encodes a ferredoxin to shuttle electrons while cross-linked to the psaD and psaE gene products, neither of which are encoded in the C. paramecium plastid genome but could conceivably be nucleus encoded.
Metabolic Shift, Codon Usage, and Increased Evolutionary Rate in C. paramecium
Increased substitution rates and base composition biases have been observed in the organellar genomes of plants and algae that have switched modes of nutrition, but it is often unclear whether a change from autotrophy to heterotrophy is the cause of these genetic changes or a result of them. Depamphilis and Palmer (1990) suggested that the latter may be true, that is, genetic changes precede the loss of photosynthesis. In cryptomonads, evidence in support of this notion comes from an analysis by Hoef-Emden et al. (2005) where an accelerated evolutionary rate was observed in plastid rbcL and nucleomorph 18S rRNA genes in both photosynthetic and nonphotosynthetic members of the genus Cryptomonas, including C. paramecium. In rbcL, rate accelerations were found to correlate with a change in codon usage from an “adaptive” pattern to a “mutational” pattern in the 2-fold degenerate NNY codons of asparagine, aspartate, histidine, phenylalanine, and tyrosine residues, that is, a shift from NNC to NNU codons in the direction of overall genome composition bias (Hoef-Emden et al. 2005). This was attributed to relaxed evolutionary constraints and reduced expression level, with the most extreme codon usage shifts and rate accelerations observed in the recently diverged heterotrophic taxa, such as C. paramecium.
We examined the codon usage of all 82 predicted plastid protein genes in C. paramecium and compared it with that seen in the photosynthetic cryptomonads R. salina and G. theta (supplementary table S2, Supplementary Material online). As is the case for both photosynthetic species, and as one might expect in a somewhat G + C-poor organellar genome (38% G + C), codons in the C. paramecium genome are generally biased toward A and T residues at degenerate sites. However, this bias is markedly less striking in C. paramecium than in the R. salina and G. theta genomes. For example, when 4-fold degenerate glycine codons are considered, 65.4% are GGA or GGU in C. paramecium (872 of 1,333 in total) compared with 85.9% GGA/GGU in G. theta (1826/2126) and 80.9% in R. salina (1843/2279; supplementary table S2, Supplementary Material online). When third codon positions as a whole are examined, the C. paramecium genome is considerably more G + C rich: 31.0% G + C compared with only 18.3% in G. theta and 23.1% in R. salina. The 2-fold degenerate codons mentioned above show the same pattern. This is most apparent in the case of histidine, where 46.0% are CAC codons in C. paramecium (205/426 in total) compared with only 27.7% in G. theta and 35.8% in R. salina, as well as with aparagine, where 40.0% of the codons are GAC in C. paramecium (328/820) compared with 16.9% in G. theta and 23.5% in R. salina (supplementary table S2, Supplementary Material online). This pattern is opposite to that seen when the rbcL gene is examined in isolation (Hoef-Emden et al. 2005) and is consistent with the higher overall G + C content of the C. paramecium plastid genome (38%) relative to G. theta (32%) and R. salina (34%). Third codon position G + C content does not vary greatly from gene to gene in C. paramecium, even when loci that might be expected to be evolving under reduced evolutionary constraints are considered. For instance, the single remaining pet gene, petF, is 27.8% G + C at the third codon position in C. paramecium, with G. theta and R. salina being 13.4% and 14.4% G + C, respectively. This compares with 27.7% and 25.7% third position G + C for the C. paramecium rbcL and rbcS genes, respectively, 31.3% G + C for the atp gene cluster (atpA, D, G, H, and I analyzed together) and 32.5% G + C for the rpl gene family taken as a whole.
