Abstract
Familial hypercholesterolemia (FH) is a condition caused by mutations in the low-density lipoprotein receptor (LDLR) gene. Expression of LDLR is highly regulated and excess receptor expression is cytotoxic. To incorporate essential gene regulation into a gene therapy vector for FH, we generated vectors in which the expression of therapeutic human LDLR gene, or luciferase reporter gene, is driven by 10 kb of human LDLR genomic DNA encompassing the promoter region including elements essential for physiologically regulated expression. Using luciferase expression and specific LDL binding and internalization assays, we have shown in vitro that the genomic promoter element confers long-term, physiologically regulated gene expression and complementation of receptor deficiency in culture for 240 cell-generations. This was demonstrated in the presence of sterols or statins, modifiers of LDLR promoter activity. In vivo, we demonstrate efficient liver-specific delivery and expression of luciferase following hydrodynamic tail-vein injection and confirm that expression from the LDLR promoter element is sensitive to statin administration. We also demonstrate long-term LDLR expression from the 10-kb promoter element up to 9 months following delivery. The vector system that we describe provides the efficient delivery, long-term expression, and physiological regulation required for a successful gene therapy intervention for FH.
Introduction
One aim of gene therapy for recessive genetic diseases is to complement the loss of function of an endogenous gene. For the treatment of some conditions, long-term expression and physiological regulation of transgene expression may prove advantageous for effective therapy. Familial hypercholesterolemia (FH) is one such condition. FH is caused by mutations in the low-density lipoprotein receptor (LDLR) and is characterized by high circulating levels of cholesterol and affects around 1:500 of the population.1 As a classic monogenic loss-of-function condition, gene therapy for FH has long been under development. Gene therapy strategies for FH have been developed by a number of research groups worldwide with the main focus being on viral delivery systems, such as retrovirus,2,3,4,5,6 adenovirus,7,8,9,10,11 and adeno-associated virus.12 These previous studies included a clinical trial3 but showed varying degrees of success with little evidence of long-term therapeutic effect. All the cited studies used classic mini-gene expression vectors where expression of LDLR was driven by strong heterologous promoters. The use of these vectors leads to an overexpression of the LDLR that results in a characteristic initial lowering of plasma cholesterol, which is not maintained in the long-term.
Expression from the LDLR genomic locus is controlled by levels of intracellular cholesterol. When intracellular cholesterol levels fall, LDLR expression is triggered by the binding of sterol response element (SRE)–binding proteins to the SREs in the promoter region.13 This increases the number of active LDL receptors on the cell surface, which bind and internalize LDL particles from the circulation. As the intracellular cholesterol stores become replete, the SRE-binding proteins become less active and expression from the LDLR locus is repressed. When the LDLR is expressed from constitutively active promoters that lack regulation elements essential for expression control by intracellular cholesterol levels, the continuous expression of LDLR and internalization of cholesterol from the extracellular space causes a toxic build up of cholesterol in the cell. This effect is seen in vitro14 and in vivo15 following delivery of LDLR expression plasmids under the control of strong viral promoters.
We have shown previously that in vitro delivery of the whole genomic locus of the LDLR leads to full and long-term physiological complementation of the LDLR deficit in Ldlr-deficient models of FH, including primary FH patient fibroblasts.16,17 Further work in vivo with the LDLR whole genomic locus within a 154-kb vector demonstrated prolonged human LDLR expression, up to 4 months after delivery. This was compared to reporter gene expression from a strong heterologous promoter, where expression was repressed 30 days after delivery.18 However, large genomic DNA transgene constructs are technically more challenging to deliver than small complementary DNA (cDNA) constructs and we found in vivo that delivery was not efficient enough to provide functional complementation in the Ldlr−/− mouse model of receptor deficiency.
In this study, we have devised a novel gene therapy approach to combine transgene delivery with classic drug treatment to improve expression in vivo. For this, we have investigated the administration of statin drugs following injection. Statins are a class of drugs known as 3-hydroxy-3-methylglutaryl coenzyme A reductase inhibitors. Statins act by inhibiting the conversion of 3-hydroxy-3-methylglutaryl coenzyme A to mevalonate, the rate-limiting step in cholesterol synthesis. The reduction in de novo synthesis of cholesterol leads to an upregulation of LDLR expression due to the overall reduction in the amount of intracellular cholesterol. Administration of statins following transgene delivery should enhance expression from the transgene, while maintaining cellular integrity by ensuring a physiological uptake of cholesterol from the extracellular space.
We have devised an expression system, which combines the advantages of the small size of a cDNA vector with the inclusion of critical regulation elements of the genomic locus. We used a portion of the complete LDLR genomic locus that contained all known essential expression control elements driving the human LDLR cDNA. This decreased the size of the vector fivefold, resulting in increased delivery efficiency, while maintaining full physiological control of functional complementation. Our vectors incorporate the highly beneficial EBNA-1 and oriP episomal maintenance elements from the Epstein–Barr virus. These elements have been shown by us and others to facilitate the retention of plasmid DNA as circular extrachromosomal plasmids in the presence of human genomic origins of replication in cis in a range of mammalian cell types.16,19,20 and also to enhance stable gene expression in vivo.19
Our second-generation gene therapy vectors contain the human LDLR cDNA controlled by 10 kb of genomic DNA from the native human LDLR locus incorporating a large promoter element of 10 kb of genomic DNA upstream from the initiating ATG codon including the three SREs, the 5′-untranslated region and a further ~8 kb of upstream sequence. We show that the vectors carrying the 10-kb LDLR promoter element confer full physiological expression on the LDLR and luciferase transgenes in vitro. In addition, we show that the gene therapy vectors containing the LDLR cDNA driven by the LDLR promoter confer long-term expression in vitro in cellular models and in vivo in mouse liver following hydrodynamic tail-vein injection. We found that statin administration does indeed increase luciferase expression from the genomic promoter element both in vitro and in vivo confirming that our vectors are capable of physiological regulation by the changing cellular milieu.
Results
Generation of advanced regulated LDLR expression vectors
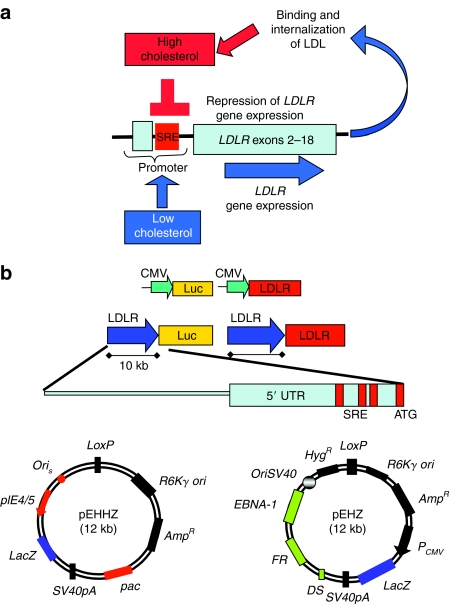
Expression from the LDLR genomic locus is sensitive to intracellular levels of cholesterol and is controlled through a negative-feedback loop (Figure 1a). The critical elements for sensitivity to cellular sterol levels are three SREs situated in the promoter region of the gene (Figure 1a). We generated a series of vectors in which expression of the firefly luciferase (Luc) cDNA or the human LDLR cDNA was placed under control of either the immediate early promoter from the cytomegalovirus (CMV) or a 10-kb piece of genomic DNA 5′ to the LDLR gene (Figure 1b). The 5′ LDLR sequence chosen is a large promoter element of 10 kb of genomic DNA upstream from the initiating ATG codon in exon 2. The 10-kb genomic DNA comprises a portion of the first coding exon (exon 2), the previous noncoding exon (exon 1), the three SREs, the 5′-untranslated region, and ~8 kb of upstream sequence, which may contain unknown further elements essential for expression in the correct physiological context. The 10-kb promoter element was excised from a bacterial artificial chromosome insert containing the human LDLR locus and placed into the Luc or LDLR expression plasmids using homologous recombination in bacteria, an efficient means of subcloning large portions of genomic DNA without the risk of PCR-generated mutagenesis (see Materials and Methods for details and primer sequences). Two plasmids containing Luciferase (pCMV-Luc, pLDLR-Luc) were constructed to test promoter regulation in vitro and in vivo using luciferase reporter gene assays. Two plasmids containing the LDLR cDNA (pCMV-LDLR, pLDLR-LDLR) were constructed to express functional LDL receptors in vitro and in vivo.
