Abstract
Human artificial chromosome (HAC) has several advantages as a gene therapy vector, including stable episomal maintenance that avoids insertional mutations and the ability to carry large gene inserts including the regulatory elements. Induced pluripotent stem (iPS) cells have great potential for gene therapy, as such cells can be generated from the individual's own tissues, and when reintroduced can contribute to the specialized function of any tissue. As a proof of concept, we show herein the complete correction of a genetic deficiency in iPS cells derived from Duchenne muscular dystrophy (DMD) model (mdx) mice and a human DMD patient using a HAC with a complete genomic dystrophin sequence (DYS-HAC). Deletion or mutation of dystrophin in iPS cells was corrected by transferring the DYS-HAC via microcell-mediated chromosome transfer (MMCT). DMD patient- and mdx-specific iPS cells with the DYS-HAC gave rise to differentiation of three germ layers in the teratoma, and human dystrophin expression was detected in muscle-like tissues. Furthermore, chimeric mice from mdx-iPS (DYS-HAC) cells were produced and DYS-HAC was detected in all tissues examined, with tissue-specific expression of dystrophin. Therefore, the combination of patient-specific iPS cells and HAC-containing defective genes represents a powerful tool for gene and cell therapies.
Introduction
Embryonic stem (ES) cells have great potential for cell therapy against genetic disorders, as such cells can contribute to the specialized function of any tissue.1,2 However, one potential problem using ES cells to treat genetic disorders is host immunorejection of the transplanted cells. Although ES cells for gene therapy may be created for a patient using the nuclear transfer technique, many ethical concerns are associated with this practice.3,4 Conversely, induced pluripotent stem (iPS) cells can be generated from the tissues of the individual with defined factors.5,6,7 Gene and cell therapies with iPS cells will thus offer advantages over nuclear transfer ES cell–mediated gene therapy with respect to ethical problems, as well as providing a genetic match with the patient and so decreasing the likelihood of immunorejection.
Homologous recombination has been used for the gene restoration of various genetic defects in ES or iPS cells using one's own genetic information.8,9 However, gene defects with unknown sites of mutation and those involving large deletions cannot be restored by homologous recombination.10 Duchenne muscular dystrophy (DMD) is caused by dysfunction of the dystrophin gene.11,12,13,14,15 As some DMD patients show a large deletion in the DMD gene, these defects cannot be restored by homologous recombination or exon-skipping approaches. Although several vectors have been developed for DMD gene therapy, no episomal vector containing the entire dystrophin genomic region has been reported, due to the extremely large size of this region (2.4 megabases).16 Human artificial chromosome (HAC) offers several advantages as gene therapy vector, including stable episomal maintenance that avoids insertional mutations and the ability to carry large gene inserts including the associated regulatory elements.17,18,19,20,21,22,23 We therefore recently developed a HAC vector containing an entire dystrophin genome for DMD gene therapy.21 In this study, we established a HAC-mediated genomic transfer system as a paradigm for the treatment of a genetic disorder such as DMD by combining patient-derived iPS cells with a HAC vector containing the normal version of a defective gene.
Results
Characterization of iPS cells from mdx mice
First, we attempted genetic correction of iPS cells derived from mdx mouse, as a model for DMD (Figure 1). The mdx-iPS cells were induced from mdx mouse embryonic fibroblasts by retroviral infection of the three factors including Klf4, Sox2, and Oct4. Transduced fibroblasts from the mdx mice gave rise to ES cell–like colonies, and these colonies were isolated based on morphological criteria. Most mdx-iPS cells were positive for ES cell markers, displayed a normal karyotype, and generated teratoma and chimeras (Supplementary Figure S1). Two randomly selected clones were used for the following experiments.
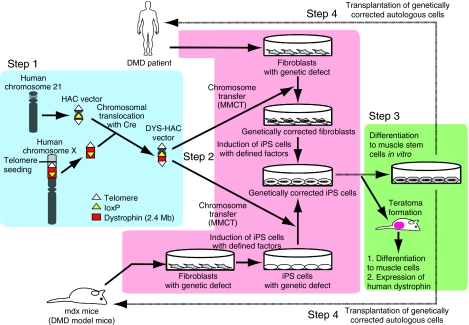
Figure 1.
Schematic diagram of the HAC vector system for iPS cell–mediated gene therapy. Human dystrophin gene was cloned into a human chromosome 21–derived HAC vector using a combination of Cre-loxP-mediated chromosomal translocation and telomere-directed chromosomal truncation (Step 1, blue background). In Step 2 (pink background), mdx mice- or DMD patient–derived iPS cells were genetically restored by transfer of the DYS-HAC vector. The DYS-HAC was transferred to mdx-iPS cells directly. However, the DYS-HAC was transferred to DMD patient–derived fibroblasts via MMCT, as we failed to directly transfer the HAC into human iPS cells, then DMD-fibroblasts (DYS-HAC) were induced into iPS cells. Inability of MMCT into human iPS and embryonic stem cells is an unsolved issue. In Step 3 (green background), differentiation to muscle cells in vivo (teratoma formation) and expression of human dystrophin in muscle cells were confirmed. Step 4 represents the future and final goal of the transplantation of genetically corrected autologous cells by elegant differentiation and implantation technologies to come in a near future (dotted line). DMD, Duchenne muscular dystrophy; HAC, human artificial chromosome; iPS, induced pluripotent stem cells; Mb, megabase; MMCT, microcell-mediated chromosome transfer.
Correction of mdx-iPS cells with DYS-HAC
We have previously developed a novel HAC vector containing full-length genomic dystrophin, which was designated as DYS-HAC.21 This DYS-HAC, which contains a visualization marker gene GFP and suicide gene TK, was transferred to the two independent mdx-iPS clones via microcell-mediated chromosome transfer (MMCT). Eight GFP+ clones were selected and examined in the following experiments (Figure 2a). PCR analysis using primers for the detection of the DYS-HAC showed that six of the eight clones contained the intact dystrophin region in the HAC (Figure 2b). Reverse transcription (RT)-PCR analysis showed expression of dystrophin from the DYS-HAC was detected in six clones with the intact genomic dystrophin, whereas enhanced green fluorescent protein (EGFP) was expressed in all the clones examined (Figure 2d). Fluorescence in situ hybridization (FISH) analyses showed that the DYS-HAC was present as an individual chromosome in the mdx-iPS cells (Figure 2c). These results show that the DYS-HAC can be transferred to mouse iPS cells at a comparable efficiency to that in mouse ES cells.21 To test the stemness of the iPS cells, RT-PCR analyses using primers for ES cell–specific genes were performed (Figure 2d). Endogenous Rex1, Nanog, Oct4, and Sox2 were expressed in all mdx-iPS (DYS-HAC) cells, comparable to the parent mdx-iPS cells and Nanog-iPS cells generated previously with four Yamanaka's factors, including c-Myc. Exogenous expressions of Klf4, Sox2, and Oct4 in most mdx-iPS (DYS-HAC) cells were lower than those of parent mdx-iPS cells, suggesting that expression of transgenes was more or less silenced after MMCT.
