Abstract
We describe two modular protocols for immunostaining and multiparameter flow cytometric analysis of major human antigen-presenting cells (dendritic cells, monocytes, B lymphocytes) in minimally manipulated whole blood. Simultaneous detection of up to eight colors is enabled by careful selection and testing of cell-subset-defining monoclonal antibodies (anchor markers) in the appropriate fluorochrome combinations, to demonstrate the quantification of surface expression levels of molecules involved in chemotaxis (e.g. CX3CR1, CCR2), adhesion (e.g. CD11b, CD62L), antigen presentation (e.g. CD83, CD86, CD209) and immune regulation (e.g. CD101) on circulating antigen-presenting cells. Each immunostaining reaction requires as little as 50–100 μl of peripheral whole blood, no density-gradient separation, and the entire procedure from preparation of reagents to flow cytometry can be completed in <5 h.
INTRODUCTION
Antigen presentation is an innate immune function that is crucial in self/non-self discrimination and in the pathogenesis of most immune-mediated diseases. The chief immune cells endowed with this function, known also as professional antigen-presenting cells (APCs), include dendritic cells (DCs), macrophages, monocytes and B lymphocytes. DCs, macrophages and monocytes, including immature monocytes, are particularly competent in antigen capturing and presentation and in activating antigen-specific T-cell responses through major histocompatibility complex class II (MHC II) molecules1-7. MHC II molecules are encoded in the highly genetically polymorphic human leukocyte antigen (HLA) region that produces multiple molecules required for antigen presentation, including HLA-DR, a MHC II molecule predominantly and abundantly expressed by APCs. Because APCs contribute crucially to the pathogenesis of a range of diseases, detailed characterization of their functional status would be valuable in devising strategies for diagnosis, immunologic monitoring and therapy and would furnish important insight into mechanisms of APC mobilization. More generally, the human immune system is poorly understood8,9, and there is an urgent need to improve our knowledge using the latest technologies.
Phenotypic characterization of antigen-presenting cells has been hampered by the large number of surface markers required to identify a subpopulation or subset, and furthermore limitations of conventional three- to four-color flow cytometers are major drawbacks1,10,11. Advantages of single-cell immunophenotyping of blood antigen-presenting cells by flow cytometry have been demonstrated and proven feasible12, offering relatively high sensitivity and statistical power compared with non-flow-based technologies. Polychromatic (>6 colors) flow cytometry increases the data-capturing throughput and enables concurrent, multiplexed analysis of cell subsets and populations in the same reaction tube (e.g. calculation of percentages or ratios of cell subsets, parallel comparison of surface expression density of a molecule between two cell populations), as well as a qualitative assessment and characterization of the functional status of cells13,14. In conjunction with the use of minimally manipulated leukocytes derived from erythrocyte-lysed fresh blood, instead of density-gradient-enriched mononuclear cells, immunophenotypic data can be captured with minimal introduction of artefacts or factors that affect immunostaining15.
Selection and rigorous testing of monoclonal antibodies and fluorochromes are crucial to successful polychromatic flow cytometry. Recently, Jansen et al. reported an eight-color immunostaining protocol for flow cytometry that uses MHC II, CD14, CD11c and CD123 with/without CD19 as cell-subset-defining markers16. Our early experience with eight-color flow cytometry using six subset-defining monoclonal antibodies concomitantly (targeting CD14, CD16, CD19, CD11c, CD123 and HLA-DR) indicated that an accurate definition of DC subsets is better achieved using the stringent Lineage cocktail 1 (Lin1)–FITC negative selection markers or antibodies (BD Biosciences) that target CD3, CD14, CD16, CD19, CD20 and CD56; these lineage markers are all undetectable on the two major DC subsets. In other words, a lack of expression of Lin1 effectively excludes T lymphocytes, CD14+ monocytes, CD14+CD16+ monocytes, CD16+ monocytes, polymorphonuclear neutrophils (PMNs), B lymphocytes and natural killer cells. Moreover, CD11c (integrin αX) and CD123 (IL-3 receptor-α), without the exclusion of lineage markers, are not DC-specific as they are expressed by a variety of myeloid and lymphoid cells.
Therefore, to maximise the capabilities of an eight-color flow cytometer (BD LSR II with blue, red and violet lasers) while preserving the accurate identification of monocyte subsets, B lymphocytes and DC subsets in addition to retaining the capacity to analyse three to four target molecules of interest, we have devised two protocols with similar workflow, Protocols A and B (Fig. 1), that have undergone testing and ascertainment. The two immunostaining protocols are modular and can be easily performed in parallel, or independently, to define the major circulating antigen-presenting cell subsets in whole blood: CD14+, CD14+CD16+ and CD16+ monocytes; CD19+HLA-DR+ B cells; and myeloid (MDCs or preDC1) and plasmacytoid (PDCs or preDC2) DCs. Performing both protocols concurrently can reduce the elapsed time from phlebotomy to immunostaining (‘arm-to-antibody’ time) and minimize any potential time-dependent in vitro confounding effects when studying innate cellular attributes that are intrinsically volatile. The use of predeveloped, standardized fluorescent beads for normalization of fluorescence intensity data as well as for periodic fine-tuning of the flow cytometer17,18 can reduce non-biological and/or instrumental variation arising from day-to-day operation of the flow cytometer, especially for multicenter and/or longitudinal immunophenotyping studies.
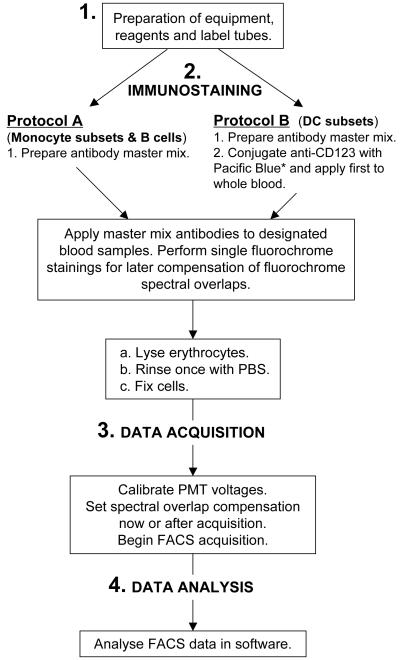
Figure 1.
Diagram illustrating the workflow for multiplexed immunophenotypic analysis of the major circulating APC subsets by flow cytometry. *Manufacturer-preconjugated anti-CD123–eFluor 450 can substitute for anti-CD123–Pacific Blue and obviate the need for manual conjugation.
Monocytes are progenitors for macrophages and certain DCs, and two principal subpopulations have been described: CD14+HLA-DR+ (‘inflammatory’) and CD16+HLA-DR+ (‘resident’) monocytes19,20. Further characterization based on their differential expression of chemokine receptors and adhesion molecules identified inflammatory CD14+ monocytes as CX3CR1lowCCR2+CD62L+CD11bhigh, and resident CD16+ monocytes as CX3CR1highCCR2−CD62L−CD11bint 2,20. CD14+ and CD16+ monocytes differ in size [~18 μm or intermediate-high forward scatter channel (FSC), and ~14 um or low-intermediate FSC, respectively], cytoplasmic granularity [relatively higher side scatter channel (SSC) and lower SSC, respectively] and in their functional capacities1,2,19,20 (see also Fig. 2a for FSC and SSC comparisons). Although less well understood, CD14+CD16+ monocytes are another subset, possibly in a transitional or intermediary state, that is expanded and active in inflammatory disorders6,21-23. Even in the absence of an exogenous stimulus, the CD14+CD16+ subset differentiates within 12 h from CD14+ monocytes, and therefore we recommend proceeding with the immunostaining within 4 h of blood donation and to rotate the blood collection tubes if possible to avoid subsequent miscalculation of monocyte subset ratios and changes in the expression levels of certain surface molecules (Fig. 3).
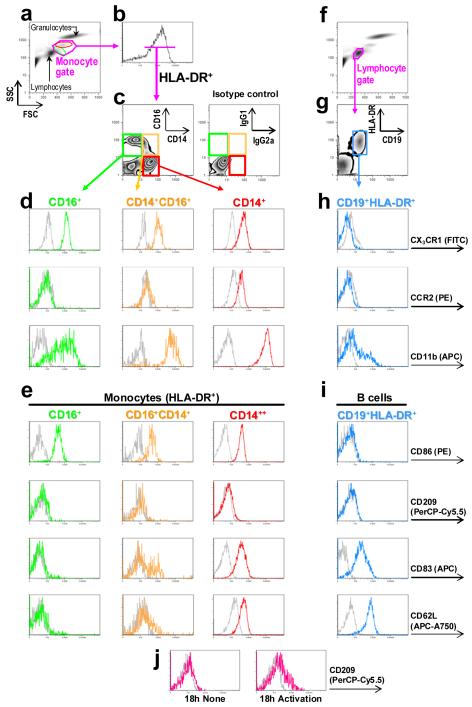
Figure 2.