Figure 1.
Design and construction of second-generation genomic DNA mini-gene expression vectors to exploit physiological regulation of expression at the low-density lipoprotein receptor (LDLR) genomic locus. (a) Expression of the LDLR is controlled by intracellular levels of cholesterol. Sterol response elements (SREs) in the promoter detect decreasing levels of intracellular cholesterol and drive increased expression from the LDLR promoter. This response is then repressed as cellular cholesterol levels rise. (b) A series of plasmids was created, which contained either the cytomegalovirus (CMV) promoter or a 10-kb piece of genomic DNA encompassing the LDLR genomic promoter and upstream regulatory elements. The promoters drive expression of either the luciferase or the LDLR cDNA. The plasmids were retrofitted with either pEHHZ [containing the HSV-1 amplicon–packaging elements and the episomal retention features from the Epstein–Barr virus (EBV)] or pEHZ (containing the EBV episomal retention elements). Both retrofitting plasmids contain LacZ as a reporter gene under the control of a CMV promoter. Plasmids: pEHHZ-CMV-Luc, pEHHZ-LDLR-Luc, pEHHZ-CMV-LDLR, pEHHZ-LDLR-LDLR, pEHZ-CMV-Luc, pEHZ-LDLR-Luc, pEHZ-0-Luc, pEHZ-CMV-LDLR, pEHZ-LDLR-LDLR. UTR, untranslated region.
For in vitro analysis of expression profiles, viral vector delivery was used. Herpes simplex virus (HSV)-1 amplicons, a highly effective viral delivery system,16,17,21 expressing the various transgene plasmids were used for transgene delivery in vitro, as this resulted in higher transduction rates in all cell types used here. The transduction rate in all cell types around 30% was seen when using a multiplicity of infection (MOI) of 10 and cells displayed no evidence of cytotoxicity following infection. To generate the HSV-1 amplicon vectors, plasmids were retrofitted using Cre/LoxP-mediated recombination with pEHHZ (Figure 1b)16 to produce pEHHZ-CMV-luc, pEHHZ-LDLR-Luc, pEHHZ-CMV-LDLR, and pEHHZ-LDLR-LDLR.
For in vivo work, nonviral hydrodynamic tail-vein injection was used to deliver plasmids, a method that we have previously shown to be highly effective at targeting LDLR expression vectors to the liver.18 The episomal maintenance elements from the Epstein–Barr virus were included in all vectors to ensure long-term vector retention and expression in vitro16 and in vivo.18 It has previously been shown that Epstein–Barr virus–retention elements conferred long-term plasmid retention and enhanced stable transgene expression in mouse liver using plasmids containing as little as 6 kb of genomic DNA in the liver.19 Plasmids were retrofitted using Cre/LoxP-mediated recombination pEHZ, (Figure 1b)18 to produce pEHZ-CMV-luc, pEHZ-LDLR-Luc, pEHZ-0-Luc (a promoter-less construct), pEHZ-CMV-LDLR, and pEHZ-LDLR-LDLR.
Both retrofitting plasmids, pEHZ and pEHHZ, contain LacZ as a reporter under the control of a CMV promoter (Figure 1b).
Luciferase expression in vitro from the pEHHZ-LDLR-Luc construct is under physiological regulation
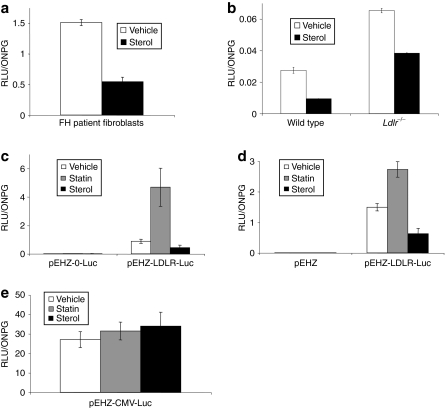
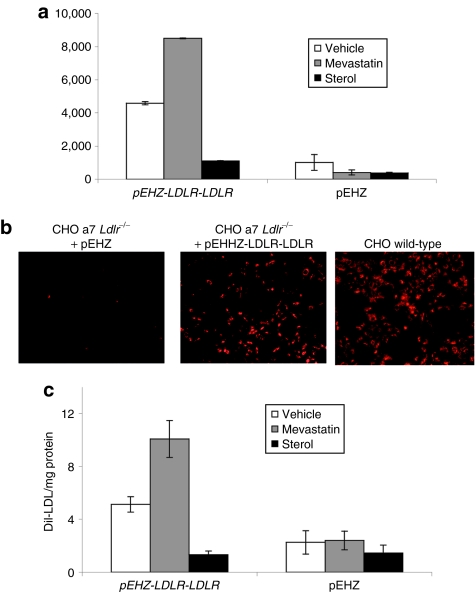
The rationale behind the vector design was to decrease the size of the expression plasmid by using those genomic regulatory elements deemed necessary for providing physiologically regulated expression of the cDNA trangene. We wished to confirm that the vectors containing the 10-kb promoter element conferred similar regulation control to the full LDLR genomic locus. We first looked at luciferase expression in primary cultures in an attempt to repeat previous work completed in primary human fibroblasts using a 135-kb plasmid containing the full LDLR genomic locus.16,17 Primary skin fibroblasts from FH patients (FH-488; Coriel Cell Repository, Camden, NJ) and fibroblasts grown from ear biopsy tissue from wild-type and Ldlr−/− mice were grown for 72 hours in media containing lipid-depleted serum (LPDS; Biomedical Technologies, Stoughton, MA) supplemented with either sterols (cholesterol and 25-hydroxycholesterol) or vehicle control (ethanol). Cells were then infected with HSV-1 amplicons expressing pEHHZ-LDLR-Luc (MOI = 10) and incubated for 48 hours before analysis of luciferase expression. All three fibroblast cell types showed robust luciferase expression that was repressed by up to 40% in the presence of sterols as expected (Figure 2a,b) demonstrating a similar sensitivity to sterols as shown by the whole genomic locus. The function of the pLDLR genomic promoter was further assessed in cell lines with an additional modifier of LDLR promoter activity, 3-hydroxy-3-methylglutaryl coenzyme A reductase inhibitors, or statins. Chinese hamster ovary (CHO) cells deficient for the Ldlr (CHO a7 Ldlr−/−) and the human hepatocarcinoma cell line Hep3b were used. Cell lines were infected with pEHHZ-LDLR-Luc HSV-1 amplicons (MOI = 10). Following transduction, the cells were incubated for 48 hours in media containing 10% lipoprotein-deficient serum (LPDS; Biomedical Technologies) and either sterols (cholesterol and 25-hydroxycholesterol), 2 µmol/l lovastatin (Merck, Nottingham, UK), or 10-µl vehicle (ethanol). At 48 hours post-transduction, cells were analyzed for luciferase activity. CHO a7 Ldlr−/− cells transduced with the pEHHZ-LDLR-Luc plasmid and incubated in lovastatin displayed a fivefold increase in luciferase expression compared to vehicle control (Figure 2d). Cells transduced with the pEHHZ-LDLR-Luc plasmid and incubated with sterols showed a 50% reduction in luciferase expression when compared to the vehicle control. A similar effect on luciferase expression from the LDLR promoter was seen in Hep3b cells (Figure 2c). To control for pleiotropic effects of statins and sterols on gene expression and luciferase activity, we transduced CHO a7 Ldlr−/− cells with the pEHHZ-CMV-Luc amplicon vector (MOI = 10) and incubated the cells for 48 hours in media containing 10% LPDS and either sterols or lovastatin. No effect of statins or sterols was seen on luciferase expression (Figure 2e) confirming that the effect of statins and sterols on luciferase expression was specific to the LDLR promoter constructs as expected.