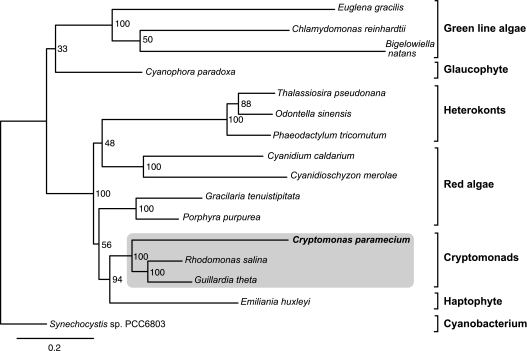
To gain further insight into the question of genome evolution and sequence divergence in nonphotosynthetic cryptomonads relative to photosynthetic species, we constructed maximum likelihood phylogenetic trees for 77 proteins encoded in all three sequenced cryptomonad genomes (i.e., G. theta, R. salina, and C. paramecium), as well as a variety of additional algae, where possible. Curiously, the cryptomonad sequences were monophyletic in only 40 of 77 trees (data not shown). Upon close examination, no convincing instances of “recent” lateral gene transfer were detected beyond the noncyanobacterial type rpl36 gene identified by Rice and Palmer (2006), which appears to be a feature of cryptomonads as a whole. Instead, the anomalous phylogenetic position of C. paramecium proteins in many of our trees appears simply to be the result of “long-branch attraction.” The C. paramecium branches were very often markedly longer than those of R. salina and/or G. theta, and in cases where a C. paramecium homolog did not branch with G. theta and R. salina, the C. paramecium sequence was particularly highly divergent. With the exception of the atp genes, there was no obvious pattern to the types of genes/proteins that were exceptionally divergent in sequence (see below). On the basis of pairwise distance calculations alone, the evolutionary divergence between C. paramecium and G. theta/R. salina was invariably greater than between G. theta and R. salina (supplementary table S1, Supplementary Material online).
We next analyzed a supermatrix of 22 broadly distributed proteins (see Materials and Methods) in order to assess the relative branching order of C. paramecium, G. theta, and R. salina, as well as overall sequence divergence. In this phylogeny (fig. 4), the three cryptomonads branched together with high statistical support, as did G. theta and R. salina to the exclusion of C. paramecium. Notably, the C. paramecium branch was more than twice as long as the R. salina and G. theta branches. The same pattern was observed when the atp synthase proteins, RNA polymerase subunits, small subunit ribosomal proteins, and large subunit ribosomal proteins were analyzed as individual supermatrices (data not shown). In the case of the atpA/B/H concatenate, the C. paramecium branch was >4 times as long as that of G. theta or R. salina. In sum, it would appear that most of the proteins encoded in the C. paramecium plastid genome—including “housekeeping” proteins involved in transcription and translation—have evolved under reduced and/or different selective constraints in C. paramecium compared with the photosynthetic cryptomonads G. theta and R. salina. Genomic data from additional nonphotosynthetic and photosynthetic members of the genus Cryptomonas will be necessary to explore this issue in a more systematic fashion.
FIG. 4.—
Maximum likelihood phylogenetic tree showing the position of the nonphotosynthetic cryptomonad Cryptomonas paramecium. The tree was constructed using RaxML from a supermatrix of 22 proteins and 5,076 unambiguously aligned amino acid positions (see Materials and Methods). Rapid bootstrap values (300 replicates) are shown. The primary and secondary plastid-containing algae are rooted with a single cyanobacterial sequence. The scale bar shows the inferred number of amino acid substitutions per site.
Genome Reduction
Why does C. paramecium retain a plastid at all? One of the first complete genes isolated from the G. theta plastid genome was for an acyl carrier protein called acpP (sometimes annotated as acpA). It is a required cofactor in the synthesis and metabolism of fatty acids (Wang and Liu 1991), and significantly, it is present in the plastid genome of C. paramecium presented here. Further evidence for fatty acid biosynthesis occurring in the C. paramecium plastid comes from the recent discovery of a nuclear gene for the plastid-targeted protein fabD in a previous small-scale genome sequence survey (Khan, Kozera, et al. 2007). FabD encodes the malonyl Co-A:ACP transcylase protein catalyzing the transfer of a malonyl moiety in fatty acid synthesis and given what is known about the essential nature of fabD and other fatty acid biosynthetic genes in other secondarily nonphotosynthetic organisms (Waller and McFadden 2005; Barbrook et al. 2006), it seems reasonable to predict that fatty acid biosynthesis is an essential plastid pathway in cryptomonads. Furthermore, the maintenance of sufB and sufC in the C. paramecium plastid genome suggests a role in iron-sulfur cluster assembly, which is also suggested to occur in the Prototheca plastid (Borza et al. 2005). The presence of the chlI gene in the nonphotosynthetic C. paramecium plastid genome may provide additional insight into the role of this magnesium chelatase component in plastid-to-nucleus signaling (Nott et al. 2006).