Flow cytometric analysis of monocyte subsets and CD19+HLA-DR+ B lymphocytes in whole blood. (a) CD16+ ‘resident’ and CD14+ ‘inflammatory’ monocytes have preponderant localizations on a FSC (linear) vs SSC (logarithmic) bivariate plot (green and red ovals, respectively), owing to their different intrinsic biophysical properties. Cells within the monocyte gate (pink enclosure) are selected for (b) expression of HLA-DR–PerCP and further separated based on (c) expression of CD14–Pacific Orange and CD16–Pacific Blue, and displayed on a quantile contour (zebra) plot. Negative controls, nonspecific mouse IgG2a and IgG1, matching the respective fluorochrome and immunoglobulin isotypes of CD14 and CD16, respectively, show no significant positivity. (d) CD16+HLA-DR+ (green), CD14+CD16+HLA-DR+ (yellow) and CD14+HLA-DR+ (red) monocyte subsets are analyzed further for their expression of CX3CR1–FITC, CCR2–PE and CD11b–APC, and (e) CD86–PE, CD209–PerCP-Cy5.5, CD83–APC and CD62L–APC-Alexa Fluor 750, as shown in one-dimensional histograms. B lymphocytes are analyzed from (f) cells localized to the lymphocyte gate that (g) express CD19 and HLA-DR. Histograms illustrate B-cell expression profiles of (h) CX3CR1, CCR2 and CD11b, and (i) CD86, CD209, CD83 and CD62L in parallel with monocytes co-immunostained in the same reactions. (j) Functionality of the anti-human CD209 antibody in PerCP-Cy5.5 is demonstrated in a separate reaction by activation of peripheral whole blood with IL-1β (10 ng/ml), TNF-α (50 ng/ml), IFN-γ (1000 U/ml; Peprotech) and ultra-pure lipopolysaccharide (100 ng/ml; InvivoGen); cells in the monocyte gate expressing CD11c and HLA-DR are shown.
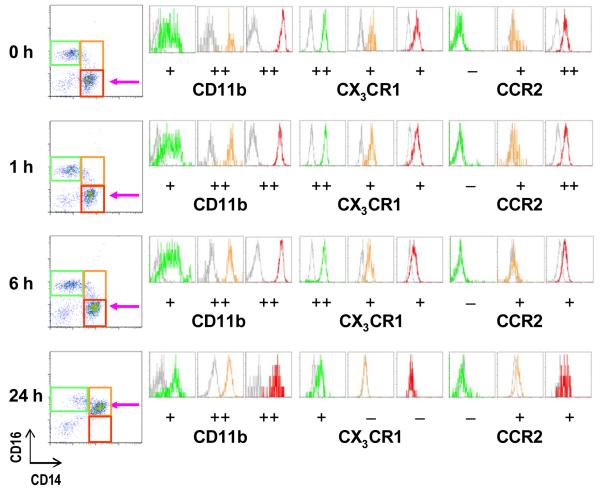
Figure 3.
A time-course experiment showing the natural differentiation or transition of CD14+ monocytes into CD14+CD16+ cells (pink arrow at 24 h) in citrated whole blood even in the absence of any stimulus. Note that CX3CR1 expression levels on all monocyte subsets decrease over time, whereas CCR2 is expressed only on CD14+ and at a low level on CD14+CD16+ monocytes but is also downregulated with time. FSC, forward scatter channel. SSC, side scatter channel. ‘-’, negative expression. ‘+’, low-to-intermediate level of expression. ‘++’, high level of expression.
MDCs and PDCs derive from different hematopoietic lineages and have divergent physiological functions7,24-29. In conjunction with positive expression of MHC II molecules such as HLA-DR and the absence of Lin1 markers, MDCs and PDCs can be further classified as Lin1−HLA-DR+CD11c+ and Lin1−HLA-DR+CD123+, respectively. CD303 (BDCA-2) can be used alternatively to identify PDCs, whereas CD1c+ (BDCA-1) and CD141+ (BDCA-3 or thrombomodulin) are markers that identify additional MDC subsets30-32. CD303 is a PDC-specific C-type lectin involved in antigen uptake, processing and presentation to T cells and regulates type I interferon release, a cell-defining attribute of PDCs33.
Neutrophils can be identified and distinguished from monocytes, macrophages and DCs by virtue of their high FSC and SSC11 – thus, the use of a different gating strategy (see also Fig. 2a) and immunophenotypic profile: CD16highCX3CR1−CD62L+CD11bhigh. In line with recent findings, neutrophils differentiate from the CX3CR1-negative lineage of myeloid progenitors that diverge from the macrophage-DC precursors early in hematopoiesis29. Investigators interested in this cell population can maximize the yield of immunophenotyping data by gating on the granulocyte cluster (Fig. 2a).
Protocol A details the use of eight fluorochrome-conjugated monoclonal antibodies (or ten parameters) to characterize in whole blood CD14+, CD14+CD16+ and CD16+ monocyte and B-cell surface expression of chemokine receptors (e.g. CX3CR1 or fractalkine receptor, CCR2), adhesion molecules (e.g. CD11b or integrin-αM, CD62L or L-selectin) and costimulatory/accessory antigen presentation molecules (e.g. CD83, CD86, CD209 or DC-SIGN).Determination of monocyte subset ratios is also shown. Protocol B takes advantage of the Lin1 immunostaining strategy focusing on the identification of CD11c+ MDCs and CD123+ PDCs, with demonstration of the alternative and/or concomitant use of the DC markers CD1c and CD303 in combination with Lin1−HLA-DR+ to ascertain the identity of MDCs and PDCs, respectively. Specific to this protocol is the use of a violet laser-excitable fluorochrome, particularly Pacific Blue (peak excitation/emission of ~410/455 nm) or an equivalent such as eFluor 450 (peak excitation/emission of ~405/450 nm), conjugated with anti-CD123 antibody that opens up other commonly used channels (e.g. PE, Alexa Fluor 700, APC-Alexa Fluor 750) for use.
Isotype control immunoglobulins are not needed for quantitative immunophenotyping18. In a recent large-scale, gene-immunophenotyping study of nearly 200 individuals that we carried out14, we eliminated the use of the isotype controls after ascertaining beforehand that the monoclonal antibodies are specific. Isotype control immunoglobulins are liable to fluctuate over time, and the use of fluorescent microspheres is a solution to the time-dependent effects or variability observed in longitudinal studies18. We recommend only the initial use of isotype-matched control immunoglobulins to establish that a target antibody is specific, then proceed to longitudinal immunophenotyping using fluorescent microspheres to normalize the acquired data.
When performing large-scale, quantitative immunophenotyping by polychromatic flow cytometry over a prolonged period (i.e. on the order of months), the investigator should have a precise hypothesis a priori, determine the power of the study and calculate the sample size required, to enable estimation of the amount of antibodies and reagents required. Apart from instrumental variation that can be remedied by using standardized fluorescent microspheres18, a batch-to-batch effect of reagents is probably another major source of variability. In a carefully planned study with a known number of immunostaining reactions, the investigator can then order an oversized batch and pool the antibodies to circumvent batch variation.
Each immunostaining reaction requires as little as 50–100 μl of fresh whole blood without prior density-gradient separation. This method provides a cost-effective, practical approach to high-throughput polychromatic immunophenotyping of the major APC subsets in whole blood and serves as a framework upon which to incorporate additional analysis parameters, depending on the flow cytometer setup and fluorochromes used.
MATERIALS
REAGENTS
Fresh whole blood from consenting healthy human donors ! CAUTION Universal precaution and work in Category 2 biological safety cabinets; where possible, use screened blood and dispose of bio-hazardous waste as per local institutional protocol. ! CAUTION All experiments using human tissue must conform to National and Institutional regulations.
Citrate phosphate dextrose adenine (CPDA)-pretreated blood collection tubes (Greiner bio-one, cat. no. 455094; Sarstedt, cat. no. 01.1610.001) ▲ CRITICAL Our testing of multiple lots of CPDA and sodium heparin tubes indicated that the former did not contain endotoxin levels above the detection limit of the assay, whereas the latter had more variable background levels of endotoxin (range <0.01 to 1.04 EU/ml, n = 10 total number of tubes tested from at least four lots); indicators of increased neutrophil degranulation, cell death and monocyte activation were noted with sodium heparin tubes.