Figure 2.
Luciferase expression in cells following transduction with pEHHZ-LDLR-Luc was sensitive to regulation by sterols and statins. (a) Familial hypercholesterolemia (FH) patient fibroblasts transduced with pEHHZ-LDLR-Luc showed a 50% reduction in luciferase expression after incubation with sterols. (b) An equivalent 50% reduction in luciferase expression in the presence of sterols was seen in wild-type or Ldlr−/− mouse primary fibroblasts transduced with pEHHZ-LDLR-Luc. (c) Luciferase expression from Chinese hamster ovary (CHO) a7 Ldlr−/− cells transduced with pEHHZ-LDLR-Luc responded to cellular stimuli in a physiologically relevant manner showing a ~50% reduction in gene expression seen with the addition of statins, and an approximately fivefold increase in expression seen with the addition of lovastatin. (d) Luciferase expression in human hepatocarcinoma Hep3b cells transduced with pEHHZ-LDLR-Luc responded to cellular stimuli in a physiologically relevant manner exhibiting a ~50% reduction in gene expression on addition of statins and an approximately twofold increase in luciferase expression on addition of mevastatin. (e) Luciferase expression from CHO a7 Ldlr−/− cells transduced with pEHHZ-CMV-Luc showed no change in luciferase expression when cells were treated with sterols or statins. In each experiment, cells were transduced at a multiplicity of infection of 10 and expression was assayed 48–72 hours post-transduction. Luciferase data are normalized to O-nitrophenyl-β-galactopyranoside (ONPG) to control for differences in vector transduction. Means are from three independent experiments each repeated in quadruplicate. Results are mean ± SD. RLU, relative light units.
Luciferase expression in vivo from the pEHZ-LDLR-Luc plasmid is robust and physiologically regulated
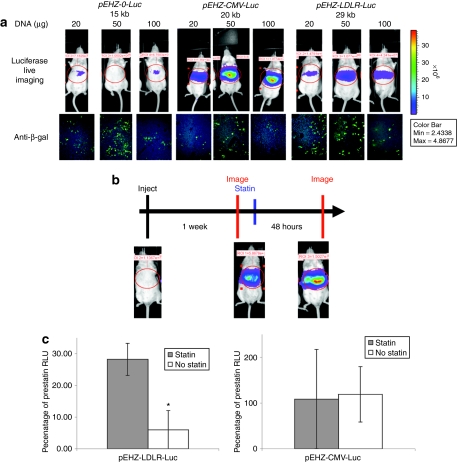
Once we confirmed that the 10-kb piece of genomic DNA encompassing the native human LDLR promoter was sufficient to confer physiological regulation of expression of luciferase in cell culture, we wished to assess whether expression would be robust and sensitive to physiological stimuli in vivo. We have shown previously that hydrodynamic tail-vein injection is an effective means of delivering large genomic plasmids (>100 kb) to the liver.18 First, we wanted to assess whether luciferase expression from the LDLR-Luc expression cassette was detectable in vivo, and second, to assess the efficacy of delivery of smaller plasmids by altering the amount of plasmid delivered in weight.
We used hydrodynamic tail-vein injection to deliver 20, 50, or 100 µg of plasmid DNA (pEHZ-0-Luc, a promoter-less luciferase construct; pEHZ-CMV-Luc; or pEHZ-LDLR-Luc) to MF-1 mice and analyzed luciferase expression and transfection efficiency at 48 hours after injection. As expected, animals injected with pEHZ-0-Luc showed negligible luciferase expression (Figure 3a). All liver sections were assessed for transfection efficiency by immunostaining for β-galactosidase (β-gal) expression and we estimated that we achieved ~10–30% transfection (Figure 3a). In the pEHZ-CMV-Luc and pEHZ-LDLR-Luc injected animals, luciferase expression was detectable following injection of all three plasmid weights. Animals injected with 50 and 100 µg of plasmid DNA showed greater luciferase expression than those injected with 20 µg (Figure 3a) although the number of transfected cells remained broadly similar across all delivery doses. This was an indication of the higher levels of efficiency seen with this technique when using smaller plasmids.
Figure 3.
Luciferase expression in vivo from pEHZ-LDLR-Luc was robust and sensitive to drug administration following hydrodynamic tail-vein injection. (a) Luciferase expression was measured at 48 hours following injection of three different amounts of DNA (20, 50, or 100 µg) of pEHZ-0-Luc, pEHZ-CMV-Luc, and pEHZ-LDLR-Luc. Robust luciferase expression was seen at 48 hours following injection of 50 and 100 µg of pEHZ-CMV-Luc and pEHZ-LDLR-Luc plasmid. Animals injected with the promoter-less construct pEHZ-0-Luc showed negligible luciferase expression. LacZ transgene expression was confirmed in the liver of each animal by immunohistochemistry. The pEHZ-LDLR-Luc construct showed a transfection of hepatocytes of ~25% at the 50-µg dose of plasmid. (b) Time-line of the statin administration experiment. Plasmid DNA was delivered to animals that were imaged 1 week later. Animals were then given an intraperitoneal (i.p.) injection of 600 mg/kg pravastatin (or saline control) and reimaged 48 hours later. Luciferase images from a mouse given pEHZ-LDLR-Luc and then treated with statin are shown. (c) Luciferase expression in vivo from the pEHZ-LDLR-Luc plasmid was robust and sensitive to statins. At 48 hours following an i.p. dose of statin or saline animals injected with pEHZ-LDLR-Luc showed a fivefold increase in luciferase levels compared to untreated animals. Animals injected with pEHZ-CMV-Luc showed no significant difference in luciferase expression following statin administration. Luciferase levels are expressed as a percentage of the luciferase levels calculated from the prestatin administration imaging. Three mice were used for each treatment group. Results are mean ± SD; *P < 0.05. RLU, relative light units.
We next wished to assess the whether luciferase expression from the pLDLR promoter element would be sensitive to in vivo delivery of agents known to increase expression from the endogenous LDLR locus. In this case, we used pravastatin (Merck), a widely used statin that has been shown to have an effect on cholesterol homeostasis in rodent models.22,23,24 MF-1 mice were injected with either 50 µg of pEHZ, pEHZ-CMV-Luc, or pEHZ-LDLR-Luc and imaged 1 week later at which point stable luciferase expression is achieved. Immediately after imaging, animals received a single intraperitoneal injection of 600 mg/kg pravastatin (100 µl), or 100 µl phosphate-buffered saline vehicle. The animals were then imaged at 48 hours after pravastatin injection (Figure 3b). As expected, animals injected with pEHZ-CMV-Luc showed no significant difference in luciferase expression between the statin injected group and the non-statin-injected group (P = 0.48, Figure 3b). However, in the mice injected with pEHZ-LDLR-Luc, the animals which were given a dose of pravastatin showed a significant, fivefold elevation of luciferase expression 48 hours poststatin delivery compared to untreated animals (P = 0.008; Figure 3b), consistent with the pravastatin administration acting to increased levels of luciferase expression from the LDLR promoter element. Overall, luciferase expression from the LDLR-Luc cassette expression is robustly detected following hydrodynamic tail-vein injection and is specifically sensitive to modulation by statins.
