Abstract
Reversible immortalization holds great potential for primary tissue expansion to develop cell-based therapies as well as for basic research. Human olfactory ensheathing glia (hOEG) are promising candidates for treating spinal cord injury and for studying extrinsic neuroregenerative mechanisms. We used lentivectors with Cre/loxP technology to achieve reversible gene transfer of BMI1, SV40 large T antigen (TAg), a short hairpin RNA against p53 (shp53), and the catalytic subunit of telomerase (TERT) in primary cultures of hOEG from human donor cadaver olfactory bulbs. Several combinations of these genes were able to immortalize hOEG, conserving their antigenic markers and neuroregenerative properties but only those transduced by BMI1/TERT did not accumulate karyotypic alterations or increase senescence marker levels. Strikingly, these were also the only cells which continued to proliferate after transgene removal by Cre recombinase delivery, whereas hOEG immortalized by shp53 or TAg in combination with TERT entered into growth arrest and died. These data support the idea that immortalization and halting senescent changes are separate processes; hOEG immortalized by BMI1/TERT can revert back to their former primary cell replicative state when deimmortalized, whereas those transduced by the other combinations depend on the presence of these transgenes to maintain their aberrant proliferative state.
Introduction
Cell-based therapy depends on the in vitro expansion of primary tissues. Most normal human somatic cells in culture conditions undergo a finite number of divisions before entering a nonreplicative state termed senescence (mortality stage 1, M1). Transduction by a number of genes can achieve extension of replicative life span but such transformed cells eventually enter crisis (mortality stage 2, M2) due to telomere shortening and are unable to continue further replication. In many cases, expression of the telomerase catalytic subunit (TERT) can bypass this M2 crisis to immortalize the cells.1 Reversible immortalization is a technique enabling the extension of cellular proliferation by the introduction of transgenes, which, after the cells have been cultured for the desired time, can be silenced or eliminated using temperature-sensitive mutants, conditional promoters, or Cre-lox technology.2,3 Recently, other cellular genetic modification approaches have been developed to retrodifferentiate primary tissues into induced pluripotent cells4 (reviewed in Lowry and Plath5) or even transdifferentiate cells of one tissue type directly into another.6,7,8 These cell reprogramming technologies have generated intense interest as potential tools for obtaining large quantities of patient-specific cell types for cell therapy applications as well as basic research tools. Although it may often be desirable to obtain cell lines with robust and unlimited growth, it is equally important that these reprogramming processes do not provoke undesired structural or functional alterations, thus requiring careful analysis of the interaction of each genetic mechanism with any given cell type.
We have used reversible immortalization to obtain cell lines derived from human olfactory ensheathing glia (hOEG), a cell type of particular interest because of its capacity to promote neuroregeneration in the central nervous system.9,10 Apart from their potential application in the clinic for cell therapy of central nervous system lesions such as spinal cord injury, hOEG cell lines would be of great utility for understanding the molecular mechanisms underlying extrinsic cues leading to neuroregeneration.
Primary hOEG can be obtained from olfactory bulbs or olfactory mucosa, but there is some difficulty in obtaining large amounts of cadaver donor tissue within a time frame permitting growth of viable cultures. Olfactory mucosa may also be obtained from live donors but these biopsies are small tissue samples. To obtain large numbers of homogeneous hOEG, we have thus investigated their reversible immortalization using Hlox lentivectors2 encoding human TERT, the SV40 large T antigen (TAg), a short hairpin RNA directed against p53 (shp53), or murine BMI1. TAg is a viral protein which binds directly to the Rb family and p53 tumor suppressor proteins to inhibit their growth-repressing activities.11 The Polycomb group transcription factor BMI1 also inhibits both p53 and Rb pathways via its repressive effect on the Ink4a/Arf tumor suppressor locus.12 Both p53 and Rb are known to play important roles in the control of cellular replication and the immortalization of various cell types by manipulation of their signal transduction cascades has been well documented. We also tested the effect of directly reducing p53 expression using a lentivector encoding shp53.13
In this study we have examined the immortalization requirements of hOEG with respect to TAg, BMI1, shp53, and TERT and how each of the transgene combinations affects senescence pathways in these cells. Importantly, we observed that although several transgene combinations are able to immortalize hOEG conserving antigenic markers and neuroregenerative function, survival of the cells after transgene removal and maintenance of unaltered karyotype occurs only when the activation of senescence pathways is prevented.
Results
Replicative life span of hOEG can be extended by combined transduction of TERT with BMI1, shp53, or TAg
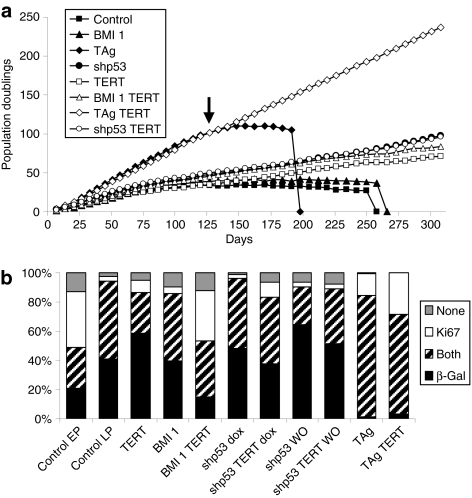
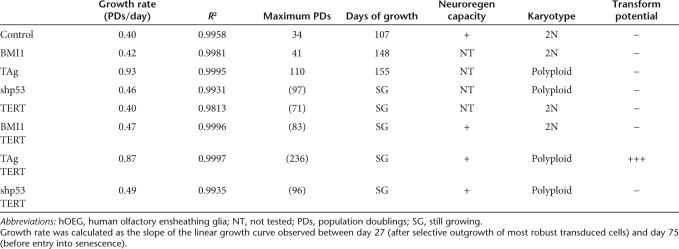
Human olfactory bulb tissue was derived from a 13-year-old female donor cadaver and vigorously growing cultures of primary olfactory ensheathing glia were established. We were able to obtain viable populations of lentivector-infected cells transduced individually by BMI1, TAg, shp53, or TERT, as well as combinations of each of the first three with TERT (Figure 1a). The expression of the different transgenes in the transduced populations was confirmed by reverse transcription–PCR (RT-PCR) or flow cytometry and their growth curves were followed over 300 days. In Figure 1a and Table 1, it can be seen that the control primary glia were able to proliferate to ~34 population doublings (PDs) during the first 107 days but thereafter enter in senescence. Entry into this state of replicative senescence (slope = 0 in the growth curve) could be delayed to 41 or 110 PDs by transduction with BMI1 or TAg, respectively. Cotransduction of BMI1 or TAg with TERT enabled the cells to evade this crisis altogether and rendered the cells immortal. Curiously, transduction of these young primary human glia with either TERT or shp53 alone or in combination also resulted in their immortalization, at least within the time span of our study. Independent of the number of PDs, senescence of untransduced OEG or those transduced only by BMI1 or TAg occurred at approximately the same time point (107–155 days) in all three cases. This supports the idea that replicative senescence is not merely the result of exhausting a predetermined number of replicative cycles, but is also due to the accumulation of environmentally imposed cellular changes over time. Finally, transduction with TAg had a marked accelerating effect on proliferation rate (Table 1), more than doubling this parameter. Transduction by shp53 provoked a smaller but also significant increase, whereas transduction by either BMI1 or TERT separately did not alter proliferation rate. Representative bright field images of these populations are shown in Supplementary Figure S1 where it can be observed that the cells most similar to the untransformed population are those immortalized by BMI1/TERT. Those immortalized by TAg/TERT exhibit a smaller, more compact morphology whereas those immortalized by shp53/TERT are of an intermediate morphology.
Figure 1.
