Abstract
Three families of proteinaceous pheromones have been described in the house mouse: androgen-binding proteins (ABPs), exocrine gland–secreting peptides (ESPs), and major urinary proteins (MUPs), each of which is thought to communicate different information. All three are encoded by large gene clusters in different regions of the mouse genome, clusters that have expanded dramatically during mouse evolutionary history. We report copy number variation among the most recently duplicated Abp genes, which suggests substantial volatility in this gene region. It appears that groups of these genes behave as low copy repeats (LCRs), duplicating as relatively large blocks of genes by nonallelic homologous recombination. An analysis of gene conversion suggested that it did not contribute to the very low or absent divergence among the paralogs duplicated in this way. We evaluated the ESP and MUP gene regions for signs of the LCR pattern but could find no compelling evidence for duplication of gene blocks of any significant size. Assessment of the entire Abp gene region with the Mouse Paralogy Browser supported the conclusion that substantial volatility has occurred there. This was especially evident when comparing strains with all or part of the Mus musculus musculus or Mus musculus castaneus Abp region. No particularly remarkable volatility was observed in the other two gene families, and we discuss the significance of this in light of the various roles proposed for the three families of mouse proteinaceous pheromones.
Keywords: house mouse, gene duplication, androgen-binding protein, pheromone, ESP, MUP
Introduction
There is a great deal of interest in mammalian communication by pheromones, and much attention has been focused on rodents. This has been especially true of the house mouse, Mus musculus, where at least three large gene families encoding proteinaceous pheromones have been described: androgen-binding proteins (ABPs), exocrine gland–secreting peptides (ESPs), and major urinary proteins (MUPs).
ABPs have been shown to mediate assortative mate selection, based on subspecies recognition that potentially limits gene exchange between subspecies where they meet (Laukaitis et al. 1997; Talley et al. 2001). There is evidence that ABP-mediated mate preference across a transect of the European mouse hybrid zone is a case of reproductive character displacement as predicted by reinforcement (Bimova et al. 2005).
ESPs constitute a newly described family of mouse proteinaceous pheromones (Kimoto et al. 2005). Female mice respond to direct facial exposure to an ESP expressed in male exorbital lacrimal glands and released into tear fluid by upregulating c-Fos and egr1 gene expression in vomeronasal sensory neurons (Kimoto et al. 2007). The same response occurs after close contact with the face or bedding of male mice, and a recombinant ESP protein stimulates electrical activity in an isolated female vomeronasal organ. The male response to similar signals is unremarkable (Kimoto et al. 2005, 2007).
The MUPs are a family of lipocalins shown to mediate female recognition of potential mates (for a review, see Hurst [2009]). For the most part, MUP genes are expressed in liver and the products are passed through the kidneys into the urine. Each adult mouse expresses a pattern of 8–14 different MUP isoforms in its urine, which is determined by its genotype and by its sex because some MUP genes show sex-limited expression (Hurst 2009). This individual recognition profile has been likened to a protein “bar code” (Robertson et al. 1996; Beynon and Hurst 2003; Armstrong et al. 2005; Cheetham et al. 2007; Logan et al. 2008). MUPs have also been implicated in male–male aggression. Chamero et al. (2007) isolated high molecular weight components of male urine that activated dissociated vomeronasal neurons and were sufficient to cause male–male aggressive behavior when painted onto previously castrated males.
The major histocompatibility complex (MHC) has also been implicated in a mechanism that reduces inbreeding by mediating assortative mate selection based on MHC genotype. It has been suggested that nine-amino acid peptide ligands bind specifically to different MHC proteins and that these ligands are released when the MHC proteins break down (for a review, see Hurst [2009]). The released ligands are filtered through the kidneys and secreted into the urine where they act as pheromones communicating information about the MHC genotype. Some researchers have pointed to difficulties with this model (Hurst 2009), and to date, the genetic basis for the putative peptide pheromones is unknown. Thus, we cannot include the MHC pheromone system in the genomic comparison reported here.
All three families of proteinaceous pheromone genes, Abps, ESPs, and MUPs, are encoded by large gene clusters in different regions of the mouse genome. The structure of the Abp linkage group is complex in part because the ABP protein is a dimer composed of an alpha subunit disulfide bridged to a beta–gamma subunit, and so two different subunit genes are required to make the subunits for a functional dimer. The Abp gene region is comprised of 64 paralogs mapping in a cluster on chromosome 7 (Laukaitis et al. 2008). Of these, 30 are Abpa genes, which encode ABP alpha subunits, and 34 are Abpbg genes, which encode the beta–gamma subunits. The majority of Abp genes occur in 27 alpha/beta–gamma pairs in a 5′-5’ orientation and numbered 1–27 (Laukaitis et al. 2008). We designate these pairs <Abpa-Abpbg> or <Abpbg-Abpa> modules, where the arrowheads point in the 3’ direction.
A group of at least 38 ESP genes, ten of which are putative pseudogenes, map in a cluster on chromosome 17 (Kimoto et al. 2007). The expressed genes encode the monomeric ESP pheromones. The gene family encoding the monomeric MUPs consists of at least 40–42 genes (there are still gaps in this region of the mouse genome), half of which are pseudogenes. All of these map in a cluster on chromosome 4 (Logan et al. 2008; Mudge et al. 2008). As in the case of the Abp region, this cluster of genes is relatively complex, in that the center of the linkage group consists of at least 15 gene pairs, each containing a gene and a pseudogene in 5′-5’ orientation (Logan et al. 2008).
Each of these gene families, Abp, ESP, and MUP, has expanded dramatically during house mouse evolutionary history (Kimoto et al. 2007; Laukaitis et al. 2008; Logan et al. 2008), and two questions that fascinate us are: What mechanism(s) is responsible for the rapid expansions of these families of pheromone genes? Has the same mechanism caused the expansion of all three gene families?