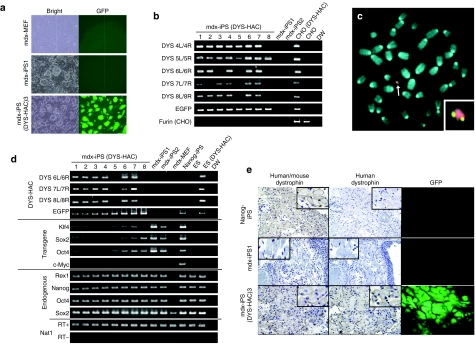
Figure 2.
Characterization of mdx-iPS with DYS-HAC. (a) Morphology of mdx-MEF, mdx-iPS, and mdx-iPS (DYS-HAC) cells. Phase-contrast (left panel) and GFP-fluorescence (right panel) micrographs are shown. (b) Genomic PCR analyses for detecting DYS-HAC in mdx-iPS cells. (c) FISH analyses for mdx-iPS (DYS-HAC) cells. An arrow indicates the DYS-HAC and the inset shows an enlarged image of the DYS-HAC. (d) RT-PCR analyses of ES cell–marker genes, four exogenous transcription factors, and human dystrophin. EGFP and Nat1 were used as internal controls. Primers for DYS 6L/6R, 7L/7R, and 8L/8R detected the isoform of dystrophin expressed in ES and iPS cells. (e) Immunohistochemical analyses of dystrophin in muscle-like tissues of each teratoma. Immunodetection of mouse and human dystrophin (left panel), immunodetection of human-specific dystrophin (middle panel), and GFP micrography (right panel) are shown. The insets show enlarged images of immunohistochemistry. Nanog-iPS- and mdx-iPS-derived teratomas were used as positive and negative controls, respectively. CHO, Chinese hamster ovary; EGFP, enhanced green fluorescent protein; GFP, green fluorescent protein; HAC, human artificial chromosome; iPS, induced pluripotent stem cells; MEF, mouse embryonic fibroblast.
To determine whether the mdx-iPS (DYS-HAC) cells have the ability to differentiate into all three embryonic germ layers (endoderm, mesoderm, and ectoderm), cells were subcutaneously injected into nude mice. Transplanted mdx-iPS (DYS-HAC) gave rise to typical teratomas (n = 10), and GFP+ tissues were detected in these teratomas (Supplementary Figure S2a,b). Histological analyses showed that tumors contained all three embryonic germ layers (Supplementary Figure S2c). FISH analyses showed that DYS-HAC was detected in 90% of cells in tumor tissues (Supplementary Figure S2d). Immunohistochemical analyses detected the expression of human dystrophin in muscle-like tissues of teratomas derived from mdx-iPS (DYS-HAC) cells, but not in those derived from mdx-iPS cells (Figure 2e). These data suggest that loss of dystrophin expression in mdx-derived muscle tissue was restored by transferring the DYS-HAC into mdx-iPS cells.
Expression in tissues from chimeric mice with DYS-HAC
In order to determine developmental potential and expression of the transferred human dystrophin in various tissues, chimeric mice were produced from mdx-iPS (DYS-HAC) (Supplementary Figure S2e). Chimeras with various forms of coat-color chimerism were obtained, and GFP+ chimeric mice were used for the following analyses. The EGFP gene driven by the CAG promoter on the DYS-HAC was expressed in all tissues examined (Figure 3a), suggesting that DYS-HAC was stably maintained in vivo. To investigate whether the tissue-specific isoform of the human dystrophin on the DYS-HAC was expressed, total RNAs from various tissues of the chimeras were analyzed by RT-PCR using three pairs of specific primers to detect human dystrophin tissue-specific transcripts. Isoforms Dp427I and Dp427m were expressed in chimeric heart and skeletal muscle, and Dp140 was expressed in chimeric brain, comparable to the human expression profile (Figure 3b). FISH analyses showed that the DYS-HAC was detected in 50% of cells at least in the brain, liver, kidney, spleen, and skeletal muscle (Figure 3c). This detection correlated with coat-color chimerism, and the DYS-HAC was present as an individual chromosome in chimeric tail fibroblasts (Supplementary Figure S2e,f). Immunohistochemical analysis using a human dystrophin–specific antibody showed human dystrophin protein localized at the sarcolemmal membrane in skeletal muscle (Figure 3d). These results suggest that isoforms of human dystrophin on the DYS-HAC were expressed in a tissue-specific manner, and the human dystrophin protein was localized at the correct locus in iPS-derived chimeric skeletal muscle. These data were consistent with the results from our previous study using chimeric mice from normal mouse ES cells with the DYS-HAC.21
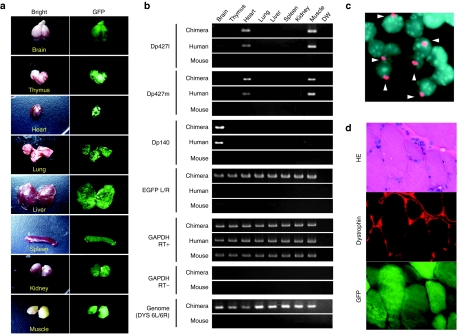
Figure 3.
Expression analysis in chimeric mice with DYS-HAC. (a) Chimeric tissues derived from mdx-iPS (DYS-HAC). Bright (left panel) and fluorescence (right panel) micrographs are shown. (b) Representative genomic PCR and RT-PCR data for detection of DYS-HAC in each chimeric tissue. (c) FISH analysis of chimeric kidney derived from mdx-iPS (DYS-HAC) cells. Digoxigenin-labeled human COT-1 DNA (red) was used to detect DYS-HAC in tissues. Chromosomal DNA was counterstained with DAPI. Arrowheads show nuclei containing the DYS-HAC. (d) Immunohistochemical analyses of dystrophin in chimeric muscle. HE staining (top panel), immunodetection of dystrophin (middle panel), and GFP micrography (bottom panel) are shown. EGFP, enhanced green fluorescent protein; GFP, green fluorescent protein; HAC, human artificial chromosome; HE, hematoxylin and eosin; iPS, induced pluripotent stem cells.