Proliferation and senescence properties of human olfactory ensheathing glia (hOEG) transduced with different combinations of immortalizing genes. (a) Growth curves of uninfected (control) hOEG and seven populations infected with the indicated combinations of immortalizing lentivectors. Cumulative population doublings were calculated as described in Materials and Methods from the cell number counted at each culture passage. The arrow indicates the point at which the cells were used for further analysis in Figures 1b and 2. (b) Cytochemical analysis for lysosomal senescence-associated β-galactosidase (SA-β-gal) activity, and the Ki67 antigen, a marker of cell proliferation. The bar graph shows the percentage of cells negative for both markers (gray), positive for only SA-β-gal (black) or only Ki67 (white) or both (striped) in primary cells (control) at passage 4 (early passage: EP) or passage 21 (late passage: LP) and populations at passage 21 which were originally transduced by the indicated lentivector combinations. Dox, treated with 1 µg/ml doxycycline; WO, without doxycycline (withdrawn after 87 days of culture, 16 passages).
Table 1.
Summary of transduced hOEG properties
A molecular proliferation/senescence profile similar to early passage hOEG is conserved only in hOEG immortalized by BMI1/TERT
We determined the percentages of proliferating and senescent cells in each of the transduced populations (Figure 1b and Supplementary Figure S2) by double staining of the cells for the cell cycle–associated protein Ki67 (immunofluorescence), and enzymatic activity for the senescence-associated marker lysosomal β-galactosidase [senescence-associated β-galactosidase (SA-β-gal)]. As seen in Figure 1b, cells could be classified as senescent quiescent (black), senescent proliferative (striped), nonsenescent proliferative (white), or nonsenescent quiescent (gray). We first compared untreated hOEG from early and late passage (LP) numbers, when these cells had already entered replicative senescence (p21, arrow in Figure 1a), corresponding to a difference of ~87 days in culture (16 culture passages or 30 PDs). As expected, an increase in the fraction of senescent cells was observed, but interestingly about half of these are senescent proliferative (striped fraction) in both cases. Significantly, this produced a marked reduction in the fraction of nonsenescent proliferative cells (white), which is indicative of the renewal capacity of the population. Examination of the hOEG populations transduced by the different transgene combinations after 21 passages (120 days in culture) showed that the combination of BMI1/TERT was surprisingly unique in maintaining a Ki67/SA-β-gal profile similar to that of early passage control hOEG, with a large fraction of nonsenescent proliferative cells. Transduction by either TERT or BMI1 alone resulted in Ki67/SA-β-gal profiles more similar to LP hOEG, with increased fractions of senescent quiescent cells. TAg transduction, either alone or in combination with TERT, provoked the highest expression levels of Ki67, but most of these cells were senescent proliferative and the quiescent fraction became negligible. It should be noted that the profile of hOEG transduced with shp53 did not change considerably with respect to the LP hOEG control population and even its combination with TERT did not greatly reduce the proportion of senescent cells. The maintenance of the proliferative fraction was due to knockdown of p53 because this was partially reverted when shp53 expression was switched off by doxycycline withdrawal.
Transduction by TERT, BMI1, shp53, and TAg induces differential profiles of molecular senescence markers
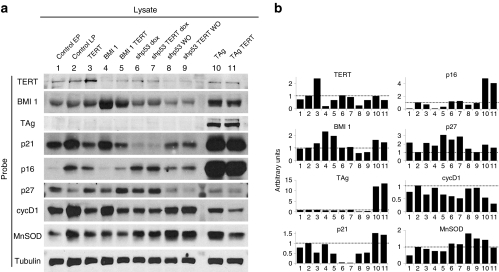
We next analyzed molecular markers of replicative senescence approximately at the same point of the growth curve (P23, arrow in Figure 1a) by western blot (Figure 2). To document the changes of these markers during replicative senescence under our culture conditions, we compared early passage primary hOEG to LP hOEG. Transgenic overexpression of TERT, BMI1, and TAg was confirmed in all of the cell populations transduced with these genes. As direct measurement of p53 levels may be uninformative due to the complex regulation of its synthesis, degradation, and function, to confirm activity of the shp53 transgene we analyzed its effect on the expression of p21, a direct downstream target of p53, which we observed to be drastically reduced, and increased again by switching off shp53 expression upon doxycycline withdrawal.
Figure 2.
Protein levels of cell cycle–associated components. Western blot analysis of protein lysates made from control human olfactory ensheathing glia (hOEG) at passage 6 (lane 1, early passage: EP), passage 23 (lane 2, late passage: LP), and passage 23 populations of hOEG originally transduced by the indicated lentivector combinations (lanes 3–11). (a) Representative blots derived from gels loaded with 30 µg of total protein per lane were probed with antibodies specific for the indicated proteins. All blots were repeated at least three times except for those probed for p21 (twice) and cyclin D1 (once). Dox, treated with 1 µg/ml doxycycline; WO, without doxycycline (withdrawn after 87 days of culture, 16 passages). (b) Graphs representing protein quantity derived by densitometry of the electrophoretic bands shown in a, normalized with respect to tubulin. Band densities of lysates from LP untransduced cells (control LP) were assigned a value of 1 (dotted line).
The senescence marker p21 (Cip1), blocks cell cycle progression through its inhibitory effect on cyclin-dependent kinases.14 In this study (Figure 2), p21 was increased in LP hOEG and, interestingly, this was reverted in cells transduced by combinations with TERT whereas BMI1 transduction appeared to have little effect on p21. As mentioned above, shp53 transduction almost eliminated p21, but cells transduced by TAg showed strikingly high p21 levels in spite of the well-documented role of TAg in inhibiting and inactivating p53.11 This agrees with previous findings indicating either the presence of a fraction of p53 not bound to TAg15 and still able to activate p21,16 or activation of p21 by a p53-independent mechanism.17
Another well-studied cyclin-dependent kinase inhibitor of a different class, p16 (INK4a),18,19 was progressively upregulated with increasing PDs in our primary hOEG (Figure 2). BMI1 was the only transgene which reduced p16 levels in hOEG down to those comparable to early passage cells, probably due to its repression of p16 expression.12 Curiously, p16 levels were also highly increased in TAg-transduced cells.
The third senescence marker which we analyzed, p27 (Kip1), is a p21-related cyclin-dependent kinase inhibitor,20,21 whose upstream regulatory events are less characterized.22 As can be seen in Figure 2, p27 levels in hOEG did not rise, but actually fell with increasing passage number of the primary cells, and its levels in the transduced populations were almost the opposite pattern of p21 levels. In TAg-transduced hOEG, p27 levels were downregulated whereas in cells transduced by BMI1, shp53, or TERT they were slightly higher than in primary cells of the same passage number. Re-activation of p53 expression by doxycycline withdrawal led to decreased p27 levels, confirming that the increased p27 expression was indeed mediated by knockdown of p53.
To gain further insight into the growth behavior of each hOEG population we next examined cyclin D1 levels, which are known to increase upon growth arrest in G0-G1 phase. Indeed, we observed high cyclin D1 levels in all nongrowing hOEG populations (Figure 2, control LP cells, and those transformed by BMI1 alone or shp53 after doxycycline withdrawal) except for hOEG transduced by TAg alone, in which a moderate level of cyclin D1 was observed. This is consistent with our results of Ki67 staining where nearly all TAg-transduced cells were proliferative. The proliferative decline of this population is a classical case of crisis where continued cell division is progressively cancelled out by increased cell death. As an indicator of cellular stress, we studied the levels of manganese superoxide dismutase (MnSOD).23 Consistent with the idea that oxidative stress plays a major role in senescence and crisis of cultured cells, we observed that all nongrowing hOEG populations showed upregulation of MnSOD except for cells transduced by BMI1 alone. Interestingly, both BMI1 and TERT either separately or together, appeared to reduce MnSOD levels, consistent with other studies showing that FoxM1c counteracts oxidative stress–induced senescence via BMI124 and that TERT protects cells from mitochondrial oxidative stress.25
Immortalization of hOEG with BMI1/TERT does not induce karyotypic changes or a transformed phenotype and maintains typical OEG antigens as well as neuroregenerative capacity
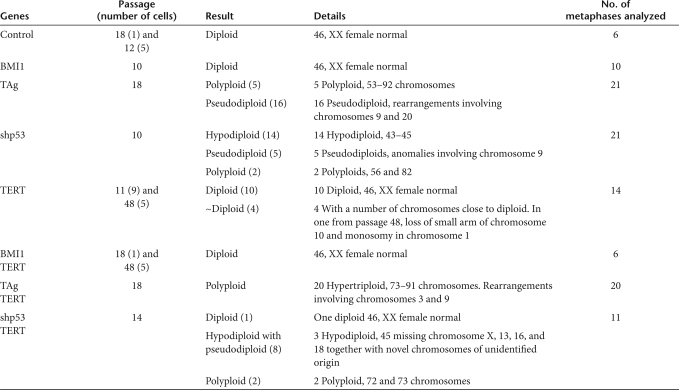
We carried out karyotype analyses of hOEG transduced by each of the transgene combinations (Table 2 and Supplementary Figure S3). The control primary cells exhibited a normal 46, XX karyotype. Cells transduced by shp53 were highly variable with polyploid, pseudodiploid, and hypodiploid karyotypes; and those transduced by TAg showed mainly polyploid and pseudodiploid karyotype. One LP (P48) cell transduced by TERT alone exhibited chromosomal aberrations. Only cells transduced by BMI1 or the combination BMI1/TERT appeared to be genetically stable and maintain the normal female karyotype, even after >300 days of culture (P48).