Functional complementation by pEHHZ-LDLR-LDLR vectors in cells lacking native LDLR
Following confirmation that the LDLR 10-kb genomic DNA promoter element was capable of driving physiologically regulated transgene expression, we next wished to assess whether the LDLR-LDLR expression cassette was capable of prolonged expression of functional LDL receptors in a physiologically relevant manner to complement LDL receptor deficiency similar to the whole locus bacterial artificial chromosome vectors.16,17
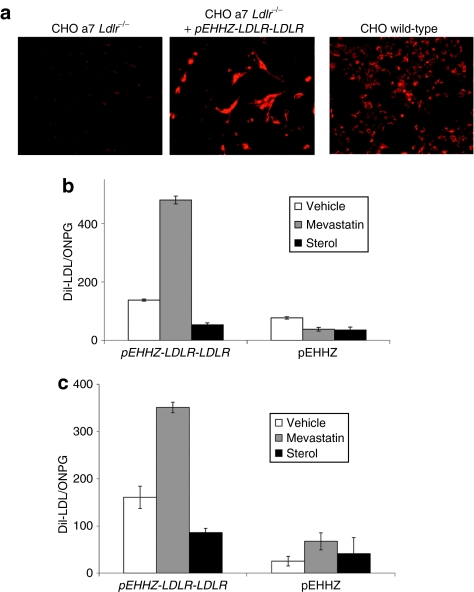
To confirm functional LDLR expression from the pLDLR-LDLR plasmid, we transduced CHO a7 Ldlr−/− cells with HSV-1 amplicons carrying the pEHHZ-LDLR-LDLR vector (MOI = 10) and measured binding and internalization of the fluorescently labeled LDL homologue, DiI-LDL. Cells transduced with the pEHHZ-LDLR-LDLR vector exhibit DiI-LDL binding similar to that seen in wild-type CHO cells (Figure 4a). We then undertook quantitative DiI-LDL analysis to measure functional LDLR binding and internalization of LDL particles. Quantitative analysis of DiI-LDL binding and internalization in CHO a7 Ldlr−/− cells and primary LDLR-deficient fibroblasts from an FH patient (FH-488; Coriel, Camden, NJ) transduced by pEHHZ-LDLR-LDLR HSV-1 amplicons showed exquisite regulation following the addition of sterols or statins. In the presence of sterols, in both cell types, binding and internalization of DiI-LDL was decreased by ~50% (Figure 4b,c) indicating a reduction in the total number of functioning LDL receptors on the cell surface consistent with a reduction in the levels of expression from the LDLR promoter element. In the presence of 2 µmol/l simvastatin (FH cells) or 10 µmol/l mevastatin (CHO a7 Ldlr−/− cells), the binding and internalization of DiI-LDL increased twofold (Figure 4b,c) consistent with statins increasing expression from the LDLR promoter element by inhibiting de novo synthesis of cholesterol. These results are consistent with the observed regulation of luciferase expression and confirm that the pLDLR-LDLR construct is able to complement the genetic deficiency of LDL receptors in a physiologically regulated manner.
Figure 4.
Functional complementation of LDLR deficiency in CHO a7 Ldlr−/− cell lines and familial hypercholesterolemia (FH) primary patient cells. (a) Incubation of Chinese hamster ovary (CHO) a7 Ldlr−/− cells with fluorescently labeled low-density lipoprotein (LDL) (DiI-LDL) after transduction with pEHHZ-LDLR-LDLR confirmed expression of functional LDL receptor (LDLR) from the plasmid. (b) Transduction of CHO a7 Ldlr−/− with pEHHZ-LDLR-LDLR resulted in functional complementation of the LDLR deficiency under physiological regulation. (c) Transduction of FH primary patient fibroblasts with pEHHZ-LDLR-LDLR led to functional complementation of LDLR deficiency under physiological regulation. Cells were transduced at a multiplicity of infection of 10 and expression was assayed 48–72 hours post-transduction. Luciferase data are normalized to O-nitrophenyl-β-galactopyranoside (ONPG) to control for differences in vector transduction. Means are from three independent experiments each repeated in quadruplicate. Results are mean ± SD.
Long-term expression and functional complementation of LDLR in vitro
Following confirmation that the 10-kb LDLR promoter element conferred physiological regulation of LDLR in vitro in the short-term, we wanted to assess whether the promoter element was capable of conferring long-term LDLR expression. CHO a7 Ldlr−/− cells were transfected with either pEHZ-LDLR-LDLR or pEHZ alone. Clonal cell lines were expanded under hygromycin selection for 80 cell-generations. Cells were then grown for 48 hours in lipid-depleted serum containing either ethanol vehicle, 12 µg/ml cholesterol and 0.6 µg/ml 25-hydroxycholesterol, or 10 µmol/l mevastatin. Quantitative analysis of DiI-LDL binding and internalization in cell lines showed that the pEHZ-LDLR-LDLR construct drove regulated LDL expression (Figure 5a). Clonal cell lines were then grown under hygromycin selection for a total of 240 cell-generations and were then assessed for LDLR expression by qualitative and quantitative analysis of DiI-LDL binding and internalization. Clonal cells expressing pEHZ-LDLR-LDLR showed binding and internalization of DiI-LDL comparable to wild-type cells (Figure 5b). As expected, clonal cell lines carrying pEHZ showed negligible DiI-LDL binding and internalization (Figure 5b). Cells were then assessed for regulated expression as described above. Cells transfected with pEHZ-LDLR-LDLR retained DiI-LDL binding and internalization comparable to that seen at 80 generations, which remained sensitive to sterols and statins (Figure 5c) indicating efficient, long-term correction of the genetic deficit in this cell line.
Figure 5.
Long-term, stable correction of low-density lipoprotein receptor (LDLR) deficiency in clonal Chinese hamster ovary (CHO) a7 Ldlr−/− cells transfected with pEHZ-LDLR-LDLR. (a) DiI-LDL binding and internalization in clonal CHO a7 Ldlr−/− cells stably transfected with pEHZ-LDLR-LDLR showed functional complementation of LDL receptor function 80 generations post-transfection. Expression of LDLR was sensitive to incubation with sterols and statins. (b) Fluorescent imaging of clonal CHO a7 Ldlr−/− cells carrying pEHZ-LDLR-LDLR 240 generations post-transfection showed binding and internalization of DiI-LDL comparable to untransfected wild-type CHO cells. (c) DiI-LDL binding and internalization in clonal CHO a7 Ldlr−/− cells stably transfected with pEHZ-LDLR-LDLR showed functional complementation of LDLR function 240 generations post-transfection. Expression of LDLR was sensitive to incubation with sterols and statins. Means are from three independent experiments each repeated in quadruplicate. Results are mean ± SD.
Long-term maintenance and expression of LDLR from the genomic promoter in vivo
Previously, we have shown that hydrodynamic tail-vein injection of a large genomic plasmid containing the full LDLR genomic locus results in long-term expression from the genomic transgene.18 Here, we wanted to investigate whether the pEHZ-LDLR-LDLR plasmid was also capable of conferring similar long-term expression from the genomic promoter element following hydrodynamic tail-vein injection in vivo. Animals were injected with either pEHZ alone (n = 6) or pEHZ-LDLR-LDLR plasmid (n = 6). One animal from each group was killed at 48 hours, 30 days, 120 days, and 270 days and the livers removed for analysis.
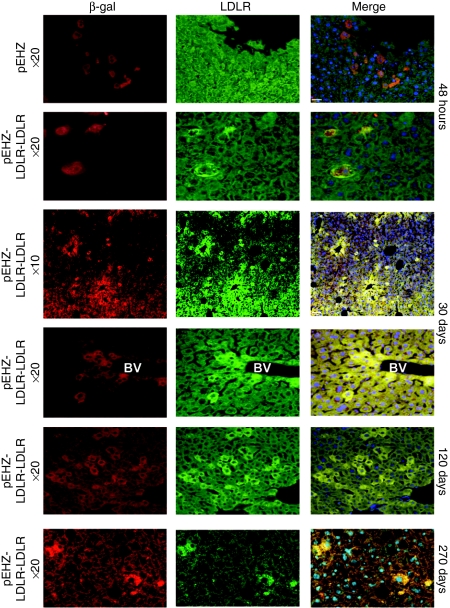
Immunohistochemistry was performed on thin sections to assess expression of human LDLR protein from the pEHZ-LDLR-LDLR plasmid. Immunostaining on liver sections taken at 48 hours from animals injected with pEHZ alone showed high expression of β-gal as expected, but no cells positive for human LDLR protein (Figure 6). Liver sections taken from animals injected with pEHZ-LDLR-LDLR displayed robust β-gal expression, which colocalizes with human LDLR protein expression (Figure 6). At 30 days, 120 days, and 270 days after gene delivery, expression of human LDLR protein that colocalizes with β-gal was still seen (Figure 6). Positive cells showed the typical sinusoidal pattern of transfection, and staining demonstrated a high level of efficiency in transfection (Figure 6). These results confirm that delivery of the pEHZ-LDLR-LDLR plasmid leads to long-term expression in vivo.