We suspect that the process responsible for this gene family expansion may have resulted in copy number variation (CNV) through recent gene birth (duplication) and death (deletion). It is also possible that some events in Abp evolutionary history, and potentially in the evolutionary histories of the other two families, have been obscured by gene conversion. In undertaking this study, it was our objective to determine the mechanism(s) by which duplication has produced the complex family of Abp genes (Emes et al. 2004 and Laukaitis et al. 2008). We also compared the ESP and MUP gene regions studied by others for evidence of the same mechanisms operating in the Abp region. We evaluated the three pheromone gene regions for signs of duplication as low copy repeats (LCRs; Lupski 1998; Stankiewicz and Lupski 2002) that have duplicated by nonallelic homologous recombination (NAHR; Shaffer and Lupski 2000; Stankiewicz and Lupski 2002), and we evaluated volatility in all three regions, using the Mouse Paralogy Browser (She et al. 2008). One of them, the Abp region, shows substantial variation among a number of mouse strains. We especially noted an extensive history of these differences in wild-derived M. m. musculus strains, in classical inbred strains with part or all of an M. m. musculus Abp region, and in one wild-derived M. m. castaneus strain. No particularly remarkable volatility was observed in the other two gene families, and we discuss the significance of this in light of the various roles proposed for the three families of mouse proteinaceous pheromones.
Materials and Methods
Identifying Sequence Similarity and Repeated Blocks of Microsatellites Interspersed among Abp Genes
The microsatellites (di-nucleotide and tri-nucleotide repeats) interspersed among the most recently duplicated Abp genes described above were studied with the UCSC browser (Karolchik et al. 2003); http://www.genome.ucsc.edu). The microsatellites were visually scanned for repeated patterns, and the areas corresponding to the repeated microsatellites were matched to the Abp linkage map to look for corresponding repeated patterns of Abp genes and pseudogenes. The complete DNA of the genome region corresponding to each of the other two mouse pheromone gene families was divided into runs of 0.5 Mb, and the microsatellites scanned for repeat patterns.
DNA sequences corresponding to the putatively repeated blocks of the Abp genes (described above) were obtained with the UCSC genome browser and aligned using DNAsis Max (Hitachi). In a similar fashion, 35-kb sequences on either side of the blocks were obtained and aligned. Maps of repeat elements in the Abp gene blocks and in the flanking regions were obtained with the UCSC genome browser and aligned with each other and with the DNA sequence alignments described above. These were used to construct figures of these alignments and maps.
Detecting Gene Conversion
Sequences were aligned with DNAsis Max (Hitachi). The program GENECONV (http://www.math.wustl.edu/∼sawyer/geneconv/) was used to search for gene conversion tracks. GENECONV seeks aligned DNA or protein segments for which a pair of sequences is sufficiently similar to suggest that gene conversion occurred. These are classified as inner or outer fragments. Inner fragments are evidence of a possible gene conversion event between ancestors of two sequences in the alignment. Outer fragments are runs of unique sites that may be evidence of past gene conversion events that originated from outside of the alignment or else from within the alignment but such that evidence of the source has been destroyed by later mutation or gene conversion (see http://www.math.wustl.edu/∼sawyer/geneconv/gconvdoc.html#AssessSig). GC content of the Abp region was determined using an online calculator provided by EnCore Biotechnology, Inc. (http://www.encorbio.com/protocols/Nuc-MW.htm).
Testing the Abp Gene Family for CNV
Multiplex ligation probe assay (MLPA) was used to search for CNV in the most recently duplicated Abp genes: a9p/a14p/a16p (Abp genes a9, a14, and a16, all of which are pseudogenes [p]) and a10/a15/a17 (none of these Abpa genes are pseudogenes). Essentially, this technique relies on specific binding between a pair of adjacent probes and target DNA. If binding is exact, a high temperature ligase joins the probes and this ligated product serves as a template for subsequent rounds of amplification with primers complementary to sequences on the probe ends (Schouten et al. 2002). Analysis was performed on a microsatellite platform. Template was the limiting component, and quantitation was based on comparing experimental peak size to the peak sizes in a panel of single-copy controls. Due to the extreme similarity among the members of set a9p, a14p, a16p, a single probe had to be used in MLPA analysis, and the same was true of set a10, a15, a17. Genomic DNA samples for this study came from Jackson Laboratory.
Protocols were modified from MRC-Holland (www.MRC-Holland.com), and probe pairs were designed to differentiate paralogs by including a minimum of five nucleotides different from their nearest match. Critically, the 5’ nucleotide of the right probe, which was phosphorylated for subsequent ligation, was unique for each probe set. Probes included a complementary region, a 2–4 bp linker segment, and a primer-binding sequence. The probe pair lengths are designed to vary, when ligated, by at least two base pairs so peaks can be easily distinguished with microsatellite analysis.
Assessing Genome Region Volatility
We used the Mouse Paralogy Browser (She et al. 2008) to assess volatility in the three pheromone family gene regions of the mouse genome (fig. 6). The coordinates for the boundaries of each gene cluster were entered, and the patterns examined for insertions and deletions. For each strain shown, the upper gray bar represents the locations of the probes. In the lower bars, blank areas represent no change from the C57BL/6 control (the mouse genome); dark and light blue bars represent insertions at P ≥ 0.9 and P ≥ 0.8, respectively; red and yellow bars represent deletions at P ≥ 0.9 and P ≥ 0.8, respectively. Black bars are insertions and deletions below those confidence levels.