Correction of DMD-iPS cells with DYS-HAC
Next, we attempted genetic correction of iPS cells derived from a DMD patient (Figure 1). We chose a DMD patient with deletion of exons 4–43 for iPS cell induction because a large deletion of this type cannot be corrected even using homologous recombination or other conventional vectors. The DYS-HAC was transferred to DMD patient–derived fibroblasts via MMCT, as we failed to directly transfer the HAC into human iPS or human ES cells due to the difficulty of cloning colonies derived from single cell following transfection or MMCT, which is an unsolved issue. The DYS-HAC was successfully transferred to DMD-fibroblasts, as shown by PCR and multiplex PCR analyses (Figure 4a and Supplementary Figure S3a). FISH analyses showed that the DYS-HAC was present as an individual chromosome in DMD patient–derived fibroblasts (data not shown). The iPS cells were generated from the DMD-fibroblasts with the DYS-HAC using a combination of lentiviral infection with mouse Slc7a1 and retroviral infection with KLF4, SOX2, OCT4, and c-MYC, as reported previously.6 Thirty GFP+ and human ES–like DMD-iPS (DYS-HAC) cells derived from three independent DMD-fibroblast (DYS-HAC) clones were selected, and randomly selected nine clones were analyzed in the following experiments. To test the stemness of iPS cells, RT-PCR analyses using primers for ES cell–specific genes were performed (Figure 4c). Endogenous REX1, NANOG, OCT4, and SOX2 were differentially expressed in iPS cells. Exogenous OCT4, KLF4, SOX2, and c-MYC were expressed in some iPS cells. These findings show that DMD-iPS (DYS-HAC) cells were comparable to DMD-iPS and normal human iPS cells. PCR and multiplex PCR analyses showed that the DYS-HAC was maintained in all examined iPS cells, comparable to parent DMD-fibroblast (DYS-HAC) clones (Supplementary Figure S3a,b). FISH analyses showed that the DYS-HAC was present as an individual chromosome in the DMD-iPS (DYS-HAC) cells (Figure 4d). To examine mitotic stability of the DYS-HAC in DMD-iPS (DYS-HAC) cells, iPS cells were cultured for about 4 months without selection. FISH analyses revealed that the DYS-HAC was independently and stably maintained in DMD-iPS (DYS-HAC) cells (Figure 4e). These data suggest that the DYS-HAC could be maintained stably during iPS generation and even after long-term culture in vitro.
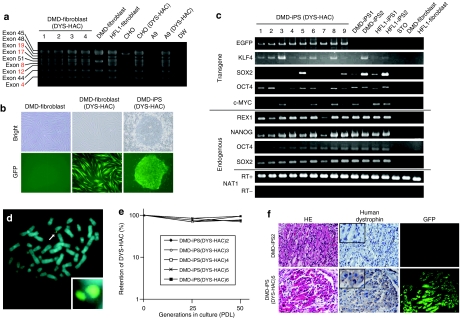
Figure 4.
Characterization of DMD-iPS with DYS-HAC. (a) Genomic multiplex PCR analyses for detecting the dystrophin genome on the DYS-HAC in DMD-fibroblast cells. Red lines show exons deleted in the DMD-fibroblast. (b) Morphology of DMD-fibroblasts, DMD-fibroblasts (DYS-HAC), and DMD-iPS (DYS-HAC) cells. Phase-contrast (top panel) and fluorescence (bottom panel) micrographs are shown. (c) RT-PCR analyses of embryonic stem cell–marker genes and four transcription factors. (d) FISH analyses for DMD-iPS (DYS-HAC) cells. An arrow indicates the DYS-HAC and the inset shows an enlarged image of the DYS-HAC. (e) Mitotic stability of the DYS-HAC in DMD-iPS (DYS-HAC) cells. (f) Immunohistochemical analyses of dystrophin in muscle-like tissues of each teratoma. HE staining (left panel), immunodetection of dystrophin (middle panel), and GFP micrography (right panel) are shown. The insets show enlarged images of immunohistochemistry. CHO, Chinese hamster ovary; DMD, Duchenne muscular dystrophy; GFP, green fluorescent protein; HAC, human artificial chromosome; HE, hematoxylin and eosin; iPS, induced pluripotent stem cells; PDL, population doubling.
To determine whether DMD-iPS (DYS-HAC) cells could differentiate into all three embryonic germ layers, cells were injected into testes of severe combined immunodeficiency mice. Transplanted DMD-iPS (DYS-HAC) gave rise to typical teratomas (n = 12), and GFP+ tissues were detected in these teratomas (Supplementary Figure S4a). Histological analyses revealed all three embryonic germ layers in all teratomas (Supplementary Figure S4b). FISH analyses showed that the DYS-HAC was detected in 90% of cells in tumor tissues (Supplementary Figure S4c). Immunohistochemical analysis showed expression of human dystrophin in muscle-like tissues of teratomas derived from DMD-iPS (DYS-HAC) cells, but not in those derived from DMD iPS cells (Figure 4f). These data were comparable to the results of the mdx-iPS (DYS-HAC) experiments mentioned above. These data suggest that loss of dystrophin expression in DMD-derived muscle tissue was restored by transferring the DYS-HAC into DMD-iPS cells.
Discussion
Previously, mouse iPS cells derived from sickle cell anemia have been corrected using the homologous recombination approach, with the model mice treated using iPS-derived hematopoietic progenitors.9 Most recently, Fanconi anemia patient–derived iPS cells were corrected using the viral vector, and the iPS cells could give rise to hematopoietic progenitors of the myeloid and erythroid lineages, showing the correction of the disease phenotype.24 However, genetic restoration by the homologous recombination and gene transfer using the conventional vectors, including viral and plasmid vector, cannot be applied in DMD patients with large deletions in the gene. The present results have shown the first complete genetic correction of a human genetic disorder, DMD with deletion of exon 4–43, using a novel HAC as an episomal vector. As Kimura et al. recently reported myogenic conversion from fibroblasts by transducing the inducible myogenic regulator, MyoD, the DMD-fibroblast (DYS-HAC) developed in this study may also represent a source for gene therapy.25
To the best of our knowledge, iPS cells have been generated from human primary fibroblast populations, but not from cloned cells.6 In this study, we could generate patient-derived genetically corrected iPS cells even from fibroblasts cloned after MMCT. Although several dystrophin isoforms cannot be expressed using previously reported vector systems, our HAC vector system containing an entire genomic dystrophin enabled isoform expressions in iPS-derived various tissues with the DYS-HAC in a tissue-specific manner. As the integrating viral vectors were used for iPS generation in this study, however, iPS induction using a nonintegrating vector system, protein introduction system, or chemical small molecules may be needed for safe gene therapy.26,27,28,29,30,31,32,33 The HAC vector may also be a promising tool for safe iPS generation: the HAC is a nonintegrating vector, and the elimination of a HAC, with a conditional centromere, from the cells can also be performed.34
In the MMCT methods, it is possible that host chromosomes (e.g., A9 and CHO) can be transferred along with the HAC at once. However, we have seldom observed the co-transfer of host chromosomes during MMCT (~1/100 clones). In this study, host chromosomes were not detected in mdx-iPS cells and DMD-fibroblasts after MMCT (data not shown).