Figure 6.
Long-term expression over 9 months of human low-density lipoprotein receptor (LDLR) protein in vivo following hydrodynamic tail-vein injection of pEHZ-LDLR-LDLR. Human LDLR protein was detected up to 270 days following delivery. Sections were co-stained with antibodies specific to β-galactosidase (red) and human LDLR (green) and counterstained with the 4′-6-diamidino-2-phenylindole nuclear stain (blue). At 48 hours after injection with pEHZ-LDLR-LDLR, livers showed colocalized staining for human LDLR and β-galactosidase. This colocalization is absent in the animal injected with pEHZ alone, which is only positive for β-galactosidase expression. At 30 days postinjection, liver sections from the animal injected with pEHZ-LDLR-LDLR showed cells expressing human LDLR protein and β-galactosidase. At ×10 magnification, the extent of transfection was estimated at 20% transfection efficiency at 30 days. pEHZ-LDLR-LDLR transgene expression was shown to persist at 120 and 270 days postinjection. Bacterial plasmid rescue assays from genomic DNA isolated from animals injected with pEHZ-LDLR-LDLR demonstrated plasmid retention as an episomal element at each time point (data not shown). BV, blood vessel.
Plasmid rescue was performed by isolating genomic DNA from livers of animals 48 hours, 30 days, 120 days, and 270 days postinjection with pEHZ-LDLR-LDLR and electroporating the DNA into bacteria. DNA prepared from eight bacterial colonies at each time point confirmed the presence of intact circular pEHZ-LDLR-LDLR plasmid at each time point (data not shown).
Discussion
The application of vectors that provide prolonged transgene expression at therapeutic levels under physiological regulation has broad potential in the field of gene therapy. Many disorders would benefit from the use of vectors in which expression is under the control of endogenous regulation elements; yet to date few studies have looked at developing this. The most elegant way of achieving physiological expression is through the use of a whole genomic locus; however, this may require vectors to deliver a very large genomic DNA transgene >100 kb, which may present technical difficulties. There are several viral systems, which can accommodate large DNA inserts, such as the HSV-1 amplicons,16,17,21,25 used here for in vitro analysis. These vector systems represent a suitable means of transgene delivery to many cell types in vitro and in vivo. However, for this study a nonviral method was used for delivery to the liver in vivo as this proved more efficient.
In our previous work developing gene therapy for FH using LDLR gene delivery, we found that the use of the whole genomic locus of the LDLR was a highly effective means of complementing the loss of function seen in this condition in vitro, by providing physiological expression of LDLR.16,17 However, when we attempted the use of the whole genomic locus in vivo, delivery efficiency of constructs >100 kb in size to a large target organ, such as the liver, became an obstacle to efficient transfection. We therefore redesigned our gene expression vectors to combine the convenient size of a cDNA vector with the physiological regulation of an entire genomic locus. The promoter region of the human LDLR locus has been the subject of extensive research and key sequences known to effect gene expression have been identified.26 We have built on this previous work and isolated a 10-kb region of genomic DNA upstream from the initiating ATG start codon that contained all the known relevant regulatory elements. This promoter element was used to drive cDNA transgenes as a genomic DNA mini-gene construct. The resulting vectors were approximately one-fifth the size of the whole genomic locus transgene vectors that we used in previous studies,16,18 i.e., 30 kb versus 154 kb.
The use of vectors containing the luciferase reporter gene driven by the 10-kb promoter element allowed us to analyze the regulation of expression from the promoter element in vivo using noninvasive live bioluminescence imaging. The ability to follow the luciferase expression profile in individual animals over time proved to be very powerful. We were able to show that the 3-hydroxy-3-methylglutaryl coenzyme A reductase inhibitor pravastatin enhanced the expression from the LDLR promoter element, a result which raises an interesting therapeutic option. By co-treating FH patients with the LDLR-LDLR gene therapy vector plus statin therapy, we could possibly enhance therapeutic outcome by decreasing intracellular cholesterol and hence increase LDLR-LDLR transgene cassette expression to enhance the cholesterol-lowering effect of the therapy. This is an important concept because we would increase expression from transduced cells without leading to a toxic build up of intracellular cholesterol, which would trigger apoptotic cell death.15 Administration of statins effectively tips the balance of cellular cholesterol homeostasis in the favor of receptor-mediated endocytosis instead of de novo synthesis. This will be investigated in future studies looking at functional complementation in Ldlr−/− mice.
In this study, we have found that a 10-kb sequence of upstream genomic DNA was sufficient to confer physiological regulation on a transgene. In this work, in vitro studies were performed in a variety of cell lines to analyze the long-term expression characteristics of the LDLR promoter element. The 10-kb LDLR genomic DNA promoter element was shown to be sensitive to substances known to affect the activity of the native promoter indicating responsiveness to changing cellular milieu similar to the expression pattern from the whole genomic locus transgene. In vitro incubation with sterols lead to a decrease in expression of either luciferase or LDLR cDNA from the LDLR promoter element. In vitro and in vivo, we observed effective upregulation of luciferase expression, typically around a fivefold increase, following administration of statins. The Epstein–Barr virus elements were included in our vectors as they have been previously shown to enhance retention in vitro and expression in vivo,16,18,19,20,27,28,29,30 although this study did not specifically investigate episomal replication. The presence of genomic DNA inserts as small as 6 kb (ref. 19) are known to also benefit expression. In our previous article, we showed expression of human LDLR protein from a genomic transgene plasmid up to 4 months following hydrodynamic tail-vein injection.18 In this study, we have extended this finding with the second-generation vectors that contain 10 kb of genomic DNA promoter sequence to 270 days. Overall, long-term expression under physiological regulation makes the transgene cassette design attractive for further gene therapy protocols for FH.
Nonviral methods for in vivo gene delivery are an attractive alternative to viral-mediated gene transfer as they offer gene transfer without the potential risks of immunogenicity and cell transformation. Here, we have used hydrodynamic tail-vein injection to deliver transgene DNA directly to the liver. Hydrodynamic tail-vein injection can be a highly efficient means of delivering DNA to the liver. Previous work has shown it to be effective when using small plasmids31,32,33,34 or large vectors with inserts >100 kb (refs. 18,35). The application of hydrodynamic tail-vein injection in small rodent models is well described18,20,33,36,37 and recent work has attempted to transfer the technology to large animal models and humans. Hydrodynamic delivery of plasmid DNA to porcine liver38,39,40 shows that gene transfection is possible in this large animal model, albeit at a level ~200-fold lower than in mice and rats, probably due to the decreased elasticity of the pig liver.39 Further improvements in the methods, employing novel surgical techniques designed to make the procedure minimally invasive,40,41 will allow the technique to move closer to the clinic. One recently published study using balloon catheter technology for hydrodynamic delivery to humans demonstrated that the technique can be applied to humans;40 however, in this study transgene expression was not demonstrated in treated patients.
Our work has developed expression plasmids that combine the efficient delivery of a small cDNA vector with the long-term physiological regulation provided by a genomic transgene. We also show that the use of drugs in addition to gene transfer can enhance transgene expression. Future work will assess the efficacy of these vectors for therapeutic intervention in animal models of FH.