FIG. 6.—
Comparison of volatility in the three pheromone gene families. The Mouse Paralogy Browser (She et al. 2008) was used to search for volatility occurring by insertion and deletion in all three gene regions. For each strain shown, the upper gray bar represents the locations of the probes. In the lower bars, blank areas represent no change from the C57BL/6 control (the mouse genome); dark and light blue bars represent insertions at P ≥ 0.9 and P ≥ 0.8, respectively; red and yellow bars represent deletions at P ≥ 0.9 and P ≥ 0.8, respectively. Black bars are insertions and deletions below those confidence levels.
Results
Blocks of Abp Genes Behave as LCRs
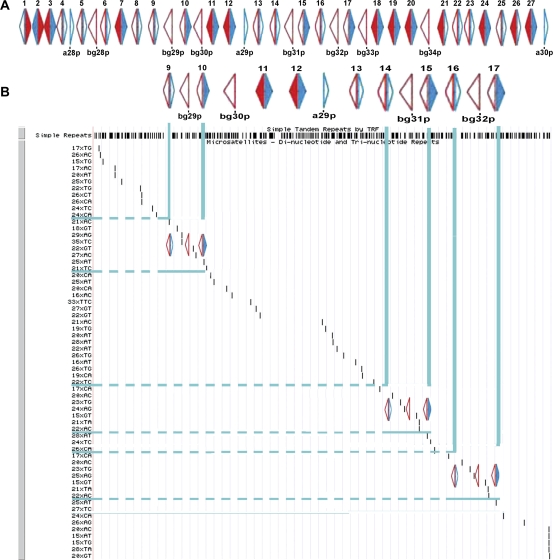
To search for duplicated blocks of <Abpbg-Abpa> modules, we studied patterns of microsatellites (di-nucleotide and tri-nucleotide repeats) in segments of the Abp region of the mouse genome where very recent duplication has occurred. We scanned these for similar or identical groups of microsatellites interspersed among the Abp genes, focusing on the region from bg9p (p=pseudogene) to a17 because a9p, a14p, a16p, and a10, a15, a17 appear to be sets of very recently duplicated paralogs (Laukaitis et al. 2008). Figure 1 shows that, with the exception of 24xAG versus 25xAG, the repeats 17xCA, 20xAC, 23xTG, 15xGT, 21xTA, and 22xAC are exactly duplicated in the segment beginning with bg14p and ending with a15 (hereinafter abbreviated 14-(31)-15) and the segment beginning with bg16p and ending with a17 (abbreviated 16-(32)-17). In the foregoing abbreviations, numbers outside parentheses represent <Abpbg-Abpa> modules, whereas those inside parentheses are single Abpbg pseudogenes falling between two modules.
FIG. 1.—
Use of microsatellite patterns to search for duplicated blocks of Abp genes. Microsatellites interspersed among the most recently duplicated Abp genes were analyzed with the UCSC genome browser. Di-nucleotide and tri-nucleotide repeats identified in the genome region were visually scanned for repeated patterns. Panel A: the complete map of Abp genes modified from Laukaitis et al. (2008). Blue arrows depict Abpa genes and red arrows depict Abpbg genes; solid filled arrows are potentially expressed genes, whereas open arrows are putative pseudogenes. <Abpbg-Abpa> modules are numbered 1–27 above the linkage map, whereas individual genes are numbered below the map. Panel B: the portion of the complete map that shows bg9p to a17 for alignment of microsatellites and Abp genes. Horizontal blue bars delineate the areas corresponding to the repeated microsatellites, and these were matched to the Abp linkage map (vertical blue bars) to look for corresponding repeated patterns of Abp genes and pseudogenes. Blocks identified in this fashion include <bg9p-a9p>-<bg29p-<bg10p-a10>, <bg14p-a14p>-<bg31p-<bg15p-a15>, and <bg16p-a16p>-<bg32p-<bg17p-a17>.
Interestingly, the most closely related Abp paralog sets (14-(31)-15 and 16-(32)-17), identified from these two nearly identical microsatellite repeat regions, have similar Abp paralog structures, two <Abpbg-Abpa> modules flanking an Abpbg pseudogene and, from left-to-right, their paralogs have identical nucleotide sequences (e.g., bg14p is identical to bg16p and a14p is identical to a16p, etc. [Laukaitis et al. 2008]). When we compared the microsatellites in the <bg9p-a9p>-<bg29p-<bg10p-a10> (abbreviated 9-(29)-10) with the 14-(31)-15 and 16-(32)-17 segments, we found a strikingly similar but not identical pattern of microsatellites.
Based on the above observation, we aligned the three segments to look for sequence identity and for similarities in other repeat elements (short interspersed nuclear elements [SINEs], long interspersed nuclear elements [LINEs], long terminal repeats [LTRs], etc.). Figure 2 shows that all three segments share an overwhelming majority of their sequences over their ∼150-kb lengths and have strikingly similar patterns of repeat elements interspersed among the Abp paralogs they contain. They also have similar patterns of single nucleotide polymorphisms. It appears that duplication of the 14-(31)-15 and 16-(32)-17 blocks was so recent as to result in essentially identical, adjacent ∼150-kb sequences in the mouse genome.
FIG. 2.—
Alignment and structure of LCRs that duplicated as blocks of <Abpbg-Abpa> modules. Panel A: alignment of the three blocks; yellow shading indicates nucleotides identical in all three segments; green indicates identity in 2 of the 3. Panel B: structures of the repeat elements (SINES, LINES, LTRs, etc.) from the UCSC genome browser.