Table 2.
Karyotype analysis
We also studied the capacity for anchorage-independent growth in soft agar of these cells (Supplementary Figure S4). Only hOEG transduced by the combination of TAg with TERT were able to form colonies, indicating a transformed phenotype for these cells. Cells transduced by all other transgene combinations including TAg and TERT separately, were incapable of anchorage-independent growth.
Immunological analysis of the immortalized hOEG populations (those transduced with BMI1/TERT, TAg/TERT, or shp53/TERT) revealed that all of these cells express most typical OEG markers with few differences with respect to the control primary cultures: all cells stained positive for neuroligin, amyloid precursor protein, nestin, S100β, diffuse cytoplasmic glial fibrillary acidic protein, and vimentin (Supplementary Figure S5). The expression of Troy, a receptor for myelin-associated inhibitors of axonal regeneration, was found to be downregulated in hOEG transduced by BMI1/TERT or TAg/TERT but not in those transduced by shp53/TERT.
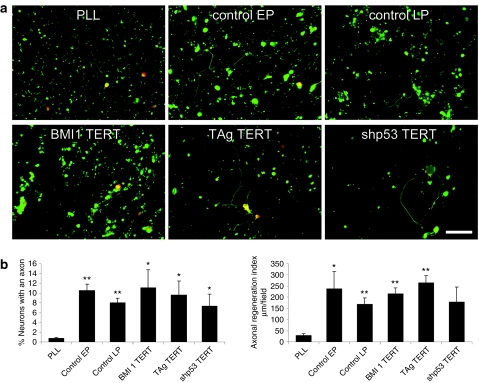
The functional properties of the immortalized hOEG were evaluated by examining their capacity to induce axonal growth in adult rat retinal ganglion neurons.26 Significantly, all of the immortalized hOEG populations exhibited neuroregenerative properties comparable to the control primary glia despite chromosomal changes in those transduced by shp53/TERT or TAg/TERT (Figure 3).
Figure 3.
Neuroregenerative capacity. (a) Representative fluorescence images of the neuroregeneration assay using rat retinal ganglion neurons showing neurons labeled with microtubule-associated protein 2 (MAP2) antibody in red and axons labeled with phosphorylated MAP1B and the high-molecular-weight neurofilament subunit proteins antibody in green. A minimum of 20 such fields were chosen at random and quantified for each treatment, and experiments were performed in triplicate (i.e., at least 60 fields analyzed for each sample). Bar = 50 µm. (b) Capacity to promote neuroregeneration in adult rat retinal ganglion neuronal cultures with respect to the percentage of neurons containing axons as well as the average length of regenerated axons. Axonal regeneration was quantified as the percentage of neurons with an axon, whereas total neuron number was determined by staining with the MAP2 antibody. Axons were defined as polarized neurites stained with antibodies against phosphorylated MAP1B and the high-molecular-weight neurofilament subunit proteins. We also determined the mean axonal length and the axonal regeneration index, a parameter defined as the axonal length/field. Graphs show mean values and bars indicate the standard deviations of three independent experiments. EP, early passage corresponds to culture passage 5 or 8; LP, late passage corresponds to culture passage 21 or 22; PLL, poly-L-lysine; the immortalized cells were used in passage 21, 22, or 23. The P values with respect to PLL are shown as: *P < 0.05 and **P < 0.01.
BMI1/TERT-transduced hOEG are viable after deimmortalization and promote neuroregeneration
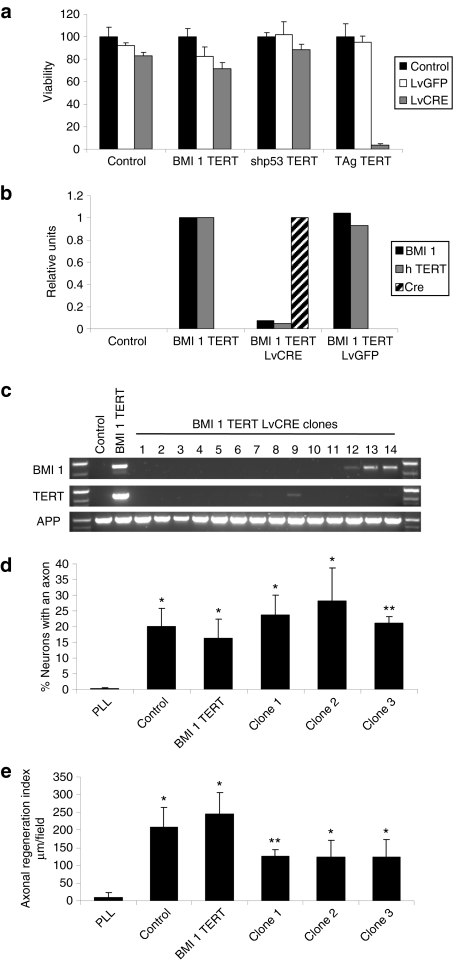
As transduction by any one of BMI1, shp53, or TAg in combination with TERT was able to immortalize hOEG at least for the duration of our study, we next evaluated the effects of “deimmortalization” of these cells—removal of the transgenes, which were all integrated as floxed cassettes in the cellular genome. One week after infection with lentivectors encoding either Cre recombinase or green fluorescent protein (GFP), most cells immortalized by BMI1/TERT or shp53/TERT were viable, although a small amount of toxicity was associated with Cre recombinase27,28,29 or the lentivector itself (Figure 4a, untransformed or BMI1/TERT-transduced cells, respectively). In contrast, the deimmortalization process was extremely toxic to cells immortalized by TAg/TERT. In the case of hOEG transduced by shp53/TERT, we were also able to eliminate expression of the shp53 transgene by doxycycline withdrawal from the culture medium. Although these cells were able to continue dividing for a few cycles, their replication rate was significantly reduced and, after ~20 days, the cells entered into crisis (Supplementary Figure S6). This was probably due to restored lethality of high p16 levels rather than to telomere shortening because the same phenomenon was observed even in those cells cotransduced by TERT.
Figure 4.