Monoclonal antibodies (see Table 1) ! CAUTION Sodium azide is a preservative present in most antibody preparations that is irritating to the skin and can be explosive when accumulated in plumbing; flush and dispose of with copious water. Even though azide is present, to avoid microbial contamination, open vials containing antibody in a laminar flow hood and remove aliquots with sterile pipette tips. ▲ CRITICAL Prior to starting a longitudinal, quantitative immunophenotyping study, perform a titration analysis to determine the saturating amount or concentration of each antibody required to ensure maximal coverage of the expressed target antigen in order to attribute any variation observed to true biological differences. This can be performed by using incremental doses of a single antibody to immunostain a donor’s blood samples under the same experimental conditions and determine the dose required to obtain a plateau of fluorescence intensity by constructing a line plot of antibody concentration vs mean (or median) fluorescence intensity, in conjunction with overlaid curves displayed histographically34. The optimal concentration is usually the one at the beginning of a saturation plateau34. For markers not requiring absolute quantitation in a study, to reduce cost, adequate staining can be often obtained with less than a saturating amount of antibody.
Isotype control immunoglobulins (see Table 1).
Zenon Pacific Blue Mouse IgG2a Labeling Kit (Invitrogen, cat. no. Z25156) – only if performing manual conjugation of Pacific Blue with purified anti-CD123 (Miltenyi Biotec, clone AC145, cat. no. 130-090-940) (see REAGENT SETUP for conjugation procedure); otherwise, not required if using a manufacturer-preconjugated Pacific Blue equivalent such as anti-CD123–eFluor 450 (eBioscience, clone 6H6, cat. no. 48-1239).
BD FACS Lysing Solution, 10x (BD Biosciences, cat. no. 349202)
BD CellFIX, 10x (BD Biosciences, cat. no. 340181)
FluoroSpheres, 6-peak calibration beads (DakoCytomation, cat. no. K0110) or 8-peak calibration beads (DakoCytomation, cat. no. K0112)
Sterile, endotoxin-free PBS, 1x (Sigma, cat. no. D8537)
Sterile, endotoxin-free distilled H2O (GIBCO Invitrogen, cat. no. 15230-089).
Table 1. Monoclonal antibodies used in Protocols A and B.
Lineage Cocktail 1 (Lin1) antibodies target CD3, CD14, CD16, CD19, CD20 and CD56
| Protocol A - Monocyte subsets & B lymphocytes | ||||||||
|---|---|---|---|---|---|---|---|---|
| Target antigen | Fluorochrome | Antibody clone | Antibody host species & isotype |
Manufacturer | Product catalog number* |
Laser used | Bandpass filters (nm) |
Long-pass dichroic mirror (nm) |
| CCR2 | PE | 48607.211 | mouse IgG2b | R&D Systems | FAB151P | blue | 575/26 | 550 |
| CD11b | APC | ICRF44 | mouse IgG1 | BioLegend | 301310 | red | 660/20 | |
| CD14† | Pacific Orange | TüK4 | mouse IgG2a | Invitrogen | MHCD1430 | violet | 585/42 | 505 |
| CD16† | Pacific Blue | 3G8 | mouse IgG1 | BioLegend | 302021 | violet | 450/50 | |
| CD19† | Alexa Fluor 700 | SJ25-C1 | mouse IgG1 | Invitrogen | MHCD1929 | red | 730/45 | 710 |
| CD62L | APC-Alexa Fluor 750 | DREG-56 | mouse IgG1 | Invitrogen | MHCD62L | red | 780/60 | 755 |
| CD83 | APC | HB15e | mouse IgG1 | BD Biosciences | 551073 | red | 660/20 | |
| CD86 | PE | FUN-1 | mouse IgG1 | BD Biosciences | 555658 | blue | 575/26 | 550 |
| CD209 | PerCP-Cy5.5 | DCN46 | mouse IgG2b | BD Biosciences | 558263 | red | 695/40 | 685 |
| CX3CR1 | FITC | 2A9-1 | rat IgG2b | MBL International | D070-4 | blue | 530/30 | 505 |
| HLA-DR | FITC | L243 | mouse IgG2a | BD Biosciences | 307604 | blue | 530/30 | 505 |
| HLA-DR† | PerCP | L243 | mouse IgG2a | BD Biosciences | 347364 | red | 695/40 | 685 |
| BD Multicolor CD83/CD86/CD209 | APC/PE/PerCP-Cy5.5 | HB15e/FUN-1/DCN46 | mouse IgG1/IgG1/IgG2b | BD Biosciencs | 334098 | (see above) | ||
| Protocol B - DC subsets | ||||||||
|---|---|---|---|---|---|---|---|---|
| Target antigen | Fluorochrome | Antibody clone | Antibody host species & isotype |
Manufacturer | Product catalog number* |
Laser used | Bandpass filters (nm) |
Long-pass dichroic mirror (nm) |
| CCR2 | PE | (see above) | ||||||
| CD1c | PE | AD5-8E7 | mouse IgG2a | Miltenyi Biotec | 130-090-508 | blue | 575/26 | 550 |
| CD11b | Alexa Fluor 700 | VIM12 | mouse IgG1 | Invitrogen | CD11b29 | red | 730/45 | 710 |
| CD11b | APC-Cy7 | ICRF44 | mouse IgG1 | BD Biosciences | 557754 | red | 780/60 | 755 |
| CD11c† | APC | S-HCL-3 | mouse IgG2b | BD Biosciences | 333144 | red | 660/20 | |
| CD62L | APC-Cy7 | DREG-56 | mouse IgG1 | eBioscience | 10-0629 | red | 780/60 | 755 |
| CD101 | PE | BB27 | mouse IgG1 | Serotec | MCA2236PE | blue | 575/26 | 550 |
| CD123† | pure** | AC145 | mouse IgG2a | Miltenyi Biotec | 130-090-940 | violet (for Pacific Blue) | 450/50 | |
| CD123†† | eFluor 450 | 6H6 | mouse IgG1 | eBioscience | 48-1239 | violet | 450/50 | |
| CD303 | APC | AC144 | mouse IgG1 | Miltenyi Biotec | 130-090-905 | red | 660/20 | |
| HLA-DR† | PerCP | (see above) | ||||||
| Lineage Cocktail 1 (Lin1)†§ | FITC | multiple | mouse IgG1/IgG2b | BD Biosciences | 340546 | blue | 530/30 | 505 |
Catalog numbers can vary in the USA, the European Union and other countries.
For conjugation with Pacific Blue using the Invitrogen Zenon Pacific Blue Mouse IgG2a Labeling Kit.
Anchor markers used in Protocols A and B.
CD123–eFluor 450 can be a substitute for CD123–Pacific Blue.
EQUIPMENT
BD LSR II flow cytometer (BD Biosciences, cat. no. 33300456)
Eppendorf Centrifuge 5417R with Fixed-Angle Rotor with Aerosol-Tight Lid
Category 2 biological safety cabinet
Sterile 1.5-ml or 2.0-ml polypropylene microcentrifuge tubes (Sarstedt), or 2-ml 96-well Polypropylene Deep-Well Plates (BD Falcon) in conjunction with adhesive PCR film (Thermo Scientific) for sealing during incubation. ! CAUTION When using 96-well polypropylene plates with adhesive sealing film, avoid contact of blood and reagents with the film
10-ml FACS tubes
BD FACSDiva software (BD Biosciences, http://www.bdbiosciences.com) for data acquisition
FlowJo software (TreeStar Inc., http://www.flowjo.com) for data analysis
REAGENT SETUP
-
Anti-CD123 monoclonal antibody
Either manually conjugate purified anti-CD123 monoclonal antibody (Miltenyi) with Pacific Blue (Invitrogen) as per manufacturer’s instructions (see also PROCEDURE step 3B) or use manufacturer-preconjugated anti-CD123–eFluor 450 (eBioscience) thereby obviating the conjugation procedure detailed in step 3Biii. Store all antibodies at 4°C.