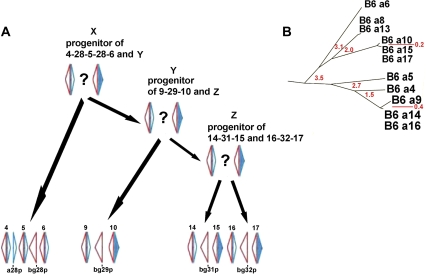
Inspection of the Abp linkage group and the intron tree published by Laukaitis et al. (2008) also suggested that a fourth block, containing <bg4p-a4p>-a28p>-<bg5p-a5p>-<bg28p-<bg6p-a6p> (abbreviated 4-(a28)-5-(bg28)-6, where all are pseudogenes), is an ancestor of the other three described above. Panel A of figure 3 shows our proposal for duplication of these four blocks, and panel B shows a portion of the intron tree excerpted from Laukaitis et al. (2008) that supports the proposed relationships in our model. We also aligned the 35-kb regions on either side of the four gene blocks and found significant sequence identity for at least 5 kb immediately adjacent to both sides of all four (SF1). We conclude that these segments have duplicated as LCRs (see Lupski 1998; Stankiewicz and Lupski 2002), probably because the similar repeats on either side of the <Abpbg-Abpa> modules make them susceptible to NAHR (Shaffer and Lupski 2000; Stankiewicz and Lupski 2002).
FIG. 3.—
Model proposed to explain the LCR relationship of the blocks of <Abpbg-Abpa> modules. Panel A: the model. The hypothetical progenitor blocks X, Y, and Z are each shown with two <Abpbg-Abpa> modules flanking a question mark. The blocks containing numbered modules and single genes represent the duplication products that now have <bg4p-a4p>, <bg9p-a9p>, <bg14p-a14p>, <bg16p-a16p> on their left flanks and <bg6p-a6p>, <bg10p-a10>, <bg15p-a15>, and <bg17p-a17> on their right flanks. We propose that three duplications, represented by three sets of black arrows, account for the NAHR-mediated duplications that produced the four related blocks of Abp genes shown. Panel B: a portion of the intron tree, modified from Laukaitis et al. (2008), shown for reference to the relationships among the Abp genes in the model (not including B6 a8 and B6 a13). B6 is an abbreviation of C57BL/6, the strain that provided the sequences for the mouse genome; red numbers are estimates of ages of duplications in MYR.
Gene Conversion Has Contributed Minimally to Similarity among Abp Paralogs
We undertook to determine whether gene conversion has contributed significantly to sequence identity in the Abp gene region. Exchange of genetic information through gene conversion or ectopic recombination between tandem gene copies following duplication can also obscure divergence because changes in one of the duplicated gene copies may be erased. Laukaitis et al. (2008) discounted the explanation that nonallelic gene conversion caused the low divergence of the Abp sequences they studied because the phylogenetic tree of ribosomal protein L23a pseudogenes frequently appears to have coduplicated with <Abpa-Abpbg> gene modules. We pursued this at a smaller scale by analyzing Abp genes with GENECONV to look for evidence of short gene conversion tracks.
Our GENECONV analysis of Abpa paralogs identified no inner (conversion between genes within alignment) and no outer (conversion with genes outside alignment) fragments that were globally significant, suggesting that there is no compelling evidence of gene conversion in Abpa paralogs. In the case of the Abpbg paralogs, the analysis identified only one inner (Abpbg26 and Abpbg34) and two outer fragments (Abpbg5p and Abpbg19) that were globally significant. We also calculated the GC content of the Abp gene region because sequences undergoing frequent gene conversion, either ectopic or allelic, are expected to become GC rich (Galtier et al. 2001, 2008). We found that the average GC content in the Abp gene region is low, about 41–42%, compared with genes undergoing gene conversion, such as ribosomal operons and transfer RNAs which have much higher GC contents (Galtier et al. 2001). Thus, it appears that gene conversion has made a minimal, but not nonexistent, contribution to the evolutionary history of the Abp gene family. It certainly has not been significant enough to have confounded the phylogenetic inference presented by Laukaitis et al. (2008), and we feel that it should not adversely affect our analysis of recently duplicated products presented here.
The Abp Gene Region Has Expanded Most Rapidly in the Center
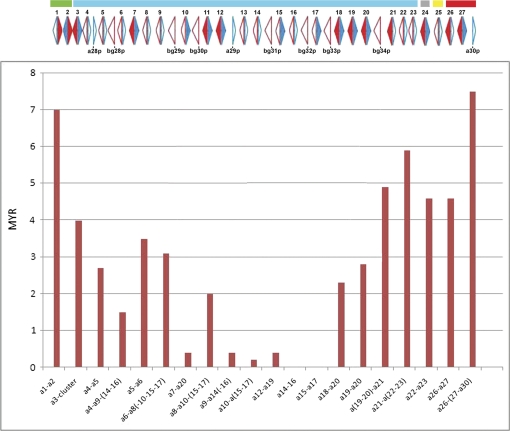
Laukaitis et al. (2008) reported evidence suggesting that expansion of the Abp gene family began in the common ancestor of the genus Mus. They presented phylogenetic evidence that the 30 Abpa genes in the mouse genome fall into five ancestral clades, and they estimated the ages of various duplications in each clade which they calculated from the age of Mus. pahari as the out-group of the genus Mus. The Abp genes in the four blocks shown in figure 3 all fall into the largest clade of the five that Laukaitis et al. (2008) described, suggesting that recent duplications by NAHR may be occurring in the interior of the Abp region. To verify this, we plotted the duplication ages on the Abp linkage map and observed that the oldest duplication events occurred at what is now the periphery of the region and that the youngest events are in the center (fig. 4). This suggests that the most recent duplication activity occurred in the interior, providing new genetic material that has pushed apart the older paralogs on the flanks. It is in this large, central clade where the most volatility seems to be occurring.