Overall, the presence of shared plastid genes and plastid-targeted proteins in a wide array of primary and secondary plastid-bearing organisms is indicative of a nonrandom retention of metabolic processes. In the case of genome structure, de Koning and Keeling (2006) suggest that nonphotosynthetic plastid genomes may be the result of convergence upon a shared set of traits. They refer to the common outcome of genome reduction, with a shift in coding strand symmetry and tRNA complement in Helicosporidium sp. (green, primary) and apicomplexans (red, secondary) as “organized reduction.” In this sense, the C. paramecium genome would seem to be something of an intermediate (along with those of E. virginiana and E. longa) as being roughly the same structure as their photosynthetic counterparts, just more reduced. Examples of heterotrophs further along the continuum toward full functionality are the nonphotosynthetic angiosperms whose plastids are still in the initial phase of losing genes through large-scale deletions and pseudogenization (Wickett et al. 2008).
Conclusion
The complete C. paramecium plastid genome presented in this report is the first red algal-derived complex plastid from a free-living organism that has lost its ability to photosynthesize. The field of comparative genomics of secondarily nonphotosynthetic plastids is in its infancy and has largely consisted of sequences from plants (Krause 2008). The addition of the C. paramecium genome to the suite of complete plastid genome sequences increases the breadth of plastid genomes sampled to date and will help to identify some common trends present in highly reduced organellar genomes. There does indeed appear to be a “structured reduction” occurring in these plastid genomes, regardless of origin or complexity of the plastid (de Koning and Keeling 2006).
Although the niche of nonphotosynthetic plastid genome analysis is expanding, there remains a wealth of information to be mined. In the cryptomonads alone, it appears that, as in land plants, a nonphotosynthetic lifestyle has evolved multiple times (Hoef-Emden 2005), and systematic investigation of diverse members of this lineage presents an opportunity to discover larger trends in genome streamlining under such conditions. Expression levels of proteins encoded in the cryptomonad plastid genome have yet to be explored but once undertaken will likely provide much valuable information on the functional significance (if any) of residual photosynthesis-related genes in newly evolved heterotrophs. Looking outside cryptomonads, the number of nonphotosynthetic plastid genomes sequenced that are of secondary or tertiary origin is still very small. As organisms with secondary plastids are abundant in the marine environment and are hugely successful colonizers of a wide variety of ecological niches (Kim and Archibald 2009), it is likely that we have barely scratched the surface of genome sequences from both parasitic and free-living nonphotosynthetic organisms. Determining what genes are maintained in nonphotosynthetic plastids may yield insight into the function(s) of some of the unidentified proteins encoded in their genomes.
A better understanding of genome reduction associated with a drastic functional shift (such as the loss of photosynthesis) may also help answer the question “can a plastid be lost once it has been acquired?” This question is central to many currently proposed hypotheses dealing with the origin and spread of secondary and tertiary plastids (Archibald 2009). The exact number of times such events have happened, and how many lineages were involved, is still unclear. Increasing our knowledge regarding the continuum of photosynthetic ability may yield clues as to whether some members of the “chromalveolate” supergroup, for example, did at one time contain a plastid. Such knowledge should ultimately contribute to a greater comprehension of the processes behind the acquisition and loss of photosynthesis—one of the most influential metabolic developments on Earth.
Supplementary Material
Supplementary tables S1 and S2 are available at Genome Biology and Evolution online (http://www.oxfordjournals.org/our_journals/gbe/).
Supplementary Material
Acknowledgments
We thank H. Khan for providing alignments of plastid proteins, R. Eveleigh and A. Roger for assistance with phylogenetic analysis, and M. Schnare for help with rRNA gene prediction. K. Hoef-Emden is also thanked for discussion of codon usage patterns in cryptomonad plastid genomes. J.M.A. acknowledges support from the Canadian Institute for Advanced Research, Integrated Microbial Biodiversity Program, as well as a Canadian Institutes of Health Research (CIHR) New Investigator award. This work was supported by an operating grant from the CIHR Regional Partnership Program, together with the Nova Scotia Health Research Foundation (ROP85016).