Furthermore, advances in efficient methods for differentiation and purification of stem cells, including ES and iPS cells, are anticipated. Barberi et al. reported engraftable skeletal myoblasts from human ES cells.35 Darabi et al. also reported the generation of functional skeletal muscle from mouse ES cells by induction of Pax3 and the treatment of mdx mice using ES cell–derived cells.36 Application of these methods to iPS cells combined with our HAC vector system may open a way to more sophisticated DMD gene therapies. Taken together, genetic correction of patient-specific iPS cells by MMCT of the DYS-HAC, efficient differentiation from iPS cells into muscle stem cells in vitro, and transplantation of genetically corrected autologous cells into the same patient are needed for the gene therapy of DMD (Figure 1). Thus, stem cells derived from multiple potential sources combined with HAC-mediated gene delivery should allow safe treatment of various genetic defects, with elegant differentiation technology to come.
Materials and Methods
Cell culture. Fibroblasts from a DMD patient (GM05169) containing deletion of exons 4–43 in the dystrophin gene were obtained from Coriell Institute (Camden, NJ). Fibroblast cells were grown in α-MEM plus 15% fetal bovine serum (FBS) and 2 mmol/l L-glutamine. DMD model mice (mdx) were obtained from Charles River (Yokohama, Japan), and mouse embryonic fibroblasts (MEFs) were isolated from 13.5 days postcoitum embryos. The mdx-MEF and PLAT-E cells were grown in Dulbecco's modified Eagle's medium (Sigma, St Louis, MO) plus 10% FBS. Chinese hamster ovary (CHO) or A9 cells containing the DYS-HAC were constructed as previously described.21 The CHO (DYS-HAC) cells were maintained in Ham's F-12 nutrient mixture (Invitrogen, Carlsbad, CA) plus 10% FBS with 8 µg/ml blasticidin S hydrochloride (Funakoshi, Tokyo, Japan). The A9 (DYS-HAC) cells were maintained in Dulbecco's modified Eagle's medium plus 10% FBS with 4 µg/ml blasticidin S hydrochloride. The parental mdx-iPS cell line and microcell hybrid clones, mdx-iPS (DYS-HAC), were maintained on mitomycin C (Sigma)–treated Jcl:ICR (CLEA Japan, Tokyo, Japan) MEFs as feeder layers in Dulbecco's modified Eagle's medium with 18% FBS (Thermo Scientific HyClone, Yokohama, Japan), 1 mmol/l sodium pyruvate (Invitrogen), 0.1 mmol/l nonessential amino acids (Invitrogen), 0.1 mmol/l 2-mercaptoethanol (Sigma), 2 mmol/l L-glutamine (Invitrogen), and 1,000 U/ml leukemia inhibitory factor (Funakoshi). Human iPS cells were maintained on SNL (STO) feeder cells in primate ES medium (ReproCell, Tokyo, Japan) supplemented with 4 ng/ml recombinant basic fibroblast growth factor (Wako, Osaka, Japan). Mouse and human iPS cells were maintained as described previously.5,6 The control iPS cell line, Nanog-iPS (APS0001, iPS-MEF-Ng-20D-17), was obtained from the RIKEN BRC Cell Bank (Tsukuba, Japan). The SNL cell line for feeder layer was obtained from the SANGER Institute (Cambridge, UK).
Generation of iPS cells. Generation of iPS cells from mdx-MEFs was performed using a retroviral system as described previously.5 Briefly, retroviruses were generated with Plat-E packaging cells. Three retroviruses containing Oct4, Klf4, and Sox2, were infected into mdx-MEFs. Four days after transfection, mdx-MEFs were replated at 3.5 × 105 cells per 100-cm dish on MEF feeder cells. The next day, medium was replaced with mouse ES cell medium. Thirteen days after infection, mouse ES–like colonies were selected up to the MEF feeder cells on 24-well plates. Generation of iPS cells from human DMD-fibroblasts was performed using the retrovirus system combined with the lentivirus system as described previously.6 Briefly, lentivirus production for Slc7a1 expression was performed using 293T cells, and lentivirus was infected into human fibroblasts. Four retroviruses containing OCT4, KLF4, SOX2 and c-MYC were infected into Slc7al-expressing human DMD-fibroblasts. The four retroviral expression vectors were obtained from Addgene (Cambridge, MA). Six days after transfection, fibroblasts were replated at 5 × 104 cells per 100-cm dish on SNL feeder cells. The next day, medium was replaced with primate ES cell medium supplemented with 4 ng/ml basic fibroblast growth factor. Thirty days after transduction, human ES–like colonies were selected up to the SNL feeder cells on 24-well plates.
MMCT. MMCT was performed as described previously.37 CHO or A9 cells containing the DYS-HAC were used as donor microcell hybrids. Briefly, mdx-iPS and DMD-fibroblast cells were fused with microcells prepared from donor hybrid CHO (DYS-HAC) or A9 (DYS-HAC) cells, and selected with blasticidin S (3 µg/ml). The transferred DYS-HAC in each line was characterized by PCR, RT-PCR, and FISH analyses.