FIG. 4.—
Ages of Abpa paralogs in MYR estimated by Laukaitis et al. (2008) were plotted below the physical map of the Abp gene region. Older paralogs plot closer to the left and right boundaries, whereas younger paralogs are in the interior of the region. Colored bars above the linkage group represent the five clades described by Laukaitis et al. (2008).
Numbers of Some Abp Genes Vary
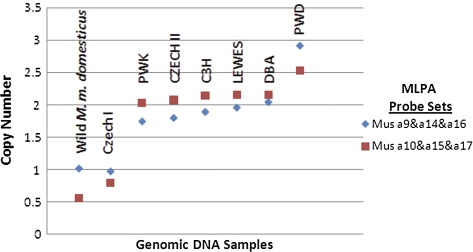
To test whether CNV could be detected in Abp genes, we selected the most recently duplicated Abpa paralogs (a9p, a14p, a16p, and a10, a15, a17) because 2 of the 3 members of each set have identical sequences in the mouse genome (Laukaitis et al. 2008). We detected 1, 2, and 3 copies of each set in various mouse inbred strains (fig. 5), suggesting that CNV of these Abp genes is common in house mice. We note that a particular copy number does not distinguish one subspecies of M. musculus from another because four different M. m. musculus wild-derived inbred strains had three different copy numbers, 1 in CZECHI, 2 in PWK and CZECHII, and 3 in PWD, whereas the wild-derived Mus musculus domesticus strain had a single copy, and the wild-derived LEWES strain had two.
FIG. 5.—
CNV of a9p/a14p/a16p, and a10/a15/a17. MLPA was used to quantitate copy number. DNA probes specific for each group of paralogs were annealed to genomic DNA and subjected to ligation. Polymerase chain reaction of the ligated products was conducted with primers specific for the tails on the probe oligomers. The graph shows the copy number for each probe set indexed to values for known single-copy genes.
These results suggest that at least some portions of the Abp region are volatile, with gene birth and death regularly occurring through duplication and/or deletion. The most striking observation in figure 5 is that CNV of a9p, a14p, a16p, and a10, a15, a17 appears to be consistently coordinated; the same number of copies of both sets appeared in each strain. This is consistent with the very similar duplication ages for the out-group paralog in each set estimated by normalizing their divergences to their divergence with their inferred Mus caroli ortholog and multiplying by the estimated divergence time of M. caroli lineages from M. m. domesticus (Laukaitis et al. 2008). This calculation provides a duplication time of 0.4 MYR for a9 and 0.2 MYR for a10 and supports our suggestion that, at least in some Abp gene regions, duplication occurred as blocks of <Abpbg-Abpa> modules rather than as individual modules. Very likely, duplication and, possibly, deletion of those blocks of genes is creating the CNV that we observe among inbred and wild-derived strains of mice in our MLPA data.
The ESP and MUP Gene Clusters Lack the Volatility That Characterizes the Abp Gene Cluster
We wished to determine whether the volatility (i.e., duplication of blocks of genes and CNV) we observed in the Abp gene region occurs in either of the other two genomic regions containing mouse pheromone genes, the ESPs and MUPs. We did not find repeated patterns of microsatellites such as those seen in the Abp region and so, in each case, we proceeded by comparing the linkage map of the gene family with the gene tree. In the case of the ESP family, mouse and rat ESPs are interspersed throughout the gene tree (Kimoto et al. 2007), leading the authors to conclude that the diversification occurred before the mouse–rat separation. They did find some small mouse-specific clades in the phylogenetic tree of ESPs, suggesting that one or more small expansions occurred in the mouse lineage. Our evaluation of their gene tree and linkage map suggested that no mechanism is needed to explain the expansion of the ESP family of genes beyond simple duplication and divergence, which can occur by slippage and unequal exchange, illegitimate recombination, or strand misalignment–realignment (Katju and Lynch 2003). NAHR, on the other hand, requires flanking repeated sequences (Shaffer and Lupski 2000; Stankiewicz and Lupski 2002).
We also compared the MUP linkage group and gene tree published by Mudge et al. (2008) for signs that groups of genes, even as few as pairs, duplicated together. The location of a gene within its clade had no correlation with its neighboring pseudogene in the linkage group, suggesting either that they had not duplicated together or that concerted evolution has obscured their joint origins. Thus, although NAHR may be operating on individual MUP genes (Mudge et al. 2008), we did not find evidence of duplication of larger gene groups of MUP genes.
To extend our search for volatility among the three families of mouse pheromone genes, we interrogated each gene region with the Mouse Paralogy Browser (She et al. 2008). Figure 6 shows the results of visualizing insertions and deletions in these three regions. The Abp region shows significant volatility with about half the strains tested having insertions and deletions over nearly the entire region. By contrast, the ESP region shows a few small deletions and insertions scattered throughout the strains tested, whereas only two strains show significant deletions in the MUP region where insertions were not seen.
Not only does the Abp gene region appear to be more volatile than the other two pheromone gene regions but also the variation seems to be skewed toward strains derived from M. m. musculus (Czech I, NZO) and strains previously shown to have part or all of an M. m. musculus Abp region (DBA/2J; Dlouhy and Karn 1984; Karn RC, unpublished data). The same is true of the strain CAST/Ei, representing the third subspecies, M. m. castaneus. Thus, it appears that much of the volatility in the Abp gene region is intersubspecific divergence over nearly the whole gene region.