EQUIPMENT SETUP
Flow cytometer
Our BD LSR II is equipped with three lasers: violet (405 nm excitation by Coherent VioFlame, 25 mW solid-state laser), blue (488 nm excitation by Coherent Sapphire, 20 mW solid-state laser) and red (633 nm excitation by JDS Uniphase 1344P, 17 mW HeNe laser). Bandpass filters 440/40 and 525/50 are configured with the violet laser; 530/30, 575/26, 695/40 and 780/60 with the blue laser; 660/20 and 730/45 and 780/60 with the red laser. The specifications of long-pass dichroic mirrors used were 505 nm, 550 nm, 685 nm, 710 nm and 755 nm (see Table 1). When frequent, in-house calibration and fine-tuning of flow cytometer as detailed by Perfetto et al.17 is not possible, regular (quarterly) evaluation and servicing of the flow cytometer by the manufacturer’s engineer should be scheduled. ! CAUTION Internal components of a flow cytometer pose serious health hazards and should not be tampered with by unqualified personnel.
PROCEDURE
Preparation of reagents (steps 1–2), 30 min
1 Label 2.0-ml microcentrifuge tubes. ▲ CRITICAL STEP 1.5-ml microcentrifuge tubes can be used if reaction volumes are scaled down proportionately. However, the yield of rare cell populations might be low. When working with large numbers of staining reactions, 2-ml 96-well Polypropylene Deep-Well Plates in conjunction with adhesive sealing film are useful; avoid contact of blood with adhesive surface. We do not recommend the use of polystyrene FACS tubes for immunostaining APCs as these cells might become activated by this surface substrate. When polypropylene FACS tubes are used, exercise care when discarding the supernatant as the pellet can be difficult to visualize. Use sterile plasticware.
2 Prepare fresh 1x BD FACS Lysing Solution and 1x BD CellFIX solution by diluting 1:10 the respective 10x solutions with sterile, endotoxin-free H2O prior to use. Keep 1x FACS Lysing Solution at 15–20°C, and CellFIX solution and 1x PBS on ice. ▲ CRITICAL STEP Keep all solutions and antibody mixes sterile.
Staining (steps 3–11), 2 h
3 Prepare the antibody master mix for immunostaining of monocyte subsets and B lymphocytes (see Protocol A) and/or DC subsets (see Protocol B).
- Antibody mix for monocyte subsets and B lymphocytes
- Add anti-human CD14–Pacific Orange, CD16–Pacific Blue, CD19–Alexa Fluor 700, and HLA–DR–PerCP (or HLA-DR–FITC) anchor antibodies to a sterile 1.5-ml microcentrifuge tube (Table 1).
- Keep mix on ice and in the dark.
- Antibody mix for DC subsets
- Add anti-human Lin1–FITC cocktail antibodies, HLA-DR–PerCP and CD11c–APC anchor antibodies to a sterile 1.5-ml microcentrifuge tube (Table 1); include anti-CD123–eFluor 450 in the mixture when substituting for CD123–Pacific Blue and skip step 3Biii.
- Keep mix on ice and in the dark.
- Manually conjugate purified anti-CD123 monoclonal antibody (Miltenyi) with Pacific Blue at 15–20°C using the Invitrogen Zenon Pacific Blue Mouse IgG2a Labeling Kit (refer to manufacturer’s instructions for details). Briefly, 1 μg (~20 μl of Miltenyi anti-human CD123, cat. no. 130-090-940) of purified anti-CD123 antibody (antibody volume not crucial if ≤20 μl is used per 100 μl of whole blood) is incubated for 5 min with 5 μl of Component A (consisting of Pacific Blue-labelled Fab fragment targeting Fc portion of anti-CD123 antibody), followed by incubation for 5 min with 5 μl of Component B (consisting of non-specific IgG that binds free Fab fragments). Anti-CD123–Pacific Blue is now conjugated and must be used within 30 min. Proceed immediately to step 4. ■ PAUSE POINT Following addition of Component A, the anti-CD123–Pacific Blue conjugate can be kept in the dark at 4°C for several weeks with the supplement of 2 mM sodium azide before proceeding to addition of Component B, according to the manufacturer’s instructions.
4 Pipet 100 μl of CPDA-treated fresh whole blood into each tube. If blood droplet is caught on the tube sidewalls, samples can be spun down gently by pressing the ‘Short’ button on the Eppendorf Centrifuge 5417R for 2 s (≤350 g).
5 Add antibodies to whole blood. For immunostaining of monocyte subsets and B lymphocytes see Protocol A. For immunostaining DC subsets see Protocol B.
- Immunostaining monocyte subsets and B lymphocytes
- Add to whole blood the appropriate volume of antibody master mix prepared above for the respective protocol. Typically, most antibodies will be added in a volume of ~10 μl so the final volume is ~180 μl; total volume to be added depends on the sum of the volume of the constituent antibodies.
-
Incubate in the dark for 30 min at 15–20°C.CRITICAL STEP During this incubation you can continue with step 6.
- Immunostaining DC subsets
- Add 25 μl of the anti-CD123–Pacific Blue antibody prepared above to 100 μl of whole blood. Skip this step and proceed to step 5Biii if using anti-CD123–eFluor 450.
-
Incubate in the dark for 10 min at 15–20°C.▲ CRITICAL STEP During this, and the subsequent, incubation one can continue with step 6.
- Add the antibody master mix prepared in step 3Bi. Typically, most antibodies will be added in a volume of ~10 μl so the final volume is 180–200 μl; total volume to be added depends on the sum of the volume of the constituent antibodies.
- Incubate in the dark for 20 min at 15–20°C.
6 While Protocol A and Protocol B antibodies are incubating with blood samples, perform single-color staining for later fluorochrome spectral overlap compensation by applying the same, single antibody that was used in the master mix for each individual fluorochrome. Incubate samples in the dark for 30 min at 15–20°C. Omit this step when using standardized fluorescence-labeled microspheres for setting spectral compensation (see also PROCEDURE – Acquisition). ▲ CRITICAL STEP Performing single-color staining for all fluorochromes each time when immunophenotyping is performed can also serve as a quality control and help to troubleshoot and gauge the quality of the antibody and the staining.
7 Following incubation of the antibody mix with blood, lyse erythrocytes by adding 2.0 ml of 1x FACS Lysing Solution per 100 μl of blood used. Incubate for 10 min at 15–20°C with inversion of tubes every 2–3 min. ▲ CRITICAL STEP Ensure cells are well resuspended for proper lysis.
8 After incubation, centrifuge at 350 g for 7 min at 15–20°C.
9 Carefully pipet off and discard supernatant. Resuspend cell pellet in 1.0 ml of cold, endotoxin-free 1x PBS for each 100 μl whole blood used and mix well.
CRITICAL STEP Perform all subsequent steps at 4°C.
10 Centrifuge at 350 g for 7 min at 4°C.
11 Carefully aspirate off and discard supernatant. Resuspend in 300 μl of 1x BD CellFIX. ■ PAUSE POINT Fixed cells can be kept at 4°C for up to 24 h prior to analysis by flow cytometry.
? TROUBLESHOOTING
Acquisition (steps 12–16), 1 h
12 Transfer fixed samples from microcentrifuge tubes to 10-ml FACS tubes.
13 Switch on flow cytometer and allow it to warm up for 30 min. Open FACSDiva software program.
14 Calibrate PMT voltages by first creating a FSC vs SSC bivariate plot in FACSDiva to visualize cell clusters, then create one-dimensional histograms for each fluorochrome used in the experiment, and set voltages by acquiring blank and fluorescent beads (DakoCytomation FluoroSpheres) gauged on histograms according to the manufacturer’s instructions. In addition, if desired, use an unstained sample of fixed cells (see Step 3 above) to establish the PMT voltages so that the majority of events (cells) contained within a gate encompassing monocyte or lymphocyte clusters (i.e. clusters that are not granulocytes, dead cells or cell debris) are in the first decade (log) of fluorescence. Benchmark against blank beads (Dako Cytomation) to monitor the autofluorescence of the target cell population on a day-to-day basis.
15 Determine fluorochrome pair compensations by first acquiring samples from single fluorochrome–antibody staining. Use bi-exponential (or ‘Logicle’) display of x-axis and y-axis when determining amount of compensation35. Set compensation now or after collection of all samples, depending on capabilities of flow cytometer and software capabilities; automatic compensation might be available on some platforms. An orderly workflow including the systematic organization of bivariate plots displaying antibody–fluorochrome pairs is essential to successful polychromatic flow cytometry (Fig. 4). For literature on flow cytometry instrument setup and fluorochrome compensation, investigators are referred to excellent reviews published by others10,33-39. ▲ CRITICAL STEP Each target antigen can have a drastically different cell-surface distribution and biophysical properties, requiring configuration of a unique set of fluorochrome compensation values. Use an abundantly expressed surface epitope when setting compensation.