Genomic PCR analyses. Genomic DNA was extracted from cell lines and chimeric tissue specimens using a genomic extraction kit (Gentra Systems, Minneapolis, MN), and PCR was performed using primers as follows. Primer pairs for the detection of the region of human dystrophin gene were as follows: DYS3L/3R [163 base pairs (bp)], 5′-AACAACTGAACAGCCGGTGGA-3′ and 5′-GGGGTGGTGGGTTGGATTTT-3′ DYS4L/4R (128 bp), 5′-GCAAGAGCAACAAAGTGGCCTA-3′ and 5′-AGCTTCTTCCAGCGTCCCTCA-3′ DYS5L/5R (132 bp), 5′-ACCTTCAGAACCGGAGGCAAC-3′ and 5′-AGGGACCCTCCTTCCATGACTC-3′ DYS6L/6R (170 bp), 5′-TGGAACGCATTTTGGGTTGTT-3′ and 5′-AAAACAATGCGCTGCCTCAAA-3′ DYS7L/7R (151 bp), 5′-TTTGCATCCTTTTGGCGTGAT-3′ and 5′-AAACTCAAGCCTGCCCCACTC-3′ DYS8L/8R (155 bp), 5′-GCTGCTAGCAATGCCACGATT-3′ and 5′-GGATGGGCTGGGAATCCATAG-3′. Primer pairs for detection of EGFP on the HAC, mouse genome (A9), and hamster genome (CHO) were as follows: EGFPL/R (479 bp), 5′-CCTGAAGTTCATCTGCACCA-3′ and 5′-TGCTCAGGTAGTGGTTGTCG-3′ Cyp3a13-5L/6R (239 bp), 5′-GCTCAGCAGGCTCAGCCCTGA-3′ and 5′-TCCAAGCCAGTAAAGGAAAGAATTAT-3′ Furin-L/R, 5′-ACTCAGAGATCCACTGCACCAGGATCCAAGGGAGG-3′ and 5′-GCTCGAGCGGCTACACCACAGACACCATTGTTGGCTACTGCTGCC-3′. Multiplex PCR analyses to detect the deletion of exons within human dystrophin were performed in accordance with the protocols of the manufacturer (Maxim Biotech, San Francisco, CA). CHO (DYS-HAC), A9 (DYS-HAC), HFL1, and HFL1-iPS were used as positive controls. mdx-iPS, DMD-fibroblasts, DMD-iPS, STO, CHO, and A9 cells were used as negative controls.
FISH analyses. FISH analyses were performed using either fixed metaphase or interphase spreads of each cell hybrid using digoxigenin-labeled (Roche, Basel, Switzerland) human COT-1 DNA (Invitrogen) and biotin-labeled BAC DNA (RP11-954B16, located in the dystrophin genomic region), essentially as described previously.37 Chromosomal DNA was counterstained with DAPI (Sigma). Images were captured using the NIS-Elements system (Nikon, Tokyo, Japan).
RT-PCR analyses. Total RNA from chimeric tissue specimens was prepared using ISOGEN (Nippon Gene, Tokyo, Japan) and purified using RNeasy columns (Qiagen, Hilden, Germany), in accordance with the manufacturer's instructions, and was then treated with RNase-free DNase I (Wako). First-strand complementary DNA (cDNA) synthesis was performed using random hexamers and SuperScript III reverse transcriptase (Invitrogen). EGFP and GAPDH were used as internal controls. Human cDNA was used as a positive control, whereas cDNA and DNA from C57BL/6 mouse tissues were used as negative controls. Total RNA from cultured cells was purified with Trizol reagent (Invitrogen) and treated with a Turbo DNA-free kit (Ambion/Applied Biosystems, Tokyo, Japan) to remove genomic DNA contamination. First-strand cDNA synthesis was undertaken using an oligo-(dT)20 primer and ReverTraAce-α (Toyobo, Osaka, Japan). In analyses of mouse iPS cells, Nanog-iPS and mouse ES (TT2) cells were used as positive controls, whereas mdx-MEF was used as a negative control. Nat1 was used as an internal control. In analyses of human iPS cells, DMD-fibroblasts were used as negative controls. Nat1 was used as an internal control. PCR was performed with cDNA using ExTaq (Takara Bio, Otsu, Japan) or AmpliTaq Gold (PerkinElmer, Waltham, MA). Amplifications were performed with an annealing temperature of 58 °C for 30–35 cycles, then amplified fragments were resolved by electrophoresis on a 2% agarose gel, followed by staining with ethidium bromide. Primer sequences were as follows: for the dystrophin isoform Dp427m, DYS 427me1L/DYS427me1R (211 bp), 5′-TTCCCCCTACAGGACTCAGA-3′ and 5′-TCTTCCCACCAAAGCATTTT-3′ for dystrophin isoform Dp427I, DYS427le1L/DYSe3R (150 bp), 5′-CTCATGATGAAAGAGAAGATGTTCAA-3′ and 5′-CTGTCAGGCCTTCGAGGA-3′ for dystrophin isoform Dp140, DYS 140e1L/DYS e45R (189 bp), 5′-TGCTGGCTGCTCTGAACTAA-3′ and 5′-GGCTTCCCAATTTTTCCTGT-3′ for EGFP, EGFPL/R (479 bp), 5′-CCTGAAGTTCATCTGCACCA-3′ and 5′-TGCTCAGGTAGTGGTTGTCG; for GAPDH (mouse and human), RPC1/2, 5′-CCATCTTCCAGGAGCGAGA-3′ and 5′-TGTCATACCAGGAAATGAGC-3′. Primers for DYS6L/6R, DYS7L/7R, and DYS8L/8R were the same as in genomic PCR analyses. RT-PCR to detect endogenous ES markers and exogenous transgenes were performed using primers as described previously.5,6 Primer pairs for the detection of transgenes of cMYC and SOX 2 were as follows: hMYC-S3148/pMXs-AS3095, 5′-CAGAGGAGGAACGAGCTAAAAC-3′ and 5′-AGACCAACTGGTAATGGTAGCG-3′ hSOX2-S691/pMXs- AS3206, 5′-GGCACCCCTGGCATGGCTCTTGGCTC-3′ and 5′-TTATCGTCGACCACTGTGCTGGCG-3′.
Generation of chimeric mice. Chimeric mice were produced from the two mdx-iPS and three mdx-iPS (DYS-HAC) cell lines. Chimera production was performed as described previously.37 Briefly, iPS cells were injected into blastocyst-stage embryos derived from ICR mice (CLEA Japan) and then transferred into pseudopregnant ICR females. Three chimeric mice showing 50% coat-color chimerism were used for expression analyses in various tissues. All chimeric mice used for analyses were 1–4 weeks old. All animal experiments were approved by the Institutional Animal Care and Use Committee of Tottori University.