Discussion
Our MLPA results suggest that CNV is common in at least part of the Abp gene region of the house mouse genome. Evaluation of the pattern of Abp paralogs and interspersed repeated elements suggests that the mechanism for this is duplication of blocks containing combinations of <Abpbg-Abpa> modules and single Abpbg paralogs. This mechanism is consistent with parts of the Abp region of the mouse genome duplicating as LCRs (Lupski 1998; Stankiewicz and Lupski 2002) by NAHR (Shaffer and Lupski 2000; Stankiewicz and Lupski 2002). Taken together, these observations lead to the conclusion that the Abp region of the mouse genome is volatile and that view is reinforced by our observations using the Mouse Paralogy Browser. Thus, gene birth and death accelerated by the ability to duplicate and delete numerous paralogs in large blocks appears to be common in the Abp region. At least for the Abp gene region, this answers our question concerning the mechanisms responsible for the rapid expansion of this pheromone gene family.
Early population surveys of alleles for the gene encoding the alpha subunit of mouse salivary ABP (originally Abpa, now designated Abpa27) showed a different a27 allele fixed in each of the three M. musculus subspecies: the a27a allele in M. m. domesticus, a27b allele in M. m. musculus, and the a27c allele in M. m. castaneus (Karn and Dlouhy 1991; Hwang et al. 1997; Karn et al. 2002). It is not known to what extent this represents the entire Abp gene region. However, the insertions and deletions in the data we obtained from the Mouse Paralogy Browser, especially in strains inbred from wild M. m. musculus (CZECHI, NZO) and M. m. castaneus (CAST/Ei), as well as those classical inbred strains with some or all the M. m. musculus subspecies Abp gene region (DBA), suggest that divergence of the Abp region of M. m. domesticus (represented by C57BL/6, the strain source of the mouse genome), M. m. musculus, and M. m. castaneus may have been accelerated by NAHR.
We did not find evidence for an LCR-type duplication of multigene segments of either the ESP gene region or the MUP gene region. We cannot, however, rule out that duplicated blocks have been obscured by homogenization from the action of concerted evolution (Mudge et al. 2008), and this answers another of our questions: whether the same mechanism caused the expansion of all three gene families. Evidently, it did not, at least not recently.
We also did not find evidence for any significant divergence of the ESP or MUP gene regions in any 1 of the 3 subspecies of M. musculus in the data we obtained from the Mouse Paralogy Browser. We conclude that there has been much less volatility, in terms of recent gene birth and death, in the evolutionary histories of these two families of pheromone genes than in the Abp family. That is consistent with the communication attributes of the three families of pheromones because ABP is the only one for which a role in mate selection based on subspecies recognition has been suggested (Laukaitis et al. 1997; Talley et al. 2001; Bimova et al. 2005). It is our contention that the contrast in volatility between the Abp gene region and the other two supports the idea of a very different communication role for mouse ABPs, one involving intersubspecific and perhaps interspecific recognition. That communication role is based on experimental evidence that both laboratory and wild mice show mating preferences for congenic targets that differ only in an Abpa allelic difference fixed in two different subspecies of mice that make secondary contact and form a hybrid zone in Central Europe (Laukaitis et al. 1997; Talley et al. 2001).
There is striking similarity between the volatility we report here for Abp genes and that reported by others for chemosensory receptor genes (especially the V1R genes; Cutler et al. 2007; Nozawa and Nei 2008). It is tempting to speculate that some of these V1R genes encode vomeronasal receptors recognizing ABP molecules, which are coevolving to provide a recognition system to reinforce subspecies and species hybridization barriers.
Did the Abp Region Undergo Two Different Phases of Expansion in the Genus Mus?
Katju and Lynch (2003) found that two-thirds of tandem gene duplicates are in inverse orientation with respect to one another, and they reviewed the models that have been proposed to produce such an orientation. The inverse (i.e., 5′-5’) orientation of Abpa and Abpbg genes in what we have termed a module fits this description. We propose that the original duplication of a single ancestral Abp gene in an early mammalian ancestor produced two paralogs in inverse adjacent order and these evolved into the original pair of Abpa and Abpbg genes. This is consistent with the most widespread Abp gene configuration in mammals (Laukaitis et al. 2008). It also seems likely that the first duplication of an <Abpa-Abpbg> module occurred in this way in the ancestor of the genus Mus because the <Abpa1-Abpbg1> … <Abpa2-Abpbg2> set, constituting one of the oldest clades, is in the inverse orientation to all the other Abp paralogs. One of the original duplicated modules was likely the ancestor of <a1-bg1>… <a2-bg2>, and the other was the ancestor of <bg26-a26p> … <bg27-a27>… <a30 (unpaired) and possibly <bg24-a24> and <bg25p-a25p> because these sets of modules are the oldest in the Abp gene group, comprising 4 of the 5 ancestral clades of genes (Laukaitis et al. 2008), and designated with colored bars at the top of fig. 4 in this paper. Very likely, the fifth clade arose later from a module that duplicated from one of these and is now represented by <bg3p-a3> and/or <bg21-a21p>.
Something apparently changed following this scenario, however, because the most recent Abp duplicates (a3-a23, a28, a29, bg3-bg23, and bg28-bg34; fig. 4) appear in direct, not inverse, order with respect to all members of their clade, and with respect to 3 of the 4 other clades (<bg24-a24>, <bg25-a25>, and <bg26-a26> … <bg27-a27> … <a30). We propose that the change involved the mechanism reported here, by which additional paralogs were generated by NAHR (Shaffer and Lupski 2000; Stankiewicz and Lupski 2002) in the manner proposed for LCRs (Lupski 1998; Stankiewicz and Lupski 2002). Rather than the relatively slower mechanism of duplication of single <Abpa-Abpbg> modules by primer slipping during DNA replication (Chen et al. 2005), blocks of multiple modules, sometimes including unpaired Abpa and Abpbg paralogs, were duplicated by NAHR, at least in the largest, most volatile Abp clade. Thus, duplication of blocks of genes is pushing the ancestral gene sets apart, leaving the more diverged sequences on the flanks (fig. 4), reminiscent of the mechanism proposed by Achaz et al. (2000) and (2001). Duplication of blocks of Abp modules on the interior of the Abp gene cluster by NAHR helps explain the very high rate of duplication (dubbed the “snowball effect” by Kondrashov F and Kondrashov A [2006]). The direct repeat nature of the most recent Abp duplicates is also consistent with duplication by NAHR, which relies on LCRs flanking the duplicating region and commonly produces direct orientation. In contrast to NAHR, most new duplicates occur in inverse orientation due to illegitimate recombination or strand misalignment–realignment (Katju and Lynch 2003). Thus, we feel that duplication of blocks of Abp modules by NAHR is the most likely scenario, at least in the youngest, largest, and most volatile clade of Abp genes.