Cell survival and neuroregenerative function after deimmortalization. (a) Viability (as measured by the MTT assay) of control human olfactory ensheathing glia (hOEG) or those immortalized by BMI1/TERT, TAg/TERT, and shp53/TERT determined 1 week after infection [multiplicity of infection (MOI): 10] with a control lentivector (LvGFP) or a lentivector expressing Cre recombinase (LvCRE). Graphs show mean values and bars indicate the standard deviations of three independent samples. (b) Quantitative reverse transcription–PCR measurement of transgene expression in control hOEG or those immortalized by BMI1/TERT determined 1 week after infection (MOI: 10) with a control lentivector (LvGFP) or a lentivector expressing Cre recombinase (LvCRE). Real-time PCR was carried out on reverse-transcribed total RNA purified from infected and control cells as described in Materials and Methods using primers specific for transgenic BMI1 (black bars), TERT (gray bars), and Cre (hatched bars). (c) PCR analysis of genomic DNA derived from control hOEG, those immortalized by BMI1/TERT, and individual clones derived from BMI1/TERT-immortalized OEG after treatment with Cre recombinase. Specific oligonucleotide pairs were used to detect transgenic BMI1 and TERT DNA whereas a third oligonucleotide pair specific for the endogenous human amyloid precursor protein was used as a control for DNA loading. (d,e) Neuroregenerative capacity of deimmortalized clones in the adult rat retinal ganglion neuron coculture assay. Graphs show means and standard deviations derived from three independent experiments represent the (d) percentage of neurons exhibiting axons and (e) the axonal regeneration index, a parameter reflecting the average length of axons regenerated. Cell passage number in these experiments was 9 for control cells and 36–37 for the rest of the cells. The P values (in d and e, with respect to PLL) are shown as: *P < 0.05 and **P < 0.01.
We focused our further studies on the hOEG transduced by BMI1/TERT because they were able to survive after deimmortalization and showed unaltered karyotype. As shown in Figure 4b, deimmortalization of these cells by infection with a Cre-encoding lentivector (but not with a control GFP-encoding lentivector) resulted in efficient Cre expression and a drastic decrease in the levels of BMI1 and TERT levels after 1 week. We have been successful in cloning these deimmortalized hOEG, and PCR analysis of genomic DNA confirmed that the BMI1 and TERT transgenes had indeed been deleted completely in 12 out of 26 clones analyzed (Figure 4c). During the 1-month period analyzed, the growth of these deimmortalized clones was similar to that of the parental immortalized population. We could also confirm that they maintained neuroregenerative capacity in vitro, showing that the number of neurons which regenerated axons (Figure 4d) by all of the hOEG populations was similar, and clearly much higher than that obtained on poly-L-lysine alone or by primary human fibroblasts (data not shown). The axonal regeneration index (Figure 4e), a parameter reflecting the mean length of regenerated axons, appeared to be slightly lower in assays with deimmortalized hOEG clones compared to those with primary and immortalized hOEG but this difference was not significant (0.05 < P < 0.06).
Discussion
The ability to obtain immortalized cells from primary cultures is often described to be due to the inhibition of replicative senescence. However, immortalization refers to indefinite cell proliferation, which may be independent of senescence, an aged state in which cells acquire molecular alterations which can modify their morphology and function. The p16Ink4a/Rb and p19Arf/p53 pathways play major roles in senescence and accumulating divisions in normal cells lead to their progressive activation. Many mechanisms of immortalization depend on preventing the activation of these pathways30,31 although this can be ineffective if the pre-existing levels of these pathways are already above a certain threshold due to the age of the cell, or conditions such as oxidative stress or oncogenic activation.32,33,34 In many cell types, senescence is also accompanied by the shortening of telomeres, but for the purpose of immortalizing cells, this can be avoided by TERT transduction.1
The primary hOEG which formed the focus of our study followed the classical replicative behavior described for human diploid cells,35 in which after a finite number of divisions, cells enter into replicative senescence (mortality stage M1) and cease to divide. The hOEG studied here were derived from a young donor (13 years) and under our culture conditions, exhibited a Hayflick limit of 34 PDs during 107 days, which could be exceeded by transduction of several immortalizing transgenes. In the cases of hOEG transduced by BMI1 or TAg, the cells eventually entered into crisis (mortality stage M2) where augmented cell death exceeded replication, leading to extinction of the culture. As has also been described for other human cell types,1 we were able to obtain immortalized cells from primary cultures of hOEG by combined transduction of TERT with BMI1, shp53, or TAg; these cells have a replicative life span well exceeding the Hayflick limit or that of cells exhibiting M2 crisis (see Figure 1a), and so at least within the duration of our study, can be considered immortalized. However, our molecular analyses reveal that most of these transgene combinations lead to indefinite cell proliferation without preventing activation of senescence markers, and genetic reversion by removal of these transgenes resulted in cell death in these cases. Only BMI1 combined with TERT was able to prevent the simultaneous rise of p16, p21, and cyclin D1 in transduced cells, and maintains a normal proliferative cell population with a nonsenescent phenotype. Significantly, BMI1/TERT transduction was also able to prolong the life span of hOEG with no changes in karyotype or transforming potential during the time span of our experiments. In hOEG transduced by TERT alone, p16, p21, and cyclin D1 levels were also maintained at low levels, but the Ki67/SA-β-gal profile of these TERT-transduced cells revealed a more senescent phenotype, and karyotypic changes were also detected in these cells. Additionally, the combination of BMI1/TERT was able to immortalize hOEG from an elderly donor36 whereas TERT alone was ineffective. BMI1 alone promoted a very efficient repression of p16 but p21 levels were increased after several passages, probably due to activation of p53 in response to classical telomere crisis and these cells died due to a cell cycle block in G1, reflected in an increase in cyclin D1 levels.
Strikingly, the hOEG population profiles in Ki67/SA-β-gal assay correlated well with the capacity of the transgene combinations to extend life span and permit viability after deimmortalization. Cells transduced by BMI1/TERT exhibit the Ki67/SA-β-gal profile most similar to that of early passage primary hOEG which is corroborated also by similar levels of p16, p21, p27, cyclin D1, and MnSOD. Importantly, it appears that this minimal perturbation of molecular pathways can maintain hOEG in “suspended senescence,” after which deimmortalization merely returns the cells to a state similar to that prior to immortalization. In contrast, incomplete repression of cellular senescence pathways results in crisis or growth arrest upon deimmortalization of hOEG transduced by TAg/TERT or shp53/TERT.
Although reversible immortalization has been used to extend life span in different primary cultures,2,3 there is little information about the behavior of the cells after deimmortalization. Our results indicate that some immortalized cells override signals blocking proliferation but do not necessarily prevent senescence progression, and deimmortalization thus cannot return these cells back to the original “young” state. Therefore, immortalization can be distinguished from the cessation of senescence, and reversible immortalization technology offers a tool for dissecting these two phenomena. Our study shows that in hOEG, transgenic expression of the single transgenes shp53, BMI1, or TAg preferentially downregulates p21, p16, or p27, respectively, which are key effectors of cellular senescence.22 The unique ability of the BMI1/TERT combination to prevent senescence progression in hOEG may be due to the fact that both p16 and p21 pathways are downregulated by these transgenes. Additionally, no karyotypic changes were noted probably due to the intact function of DNA damage sensor systems such as those involving p53.
It is important to note that regardless of their chromosomal alterations, the cells immortalized by TERT in combination with either TAg or shp53 conserve antigens typical of OEG as well as neuroregenerative function. Thus, these easy-to-grow cell lines can serve as useful models for studying hOEG-mediated neuroregeneration and perhaps even as therapeutic tools if they can be shown to be genetically stable and exhibit no tumorigenic potential.37
Human embryonic stem cells represent a potential source of all cell types for cell therapy, but ethical issues concerning their use must be resolved. Although adult stem cells or lineage-restricted precursors can be obtained from various somatic niches by ethically approved procedures, their use depends on the development of efficient methods for directing their differentiation into desired cell types. Recent studies show that even mature somatic tissues can be reprogrammed to generate induced pluripotent stem cells for large-scale expansion in culture but their subsequent differentiation into the desired tissues is not a trivial task. Our present molecular characterization of reversibly immortalized hOEG indicates that reprogramming of proliferation alone can be achieved without alteration of senescence state or other cellular properties, thus bypassing the need for retrodifferentiation and the subsequent chore of differentiating the cells to restricted lineages necessary for the use of stem cells. This important tool for cell therapy also opens the possibility for autotransplantation, which would greatly aid initial clinical trials to assess efficacy by eliminating graft rejection problems.
Materials and Methods
Detailed information regarding special reagents and antibodies can be obtained in the Supplementary Materials and Methods section.