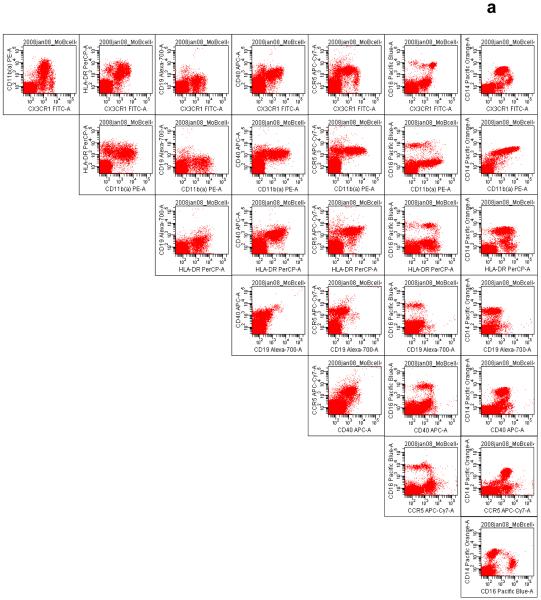
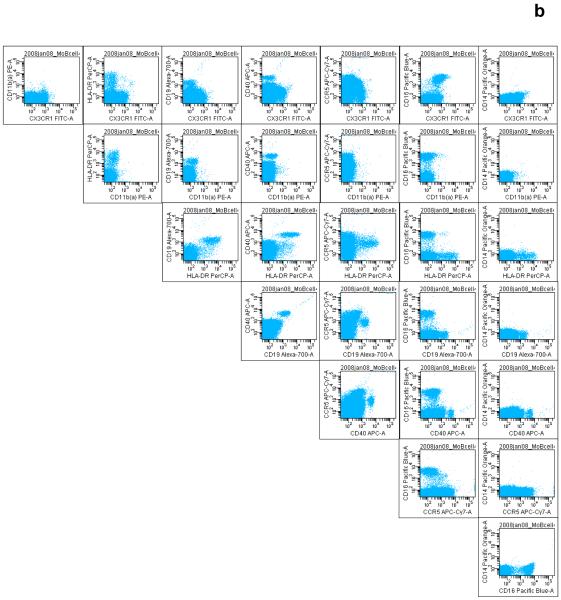
Figure 4.
The systematic display of antibody–fluorochrome pairs created in the Worksheet window of the BD FACSDiva software is useful in organizing the large number of bivariate plots, for visualizing the correct spectral overlap compensation, quality control and for discovering novel cell subsets. This mode of display is also useful for visualizing single-color staining and for ascertaining fluorochrome ‘spillover’. Acquisition events or cells captured in the (a) monocyte gate and (b) lymphocyte gate are shown. Antibodies demonstrated include those targeting CX3CR1–FITC, activated CD11b (CD11b(a))–PE, HLA-DR–PerCP, CD19–Alexa Fluor 700, CD40-APC, CCR5–APC-Cy7, CD16–Pacific Blue, and CD14–Pacific Orange (see Table 1 for details).
16 Begin acquisition of data.
? TROUBLESHOOTING
Analysis (steps 17), 1 h
17 After acquisition, export data in the *.fcs format and analyze using the FlowJo software (see also ANTICIPATED RESULTS and Figures; for latest information on cross-platform compatibility of FlowJo files, check the FlowJo website.). For monocytes follow option A. For B lymphocytes follow option B. For DC subsets, follow option C.
- Monocytes
- Select HLA-DR+ cells (Fig. 2b, within pink gate), and use a quantile contour (zebra) plot to display expression of CD14–Pacific Orange vs. CD16–Pacific Blue.
- Gate on discrete clusters of CD16+ (resident), CD14+CD16+, CD14+ (inflammatory) monocytes (Fig. 2c). Leave FITC, PE, APC and APC-Alexa Fluor 750 channels open for quantification of antigen expression levels displayed in a one-dimensional histogram. When HLA-DR–FITC is used, the PerCP or PerCP-Cy5.5 channel can be used for quantification, as demonstrated with CD209–PerCP-Cy5.5 (Fig. 2h).
- B lymphocytes
- Place gate around the lymphocyte cluster on the FSC vs SSC plot (Fig. 2e), drill down on this population and create a CD19–Alexa Fluor 700 vs HLA-DR–PerCP (Fig. 2f). HLA-DR-expressing B cells are CD19+HLA-DR+. PE, APC, APC-Alexa Fluor 750 and FITC or PerCP/PerCP-Cy5.5 are fluorochrome channels left available for quantification of antigen expression levels, and are displayed histographically (Fig. 2g). PerCP or PerCP-Cy5.5 can be used as the quantification channel when HLA-DR–FITC instead of HLA-DR–PerCP is used (Fig. 2i).
- DC subsets
- Analyze ungated cells on a Lin1–FITC vs HLA-DR–PerCP bivariate plot. DCs are Lin1−HLA-DR+ (Fig. 5b, pale-blue box).
- Using a CD11c–APC vs CD123–Pacific Blue plot, calculate the MDC:PDC ratio or percentage (Fig. 5c).
-
To determine the expression levels of target molecules for both DC subsets, create two-dimensional scatter plots from the Lin1−HLA-DR+ cluster (Fig. 5b, pale-blue box), generating CD11c vs HLA-DR (for CD11c+HLA-DR+ MDCs) and CD123 vs HLA-DR (CD123+HLA-DR+ PDCs) plots (Fig. 5d), from which histograms displaying expression levels of molecules in the PE, APC-Alexa Fluor 750 and Alexa Fluor 700 channels can be created (Fig. 5e and 5f).? TROUBLESHOOTING
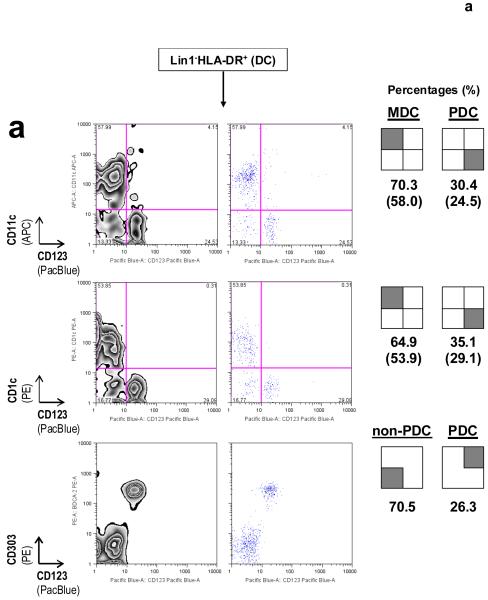
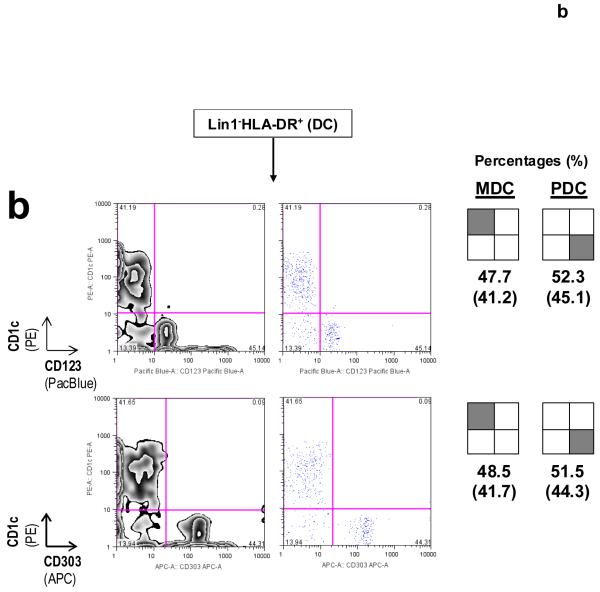
Figure 5.