The Innovation, Amplification, and Divergence Model
It is tempting to speculate that the massive Abp gene duplication that occurred in the genus Mus may have followed a model variously known as adaptive amplification (Hendrickson et al. 2002; Roth and Andersson 2004), adaptive radiation (Francino 2005) or innovation, amplification, and divergence (Bergthorsson et al. 2007); for a review, see Conant and Wolfe (2008). In essence, duplications are selected in certain genes with weak but beneficial functions (i.e., Abp genes with effects on mouse behavior). The tandem array of duplicates maintained by this selection are subject to a high rate of beneficial mutations because of the increased number of mutational targets; again in this case, NAHR has rapidly produced a large set of Abp mutational targets. Once such mutations have been fixed, selection to maintain the duplicated array is reduced and the extra copies are lost, leaving only the ancestral gene and the new duplicate with a novel function. For example, in the Abp system, duplicates such as bg4p, a4p, bg5p, a5p, bg6p, a6p, bg9p, a9p, bg10p, bg14p, a14p, bg15p, bg16p, a16p, and bg17p were silenced somewhere on the way to a10, a15, and a17 being fixed. It is particularly interesting that the progenitors continued to duplicate after being silenced because they were included in the block of genes being copied by NAHR.
Supplementary Material
Supplementary figure 1 is available at Genome Biology and Evolution online (http://www.oxfordjournals.org/our_journals/gbe/).
Supplementary Material
Acknowledgments
The authors wish to thank Erin Kelleher for advice on using the GENECONV program. We are also grateful to Chris Ponting for helpful suggestions and to Evan Eichler for assistance with the Mouse Paralogy Browser. This work was supported by a Senior Postdoctoral Fellowship (5F33HD055016-02 to R.C.K.) from the National Institute of Child Health and Human Development (NICHD). The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the NICHD.
References
- Achaz G, Coissac E, Viari A, Netter P. Analysis of intrachromosomal duplications in yeast Saccharomyces cerevisiae: a possible model for their origin. Mol Biol Evol. 2000;17:1268–1275. doi: 10.1093/oxfordjournals.molbev.a026410. [DOI] [PubMed] [Google Scholar]
- Achaz G, Netter P, Coissac E. Study of intrachromosomal duplications among the eukaryotic genomes. Mol Biol Evol. 2001;18:2280–2288. doi: 10.1093/oxfordjournals.molbev.a003774. [DOI] [PubMed] [Google Scholar]
- Armstrong S, Robertson D, Cheetham S, Hurst J, Beynon R. Structural and functional differences in isoforms of the major urinary proteins: a male-specific protein that preferentially binds a male pheromone. Biochem J. 2005;391:343–350. doi: 10.1042/BJ20050404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergthorsson U, Andersson D, Roth J. Ohno's dilemma: evolution of new genes under continuous selection. Proc Natl Acad Sci USA. 2007;104:1704–1709. doi: 10.1073/pnas.0707158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beynon R, Hurst J. Multiple roles of major urinary proteins in the house mouse, Mus domesticus. Biochem Soc Trans. 2003;31:142–146. doi: 10.1042/bst0310142. [DOI] [PubMed] [Google Scholar]
- Bimova B, Karn R, Pialek J. The role of salivary androgen-binding protein in reproductive isolation between two subspecies of house mouse: Mus musculus musculus and Mus musculus domesticus. Biol J Linn Soc Lond. 2005;84:349–361. [Google Scholar]
- Chamero P, et al. Identification of protein pheromones that promote aggressive behaviour. Nature. 2007;450:899–902. doi: 10.1038/nature05997. [DOI] [PubMed] [Google Scholar]
- Cheetham S, et al. The genetic basis of individual recognition signals in the mouse. Curr Biol. 2007;17:1771–1777. doi: 10.1016/j.cub.2007.10.007. [DOI] [PubMed] [Google Scholar]
- Chen JM, Chuzhanova N, Stenson PD, Ferec C, Cooper DN. Meta-analysis of gross insertions causing human genetic disease: novel mutational mechanisms and the role of replication slippage. Hum Mutat. 2005;25:207–221. doi: 10.1002/humu.20133. [DOI] [PubMed] [Google Scholar]
- Conant G, Wolfe K. Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet. 2008;9:938–950. doi: 10.1038/nrg2482. [DOI] [PubMed] [Google Scholar]
- Cutler G, Marshall L, Chin N, Baribault H, Kassner P. Significant gene content variation characterizes the genomes of inbred mouse strains. Genome Res. 2007;17:1743–1754. doi: 10.1101/gr.6754607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dlouhy SR, Karn RC. Multiple gene action determining a mouse salivary protein phenotype: identification of the structural gene for androgen binding protein (Abp) Biochem Genet. 1984;22:657–667. doi: 10.1007/BF00485851. [DOI] [PubMed] [Google Scholar]
- Emes RD, et al. Comparative evolutionary genomics of androgen-binding protein genes. Genome Res. 2004;14:1516–1529. doi: 10.1101/gr.2540304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francino M. An adaptive radiation model for the origin of new gene functions. Nat Genet. 2005;37:573–577. doi: 10.1038/ng1579. [DOI] [PubMed] [Google Scholar]