Primary culture of human OEG. Olfactory bulbs were obtained from a 13-year-old female cadaver donor following the legally and ethically approved protocols of the Tissue Bank for Neurological Research (Madrid). We used a previously described method26 with modifications. Briefly, at ~9 hours postmortem, superficial layers from olfactory bulbs were dissected out and the tissue dissociated. After 15-minute incubation with 0.1% trypsin in Hank's buffered salt solution, digestion was stopped with 20% fetal calf serum (FCS) in Hank's buffered salt solution, and the cells were collected by centrifugation and dissociated by several passes through Pasteur pipettes. OEG were resuspended and cultured in minimal essential medium: Dulbecco's modified Eagle medium:F12 (1:1), 10% FCS, 2 mmol/l glutamine, 20 µg/ml pituitary extract, 2 µmol/l forskolin, and 50 µg/ml primocin.
Lentivector production and titration. Pseudotyped lentivectors were produced using reagents and protocols from Didier Trono (for detailed protocol, see http://tronolab.epfl.ch/page58122.html) by transient cotransfection of 5 µg of the corresponding lentivector plasmid, 5 µg of the packaging plasmid pCMVdR8.74,38 and 2 µg of the vesicular stomatitis virus G envelope protein plasmid pMD2G (Addgene plasmid 12259) in 10-cm plates of subconfluent 293T cells using Lipofectamine Plus reagent following instructions of the supplier (Invitrogen, Carlsbad, CA). Lentivectors expressing immortalizing genes were pLOX-CWBmi1 (Addgene plasmid 12240), pLOX-TERT-iresTK (Addgene plasmid 12245), and pLOX-Ttag-iresTK (Addgene plasmid 12246) which respectively encode mouse BMI1 or TERT coupled to herpes simplex virus-1 thymidine kinase (TK) gene via the encephalomyocarditis virus internal ribosome entry site, and TAg also linked to internal ribosome entry site-TK.2 The vector pLVUHshp53-tTR-KRAB (Addgene plasmid 11649) encodes a short hairpin RNA directed against p53 under the control of an H1 promoter that is regulated by tetracycline.13 To express Cre recombinase we used pLOX-CW-CRE,39 a lentivector which transduces cells with a self-excising CRE transgene, thus avoiding genotoxicity due to sustained Cre expression. The pRRLSIN.cPPT.PGK-GFP.WPRE vector (Addgene plasmid 12252) which expresses enhanced GFP was used as a control vector. Lentivectors were titered on target cells (hOEG) with serial dilutions of the vector supernatants and the number of transduced cells determined 48 hours postinfection by flow cytometry (FACSCalibur, BD Biosciences, San Diego, CA) using antibodies directed against the corresponding transgene.
Immortalization and deimmortalization of hOEG. Primary hOEG cultures in passage 2 were seeded in 24-well plates and infected with various combinations of lentivectors encoding BMI1, TAg, shp53, and TERT at a multiplicity of infection of between 2 and 10. After reaching confluence, the cells were counted and thereafter cultured on 10-cm plates; PDs were calculated as PDs = log (Nf/N0)/log 2, where Nf is the final cell count and N0 is the initial number of cells seeded (105 cells).
Genetic reversion of immortalized hOEG was carried out by infecting the cells with lentivectors encoding either Cre recombinase or enhanced GFP at a multiplicity of infection of 10. Deimmortalized cells were analyzed for viability using the MTT (200 µg/ml) assay40 1 week after infection, or plated out at high dilution (103 cells per 10-cm plate) to obtain single colonies for clonal analysis by PCR of genomic DNA.
Flow cytometry analysis. Cells were detached by trypsin treatment, fixed by 10-minute incubation in 2% paraformaldehyde (PFA) in phosphate-buffered saline (PBS), and washed and maintained in PBS at 4 °C until analyzed. Immediately prior to analysis, cells were permeabilized by 2-minute treatment with PBS + 0.5% Tween 20, followed by blocking with PBS 0.5% + Tween 20 + 10% FCS for 15 minutes. The cells were then incubated with the primary antibody diluted in blocking solution for 20 minutes. After washing twice with PBS + 0.5% Tween 20, cells were incubated for 10 minutes with the secondary antibodies. After washing again, flow cytometry analysis was performed (FACSCalibur, BD Biosciences).
Immunocytochemistry. Cells were grown on sterile glass coverslips and fixed with 4% PFA in PBS. After blocking with PBS containing 1% FCS and 0.1% Triton X-100 for 30 minutes, cells were washed with PBS and stained by indirect immunofluorescence using the antibodies described in Materials and Materials as outlined in the text. Samples were mounted in Fluoromount G and were observed in an Axiovert 200 (Zeiss, Oberkochen, Germany) fluorescence microscope.
Cocultures of postmitotic rat retinal ganglion neurons with OEG. Extension of neurites by postmitotic rat retinal ganglion neurons was used as a culture model of axonal regeneration.26 Briefly, retinal tissue was extracted from 2-month-old adult (P60) rats and digested with papain (20 U/ml) in the presence of 50 µmol/l of the N-methyl-D-aspartic acid receptor inhibitor DL-2-amino-5-phosphonovaleric acid. The cell suspension was then plated either on 10 µg/ml poly-L-lysine–treated coverslips or OEG monolayers. The cultures were maintained at 37 °C with 5% CO2 in serum-free Neurobasal medium supplemented with B-27 12.5 mmol/l KCl for 96 hours before fixing with 4% PFA.
Quantification of axon regeneration. Preparations were quantified in a blinded manner by counting axons under a ×40 objective of an inverted Axiovert 200 microscope. A minimum of 20 randomly chosen fields were quantified for each treatment and experiments were performed in triplicate. Axonal regeneration was quantified as the percentage of neurons with an axon, whereas total neuron number was determined by staining with the microtubule-associated protein 2 antibody. Axons were defined as polarized neurites stained with antibodies against phosphorylated microtubule-associated protein 1B and the high-molecular-weight neurofilament subunit proteins. We also determined the mean axonal length and the axonal regeneration index, a parameter defined as the axonal length/field. Quantitative image analysis was performed using the NeuronJ plugin (E. Meijering) of the ImageJ program (Wayne Rasband, National Institutes of Health).
Senescence versus proliferation assay. Cells were grown over sterile coverslips and fixed by 10-minute treatment with 4% PFA in PBS at 4 °C. Cells were incubated overnight with SA-β-gal staining solution: 1 mg/ml of X-gal (5-bromo-4-chloro-3-indolyl β-D-galactoside), 40 mmol/l citric acid/sodium phosphate pH 6.0, 5 mmol/l potassium ferrocyanide, 5 mmol/l potassium ferricyanide, 150 mmol/l NaCl, 2 mmol/l MgCl2, at pH 4.0. Cells were then washed with H2O, permeabilized by a 10-minute incubation with 0.1% sodium dodecyl sulfate in PBS, blocked by 30-minute incubation in PBS containing 1% FCS and 0.1% Triton X-100, after which the cells were incubated for 1 hour with Ki67 antibody followed by anti-mouse Alexa-488-labeled secondary antibody. Finally, cell nuclei were stained with To-Pro-3 to obtain total cell counts. Samples were observed in an Axiovert 200 microscope and 10–18 random fields at ×40 magnification were photographed in both bright field and fluorescence modes. To aid in scoring of the senescence and proliferative status of the cells, the bright field (blue over white background) SA-β-gal images were transformed into green over black background images using ImageJ software and merged with the corresponding Ki67 and To-Pro-3 images in the red and blue channels, respectively (Supplementary Figure S2).
Protein expression pattern by western blot. The cells were collected, washed, and resuspended in lysis buffer (50 mmol/l Tris–HCl pH 7.5, 300 mmol/l NaCl, 0.5% sodium dodecyl sulfate, and 1% Triton X-100) and incubated for 15 minutes at 95 °C. Protein concentration of the extracts was measured using the Dc protein assay kit (Bio-Rad, Hercules, CA) and 30 µg of each cell extract was resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis in 12% polyacrylamide gels. After electrophoresis, proteins were transferred to nitrocellulose membranes by western blot and immunoreactive proteins were visualized using an enhanced chemiluminescence detection kit (Pierce, Rockford, IL) following instructions of the supplier. All blots were repeated at least three times except for those probed for p21 (twice) and cyclin D1 (once). Because some of the proteins shown in Figure 2 have very similar molecular masses, the western blot images are derived from two duplicate membranes of the same samples. Protein bands were quantified by densitometry using Quantity One software (Bio-Rad) and values were normalized with respect to tubulin values of the same lane on the same membrane. Numbers in the graph in Figure 2b represent the ratio between the value of each sample with respect to LP untransduced cells (control LP).