Teratoma formation and histology. To produce teratomas, 2 × 106 mdx-iPS (DYS-HAC), mdx-iPS, and mdx-fibroblast cells were subcutaneously injected into CD-1 (ICR)-nu mice (Charles River), and 1 × 106 DMD-iPS (DYS-HAC), DMD-iPS, and DMD-fibroblast (DYS-HAC) cells were injected into testes of severe combined immunodeficiency mice (Charles River). After 5–6 weeks in the mdx-iPS series and 9–13 weeks in the DMD-iPS series, resected teratomas were fixed in 20% formalin and processed for paraffin sectioning, then stained with hematoxylin and eosin.
Immunohistochemical analyses. Formalin-fixed and paraffin-embedded specimens and OCT (optimal cutting temperature) compound–embedded frozen specimens were used for immunohistochemistry. Primary antibodies used in this study were as follows: rabbit polyclonal antibody against both human and mouse dystrophin (1:100; Lab Vision, Fremont, CA); and mouse monoclonal antibody against human dystrophin (MANDYS106, diluted 1:4; a gift from Glenn E. Morris, Keele University, Shropshire, UK). Immunoreaction was developed by using the SAB (streptavidin–biotin) peroxidase complex method with the Histofine SAB-PO Kit (Nichirei, Tokyo, Japan) for rabbit polyclonal antibody and the Histofine Mouse Stain Kit (Nichirei) for mouse monoclonal antibody, or visualized with Alexa Fluor 555 goat anti-mouse conjugate (diluted 1:1,000; Molecular Probes, Eugene, OR).
SUPPLEMENTARY MATERIALFigure S1. Characterisation of mdx-iPS. (a) RT-PCR analyses of ES cell-marker genes and four transcription factors. Parental mdx-MEF and Nanog-iPS cells were used as negative and positive controls, respectively. Nat1 was used as an internal control. (b, c) Teratoma derived from the mdx-iPS. (d) Histological analyses of teratomas derived from mdx-iPS cells (haematoxylin and eosin stain). Five weeks after cell transplantation, structures originating from all three germ layers were found in the teratoma. Sq: squamous epithelia (ectodermal derivative); Sk: skeletal muscle (mesodermal derivative); Gr: glandular epithelia (endodermal derivative); Ci: ciliated epithelia (endodermal derivative).Figure S2. Histological and FISH analyses in mdx-iPS (DYS-HAC)-derived teratoma and chimera. (a, b) Teratoma derived from mdx-iPS(DYS-HAC). (b) Bright (top panel) and fluorescence (bottom panel) micrographs are shown. (c) Histological analyses of teratomas derived from the mdx-iPS (DYS-HAC) cells (haematoxylin and eosin stain). Five weeks after cell transplantation, structures originating from all three germ layers were observed in the teratoma. Sq: squamous epithelia (ectodermal derivative); Sk: skeletal muscle (mesodermal derivative); Gr: glandular epithelia (endodermal derivative). (d,f) FISH analysis of teratoma and chimeric tail fibroblasts derived from mdx-iPS (DYS-HAC) cells. Digoxigenin-labelled human COT-1 DNA (red) (d,f) and the biotin-labelled RP11-954B16 (green) (f) were used to detect the DYS-HAC in tissues. Chromosomal DNA was counterstained with DAPI. An arrow indicates the DYS-HAC and the inset shows an enlarged image of the DYS-HAC. (e) Chimeric mice from mdx-iPS (DYS-HAC) cells.Figure S3. Genomic PCR analyses in DMD-fibroblast (DYS-HAC) and DMD-iPS (DYS-HAC) cells. (a) Genomic PCR analyses for detecting the DYS-HAC in DMD-fibroblast cells. CHO (DYS-HAC), A9 (DYS-HAC) and HFL1 fibroblast were used as positive controls. DMD-fibroblast, CHO and A9 cells were used as negative controls. Primers of Furin and Cyp3a13 were used to detect CHO and A9 genome, respectively. (b) Genomic PCR and multiplex PCR (M-PCR) analyses for detecting the DYS-HAC in DMD-iPS cells. HFL1-iPS1, HFL1-iPS2 and HFL1 fibroblasts were used as positive controls. DMD-iPS1, DMD-iPS2 and DMD fibroblasts were used as negative controls.Figure S4. Histological and FISH analyses in DMD-iPS (DYS-HAC)-derived teratoma. (a) Teratoma derived from DMD-iPS and DMD-iPS (DYS-HAC) cells. Bright and (top panel) and fluorescence (bottom panel) micrographs are shown. (b) Histological analyses of teratomas derived from DMD-iPS (DYS-HAC) cells (haematoxylin and eosin stain). Thirteen weeks after cell transplantation, structures originating from all three germ layers were observed in the teratoma. Endoderm (glandular epithelia, left panel), mesoderm (skeletal muscle, middle panel), ectoderm (melanin-producing cells, right panel). (c) FISH analysis of teratoma derived from DMD-iPS (DYS-HAC) cells. The biotin-labelled RP11-954B16 (green) was used to detect the DYS-HAC in tissues. Chromosomal DNA was counterstained with DAPI. Arrowheads show nuclei containing the DYS-HAC.
Supplementary Material
Characterisation of mdx-iPS. (a) RT-PCR analyses of ES cell-marker genes and four transcription factors. Parental mdx-MEF and Nanog-iPS cells were used as negative and positive controls, respectively. Nat1 was used as an internal control. (b, c) Teratoma derived from the mdx-iPS. (d) Histological analyses of teratomas derived from mdx-iPS cells (haematoxylin and eosin stain). Five weeks after cell transplantation, structures originating from all three germ layers were found in the teratoma. Sq: squamous epithelia (ectodermal derivative); Sk: skeletal muscle (mesodermal derivative); Gr: glandular epithelia (endodermal derivative); Ci: ciliated epithelia (endodermal derivative).
Histological and FISH analyses in mdx-iPS (DYS-HAC)-derived teratoma and chimera. (a, b) Teratoma derived from mdx-iPS(DYS-HAC). (b) Bright (top panel) and fluorescence (bottom panel) micrographs are shown. (c) Histological analyses of teratomas derived from the mdx-iPS (DYS-HAC) cells (haematoxylin and eosin stain). Five weeks after cell transplantation, structures originating from all three germ layers were observed in the teratoma. Sq: squamous epithelia (ectodermal derivative); Sk: skeletal muscle (mesodermal derivative); Gr: glandular epithelia (endodermal derivative). (d,f) FISH analysis of teratoma and chimeric tail fibroblasts derived from mdx-iPS (DYS-HAC) cells. Digoxigenin-labelled human COT-1 DNA (red) (d,f) and the biotin-labelled RP11-954B16 (green) (f) were used to detect the DYS-HAC in tissues. Chromosomal DNA was counterstained with DAPI. An arrow indicates the DYS-HAC and the inset shows an enlarged image of the DYS-HAC. (e) Chimeric mice from mdx-iPS (DYS-HAC) cells.