- Galtier N, Duret L, Glemin S, Ranwez V. GC-biased gene conversion promotes the fixation of deleterious amino acid changes in primates. Trends Genet. 2008;25:1–5. doi: 10.1016/j.tig.2008.10.011. [DOI] [PubMed] [Google Scholar]
- Galtier N, Piganeau G, Mouchiroud D, Duret L. GC-content evolution in mammalian genomes: the biased gene conversion hypothesis. Genetics. 2001;159:907–911. doi: 10.1093/genetics/159.2.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson H, Slechta E, Bergthorsson U, Andersson D, Roth J. Amplification mutagenesis: evidence that ‘directed’ adaptive mutation and general hypermutability result from growth with a selected gene amplification. Proc Natl Acad Sci USA. 2002;99:2164–2169. doi: 10.1073/pnas.032680899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst J. Female recognition and assessment of males through scent. Behav Brain Res. 2009;200:295–303. doi: 10.1016/j.bbr.2008.12.020. [DOI] [PubMed] [Google Scholar]
- Hwang JM, Hofstetter JR, Bonhomme F, Karn RC. The microevolution of mouse salivary androgen-binding protein (ABP) paralleled subspeciation of Mus musculus. J Hered. 1997;88:93–97. doi: 10.1093/oxfordjournals.jhered.a023083. [DOI] [PubMed] [Google Scholar]
- Karn RC, Dlouhy SR. Salivary androgen-binding protein variation in Mus and other rodents. J Hered. 1991;82:453–458. doi: 10.1093/oxfordjournals.jhered.a111127. [DOI] [PubMed] [Google Scholar]
- Karn RC, Orth A, Bonhomme F, Boursot P. The complex history of a gene proposed to participate in a sexual isolation mechanism in house mice. Mol Biol Evol. 2002;19:462–471. doi: 10.1093/oxfordjournals.molbev.a004102. [DOI] [PubMed] [Google Scholar]
- Karolchik D, et al. The UCSC genome browser database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katju V, Lynch M. The structure and early evolution of recently arisen gene duplicates in the Caenorhabditis elegans genome. Genetics. 2003;165:1793–1803. doi: 10.1093/genetics/165.4.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimoto H, Haga S, Sato K, Touhara K. Sex-specific peptides from exocrine glands stimulate mouse vomeronasal sensory neurons. Nature. 2005;437:898–901. doi: 10.1038/nature04033. [DOI] [PubMed] [Google Scholar]
- Kimoto H, et al. Sex- and strain-specific expression and vomeronasal activity of mouse ESP family peptides. Curr Biol. 2007;17:1879–1884. doi: 10.1016/j.cub.2007.09.042. [DOI] [PubMed] [Google Scholar]
- Kondrashov F, Kondrashov A. Role of selection in fixation of gene duplications. J Theor Biol. 2006;239:141–151. doi: 10.1016/j.jtbi.2005.08.033. [DOI] [PubMed] [Google Scholar]
- Laukaitis C, Critser ES, Karn RC. Salivary androgen-binding protein mediates sexual isolation in Mus musculus. Evolution. 1997;51:2000–2005. doi: 10.1111/j.1558-5646.1997.tb05121.x. [DOI] [PubMed] [Google Scholar]
- Laukaitis CM, et al. Rapid bursts of androgen-binding protein (Abp) gene duplication occurred independently in diverse mammals. BMC Evol Biol. 2008;8:46–62. doi: 10.1186/1471-2148-8-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan DW, Marton TF, Stowers L. Species specificity in major urinary proteins by parallel evolution. PLoS ONE. 2008;3:e3280. doi: 10.1371/journal.pone.0003280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski J. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14:417–422. doi: 10.1016/s0168-9525(98)01555-8. [DOI] [PubMed] [Google Scholar]
- Mudge J, Armstrong S, McLaren K, Beynon R, Hurst J. Species specificity in major urinary proteins by parallel evolution. Genome Biol. 2008;9:R91. doi: 10.1186/gb-2008-9-5-r91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa M, Nei M. Genomic drift and copy number variation of chemosensory receptor genes in humans and mice. Cytogenet Genome Res. 2008;123:263–269. doi: 10.1159/000184716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson D, Cox K, Gaskell S, Evershed R, Beynon R. Molecular heterogeneity in the major urinary proteins of the house mouse Mus musculus. Biochem J. 1996;316(Pt 1):265–272. doi: 10.1042/bj3160265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth J, Andersson D. Adaptive mutation: how growth under selection stimulates Lac(+) reversion by increasing target copy number. J Bacteriol. 2004;186:4855–4860. doi: 10.1128/JB.186.15.4855-4860.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schouten JP, et al. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer L, Lupski J. Molecular mechanisms for constitutional chromosomal rearrangements in humans. Ann Rev Genet. 2000;34:297–329. doi: 10.1146/annurev.genet.34.1.297. [DOI] [PubMed] [Google Scholar]
- She X, Cheng Z, Zollner S, Church D, Eichler E. Mouse segmental duplication and copy number variation. Nat Genet. 2008;40:909–914. doi: 10.1038/ng.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski J. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- Talley HM, Laukaitis CM, Karn RC. Female preference for male saliva: implications for sexual isolation of Mus musculus subspecies. Evolution. 2001;55:631–634. doi: 10.1554/0014-3820(2001)055[0631:fpfmsi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.