Materials and Methods
Vector construction. The backbone vector used for the generation of all four plasmid vectors was p1009, a kanamycin-resistant, LoxP-containing vector generated by ligating kanamycin resistance from pET-9A into pGL3-basic (Promega, Southampton, UK). A LoxP site was added to the resulting vector by ligating oligolinkers containing BglI and EcoRI sites (Linker sequence: cggcataacttcgtataatgtatgctatacgaagttatgaattcgccgcct). A p1009/CMV intermediate plasmid was generated by ligating a BglII/PacI fragment containing pCMV and the human polyadenylation signal BGHpA into p1009. The LDLR cDNA was ligated into this vector from pHGCX-LDLR using a NotI/AflII digest to generate pCMV-LDLR. pCMV-Luc was generated by PCR amplification of a NotI/XbaI-flanked luciferase cDNA fragment and ligated into p1009/CMV. To create vectors containing the 10-kb genomic promoter element, we designed homologous recombination primers to amplify the backbone vectors, pCMV-LDLR or pCMV-Luc, using 55 bp of homology to target sequences in bacterial artificial chromosome 164O19 clone (Genbank ACO 11485) and 25 bp of primer sequence to amplify all vector components except the CMV promoter. The forward primers amplified pCMV-LDLR or pCMV-Luc from the start codons of either LDLR or luciferase (base pairs in lower case are 55-bp homology arms and have homology to LDLR genomic sequence upstream of the LDLR start codon. LDLR primer: 5′-aggacacagcaggtcgtgatccgggtcgggacactgcctggcagaggctgcgagcATGGGGCCCTGGGGCTGGAAATTGCG-3′, Luciferase primer: 5′-aggacacagcaggtcgtgatccgggtcgggacactgcctggcagaggctgcgagcATGGAAGACGCCAAAAACATAAAGA-3′). We designed a single reverse primer that contained a 55-bp homology arm situated 10-kb downstream of the LDLR start codon (reverse primer sequence; 5′-cgcctcatcttcccagtgttgggattacaggtgtgagccaccatgcccggccatactcggtacctatcgatagagaaatg-3′). Long-range PCR was performed on pCMV-LDLR and pCMV-Luc plasmids using BioXact-long polymerase (Bioline, London, UK). PCR products were column purified using PCR purification columns (Qiagen, Crawley, UK) and electroporated into bacteria containing LDLR bacterial artificial chromosome clone (164019 Genbank ACO 11485) and rec/ET plasmid pSC101. All plasmids were then retrofitted using Cre/LoxP recombination with pEHZ or pEHHZ.
Tissue culture. CHO wild-type and CHO a7 Ldlr−/− cells were grown in HAMS F12 media supplemented with 10% fetal bovine serum (FBS), L-glutamine, penicillin (100 U/ml), and streptomycin (100 U/ml). Clonal CHO cell lines were obtained through transfection with pEHZ-LDLR or pEHZ alone using 1 µl lipofectamine (Invitrogen, Paisley, UK) per microgram of plasmid DNA. Cells were expanded in the presence of 500 µg/ml hygromycin. Hep3b cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% FBS, L-glutamine, penicillin (100 U/ml), and streptomycin (100 U/ml). FH patient cell lines were obtained from The National Institute of General Medical Sciences Coriell Cell Repository (Camden, NJ). Human fibroblasts were cultured in Eagle's modified medium, with Earle's salts, supplemented with 15% FBS, L-glutamine, penicillin (100 U/ml), and streptomycin (100 U/ml). Primary mouse fibroblasts were obtained from ear biopsy tissue as described.42 Cells were grown in Dulbecco's modified Eagle's medium supplemented with 10 % FBS, non essential amino acids, L-glutamine, penicillin (100 U/ml), and streptomycin (100 U/ml).
HSV-1 amplicon production and infection. HSV-1 amplicons were produced using an improved HSV-1 helper virus–free system as previously described.21 Briefly, supernatant from nine 6-cm dishes was concentrated for 1 hour at 78,000 g over a 30% sucrose cushion and resuspended in 250 µl OptiMEM (Invitrogen) to give a stock of 5 × 108 transducing units/ml. For amplicon infection 5 × 104 (CHO, Hep3b) or 2.5 × 104 (FH-488, primary mouse fibroblasts) cells were plated per well of a 24-well dish. At 24–48 hours after plating, cells were infected with an HSV-1 amplicon at a typical MOI of 10. Amplicon particles were diluted in 200 µl of OptiMEM and added to plated cells in 24-well plates. Plates were then centrifuged 1,500 g for 15 minutes followed by 45 minutes at 700 g. The transduction medium was then replaced with medium containing FBS and/or sterols and statins.
In vitro Luciferase expression analysis. Lipoprotein-deficient FBS (LPDS) was purchased from Biomedical Technologies. Luciferase expression was analyzed using a Dynex Mix Luciferase plate-reader (LabMode, Chantilly, VA). Cells were lysed in Luciferase lysis buffer (25 mmol/l Tris–PO4 pH 7.8, 0.2 mmol/l 1,2-diaminocyclohexanetetraacetic acid, 1:10 glycerol, 1:100 Triton X-100, 2 µmol/l dithiothretol) for 20 minutes. Lysate was transferred to 96-well assay plates and was analyzed through the addition of luciferin (0.3 mg/ml; Caliper Life Sciences, Hopkinton, MA) and ATP (200 mg/ml) in assay buffer (15 mmol/l MgSO4, 15 mmol/l KPO4 pH 7.8, 0.04 mmol/l ethylene glycol tetraacetic acid pH 7.8, 2 µmol/l dithiothretol). Relative light units were normalized to β-gal expression via O-nitrophenyl-β-galactopyranoside assay. O-nitrophenyl-β-galactopyranoside assay buffer (6 mmol/l Na2HPO4, 4 mmol/l Na2H2PO4, 10 mmol/l KCl, 1 mmol/l MgSO4, 20 mg/ml O-nitrophenyl-β-galactopyranoside, 2 µmol/l dithiothretol, and 50 µl β-mercaptoethanol; 100 µl) was added to 3 µl of cell lysate and incubated until development of yellow color. The reaction was stopped using 50 mmol/l Na2CO3 (50 µl) and absorbance read at OD 460. To investigate the effect of sterols and statins on luciferase expression, the medium was supplemented with LPDS and either 12 µg/ml cholesterol and 0.6 µg/ml 25-hydroxycholesterol or appropriate volumes of statins all dissolved in ethanol. An equal volume of ethanol was added to control wells to act as a vehicle-only control.
In vitro LDLR expression analysis. Human LDL and DiI-LDL were purchased from AbD Serotec (Kidlington, Oxford). Quantitative analysis of LDLR function by measuring DiI-LDL fluorescence was performed as described17 with modified lysis buffer (25 mmol/l Tris–PO4 pH 7.8, 0.2 mmol/l 1,2-diaminocyclohexanetetraacetic acid, 1:10 glycerol, 1:100 Triton X-100, 2 µM dithiothretol). DiI fluorescence levels in the cell lysate were analyzed using a Fluostar spectrofluorophotometer plate-reader at excitation and emission wavelengths of 520 and 580 nm, respectively, and total protein content determined either using bicinchoninic acid solution (Sigma, Dorset, UK) for clonal cell lines or β-gal expression was determined using O-nitrophenyl-β-galactopyranoside assay. Nonspecific binding was determined in the presence of a 50-fold excess of unlabeled LDL and subtracted from total binding to give specific binding. To investigate the effect of sterols and statins on LDLR expression, the medium was supplemented using sterols and statins as described above.
Animals. Female MF-1 mice, an outbred strain used previously18 were obtained from Harlan, UK and housed on a 12:12 light–dark cycle and fed ad libitum. All animal procedures were conducted in accordance with the Animals (Scientific Procedures) Act 1986, and after appropriate ethical review.