Anchorage-independent growth. Cells which had been grown in culture for 175 days were seeded in 12-multiwell plates at 104 cells/well in 0.35% agar in Dulbecco's modified Eagle medium over a 0.7% agar layer. Plates were incubated for 23 days, after which time colonies were large enough to be visualized by staining with MTT (200 µg/ml) for 6 hours and photographed using a flatbed scanner. Soft agar assays were performed in triplicate.
Karyotype analysis. Chromosomal studies were performed at the Genetics Department of the Hospital “La Paz” in Madrid. Metaphase spreads were prepared from cells treated with 100 ng/ml colcemid for 6 hours and analyzed by standard protocols for high-resolution GTL banding.41
RT-PCR analysis. We used specific primers for the different genes and the corresponding transgenes: BMI1 cagcaatgactgtgatgc (forward) and aatccagaggttgattatcg (reverse, exclusively specific for transgenic BMI1); TERT gtatggctgcgtggtgaacttgc (forward) and tgccaaaagacggcaatatggtg (reverse, exclusively specific for transgenic TERT); and TAg taaagcattgcctggaacgcagt (forward) and aggactgaggggcctgaaatgag (reverse).
Total RNA was purified using RNeasy Mini Kit (Qiagen, Hilden, Germany) following commercial guidelines. Purified total RNA (1–2 mg) of each cell line was reverse transcribed with the specific reverse primers using AMV reverse transcriptase (Promega, Madison, WI) following the manufacturer's instructions. Three microliters of each RT reaction were then amplified by PCR using the following protocol: 35 cycles of 95 °C for 1 minute, 54 °C for 1 minute, and 72 °C for 2 minutes + 0.2 second/cycle.
Quantitative RT-PCR. PCR primers were designed with the Primer Express software (Applied Biosystems, Foster City, CA) and purchased from Metabion (Martinsried, Germany) to specifically amplify the TERT transgene: cttcaagaccatcctggactgat (forward) and caatatggtggaaaataaccggaa (reverse); the BMI1 transgene: ggtcatcagcaacttcatctggt (forward) and ctttcacaaattttgtaatccagagg (reverse); and CRE: agaaaaaatggtgttgccgc (forward) and gccagttgatagctggctgg (reverse).
Total RNA was purified using RNeasy Mini Kit (Qiagen, Hilden, Germany) following commercial guidelines. For RT-PCR, 0.5 mg RNA was reverse transcribed using the High Capacity Reverse Transcription kit (Applied Biosystems, Foster City, CA) following the supplier's guidelines with the conditions: 10 minutes at 25 °C, 120 minutes at 37 °C, and 5 seconds at 85 °C. Thereafter, 0.5 µl of each reverse-transcribed sample (10 ng/µl) was added to either 5 µl 2× FastStart TaqMan MasterMix (Roche, Basel, Switzerland) and 0.5 µl 20× specific Taqman Assay (ABI, Applied Biosystems) for 18S rRNA assay or 5 µl 2× SYBR Green MasterMix (ABI) and 250 nmol/l oligonucleotides for transgene detection in a total volume of 10 µl. We used universal conditions for real-time PCR: 2 minutes at 50 °C, 10 minutes at 95 °C and 40 cycles of 15 seconds at 95 °C, and 1 minute at 60 °C in a ABI 7900HT thermocycler (Applied Biosystems).
Detection of transgenes from genomic DNA. Specific primers were designed to detect the lentivector-encoded transgenes: BMI1: ttctcctcggtcttcattgg (reverse), TERT: gaaggccagcacgttcttcg (reverse), and forward for both transgenes: tgaaccgtcagatcgcctgg. As a control endogenous gene to assess template DNA, specific primers that recognized human amyloid precursor protein were used: tgccaaaattccatatggacg (forward) and gggatttgccaagcagcatat (reverse). For PCR, GoTaq Flexi DNA Polymerase (Promega) was used following the supplier's instructions with the following modifications: for detection of BMI1 and TERT, the conditions were 500 ng of genomic DNA, 1 mol/l betaine, 5% dimethyl sulfoxide, and 2 mmol/l MgCl2, and the protocol was 5 minutes at 94 °C followed by 30 cycles of 1 minute at 94 °C, 1 minute at 60 °C, and 1 minute at 72 °C, followed finally by 10 minutes at 72 °C; for amyloid precursor protein, the conditions were 100 ng of genomic DNA and 2 mmol/l MgCl2, and the protocol was 1 minute at 94 °C, followed by 35 cycles of 30 seconds at 94 °C, 30 seconds at 55 °C, and 1 minute at 72 °C, followed finally by 10 minutes at 72 °C. PCRs were resolved in 1.5% agarose gels.
Statistical analysis. Statistical comparison of the data sets was performed by the Student's t-test. The differences are given with their corresponding statistical significance or P value, which is the probability that the difference occurred merely by chance under the null hypothesis.
SUPPLEMENTARY MATERIALFigure S1. Bright field images of the different hOEG populations. First row: representative bright field images of primary hOEG cells (control) after 10 culture passages and populations immortalized with BMI1/TERT, TAg/TERT or shp53/TERT after 23 culture passages are shown. Second row: representative images of BMI1/TERT transduced cells after deimmortalization by infection with LvCRE (MOI: 10) showing the whole population (2 weeks after the infection) as well as deimmortalized clones. Scale bar, 20 μm.Figure S2. Simultaneous detection of senescence versus proliferation markers. First column shows representative lysosomal senescence-associated β-galactosidase (SA-β-Gal) activity staining using X-gal in bright field images of immortalized hOEG. Second column shows the same SA-β-Gal pattern transformed into green over black background using ImageJ software. Third and fourth columns show representative fluorescence images of Ki67 immunostaining and nuclear marking with To-Pro-3, respectively. Fifth column shows the merge of the three preceding columns. EP, early passage (passage 4); LP, late passage (passage 21), the rest of the populations were studied at passage 21; dox, treated with 1 μg/ml doxycycline; WO, without doxycycline, which was withdrawn after 87 days of culture, 16 passages.Figure S3. Karyotype analysis. Representative high-resolution GTL-banding metaphase spreads prepared from control (female) hOEG or those immortalized by BMI1/TERT, TAg/TERT and shp53/TERT. Quantitative data are given in Table 2. Arrows indicate important deletions. Primary cells (control) were analyzed in cell passage 12 and immortalized populations in cell passage 18 (BMI1 TERT and TAg TERT) and 14 (shp53 TERT).Figure S4. Anchorage-independent growth. Colony formation assay in soft agar of hOEG populations transduced by lentivectors encoding the indicated transgenes. Positive controls were performed by using the two tumor cell lines MDA-MB (mammary adenocarcinoma) and U-87 MG (glioblastoma). Living cells were stained using MTT and plate images were obtained using a flatbed scanner.Figure S5. OEG markers present on immortalized hOEG populations. Parallel cultures of unmodified primary hOEG, or populations immortalized by lentivectors encoding BMI1/TERT, TAg/TERT or shp53/TERT were fixed and stained by indirect immunofluorescence for the presence of neuroligin, amyloid precursor protein (APP), nestin, troy, S100β, glial fibrillary acidic protein (GFAP) and vimentin.Figure S6. Doxycycline-dependent behavior of shp53-transformed populations. Growth curve of hOEG transduced either by a lentivector encoding a tetracycline-inducible shp53 alone (blue), or in combination with a lentivector encoding TERT (red). The cells were grown in the presence of 1 μg/ml doxycycline (closed symbols, dox) to maintain the expression of the shp53 transgene. At day 87 (arrow), in half of each culture, the effect of eliminating shp53 expression was studied by withdrawal of doxycycline from the medium (open symbols, WO). Cumulative population doublings were calculated as described in the Materials and Methods section from the cell number counted at each culture passage.Materials and Methods.