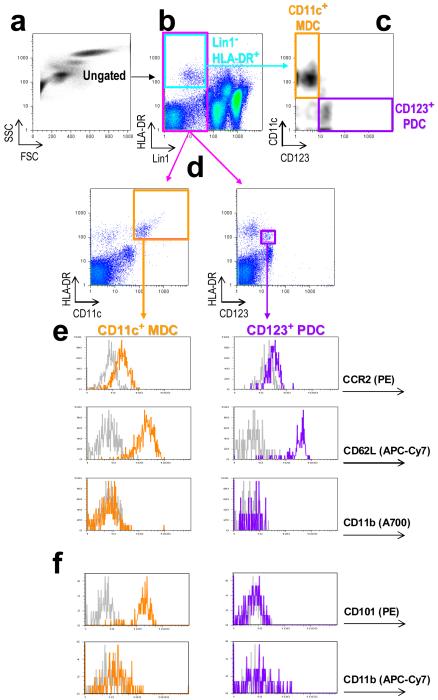
Analysis of two major DC subsets in whole blood. (a) Ungated cells that are (b) Lin1−HLA-DR+ (within pale-blue box) are analysed on a (c) bivariate density plot showing CD11c vs CD123 expression levels to allow for identification of MDCs (within orange box) and PDCs (within purple box), respectively, and for determination of DC subsets ratio or percentages. To profile expression levels of specific surface antigens on DC subsets, (b) Lin1− cells (within pink box) are gated on and further analysed for (d) CD11c+HLA-DR+ and CD123c+HLA-DR+ expression, corresponding to MDCs (orange) and PDCs (purple), respectively. Histograms show (e) expression levels of CCR2–PE, CD62L–APC-Cy7 and CD11b–Alexa Fluor 700, and (f) of CD101–PE and CD11b–APC-Cy7.
TROUBLESHOOTING
See Table 2 for troubleshooting guidance.
Table 2. Troubleshooting table.
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 3, 14, 15, 17 | Fluorochrome ‘spillover’ (i.e. non-specific fluorescence detected in a neighboring channel, e.g. false-positive events in the PE channel attributable to FITC fluorescence) |
|
Check fluorescence spectrum viewer (http://bdbiosciences.com/spectra or http://www.invitrogen.com/site/us/en/home/support/Research-Tools/Fluorescence-SpectraViewer.html). Examine single- color staining profiles and perform ‘fluorescence minus one’ or FMO36 control stainings to identify the culprit fluorochrome. Knowing the distribution of antigens and epitopes targeted by antibodies, and having a proper understanding of fluorochromes, their selection and instrument setup and operation are crucial for polychromatic flow cytometry10,34-39. Refer also to instrument user’s guide provided by the flow cytometer manufacturer. |
| 3B, 17 | Dull staining of manually conjugated Pacific Blue antibody (e.g. anti-CD123) using the Invitrogen Zenon labeling kit |
i. Suboptimal antibody: conjugate ratio. | Optimize amount of Component A used, and titrate antibody levels. Component B (blocking immunoglobulin) must also be adjusted stoichiometrically (refer to manufacturer’s instructions). Alternatively, a Pacific Blue equivalent such as eFluor 450 can be used. |
| 7, 11, 17 | Lack of consistency in phenotype readout. |
|
Allow samples to stabilize in fixative for at least 1 h after immunostaining. Analyze at a fixed time interval (e.g. 16 h after cell fixation). Avoid analysing later than 24 h as fluorescence intensity or tandem dyes might begin to degrade. Use freshly prepared standardized reagents (e.g. BD FACS lysing solution, CellFIX). |
| 7, 17 | Odd or variable FSC vs SSC bivariate profiles or large amounts of debris, including dead cells. |
|
Stop incubation promptly at 10 min, and invert tubes less vigorously. |
| 9, 11, 17 | Low cell yield. |
|
Pipet off supernatant carefully if using microcentrifuge tubes, or pour away supernatant gently when using FACS tubes. Consider scaling up blood volumes and staining reagents. |
| 14, 17 | Progressive shifting of acquisition events on FSC vs SSC bivariate plot over days and sometimes during a data collection session. This can |
|
Ferromagnetic valves are often used to regulate pressures and buffer or waste flow within certain flow cytometers. Use of magnetic bead- based cell sorting and subsequent analysis on such flow cytometers can cause valves to malfunction, leading to increased internal pressure within the flow cell. Discontinue the use of |
ANTICIPATED RESULTS
Protocol A
The gating of monocytes on a FSC vs SSC bivariate plot is important for identifying the monocyte subsets that have different sizes and granularity1,2. CD16+ monocytes have lower FSC and SSC, and are situated close to the lymphocyte cluster (Fig. 2a, green oval), whereas CD14+ monocytes, owing to their larger numbers, size (higher FSC) and higher cytoplasmic granularity (higher SSC), cluster near granulocytes (Fig. 2a, red oval). The former subset can be missed or its percentages underestimated if the gating is too conservatively placed on only the latter subpopulation. Within the monocyte gate, HLA-DR+ cells (Fig. 2b) are separated based on expression of CD14 and CD16 and are displayed on a quantile contour (zebra) plot that can also be used to calculate relative ratios or percentages of CD16+ (Fig. 2c, green box), CD14+CD16+ (Fig. 2c, yellow box) and CD14+ (Fig. 2c, red box) subsets. We have found the zebra display useful in guiding the gating of monocyte subsets. Histograms showing expression profiles of the monocyte subsets are consistent with those reported previously: CD16+HLA-DR+ monocytes are CX3CR1highCCR2− CD11blowCD62L− (Fig. 2d, green tracings), and CD14+HLA-DR+ monocytes are CX3CR1lowCCR2+CD11bhighCD62L+ (Fig. 2d, red tracings)20. CD19+HLA-DR+ B cells do not express CX3CR1 or CCR2, but the majority do express CD62L. A recent study reported that ~34% (19–44%) of B cells in peripheral blood of healthy human donors express CD11b on the cell surface, and that CD11b mediates homing of memory B cell subsets, in contrast to naïve cells40. Accumulating evidence supports the divergent lineage and functions of CD11b+ and CD11b−/low B cells40-42. Another analysis of monocyte subsets and CD19+HLA-DR+ B cells is demonstrated by CD86–PE, CD83–APC, CD209–PerCP-Cy5.5 through use of HLA-DR–FITC instead of HLA-DR–PerCP (Fig. 2). Although the expression of CD209 is negligible or undetectable on resting, circulating monocytes and HLA-DR+CD19+ B cells from a healthy blood donor (Fig. 2h and 2i), the functionality of this fluorochrome-labeled antibody is demonstrated by its increased expression on CD11c+HLA-DR+ cells in the monocyte gate following cytokine activation (Fig. 2j). We advise immunostaining within 4 h of blood donation, as changes in the monocyte subset dynamics occur with time (Fig. 3). Protocol A utilizes ten parameters or eight fluorochromes (FITC, PE, PerCP or PerCP-Cy5.5, Alexa Fluor 700, APC, APC-Alexa Fluor 750, Pacific Orange and Pacific Blue).
Protocol B
This protocol is a modification of the four-color staining reported by Willmann et al.12 and others. Because four fluorochrome channels (typically, Lin1–FITC, HLA-DR–PerCP, CD11c–APC and CD123–PE) are required for separating MDCs and PDCs, this limitation precludes the use of antibodies of interest that are commonly available in PE and APC. Using a three-laser flow cytometer (BD LSR II) and a violet-laser-excitable fluorochrome (e.g. anti-CD123 conjugated with Pacific Blue), we were able to maximize immunostaining combinations and attain seven-color multiplexed analysis of two major blood DC subsets; there exist future opportunities for robust eight-color analyses with the increased availability of monoclonal antibodies coupled with new fluorochromes taking advantage of the violet laser such as eFluor 605 and eFluor 650 monoclonal antibodies (eBioscience). In this protocol, Lin1 and HLA-DR are indispensable for identifying DCs (Fig. 5a and 5b) that can be subclassified into MDCs (Lin1−HLA-DR+CD11c+) and PDCs (Lin1− HLA-DR+CD123+) (Fig. 5d). CD1c and CD303 are alternative markers that can identify Lin1− HLA-DR+ MDC and PDC, respectively, (Fig. 6a and 6b) but should not be regarded as equivalent to CD11c and CD123 in view of the tremendous DC heterogeneity26,27,30-32,44. MDC:PDC ratios or percentages (Fig. 5c) have been reported to alter in disease states; however, gating strategies and the use of DC-defining markers can vary between studies45,46. The CD11c–CD123 bivariate plot of Lin1−HLA-DR+ cells (Fig. 5c and Fig. 5a, top row) is a common strategy used to simultaneously display ratios between the two DC subsets44,45. For quantification of expression levels (i.e. in mean or median fluorescence intensity units), we find that the CD11c+HLA-DR+ and CD123+HLA-DR+ plots (Fig. 5d) generated from Lin1− cells (Fig. 5b, pink box) are preferable as they provide a clear visualization of DC clusters (Fig. 5d, orange and purple boxes) and other cells or events; this is also the method demonstrated in the product sheet provided by the manufacturer of the Lin1 mixture antibodies (BD Biosciences). Protocol B has the capacity for at least nine parameters or seven fluorochromes (FITC, PE, PerCP, APC, APC-Alexa Fluor 750, Alexa Fluor 700, Pacific Blue). Using CD101 expression profiling of circulating DCs as an example (Fig. 5f), we have concurrently identified positive CD101 surface expression on MDCs but non-expression on PDCs as a novel strategy for differentiating the two DC subsets, findings that might have implications also in their differential immunomodulatory functions31,47-50,52.