References
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archibald JM. Nucleomorph genomes: structure, function, origin and evolution. Bioessays. 2007;29:392–402. doi: 10.1002/bies.20551. [DOI] [PubMed] [Google Scholar]
- Archibald JM. The puzzle of plastid evolution. Curr Biol. 2009;19:R81–R88. doi: 10.1016/j.cub.2008.11.067. [DOI] [PubMed] [Google Scholar]
- Barbrook AC, Howe CJ, Purton S. Why are plastid genomes retained in non-photosynthetic organisms? Trends Plant Sci. 2006;11:101–108. doi: 10.1016/j.tplants.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Borza T, Popescu CE, Lee RW. Multiple metabolic roles for the nonphotosynthetic plastid of the green alga Prototheca wickerhamii. Eukaryot Cell. 2005;4:253–261. doi: 10.1128/EC.4.2.253-261.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning AP, Keeling PJ. The complete plastid genome sequence of the parasitic green alga Helicosporidium sp. is highly reduced and structured. BMC Biol. 2006;4:12. doi: 10.1186/1741-7007-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depamphilis CW, Palmer JD. Loss of photosynthetic and chlororespiratory genes from the plastid genome of a parasitic flowering plant. Nature. 1990;348:337–339. doi: 10.1038/348337a0. [DOI] [PubMed] [Google Scholar]
- Douglas SE, Murphy CA, Spencer DF, Gray MW. Cryptomonad algae are evolutionary chimaeras of two phylogenetically distinct unicellular eukaryotes. Nature. 1991;350:148–151. doi: 10.1038/350148a0. [DOI] [PubMed] [Google Scholar]
- Douglas SE, Penny SL. The plastid genome of the cryptophyte alga, Guillardia theta: complete sequence and conserved synteny groups confirm its common ancestry with red algae. J Mol Evol. 1999;48:236–244. doi: 10.1007/pl00006462. [DOI] [PubMed] [Google Scholar]
- Douglas SE, et al. The highly reduced genome of an enslaved algal nucleus. Nature. 2001;410:1091–1096. doi: 10.1038/35074092. [DOI] [PubMed] [Google Scholar]
- Gockel G, Hachtel W. Complete gene map of the plastid genome of the nonphotosynthetic euglenoid flagellate Astasia longa. Protist. 2000;151:347–351. doi: 10.1078/S1434-4610(04)70033-4. [DOI] [PubMed] [Google Scholar]
- Gould SB, Waller RF, McFadden GI. Plastid evolution. Annu Rev Plant Biol. 2008;59:491–517. doi: 10.1146/annurev.arplant.59.032607.092915. [DOI] [PubMed] [Google Scholar]
- Graham LE, Wilcox LW. Algae. Upper Saddle River (NJ): Prentice-Hall; 2000. [Google Scholar]
- Grigoriev A. Analyzing genomes with cumulative skew diagrams. Nucleic Acids Res. 1998;26:2286–2290. doi: 10.1093/nar/26.10.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallick RB, et al. Complete sequence of Euglena gracilis chloroplast DNA. Nucleic Acids Res. 1993;21:3537–3544. doi: 10.1093/nar/21.15.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauth AM, Maier UG, Lang BF, Burger G. The Rhodomonas salina mitochondrial genome: bacteria-like operons, compact gene arrangement and complex repeat region. Nucleic Acids Res. 2005;33:4433–4442. doi: 10.1093/nar/gki757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoef-Emden K. Multiple independent losses of photosynthesis and differing evolutionary rates in the genus Cryptomonas (Cryptophyceae): combined phylogenetic analyses of DNA sequences of the nuclear and the nucleomorph ribosomal operons. J Mol Evol. 2005;60:183–195. doi: 10.1007/s00239-004-0089-5. [DOI] [PubMed] [Google Scholar]
- Hoef-Emden K. Molecular phylogeny of the phycocyanin-containing cryptophytes: evolution of biliproteins and geographical distribution. J Phycol. 2008;44:985–993. doi: 10.1111/j.1529-8817.2008.00530.x. [DOI] [PubMed] [Google Scholar]
- Hoef-Emden K, Marin B, Melkonian M. Nuclear and nucleomorph SSU rDNA phylogeny in the Cryptophyta and the evolution of cryptophyte diversity. J Mol Evol. 2002;55:161–179. doi: 10.1007/s00239-002-2313-5. [DOI] [PubMed] [Google Scholar]