Supplementary Material
Bright field images of the different hOEG populations. First row: representative bright field images of primary hOEG cells (control) after 10 culture passages and populations immortalized with BMI1/TERT, TAg/TERT or shp53/TERT after 23 culture passages are shown. Second row: representative images of BMI1/TERT transduced cells after deimmortalization by infection with LvCRE (MOI: 10) showing the whole population (2 weeks after the infection) as well as deimmortalized clones. Scale bar, 20 μm.
Simultaneous detection of senescence versus proliferation markers. First column shows representative lysosomal senescence-associated β-galactosidase (SA-β-Gal) activity staining using X-gal in bright field images of immortalized hOEG. Second column shows the same SA-β-Gal pattern transformed into green over black background using ImageJ software. Third and fourth columns show representative fluorescence images of Ki67 immunostaining and nuclear marking with To-Pro-3, respectively. Fifth column shows the merge of the three preceding columns. EP, early passage (passage 4); LP, late passage (passage 21), the rest of the populations were studied at passage 21; dox, treated with 1 μg/ml doxycycline; WO, without doxycycline, which was withdrawn after 87 days of culture, 16 passages.
Karyotype analysis. Representative high-resolution GTL-banding metaphase spreads prepared from control (female) hOEG or those immortalized by BMI1/TERT, TAg/TERT and shp53/TERT. Quantitative data are given in Table 2. Arrows indicate important deletions. Primary cells (control) were analyzed in cell passage 12 and immortalized populations in cell passage 18 (BMI1 TERT and TAg TERT) and 14 (shp53 TERT).
Anchorage-independent growth. Colony formation assay in soft agar of hOEG populations transduced by lentivectors encoding the indicated transgenes. Positive controls were performed by using the two tumor cell lines MDA-MB (mammary adenocarcinoma) and U-87 MG (glioblastoma). Living cells were stained using MTT and plate images were obtained using a flatbed scanner.
OEG markers present on immortalized hOEG populations. Parallel cultures of unmodified primary hOEG, or populations immortalized by lentivectors encoding BMI1/TERT, TAg/TERT or shp53/TERT were fixed and stained by indirect immunofluorescence for the presence of neuroligin, amyloid precursor protein (APP), nestin, troy, S100β, glial fibrillary acidic protein (GFAP) and vimentin.
Doxycycline-dependent behavior of shp53-transformed populations. Growth curve of hOEG transduced either by a lentivector encoding a tetracycline-inducible shp53 alone (blue), or in combination with a lentivector encoding TERT (red). The cells were grown in the presence of 1 μg/ml doxycycline (closed symbols, dox) to maintain the expression of the shp53 transgene. At day 87 (arrow), in half of each culture, the effect of eliminating shp53 expression was studied by withdrawal of doxycycline from the medium (open symbols, WO). Cumulative population doublings were calculated as described in the Materials and Methods section from the cell number counted at each culture passage.
Acknowledgments
We are grateful for advice and technical aid provided by the Microscopy service of the Centro de Biología Molecular “Severo Ochoa” and Ricardo Ramos from the Universidad Autónoma de Madrid Interdepartmental Research Services (SIDI). This work was supported by grants from Noscira S.A. and the Fundación Marcelino Botín. F.L. held Ramón y Cajal and I3 research incorporation contracts from the Spanish Ministry of Science.
REFERENCES
- Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, et al. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- Salmon P, Oberholzer J, Occhiodoro T, Morel P, Lou J., and , Trono D. Reversible immortalization of human primary cells by lentivector-mediated transfer of specific genes. Mol Ther. 2000;2:404–414. doi: 10.1006/mthe.2000.0141. [DOI] [PubMed] [Google Scholar]
- Westerman KA., and , Leboulch P. Reversible immortalization of mammalian cells mediated by retroviral transfer and site-specific recombination. Proc Natl Acad Sci USA. 1996;93:8971–8976. doi: 10.1073/pnas.93.17.8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., and , Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Lowry WE., and , Plath K. The many ways to make an iPS cell. Nat Biotechnol. 2008;26:1246–1248. doi: 10.1038/nbt1108-1246. [DOI] [PubMed] [Google Scholar]
- Feng R, Desbordes SC, Xie H, Tillo ES, Pixley F, Stanley ER, et al. PU.1 and C/EBPalpha/beta convert fibroblasts into macrophage-like cells. Proc Natl Acad Sci USA. 2008;105:6057–6062. doi: 10.1073/pnas.0711961105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H, Tapscott SJ, Davis RL, Thayer MJ, Adam MA, Lassar AB, et al. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proc Natl Acad Sci USA. 1989;86:5434–5438. doi: 10.1073/pnas.86.14.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Brown J, Kanarek A, Rajagopal J., and , Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455:627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisman G., and , Li Y. Repair of neural pathways by olfactory ensheathing cells. Nat Rev Neurosci. 2007;8:312–319. doi: 10.1038/nrn2099. [DOI] [PubMed] [Google Scholar]
- Moreno-Flores MT, Díaz-Nido J, Wandosell F., and , Avila J. Olfactory ensheathing glia: drivers of axonal regeneration in the central nervous system. J Biomed Biotechnol. 2002;2:37–43. doi: 10.1155/S1110724302000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali SH., and , DeCaprio JA. Cellular transformation by SV40 large T antigen: interaction with host proteins. Semin Cancer Biol. 2001;11:15–23. doi: 10.1006/scbi.2000.0342. [DOI] [PubMed] [Google Scholar]
- Park IK, Morrison SJ., and , Clarke MF. Bmi1, stem cells, and senescence regulation. J Clin Invest. 2004;113:175–179. doi: 10.1172/JCI20800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulc J, Wiznerowicz M, Sauvain MO, Trono D., and , Aebischer P. A versatile tool for conditional gene expression and knockdown. Nat Methods. 2006;3:109–116. doi: 10.1038/nmeth846. [DOI] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ, Keyomarsi K, Dynlacht B, Tsai LH, Zhang P, et al. Inhibition of cyclin-dependent kinases by p21. Mol Biol Cell. 1995;6:387–400. doi: 10.1091/mbc.6.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Zhou S, Chen X, Guo L, Li Z, Hu D, et al. The activation of p53 mediated by Epstein-Barr virus latent membrane protein 1 in SV40 large T-antigen transformed cells. FEBS Lett. 2008;582:755–762. doi: 10.1016/j.febslet.2008.01.031. [DOI] [PubMed] [Google Scholar]
- O'Neill FJ, Hu Y, Chen T., and , Carney H. Identification of p53 unbound to T-antigen in human cells transformed by simian virus 40 T-antigen. Oncogene. 1997;14:955–965. doi: 10.1038/sj.onc.1200913. [DOI] [PubMed] [Google Scholar]
- Markovics JA, Carroll PA, Robles MT, Pope H, Coopersmith CM., and , Pipas JM. Intestinal dysplasia induced by simian virus 40 T antigen is independent of p53. J Virol. 2005;79:7492–7502. doi: 10.1128/JVI.79.12.7492-7502.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara E, Smith R, Parry D, Tahara H, Stone S., and , Peters G. Regulation of p16CDKN2 expression and its implications for cell immortalization and senescence. Mol Cell Biol. 1996;16:859–867. doi: 10.1128/mcb.16.3.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huschtscha LI., and , Reddel RR. p16(INK4a) and the control of cellular proliferative life span. Carcinogenesis. 1999;20:921–926. doi: 10.1093/carcin/20.6.921. [DOI] [PubMed] [Google Scholar]
- Polyak K, Lee MH, Erdjument-Bromage H, Koff A, Roberts JM, Tempst P, et al. Cloning of p27Kip1, a cyclin-dependent kinase inhibitor and a potential mediator of extracellular antimitogenic signals. Cell. 1994;78:59–66. doi: 10.1016/0092-8674(94)90572-x. [DOI] [PubMed] [Google Scholar]