Figure 6.
DC subset-defining markers for identifying MDCs and PDCs as described in Protocol B. (a) CD11c and CD123, in conjunction with Lin1−HLA-DR+, are surface markers that are commonly used to identify MDCs and PDCs, respectively. CD1c+ (or BDCA-1+) MDCs might represent a major subset of CD11c+ MDCs, as reported previously30-32. Co-staining for CD123 and CD303 (BDCA-2) on Lin1−HLA-DR+ cells simultaneously identifies PDCs in the same staining reaction43; numbers in brackets represent raw percentages for non-PDCs (70.5%) and Lin1−HLA-DR+CD123+CD303+CD11c− PDCs (26.3%). Concordance between Lin1−HLA-DR+CD123+ and Lin1−HLA-DR+CD303+ cells is typically in the range 0.971 to 0.989 (mean 0.981, n = 6). (b) Immunostaining from another blood donor illustrates a variation in the MDC:PDC ratio or percentages. CD1c and CD303 can be used as alternative markers to identify specific Lin1−HLA-DR+ MDC and PDC subsets, respectively, and in determining subset ratios or percentages. Numbers inside brackets represent percentages calculated from all four quadrants, whereas percentages outside brackets are normalized for MDCs and PDCs only. Data are representative of at least three independent donors.
Surface markers useful for the identification of circulating APC subsets are summarized in Table 3. As the complexity of polychromatic flow cytometry increases, multidimensional analyses of the data by computational modelling might one day replace manual gating and increase the throughput and objectivity of analyses53. Comprehensive information on flow cytometer setup, fluorochromes, spectral overlap compensations, fluorescent bead normalization for longitudinal immunophenotyping studies, operational pitfalls and other useful technical information are detailed elsewhere10,18,34-39. Further links to major manufactures of flow cytometers, accessories and fluorochrome-conjugated antibodies can be found at the end of an article by Perfetto et al.11.
Table 3. Physical properties and differential expression profile of surface phenotypes on CD16+, CD14+CD16+ and CD14+ monocytes (Mo), CD19+HLA-DR+ B cells, CD11c+ MDCs, CD123+ PDCs and CD16hiCX3CR1− neutrophils.
| Parameters | CD16+ Mo | CD16+CD14+ Mo | CD14++ Mo | CD19+HLA-DR+ B lymphocytes |
CD11c+ MDC | CD123+ PDC | CD16hi Neutrophils |
|---|---|---|---|---|---|---|---|
|
| |||||||
| FSC | low-int | low-int to high-int | high-int | low | low to int | low | high |
| SSC | int | int | int | low | int | low to int | high |
| HLA-DR | pos | pos | pos | pos | pos | pos | neg* |
| CX3CR1 | high | int | low | neg | n.d. | neg** | neg |
| CCR2 | neg | int | high | neg | high | int | neg |
| CD11b | int | int | high | neg to int | low or neg | neg | pos |
| CD62L | low or neg | low | pos | int | high | high | pos |
| CD101 | pos† | pos† | pos† | pos† | pos | neg | pos† |
ACKNOWLEDGEMENTS
We thank the Wellcome Trust, the Juvenile Diabetes Research Foundation (JDRF) International, and the National Institute for Health Research Cambridge Biomedical Research Centre for funding. The Cambridge Institute for Medical Research is a recipient of the Wellcome Trust Strategic Award (079895). E.F. was funded by the JDRF and the Prince Philip Graduate Exhibition from the Cambridge Overseas Trust. L.S.W. is a Wellcome Trust Principal Research Fellow.
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
REFERENCES
- 1.Ziegler-Heitbrock HWL. Definition of human blood monocytes. J. Leukoc. Biol. 2000;67:603–606. doi: 10.1002/jlb.67.5.603. [DOI] [PubMed] [Google Scholar]
- 2.Grage-Griebenow E, Flad HD, Ernst M. Heterogeneity of human peripheral blood monocyte subsets. J. Leukoc. Biol. 2001;69:11–20. [PubMed] [Google Scholar]
- 3.Steinman RM, Nussenzweig MC. Avoiding horror autotoxicus: the importance of dendritic cells in peripheral T cell tolerance. Proc. Natl. Acad. Sci. U.S.A. 2002;99:351–358. doi: 10.1073/pnas.231606698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al-Daccak R, Mooney N, Charron D. MHC class II signaling in antigen-presenting cells. Curr. Opin. Immunol. 2004;16:108–113. doi: 10.1016/j.coi.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 5.Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat. Rev. Immunol. 2005;5:953–964. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- 6.Tacke F, et al. Immature monocytes acquire antigens from other cells in the bone marrow and present them to T cells after maturing in the periphery. J. Exp. Med. 2006;203:583–597. doi: 10.1084/jem.20052119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reis e Sousa C. Dendritic cells in a mature age. Nat. Rev. Immunol. 2006;6:476–483. doi: 10.1038/nri1845. [DOI] [PubMed] [Google Scholar]
- 8.Davis MM. A prescription for human immunology. Immunity. 2008;29:835–838. doi: 10.1016/j.immuni.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayday AC, Peakman M. The habitual, diverse and surmountable obstacles to human immunology research. Nat. Immunol. 2008;9:575–580. doi: 10.1038/ni0608-575. [DOI] [PubMed] [Google Scholar]
- 10.Baumgarth N, Roederer M. A practical approach to multicolor flow cytometry for immunophenotyping. J. Immunol. Methods. 2000;243:77–97. doi: 10.1016/s0022-1759(00)00229-5. [DOI] [PubMed] [Google Scholar]
- 11.Perfetto SP, Chattopadhyay PK, Roederer M. Seventeen-colour flow cytometry: unravelling the immune system. Nat. Rev. Immunol. 2004;4:648–655. doi: 10.1038/nri1416. [DOI] [PubMed] [Google Scholar]
- 12.Willmann K, Dunne JF. A flow cytometric immune function assay for human peripheral blood dendritic cells. J. Leukoc. Biol. 2000;67:536–544. [PubMed] [Google Scholar]
- 13.Seder RA, Darrah PA, Roederer M. T-cell quality in memory and protection: implications for vaccine design. Nat. Rev. Immunol. 2008;8:247–258. doi: 10.1038/nri2274. [DOI] [PubMed] [Google Scholar]
- 14.Dendrou CA, et al. Gene-to-cell-specific protein phenotype for the autoimmune locus IL2RA using fresh blood from a genotype-selectable bioresource. Nat. Genet. 2009;41:1011–1015. doi: 10.1038/ng.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamul KR, Schmitz JL, Kane K, Folds JD. Comparison of the effects of Ficoll-Hypaque separation and whole blood lysis on results of immunophenotypic analysis of blood and bone marrow samples from patients with hematologic malignancies. Clin. Diagn. Lab. Immunol. 1995;2:337–342. doi: 10.1128/cdli.2.3.337-342.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jansen K, et al. Polychromatic flow cytometric high-throughput assay to analyze the innate immune response to Toll-like receptor stimulation. J. Immunol. Methods. 2008;336:183–192. doi: 10.1016/j.jim.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perfetto SP, Ambrozak D, Nguyen R, Chattopadhyay P, Roederer M. Quality assurance for polychromatic flow cytometry. Nat. Protocols. 2006;1:1522–1530. doi: 10.1038/nprot.2006.250. [DOI] [PubMed] [Google Scholar]
- 18.Dendrou CA, et al. Fluorescence intensity normalisation: correcting for time effects in large-scale flow cytometric analysis. Advances in Bioinformatics. 2009 doi: 10.1155/2009/476106. doi:10.1155/2009/476106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Passlick B, Flieger D, Ziegler-Heitbrock HWL. Identification and characterization of a novel monocyte subpopulation in human peripheral blood. Blood. 1989;74:2527–2534. [PubMed] [Google Scholar]