Genomic PCR analyses in DMD-fibroblast (DYS-HAC) and DMD-iPS (DYS-HAC) cells. (a) Genomic PCR analyses for detecting the DYS-HAC in DMD-fibroblast cells. CHO (DYS-HAC), A9 (DYS-HAC) and HFL1 fibroblast were used as positive controls. DMD-fibroblast, CHO and A9 cells were used as negative controls. Primers of Furin and Cyp3a13 were used to detect CHO and A9 genome, respectively. (b) Genomic PCR and multiplex PCR (M-PCR) analyses for detecting the DYS-HAC in DMD-iPS cells. HFL1-iPS1, HFL1-iPS2 and HFL1 fibroblasts were used as positive controls. DMD-iPS1, DMD-iPS2 and DMD fibroblasts were used as negative controls.
Histological and FISH analyses in DMD-iPS (DYS-HAC)-derived teratoma. (a) Teratoma derived from DMD-iPS and DMD-iPS (DYS-HAC) cells. Bright and (top panel) and fluorescence (bottom panel) micrographs are shown. (b) Histological analyses of teratomas derived from DMD-iPS (DYS-HAC) cells (haematoxylin and eosin stain). Thirteen weeks after cell transplantation, structures originating from all three germ layers were observed in the teratoma. Endoderm (glandular epithelia, left panel), mesoderm (skeletal muscle, middle panel), ectoderm (melanin-producing cells, right panel). (c) FISH analysis of teratoma derived from DMD-iPS (DYS-HAC) cells. The biotin-labelled RP11-954B16 (green) was used to detect the DYS-HAC in tissues. Chromosomal DNA was counterstained with DAPI. Arrowheads show nuclei containing the DYS-HAC.
Acknowledgments
We thank Glenn E. Morris (Keele University, Shropshire, UK) for providing the MONDYS106 antibody; Toshio Kitamura for providing the retroviral system; Shigeko Masuda and Yumako Miura for technical assistance; and Motonobu Katoh and Tetsuya Ohbayashi for critical discussions. This study was supported in part by JST (Japan Science and Technology Agency), CREST (Core Research for Evolutional Science and Technology) (M.O.), and the 21st Century COE (Centers of Excellence) program from the Ministry of Education, Culture, Sports, Science and Technology of Japan (M.O.).
REFERENCES
- Evans MJ., and , Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Dennis C. Cloning: mining the secrets of the egg. Nature. 2006;439:652–655. doi: 10.1038/439652a. [DOI] [PubMed] [Google Scholar]
- Hall VJ, Stojkovic P., and , Stojkovic M. Using therapeutic cloning to fight human disease: a conundrum or reality. Stem Cells. 2006;24:1628–1637. doi: 10.1634/stemcells.2005-0592. [DOI] [PubMed] [Google Scholar]
- Takahashi K., and , Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, et al. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rideout WM, 3rd, Hochedlinger K, Kyba M, Daley GQ., and , Jaenisch R. Correction of a genetic defect by nuclear transplantation and combined cell and gene therapy. Cell. 2002;109:17–27. doi: 10.1016/s0092-8674(02)00681-5. [DOI] [PubMed] [Google Scholar]
- Hanna J, Wernig M, Markoulaki S, Sun CW, Meissner A, Cassady JP, et al. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science. 2007;318:1920–1923. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]
- Odom GL, Gregorevic P., and , Chamberlain JS. Viral-mediated gene therapy for the muscular dystrophies: successes, limitations and recent advances. Biochim Biophys Acta. 2007;1772:243–262. doi: 10.1016/j.bbadis.2006.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig M, Monaco AP., and , Kunkel LM. The complete sequence of dystrophin predicts a rod-shaped cytoskeletal protein. Cell. 1988;53:219–228. doi: 10.1016/0092-8674(88)90383-2. [DOI] [PubMed] [Google Scholar]
- Perloff JK, de Leon AC., Jr, and , O'Doherty D. The cardiomyopathy of progressive muscular dystrophy. Circulation. 1966;33:625–648. doi: 10.1161/01.cir.33.4.625. [DOI] [PubMed] [Google Scholar]
- Chelly J, Hamard G, Koulakoff A, Kaplan JC, Kahn A., and , Berwald-Netter Y. Dystrophin gene transcribed from different promoters in neuronal and glial cells. Nature. 1990;344:64–65. doi: 10.1038/344064a0. [DOI] [PubMed] [Google Scholar]
- Zhao J, Uchino M, Yoshioka K, Miyatake M., and , Miike T. Dystrophin in control and mdx retina. Brain Dev. 1991;13:135–137. doi: 10.1016/s0387-7604(12)80123-9. [DOI] [PubMed] [Google Scholar]
- Byers TJ, Kunkel LM., and , Watkins SC. The subcellular distribution of dystrophin in mouse skeletal, cardiac, and smooth muscle. J Cell Biol. 1991;115:411–421. doi: 10.1083/jcb.115.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig M, Hoffman EP, Bertelson CJ, Monaco AP, Feener C., and , Kunkel LM. Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell. 1987;50:509–517. doi: 10.1016/0092-8674(87)90504-6. [DOI] [PubMed] [Google Scholar]
- Katoh M, Ayabe F, Norikane S, Okada T, Masumoto H, Horike S, et al. Construction of a novel human artificial chromosome vector for gene delivery. Biochem Biophys Res Commun. 2004;321:280–290. doi: 10.1016/j.bbrc.2004.06.145. [DOI] [PubMed] [Google Scholar]
- Kazuki Y, Hoshiya H, Kai Y, Abe S, Takiguchi M, Osaki M, et al. Correction of a genetic defect in multipotent germline stem cells using a human artificial chromosome. Gene Ther. 2008;15:617–624. doi: 10.1038/sj.gt.3303091. [DOI] [PubMed] [Google Scholar]
- Basu J., and , Willard HF. Artificial and engineered chromosomes: non-integrating vectors for gene therapy. Trends Mol Med. 2005;11:251–258. doi: 10.1016/j.molmed.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Oshimura M., and , Katoh M. Transfer of human artificial chromosome vectors into stem cells. Reprod Biomed Online. 2008;16:57–69. doi: 10.1016/s1472-6483(10)60557-3. [DOI] [PubMed] [Google Scholar]