Hydrodynamic delivery and imaging. Animals weighing 25–30 g received hydrodynamic tail-vein injections of plasmid DNA as described.18 Animals that received luciferase plasmids were imaged using an IVIS 100 luciferase imaging camera (Caliper Life Sciences). Before imaging, animals were anesthetized with isoflurane and given a 100 µl intraperitoneal injection of a 15 mg/ml luciferin solution (in phosphate-buffered saline; Caliper Life Sciences). Animals were placed inside the chamber and anesthesia maintained. Following a 4-minute incubation period, images were taken and luciferase expression quantified using LivingImage software (Caliper Life Sciences). As appropriate, animals then received 600 mg/kg pravastatin dissolved in phosphate-buffered saline as a 100 µl intraperitoneal injection, or 100 µl of phosphate-buffered saline alone. Animals that received LDLR plasmids were allowed to recover and left for the appropriate amount of time before sacrifice.
Plasmid rescue. Frozen liver tissue was homogenized in genomic lysis buffer (0.6% sodium dodecyl sulfate, 100 mmol/l NaCl, 50 mmol/l Tris pH 8, 20 mmol/l EDTA) and incubated with 100 µl proteinase K overnight at 37 °C. Genomic DNA was then purified with phenol/chloroform extraction and precipitated with 100% ethanol. DNA pellets were left to air dry and resuspended in 200 µl Tris–EDTA. To assess the presence of circular plasmid, DNA (1 µl) was electroporated into DH10B bacteria and resulting colonies grown overnight in 1.5 ml culture medium. Plasmid DNA was purified using alkaline lysis and DNA analyzed by restriction enzyme digestion.
Histochemical tissue analysis. Livers were fixed by perfusion fixation in paraformaldehyde and embedded in paraffin tissue blocks. Immunohistochemistry for β-gal expression and colocalization of human LDLR with β-gal on thin (5 µm) sections was undertaken as described previously.18
Acknowledgments
We acknowledge the technical input of Fionnadh Carrol in the use of the luciferase imaging equipment, Yasser El Sherbini for technical assistance in amplicon transduction, and Angeliki Biba for the construction of the p1009 plasmid. This work was supported by the British Heart Foundation and the Medical Research Council (UK).
REFERENCES
- Tolleshaug H, Hobgood KK, Brown MS., and , Goldstein JL. The LDL receptor locus in familial hypercholesterolemia: multiple mutations disrupt transport and processing of a membrane receptor. Cell. 1983;32:941–951. doi: 10.1016/0092-8674(83)90079-x. [DOI] [PubMed] [Google Scholar]
- Chowdhury JR, Grossman M, Gupta S, Chowdhury NR, Baker JR., Jr, and , Wilson JM. Long-term improvement of hypercholesterolemia after ex vivo gene therapy in LDLR-deficient rabbits. Science. 1991;254:1802–1805. doi: 10.1126/science.1722351. [DOI] [PubMed] [Google Scholar]
- Grossman M, Rader DJ, Muller DW, Kolansky DM, Kozarsky K, Clark BJ, 3rd, et al. A pilot study of ex vivo gene therapy for homozygous familial hypercholesterolaemia. Nat Med. 1995;1:1148–1154. doi: 10.1038/nm1195-1148. [DOI] [PubMed] [Google Scholar]
- Miyanohara A, Sharkey MF, Witztum JL, Steinberg D., and , Friedmann T. Efficient expression of retroviral vector-transduced human low density lipoprotein (LDL) receptor in LDL receptor-deficient rabbit fibroblasts in vitro. Proc Natl Acad Sci USA. 1988;85:6538–6542. doi: 10.1073/pnas.85.17.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JM, Johnston DE, Jefferson DM., and , Mulligan RC. Correction of the genetic defect in hepatocytes from the Watanabe heritable hyperlipidemic rabbit. Proc Natl Acad Sci USA. 1988;85:4421–4425. doi: 10.1073/pnas.85.12.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankkonen HM, Vähäkangas E, Marr RA, Pakkanen T, Laurema A, Leppänen P, et al. Long-term lowering of plasma cholesterol levels in LDL-receptor-deficient WHHL rabbits by gene therapy. Mol Ther. 2004;9:548–556. doi: 10.1016/j.ymthe.2004.01.015. [DOI] [PubMed] [Google Scholar]
- Ishibashi S, Brown MS, Goldstein JL, Gerard RD, Hammer RE., and , Herz J. Hypercholesterolemia in low density lipoprotein receptor knockout mice and its reversal by adenovirus-mediated gene delivery. J Clin Invest. 1993;92:883–893. doi: 10.1172/JCI116663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozarsky KF, Jooss K, Donahee M, Strauss JF., 3rd, and , Wilson JM. Effective treatment of familial hypercholesterolaemia in the mouse model using adenovirus-mediated transfer of the VLDL receptor gene. Nat Genet. 1996;13:54–62. doi: 10.1038/ng0596-54. [DOI] [PubMed] [Google Scholar]
- Kozarsky KF, McKinley DR, Austin LL, Raper SE, Stratford-Perricaudet LD., and , Wilson JM. In vivo correction of low density lipoprotein receptor deficiency in the Watanabe heritable hyperlipidemic rabbit with recombinant adenoviruses. J Biol Chem. 1994;269:13695–13702. [PubMed] [Google Scholar]
- Li J, Fang B, Eisensmith RC, Li XH, Nasonkin I, Lin-Lee YC, et al. In vivo gene therapy for hyperlipidemia: phenotypic correction in Watanabe rabbits by hepatic delivery of the rabbit LDL receptor gene. J Clin Invest. 1995;95:768–773. doi: 10.1172/JCI117725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura S, Merched A, Nour E, Dieker C, Oka K., and , Chan L. Low-density lipoprotein receptor gene therapy using helper-dependent adenovirus produces long-term protection against atherosclerosis in a mouse model of familial hypercholesterolemia. Gene Ther. 2004;11:1540–1548. doi: 10.1038/sj.gt.3302310. [DOI] [PubMed] [Google Scholar]
- Lebherz C, Gao G, Louboutin JP, Millar J, Rader D., and , Wilson JM. Gene therapy with novel adeno-associated virus vectors substantially diminishes atherosclerosis in a murine model of familial hypercholesterolemia. J Gene Med. 2004;6:663–672. doi: 10.1002/jgm.554. [DOI] [PubMed] [Google Scholar]
- Südhof TC, Van der Westhuyzen DR, Goldstein JL, Brown MS., and , Russell DW. Three direct repeats and a TATA-like sequence are required for regulated expression of the human low density lipoprotein receptor gene. J Biol Chem. 1987;262:10773–10779. [PubMed] [Google Scholar]
- Heeren J, Steinwaerder DS, Schnieders F, Cichon G, Strauss M., and , Beisiegel U. Nonphysiological overexpression of low-density lipoprotein receptors causes pathological intracellular lipid accumulation and the formation of cholesterol and cholesteryl ester crystals in vitro. J Mol Med. 1999;77:735–743. doi: 10.1007/s001099900045. [DOI] [PubMed] [Google Scholar]
- Cichon G, Willnow T, Herwig S, Uckert W, Löser P, Schmidt HH, et al. Non-physiological overexpression of the low density lipoprotein receptor (LDLr) gene in the liver induces pathological intracellular lipid and cholesterol storage. J Gene Med. 2004;6:166–175. doi: 10.1002/jgm.473. [DOI] [PubMed] [Google Scholar]
- Wade-Martins R, Saeki Y., and , Chiocca EA.2003Infectious delivery of a 135-kb LDLR genomic locus leads to regulated complementation of low-density lipoprotein receptor deficiency in human cells Mol Ther 7(5 Pt 1): 604–612. [DOI] [PubMed] [Google Scholar]