- 20.Geissmann F, Jung S, Littman DR. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity. 2003;19:71–82. doi: 10.1016/s1074-7613(03)00174-2. [DOI] [PubMed] [Google Scholar]
- 21.Fingerle G, et al. The novel subset of CD14+/CD16+ blood monocytes is expanded in sepsis patients. Blood. 1993;82:3170–3176. [PubMed] [Google Scholar]
- 22.Kawanaka N, et al. CD14+,CD16+ blood monocytes and joint inflammation in rheumatoid arthritis. Arthritis. Rheum. 2002;46:2578–2586. doi: 10.1002/art.10545. [DOI] [PubMed] [Google Scholar]
- 23.Belge KU, et al. The proinflammatory CD14+CD16+DR++ monocytes are a major source of TNF. J. Immunol. 2002;168:3536–3542. doi: 10.4049/jimmunol.168.7.3536. [DOI] [PubMed] [Google Scholar]
- 24.Spits H, Couwenberg F, Bakker AQ, Weijer K, Uittenbogaart CH. Id2 and Id3 inhibit development of CD34(+) stem cells into predendritic cell (pre-DC)2 but not into pre-DC1. Evidence for a lymphoid origin of pre-DC2. J. Exp. Med. 2000;192:1775–1784. doi: 10.1084/jem.192.12.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu YJ, Kanzler H, Soumelis V, Gilliet M. Dendritic cell lineage, plasticity and cross-regulation. Nat. Immunol. 2001;2:585–589. doi: 10.1038/89726. [DOI] [PubMed] [Google Scholar]
- 26.Liu YJ. IPC: professional type 1 interferon-producing cells and plasmacytoid dendritic cell precursors. Annu. Rev. Immunol. 2005;23:275–306. doi: 10.1146/annurev.immunol.23.021704.115633. [DOI] [PubMed] [Google Scholar]
- 27.Münz C, Steinman RM, Fujii S. Dendritic cell maturation by innate lymphocytes: coordinated stimulation of innate and adaptive immunity. J. Exp. Med. 2005;202:203–207. doi: 10.1084/jem.20050810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Banchereau J, Pascual V. Type I interferon in systemic lupus erythematosus and other autoimmune diseases. Immunity. 2006;25:383–392. doi: 10.1016/j.immuni.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 29.Fogg D, et al. A clonogenic bone marrow progenitor specific for macrophages and dendritic cells. Science. 2006;311:83–87. doi: 10.1126/science.1117729. [DOI] [PubMed] [Google Scholar]
- 30.Dzionek A, et al. BDCA-2, BDCA-3, and BDCA-4: three markers for distinct subsets of dendritic cells in human peripheral blood. J. Immunol. 2000;165:6037–6046. doi: 10.4049/jimmunol.165.11.6037. [DOI] [PubMed] [Google Scholar]
- 31.MacDonald KP, et al. Characterization of human blood dendritic cell subsets. Blood. 2002;100:4512–4520. doi: 10.1182/blood-2001-11-0097. [DOI] [PubMed] [Google Scholar]
- 32.Bonasio R, von Andrian UH. Generation, migration and function of circulating dendritic cells. Curr. Opin. Immunol. 2006;18:503–511. doi: 10.1016/j.coi.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 33.Dzionek A, et al. BDCA-2, a novel plasmacytoid dendritic cell-specific type II C-type lectin, mediates antigen capture and is a potent inhibitor of interferon alpha/beta induction. J. Exp. Med. 2001;194:1823–1834. doi: 10.1084/jem.194.12.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Radbruch A. Flow Cytometry and Cell Sorting. Edn 2. Springer-Verlag; Berlin, Heidelberg, Germany: 2000. Immunofluorescence: basic considerations. Chapter 3. [Google Scholar]
- 35.Herzenberg LA, Tung J, Moore WA, Herzenberg LA, Parks DR. Interpreting flow cytometry data: a guide for the perplexed. Nat. Immunol. 2006;7:681–685. doi: 10.1038/ni0706-681. [DOI] [PubMed] [Google Scholar]
- 36.Roederer M. Spectral compensation for flow cytometry: visualization artefacts, limitations, and caveats. Cytometry. 2001;45:194–205. doi: 10.1002/1097-0320(20011101)45:3<194::aid-cyto1163>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 37.Shapiro HM. Practical Flow Cytometry. Edn 4. John Wiley & Sons, Inc.; Hoboken, New Jersey, USA: 2003. Parameters and Probes. Chapter 7. [Google Scholar]
- 38.Maecker HT, Frey T, Nomura LE, Trotter J. Selecting fluorochrome conjugates for maximum sensitivity. Cytometry A. 2004;62A:169–173. doi: 10.1002/cyto.a.20092. [DOI] [PubMed] [Google Scholar]
- 39.Maecker HT, Trotter J. Flow cytometry controls, instrument setup, and the determination of positivity. Cytometry A. 2006;69A:1037–1042. doi: 10.1002/cyto.a.20333. [DOI] [PubMed] [Google Scholar]
- 40.Kawai K, et al. CD11b-mediated migratory property of peripheral blood B cells. J. Allergy Clin. Immunol. 2005;116:192–197. doi: 10.1016/j.jaci.2005.03.021. [DOI] [PubMed] [Google Scholar]
- 41.Montecino-Rodriguez E, Leathers H, Dorshkind K. Identification of a B-1 B cell-specified progenitor. Nat. Immunol. 2006;7:293–301. doi: 10.1038/ni1301. [DOI] [PubMed] [Google Scholar]
- 42.Arnold LW, Pennell CA, McCray SK, Clarke SH. Development of B-1 cells: segregation of phosphatidylcholine-specific B cells to the B-1 population occurs after immunoglobulin gene expression. J. Exp. Med. 1994;179:1585–1595. doi: 10.1084/jem.179.5.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nestle FO, et al. Plasmacytoid predendritic cells initiate psoriasis through interferon-alpha production. J. Exp. Med. 2005;202:135–143. doi: 10.1084/jem.20050500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Summers KL, et al. Reduced IFN-alpha secretion by blood dendritic cells in human diabetes. Clin. Immunol. 2006;121:81–89. doi: 10.1016/j.clim.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 45.Peng R, Li Y, Brezner K, Litherland S, Clare-Salzler MJ. Abnormal peripheral blood dendritic cell populations in type 1 diabetes. Ann. N. Y. Acad. Sci. 2003;1005:222–225. doi: 10.1196/annals.1288.031. [DOI] [PubMed] [Google Scholar]
- 46.Allen JS, et al. Plasmacytoid dendritic cells are proportionally expanded at diagnosis of type 1 diabetes and enhance islet autoantigen presentation to T-cells through immune complex capture. Diabetes. 2009;58:138–145. doi: 10.2337/db08-0964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ochando JC, et al. Alloantigen-presenting plasmacytoid dendritic cells mediate tolerance to vascularized grafts. Nat. Immunol. 2006;7:652–662. doi: 10.1038/ni1333. [DOI] [PubMed] [Google Scholar]
- 48.Shao L, Jacobs AR, Johnson VV, Mayer L. Activation of CD8+ regulatory T cells by human placental trophoblasts. J. Immunol. 2005;174:7539–7547. doi: 10.4049/jimmunol.174.12.7539. [DOI] [PubMed] [Google Scholar]
- 49.Allez M, Brimnes J, Dotan I, Mayer L. Expansion of CD8+ T cells with regulatory function after interaction with intestinal epithelial cells. Gastroenterology. 2002;123:1516–1526. doi: 10.1053/gast.2002.36588. [DOI] [PubMed] [Google Scholar]
- 50.Bouloc A, Bagot M, Delaire S, Bensussan A, Boumsell L. Triggering CD101 molecule on human cutaneous dendritic cells inhibits T cell proliferation via IL-10 production. Eur. J. Immunol. 2000;30:3132–3139. doi: 10.1002/1521-4141(200011)30:11<3132::AID-IMMU3132>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 51.Ashtekar AR, Saha B. Poly’s plea: membership to the club of APCs. Trends Immunol. 2003;24:485–490. doi: 10.1016/s1471-4906(03)00235-7. [DOI] [PubMed] [Google Scholar]
- 52.Rivas A, et al. V7, a novel leukocyte surface protein that participates in T cell activation. I. Tissue distribution and functional studies. J. Immunol. 1995;154:4423–4433. [PubMed] [Google Scholar]
- 53.Pyne S, et al. Automated high-dimensional flow cytometric data analysis. Proc. Natl. Acad. Sci. U. S. A. 2009;106:8519–8524. doi: 10.1073/pnas.0903028106. [DOI] [PMC free article] [PubMed] [Google Scholar]