- Hoshiya H, Kazuki Y, Abe S, Takiguchi M, Kajitani N, Watanabe Y, et al. A highly stable and nonintegrated human artificial chromosome (HAC) containing the 2.4 Mb entire human dystrophin gene. Mol Ther. 2009;17:309–317. doi: 10.1038/mt.2008.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor TP., and , Crystal RG. Genetic medicines: treatment strategies for hereditary disorders. Nat Rev Genet. 2006;7:261–276. doi: 10.1038/nrg1829. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- Raya A, Rodríguez-Pizà I, Guenechea G, Vassena R, Navarro S, Barrero MJ, et al. Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells. Nature. 2009;460:53–59. doi: 10.1038/nature08129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura E, Han JJ, Li S, Fall B, Ra J, Haraguchi M, et al. Cell-lineage regulated myogenesis for dystrophin replacement: a novel therapeutic approach for treatment of muscular dystrophy. Hum Mol Genet. 2008;17:2507–2517. doi: 10.1093/hmg/ddn151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller LU, Daley GQ., and , Williams DA. Upping the ante: recent advances in direct reprogramming. Mol Ther. 2009;17:947–953. doi: 10.1038/mt.2009.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Nagaya M, Utikal J, Weir G., and , Hochedlinger K. Induced pluripotent stem cells generated without viral integration. Science. 2008;322:945–949. doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita K, Nakagawa M, Hyenjong H, Ichisaka T., and , Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, Hämäläinen R, et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II, et al. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Wei W, Zhu S, Zhu J, Shi Y, Lin T, et al. Generation of rat and human induced pluripotent stem cells by combining genetic reprogramming and chemical inhibitors. Cell Stem Cell. 2009;4:16–19. doi: 10.1016/j.stem.2008.11.014. [DOI] [PubMed] [Google Scholar]
- Kaji K, Norrby K, Paca A, Mileikovsky M, Mohseni P., and , Woltjen K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771–775. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Wu S, Joo JY, Zhu S, Han DW, Lin T, et al. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell. 2009;4:381–384. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano M, Cardinale S, Noskov VN, Gassmann R, Vagnarelli P, Kandels-Lewis S, et al. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev Cell. 2008;14:507–522. doi: 10.1016/j.devcel.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberi T, Bradbury M, Dincer Z, Panagiotakos G, Socci ND., and , Studer L. Derivation of engraftable skeletal myoblasts from human embryonic stem cells. Nat Med. 2007;13:642–648. doi: 10.1038/nm1533. [DOI] [PubMed] [Google Scholar]
- Darabi R, Gehlbach K, Bachoo RM, Kamath S, Osawa M, Kamm KE, et al. Functional skeletal muscle regeneration from differentiating embryonic stem cells. Nat Med. 2008;14:134–143. doi: 10.1038/nm1705. [DOI] [PubMed] [Google Scholar]
- Tomizuka K, Yoshida H, Uejima H, Kugoh H, Sato K, Ohguma A, et al. Functional expression and germline transmission of a human chromosome fragment in chimaeric mice. Nat Genet. 1997;16:133–143. doi: 10.1038/ng0697-133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Characterisation of mdx-iPS. (a) RT-PCR analyses of ES cell-marker genes and four transcription factors. Parental mdx-MEF and Nanog-iPS cells were used as negative and positive controls, respectively. Nat1 was used as an internal control. (b, c) Teratoma derived from the mdx-iPS. (d) Histological analyses of teratomas derived from mdx-iPS cells (haematoxylin and eosin stain). Five weeks after cell transplantation, structures originating from all three germ layers were found in the teratoma. Sq: squamous epithelia (ectodermal derivative); Sk: skeletal muscle (mesodermal derivative); Gr: glandular epithelia (endodermal derivative); Ci: ciliated epithelia (endodermal derivative).
Histological and FISH analyses in mdx-iPS (DYS-HAC)-derived teratoma and chimera. (a, b) Teratoma derived from mdx-iPS(DYS-HAC). (b) Bright (top panel) and fluorescence (bottom panel) micrographs are shown. (c) Histological analyses of teratomas derived from the mdx-iPS (DYS-HAC) cells (haematoxylin and eosin stain). Five weeks after cell transplantation, structures originating from all three germ layers were observed in the teratoma. Sq: squamous epithelia (ectodermal derivative); Sk: skeletal muscle (mesodermal derivative); Gr: glandular epithelia (endodermal derivative). (d,f) FISH analysis of teratoma and chimeric tail fibroblasts derived from mdx-iPS (DYS-HAC) cells. Digoxigenin-labelled human COT-1 DNA (red) (d,f) and the biotin-labelled RP11-954B16 (green) (f) were used to detect the DYS-HAC in tissues. Chromosomal DNA was counterstained with DAPI. An arrow indicates the DYS-HAC and the inset shows an enlarged image of the DYS-HAC. (e) Chimeric mice from mdx-iPS (DYS-HAC) cells.
Genomic PCR analyses in DMD-fibroblast (DYS-HAC) and DMD-iPS (DYS-HAC) cells. (a) Genomic PCR analyses for detecting the DYS-HAC in DMD-fibroblast cells. CHO (DYS-HAC), A9 (DYS-HAC) and HFL1 fibroblast were used as positive controls. DMD-fibroblast, CHO and A9 cells were used as negative controls. Primers of Furin and Cyp3a13 were used to detect CHO and A9 genome, respectively. (b) Genomic PCR and multiplex PCR (M-PCR) analyses for detecting the DYS-HAC in DMD-iPS cells. HFL1-iPS1, HFL1-iPS2 and HFL1 fibroblasts were used as positive controls. DMD-iPS1, DMD-iPS2 and DMD fibroblasts were used as negative controls.
Histological and FISH analyses in DMD-iPS (DYS-HAC)-derived teratoma. (a) Teratoma derived from DMD-iPS and DMD-iPS (DYS-HAC) cells. Bright and (top panel) and fluorescence (bottom panel) micrographs are shown. (b) Histological analyses of teratomas derived from DMD-iPS (DYS-HAC) cells (haematoxylin and eosin stain). Thirteen weeks after cell transplantation, structures originating from all three germ layers were observed in the teratoma. Endoderm (glandular epithelia, left panel), mesoderm (skeletal muscle, middle panel), ectoderm (melanin-producing cells, right panel). (c) FISH analysis of teratoma derived from DMD-iPS (DYS-HAC) cells. The biotin-labelled RP11-954B16 (green) was used to detect the DYS-HAC in tissues. Chromosomal DNA was counterstained with DAPI. Arrowheads show nuclei containing the DYS-HAC.