- Hoef-Emden K, Melkonian M. Revision of the genus Cryptomonas (Cryptophyceae): a combination of molecular phylogeny and morphology provides insights into a long-hidden dimorphism. Protist. 2003;154:371–409. doi: 10.1078/143446103322454130. [DOI] [PubMed] [Google Scholar]
- Hoef-Emden K, Tran HD, Melkonian M. Lineage-specific variations of congruent evolution among DNA sequences from three genomes, and relaxed selective constraints on rbcL in Cryptomonas (Cryptophyceae) BMC Evol Biol. 2005;5:56. doi: 10.1186/1471-2148-5-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan H, et al. Retrotransposons and tandem repeat sequences in the nuclear genomes of cryptomonad algae. J Mol Evol. 2007;64:223–236. doi: 10.1007/s00239-006-0088-9. [DOI] [PubMed] [Google Scholar]
- Khan H, et al. Plastid genome sequence of the cryptophyte alga Rhodomonas salina CCMP1319: lateral transfer of putative DNA replication machinery and a test of chromist plastid phylogeny. Mol Biol Evol. 2007;24:1832–1842. doi: 10.1093/molbev/msm101. [DOI] [PubMed] [Google Scholar]
- Kim E, Archibald JM. Diversity and evolution of plastids and their genomes. In: Aronsson H, Sandelius AS, editors. The chloroplast-interactions with the environment. Berlin (Germany): Springer-Verlag; 2009. pp. 1–39. [Google Scholar]
- Kim E, et al. Complete sequence and analysis of the mitochondrial genome of Hemiselmis andersenii CCMP644 (Cryptophyceae) BMC Genomics. 2008;9:215. doi: 10.1186/1471-2164-9-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauf U, Hachtel W. The genes encoding subunits of ATP synthase are conserved in the reduced plastid genome of the heterotrophic alga Prototheca wickerhamii. Mol Genet Genomics. 2002;267:492–497. doi: 10.1007/s00438-002-0681-6. [DOI] [PubMed] [Google Scholar]
- Krause K. From chloroplasts to “cryptic” plastids: evolution of plastid genomes in parasitic plants. Curr Genet. 2008;54:111–121. doi: 10.1007/s00294-008-0208-8. [DOI] [PubMed] [Google Scholar]
- Lane CE, Archibald JM. Novel nucleomorph genome architecture in the cryptomonad genus Hemiselmis. J Eukaryot Microbiol. 2006;53:515–521. doi: 10.1111/j.1550-7408.2006.00135.x. [DOI] [PubMed] [Google Scholar]
- Lane CE, et al. Insight into the diversity and evolution of the cryptomonad nucleomorph genome. Mol Biol Evol. 2006;23:856–865. doi: 10.1093/molbev/msj066. [DOI] [PubMed] [Google Scholar]
- Lane CE, et al. Nucleomorph genome of Hemiselmis andersenii reveals complete intron loss and compaction as a driver of protein structure and function. Proc Natl Acad Sci USA. 2007;104:19908–19913. doi: 10.1073/pnas.0707419104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier UG, Hofmann CJ, Eschbach S, Wolters J, Igloi GL. Demonstration of nucleomorph-encoded eukaryotic small subunit ribosomal RNA in cryptomonads. Mol Gen Genet. 1991;230:155–160. doi: 10.1007/BF00290663. [DOI] [PubMed] [Google Scholar]
- Mazumdar J, Emma HW, Masek K, Hunter CA, Striepen B. Apicoplast fatty acid synthesis is essential for organelle biogenesis and parasite survival in Toxoplasma gondii. Proc Natl Acad Sci USA. 2006;103:13192–13197. doi: 10.1073/pnas.0603391103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden GI. Second-hand chloroplasts: evolution of cryptomonad algae. In: Callow JA, editor. Advances in botanical research. London: Academic Press Limited; 1993. pp. 189–230. [Google Scholar]
- McFadden GI. Chloroplast origin and integration. Plant Physiol. 2001;125:50–53. doi: 10.1104/pp.125.1.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden GI, Gilson PR, Hill DRA. Goniomonas: rRNA sequences indicate that this phagotrophic flagellate is a close relative of the host component of cryptomonads. Eur J Phycol. 1994;29:29–32. [Google Scholar]
- Moore RB, et al. A photosynthetic alveolate closely related to apicomplexan parasites. Nature. 2008;452:900. doi: 10.1038/nature06635. [DOI] [PubMed] [Google Scholar]