- Toyoshima H., and , Hunter T. p27, a novel inhibitor of G1 cyclin-Cdk protein kinase activity, is related to p21. Cell. 1994;78:67–74. doi: 10.1016/0092-8674(94)90573-8. [DOI] [PubMed] [Google Scholar]
- Bringold F., and , Serrano M. Tumor suppressors and oncogenes in cellular senescence. Exp Gerontol. 2000;35:317–329. doi: 10.1016/s0531-5565(00)00083-8. [DOI] [PubMed] [Google Scholar]
- Macmillan-Crow LA., and , Cruthirds DL. Invited review: manganese superoxide dismutase in disease. Free Radic Res. 2001;34:325–336. doi: 10.1080/10715760100300281. [DOI] [PubMed] [Google Scholar]
- Li SK, Smith DK, Leung WY, Cheung AM, Lam EW, Dimri GP, et al. FoxM1c counteracts oxidative stress-induced senescence and stimulates Bmi-1 expression. J Biol Chem. 2008;283:16545–16553. doi: 10.1074/jbc.M709604200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed S, Passos JF, Birket MJ, Beckmann T, Brings S, Peters H, et al. Telomerase does not counteract telomere shortening but protects mitochondrial function under oxidative stress. J Cell Sci. 2008;121:1046–1053. doi: 10.1242/jcs.019372. [DOI] [PubMed] [Google Scholar]
- Moreno-Flores MT, Lim F, Martín-Bermejo MJ, Díaz-Nido J, Avila J., and , Wandosell F. Immortalized olfactory ensheathing glia promote axonal regeneration of rat retinal ganglion neurons. J Neurochem. 2003;85:861–871. doi: 10.1046/j.1471-4159.2003.01729.x. [DOI] [PubMed] [Google Scholar]
- Loonstra A, Vooijs M, Beverloo HB, Allak BA, van Drunen E, Kanaar R, et al. Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proc Natl Acad Sci USA. 2001;98:9209–9214. doi: 10.1073/pnas.161269798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver DP., and , Livingston DM. Self-excising retroviral vectors encoding the Cre recombinase overcome Cre-mediated cellular toxicity. Mol Cell. 2001;8:233–243. doi: 10.1016/s1097-2765(01)00295-7. [DOI] [PubMed] [Google Scholar]
- Baba Y, Nakano M, Yamada Y, Saito I., and , Kanegae Y. Practical range of effective dose for Cre recombinase-expressing recombinant adenovirus without cell toxicity in mammalian cells. Microbiol Immunol. 2005;49:559–570. doi: 10.1111/j.1348-0421.2005.tb03753.x. [DOI] [PubMed] [Google Scholar]
- Lundberg AS, Hahn WC, Gupta P., and , Weinberg RA. Genes involved in senescence and immortalization. Curr Opin Cell Biol. 2000;12:705–709. doi: 10.1016/s0955-0674(00)00155-1. [DOI] [PubMed] [Google Scholar]
- Carnero A, Hudson JD, Price CM., and , Beach DH. p16INK4A and p19ARF act in overlapping pathways in cellular immortalization. Nat Cell Biol. 2000;2:148–155. doi: 10.1038/35004020. [DOI] [PubMed] [Google Scholar]
- Beauséjour CM, Krtolica A, Galimi F, Narita M, Lowe SW, Yaswen P, et al. Reversal of human cellular senescence: roles of the p53 and p16 pathways. EMBO J. 2003;22:4212–4222. doi: 10.1093/emboj/cdg417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J., and , d'Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8:729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- Fridman AL., and , Tainsky MA. Critical pathways in cellular senescence and immortalization revealed by gene expression profiling. Oncogene. 2008;27:5975–5987. doi: 10.1038/onc.2008.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayflick L. The limited in vitro lifetime of human diploid cell strains. Exp Cell Res. 1965;37:614–636. doi: 10.1016/0014-4827(65)90211-9. [DOI] [PubMed] [Google Scholar]
- Lim F, Martín-Bermejo MJ, García-Escudero V, Gallego-Hernández MT, García-Gómez A, Rábano A, et al. Reversibly immortalized human olfactory ensheathing glia from an elderly donor maintain neuroregenerative capacity Gliain the press). [DOI] [PubMed]
- Kondziolka D, Steinberg GK, Wechsler L, Meltzer CC, Elder E, Gebel J, et al. Neurotransplantation for patients with subcortical motor stroke: a phase 2 randomized trial. J Neurosurg. 2005;103:38–45. doi: 10.3171/jns.2005.103.1.0038. [DOI] [PubMed] [Google Scholar]
- Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D, et al. A third-generation lentivirus vector with a conditional packaging system. J Virol. 1998;72:8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cudré-Mauroux C, Occhiodoro T, König S, Salmon P, Bernheim L., and , Trono D. Lentivector-mediated transfer of Bmi-1 and telomerase in muscle satellite cells yields a duchenne myoblast cell line with long-term genotypic and phenotypic stability. Hum Gene Ther. 2003;14:1525–1533. doi: 10.1089/104303403322495034. [DOI] [PubMed] [Google Scholar]
- Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- Seabright M. A rapid banding technique for human chromosomes. Lancet. 1971;2:971–972. doi: 10.1016/s0140-6736(71)90287-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bright field images of the different hOEG populations. First row: representative bright field images of primary hOEG cells (control) after 10 culture passages and populations immortalized with BMI1/TERT, TAg/TERT or shp53/TERT after 23 culture passages are shown. Second row: representative images of BMI1/TERT transduced cells after deimmortalization by infection with LvCRE (MOI: 10) showing the whole population (2 weeks after the infection) as well as deimmortalized clones. Scale bar, 20 μm.
Simultaneous detection of senescence versus proliferation markers. First column shows representative lysosomal senescence-associated β-galactosidase (SA-β-Gal) activity staining using X-gal in bright field images of immortalized hOEG. Second column shows the same SA-β-Gal pattern transformed into green over black background using ImageJ software. Third and fourth columns show representative fluorescence images of Ki67 immunostaining and nuclear marking with To-Pro-3, respectively. Fifth column shows the merge of the three preceding columns. EP, early passage (passage 4); LP, late passage (passage 21), the rest of the populations were studied at passage 21; dox, treated with 1 μg/ml doxycycline; WO, without doxycycline, which was withdrawn after 87 days of culture, 16 passages.
Karyotype analysis. Representative high-resolution GTL-banding metaphase spreads prepared from control (female) hOEG or those immortalized by BMI1/TERT, TAg/TERT and shp53/TERT. Quantitative data are given in Table 2. Arrows indicate important deletions. Primary cells (control) were analyzed in cell passage 12 and immortalized populations in cell passage 18 (BMI1 TERT and TAg TERT) and 14 (shp53 TERT).
Anchorage-independent growth. Colony formation assay in soft agar of hOEG populations transduced by lentivectors encoding the indicated transgenes. Positive controls were performed by using the two tumor cell lines MDA-MB (mammary adenocarcinoma) and U-87 MG (glioblastoma). Living cells were stained using MTT and plate images were obtained using a flatbed scanner.
OEG markers present on immortalized hOEG populations. Parallel cultures of unmodified primary hOEG, or populations immortalized by lentivectors encoding BMI1/TERT, TAg/TERT or shp53/TERT were fixed and stained by indirect immunofluorescence for the presence of neuroligin, amyloid precursor protein (APP), nestin, troy, S100β, glial fibrillary acidic protein (GFAP) and vimentin.
Doxycycline-dependent behavior of shp53-transformed populations. Growth curve of hOEG transduced either by a lentivector encoding a tetracycline-inducible shp53 alone (blue), or in combination with a lentivector encoding TERT (red). The cells were grown in the presence of 1 μg/ml doxycycline (closed symbols, dox) to maintain the expression of the shp53 transgene. At day 87 (arrow), in half of each culture, the effect of eliminating shp53 expression was studied by withdrawal of doxycycline from the medium (open symbols, WO). Cumulative population doublings were calculated as described in the Materials and Methods section from the cell number counted at each culture passage.