- Lufino MM, Manservigi R., and , Wade-Martins R. An S/MAR-based infectious episomal genomic DNA expression vector provides long-term regulated functional complementation of LDLR deficiency. Nucleic Acids Res. 2007;35:e98. doi: 10.1093/nar/gkm570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbitt OC, Harbottle RP, Waddington SN, Bursill CA, Coutelle C, Channon KM, et al. Delivery and long-term expression of a 135 kb LDLR genomic DNA locus in vivo by hydrodynamic tail vein injection. J Gene Med. 2007;9:488–497. doi: 10.1002/jgm.1041. [DOI] [PubMed] [Google Scholar]
- Sclimenti CR, Neviaser AS, Baba EJ, Meuse L, Kay MA., and , Calos MP. Epstein-Barr virus vectors provide prolonged robust factor IX expression in mice. Biotechnol Prog. 2003;19:144–151. doi: 10.1021/bp0200907. [DOI] [PubMed] [Google Scholar]
- Stoll SM, Sclimenti CR, Baba EJ, Meuse L, Kay MA., and , Calos MP. Epstein-Barr virus/human vector provides high-level, long-term expression of α1-antitrypsin in mice. Mol Ther. 2001;4:122–129. doi: 10.1006/mthe.2001.0429. [DOI] [PubMed] [Google Scholar]
- Saeki Y, Fraefel C, Ichikawa T, Breakefield XO., and , Chiocca EA. Improved helper virus-free packaging system for HSV amplicon vectors using an ICP27-deleted, oversized HSV-1 DNA in a bacterial artificial chromosome. Mol Ther. 2001;3:591–601. doi: 10.1006/mthe.2001.0294. [DOI] [PubMed] [Google Scholar]
- Koga T, Shimada Y, Kuroda M, Tsujita Y, Hasegawa K., and , Yamazaki M. Tissue-selective inhibition of cholesterol synthesis in vivo by pravastatin sodium, a 3-hydroxy-3-methylglutaryl coenzyme A reductase inhibitor. Biochim Biophys Acta. 1990;1045:115–120. doi: 10.1016/0005-2760(90)90139-o. [DOI] [PubMed] [Google Scholar]
- Bisgaier CL, Essenburg AD, Auerbach BJ, Pape ME, Sekerke CS, Gee A, et al. Attenuation of plasma low density lipoprotein cholesterol by select 3-hydroxy-3-methylglutaryl coenzyme A reductase inhibitors in mice devoid of low density lipoprotein receptors. J Lipid Res. 1997;38:2502–2515. [PubMed] [Google Scholar]
- Johnson J, Carson K, Williams H, Karanam S, Newby A, Angelini G, et al. Plaque rupture after short periods of fat feeding in the apolipoprotein E-knockout mouse: model characterization and effects of pravastatin treatment. Circulation. 2005;111:1422–1430. doi: 10.1161/01.CIR.0000158435.98035.8D. [DOI] [PubMed] [Google Scholar]
- Hibbitt OC., and , Wade-Martins R. Delivery of large genomic DNA inserts >100 kb using HSV-1 amplicons. Curr Gene Ther. 2006;6:325–336. doi: 10.2174/156652306777592054. [DOI] [PubMed] [Google Scholar]
- Pak YK. Serum response element-like sequences of the human low density lipoprotein receptor promoter: possible regulation sites for sterol-independent transcriptional activation. Biochem Mol Biol Int. 1996;38:31–36. [PubMed] [Google Scholar]
- Wang S., and , Vos JM. A hybrid herpesvirus infectious vector based on Epstein-Barr virus and herpes simplex virus type 1 for gene transfer into human cells in vitro and in vivo. J Virol. 1996;70:8422–8430. doi: 10.1128/jvi.70.12.8422-8430.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade-Martins R, Frampton J., and , James MR. Long-term stability of large insert genomic DNA episomal shuttle vectors in human cells. Nucleic Acids Res. 1999;27:1674–1682. doi: 10.1093/nar/27.7.1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade-Martins R, White RE, Kimura H, Cook PR., and , James MR. Stable correction of a genetic deficiency in human cells by an episome carrying a 115 kb genomic transgene. Nat Biotechnol. 2000;18:1311–1314. doi: 10.1038/82444. [DOI] [PubMed] [Google Scholar]
- Oehmig A, Fraefel C, Breakefield XO., and , Ackermann M. Herpes simplex virus type 1 amplicons and their hybrid virus partners, EBV, AAV, and retrovirus. Curr Gene Ther. 2004;4:385–408. doi: 10.2174/1566523043346129. [DOI] [PubMed] [Google Scholar]
- Liu F, Song Y., and , Liu D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 1999;6:1258–1266. doi: 10.1038/sj.gt.3300947. [DOI] [PubMed] [Google Scholar]
- Zhang G, Budker V., and , Wolff JA. High levels of foreign gene expression in hepatocytes after tail vein injections of naked plasmid DNA. Hum Gene Ther. 1999;10:1735–1737. doi: 10.1089/10430349950017734. [DOI] [PubMed] [Google Scholar]
- Zhang G, Song YK., and , Liu D. Long-term expression of human α1-antitrypsin gene in mouse liver achieved by intravenous administration of plasmid DNA using a hydrodynamics-based procedure. Gene Ther. 2000;7:1344–1349. doi: 10.1038/sj.gt.3301229. [DOI] [PubMed] [Google Scholar]
- Zhang G, Vargo D, Budker V, Armstrong N, Knechtle S., and , Wolff JA. Expression of naked plasmid DNA injected into the afferent and efferent vessels of rodent and dog livers. Hum Gene Ther. 1997;8:1763–1772. doi: 10.1089/hum.1997.8.15-1763. [DOI] [PubMed] [Google Scholar]
- Magin-Lachmann C, Kotzamanis G, D'Aiuto L, Cooke H, Huxley C., and , Wagner E. In vitro and in vivo delivery of intact BAC DNA—comparison of different methods. J Gene Med. 2004;6:195–209. doi: 10.1002/jgm.481. [DOI] [PubMed] [Google Scholar]
- Ajuf'ev BN, Dizhe EB, Efremov AM, Mogilenko DA, Oleinikova GN, Lapikov IA, et al. Hydrodynamics-based transfer of human apolipoprotein A-I gene into mice: study of factors involving an efficacy and duration of the transferred gene expression in animals' liver. Mol Biol (Mosk) 2004;38:1076–1084. [PubMed] [Google Scholar]
- Zhang X, Dong X, Sawyer GJ, Collins L., and , Fabre JW. Regional hydrodynamic gene delivery to the rat liver with physiological volumes of DNA solution. J Gene Med. 2004;6:693–703. doi: 10.1002/jgm.595. [DOI] [PubMed] [Google Scholar]
- Yoshino H, Hashizume K., and , Kobayashi E. Naked plasmid DNA transfer to the porcine liver using rapid injection with large volume. Gene Ther. 2006;13:1696–1702. doi: 10.1038/sj.gt.3302833. [DOI] [PubMed] [Google Scholar]
- Fabre JW, Grehan A, Whitehorne M, Sawyer GJ, Dong X, Salehi S, et al. Hydrodynamic gene delivery to the pig liver via an isolated segment of the inferior vena cava. Gene Ther. 2008;15:452–462. doi: 10.1038/sj.gt.3303079. [DOI] [PubMed] [Google Scholar]
- Khorsandi SE, Bachellier P, Weber JC, Greget M, Jaeck D, Zacharoulis D, et al. Minimally invasive and selective hydrodynamic gene therapy of liver segments in the pig and human. Cancer Gene Ther. 2008;15:225–230. doi: 10.1038/sj.cgt.7701119. [DOI] [PubMed] [Google Scholar]
- Aliño SF, Herrero MJ, Noguera I, Dasí F., and , Sánchez M. Pig liver gene therapy by noninvasive interventionist catheterism. Gene Ther. 2007;14:334–343. doi: 10.1038/sj.gt.3302873. [DOI] [PubMed] [Google Scholar]
- Kulnane LS, Lehman EJ, Hock BJ, Tsuchiya KD., and , Lamb BT. Rapid and efficient detection of transgene homozygosity by FISH of mouse fibroblasts. Mamm Genome. 2002;13:223–226. doi: 10.1007/s00335-001-2128-5. [DOI] [PubMed] [Google Scholar]