- Nott A, Jung H-S, Koussevitzky S, Chory J. Plastid-to-nucleus retrograde signaling. Annu Rev Plant Biol. 2006;57:739–759. doi: 10.1146/annurev.arplant.57.032905.105310. [DOI] [PubMed] [Google Scholar]
- Obornik M, Janouskovec J, Chrudimsky T, Lukes J. Evolution of the apicoplast and its host: to autotrophy and back again. Int J Parasitol. 2008;39:1–12. doi: 10.1016/j.ijpara.2008.07.010. [DOI] [PubMed] [Google Scholar]
- Oudot-Le Secq MP, et al. Chloroplast genomes of the diatoms Phaeodactylum tricornutum and Thalassiosira pseudonana: comparison with other plastid genomes of the red lineage. Mol Genet Genomics. 2007;277:427–439. doi: 10.1007/s00438-006-0199-4. [DOI] [PubMed] [Google Scholar]
- Palmer JD. The symbiotic birth and spread of plastids: how many times and whodunnit? J Phycol. 2003;39:4–11. [Google Scholar]
- Reyes-Prieto A, Weber AP, Bhattacharya D. The origin and establishment of the plastid in algae and plants. Annu Rev Genet. 2007;41:147–168. doi: 10.1146/annurev.genet.41.110306.130134. [DOI] [PubMed] [Google Scholar]
- Rice DW, Palmer JD. An exceptional horizontal gene transfer in plastids: gene replacement by a distant bacterial paralog and evidence that haptophyte and cryptophyte plastids are sisters. BMC Biol. 2006;4:31. doi: 10.1186/1741-7007-4-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers MB, Gilson PR, Su V, McFadden GI, Keeling PJ. The complete chloroplast genome of the chlorarachniophyte Bigelowiella natans: evidence for independent origins of chlorarachniophyte and euglenid secondary endosymbionts. Mol Biol Evol. 2007;24:54–62. doi: 10.1093/molbev/msl129. [DOI] [PubMed] [Google Scholar]
- Rutherford K, et al. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- Sanchez Puerta MV, Bachvaroff TR, Delwiche CF. The complete plastid genome sequence of the haptophyte Emiliania huxleyi: a comparison to other plastid genomes. DNA Res. 2005;12:151–156. doi: 10.1093/dnares/12.2.151. [DOI] [PubMed] [Google Scholar]
- Shalchian-Tabrizi K, et al. Diversification of unicellular eukaryotes: cryptomonad colonizations of marine and fresh waters inferred from revised 18S rRNA phylogeny. Environ Microbiol. 2008;10:2635–2644. doi: 10.1111/j.1462-2920.2008.01685.x. [DOI] [PubMed] [Google Scholar]
- Simpson CL, Stern DB. The treasure trove of algal chloroplast genomes. Surprises in architecture and gene content, and their functional implications. Plant Physiol. 2002;129:957–966. doi: 10.1104/pp.010908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Stoebe B, Kowallik KV. Gene-cluster analysis in chloroplast genomics. Trends Genet. 1999;15:344–347. doi: 10.1016/s0168-9525(99)01815-6. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Heyden S, Chao EE, Cavalier-Smith T. Genetic diversity of goniomonads: an ancient divergence between marine and freshwater species. Eur J Phycol. 2004;39:343–350. [Google Scholar]
- Waller RF, McFadden GI. The apicoplast: a review of the derived plastid of apicomplexan parasites. Curr Issues Mol Biol. 2005;7:57–79. [PubMed] [Google Scholar]
- Wang SG, Liu XQ. The plastid genome of Cryptomonas f encodes an hsp70-like protein, a histone-like protein, and an acyl carrier protein. Proc Natl Acad Sci USA. 1991;88:10783–10787. doi: 10.1073/pnas.88.23.10783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickett NJ, et al. Functional gene losses occur with minimal size reduction in the plastid genome of the parasitic liverwort Aneura mirabilis. Mol Biol Evol. 2008;25:393–401. doi: 10.1093/molbev/msm267. [DOI] [PubMed] [Google Scholar]
- Wilson RJMI, et al. Complete gene map of the plastid-like DNA of the malaria parasite Plasmodium falciparum. J Mol Biol. 1996;261:155–172. doi: 10.1006/jmbi.1996.0449. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.