Abstract
Several recent studies have focused on the evolution of recently duplicated genes in Drosophila. Currently, however, little is known about the evolutionary forces acting upon duplications that are segregating in natural populations. We used a high-throughput, paired-end sequencing platform (Illumina) to identify structural variants in a population sample of African D. melanogaster. Polymerase chain reaction and sequencing confirmation of duplications detected by multiple, independent paired-ends showed that paired-end sequencing reliably uncovered the break points of structural rearrangements and allowed us to identify a number of tandem duplications segregating within a natural population. Our confirmation experiments show that rates of confirmation are very high, even at modest coverage. Our results also compare well with previous studies using microarrays (Emerson J, Cardoso-Moreira M, Borevitz JO, Long M. 2008. Natural selection shapes genome wide patterns of copy-number polymorphism in Drosophila melanogaster. Science. 320:1629–1631. and Dopman EB, Hartl DL. 2007. A portrait of copy-number polymorphism in Drosophila melanogaster. Proc Natl Acad Sci U S A. 104:19920–19925.), which both gives us confidence in the results of this study as well as confirms previous microarray results.
We were also able to identify whole-gene duplications, such as a novel duplication of Or22a, an olfactory receptor, and identify copy-number differences in genes previously known to be under positive selection, like Cyp6g1, which confers resistance to dichlorodiphenyltrichloroethane. Several “hot spots” of duplications were detected in this study, which indicate that particular regions of the genome may be more prone to generating duplications. Finally, population frequency analysis of confirmed events also showed an excess of rare variants in our population, which indicates that duplications segregating in the population may be deleterious and ultimately destined to be lost from the population.
Keywords: Drosophila, structural variation, mutation, polymorphism, high-throughput sequencing
Introduction
The study of the evolution of recently duplicated genes has provided valuable insights into the evolution of novel functions (reviewed in Long et al. 2003). Recent experimental and computational studies have documented several “new genes” in Drosophila species (e.g., Long and Langley 1993; Wang et al. 2000, 2002, 2004; Betran et al. 2002, 2006; Betran and Long 2003; Jones et al. 2005; Loppin et al. 2005; Arguello et al. 2006; Fan and Long 2007; Fiston-Lavier et al. 2007; Shih and Jones 2008; Zhou et al. 2008). Many of these studies have documented a history of positive selection early in the evolution of the duplicated coding region. The best-studied case is the jingwei gene, which encodes a fusion of the alcohol dehydrogenase (Adh) protein with the N-terminus of a testis-specific protein (Long and Langley 1993; Wang et al. 2000), where the Adh-derived portion has evolved different biochemical properties than the parental copy (Zhang et al. 2004).
The identification of new genes in Drosophila has primarily led to the discovery of fixed duplication events between species. These duplications are therefore the evolutionarily successful mutations that have become established as differences between species. By contrast, there are little data available on the evolutionary dynamics of duplications segregating within natural populations. Such mutations may be quite relevant to heritable phenotypic variation, as suggested by recent results from human genetics associating so-called “copy-number variants” with several complex diseases (Sharp et al. 2006; Sebat et al. 2007; reviewed in Kondrashov FA and Kondrashov AS 2006). In Drosophila, early reports of polymorphic gene duplications included glycerol 3 phosphate dehydrogenase (Takano et al. 1989), metallothionein (Maroni et al. 1987; Lange et al. 1990), and urate oxidase (Lootens et al. 1993). Kern and Begun (2008) recently described a whole-gene deletion polymorphism, where the absence allele is associated with large deletions of telomeric DNA (Kern and Begun 2008).
Over the last decade, understanding role of copy-number variants and structural variants in a variety of species such as Drosophila (Dopman and Hartl 2007; Emerson et al. 2008), mice (Graubert et al. 2007), Caenorhabditis elegans (Maydan et al. 2007), humans (Conrad et al. 2009; reviewed in Zhang et al. 2009), yeast (Doniger et al. 2008; Stambuk et al. 2009), dogs (Chen et al. 2009), and pigs (Fadista et al. 2008) has improved, although it is far from complete. The results of these studies have had many similarities such as evidence for selection on variants, copy number or structural variants of known genes contributing to important phenotypes, and the proportion of genetic variation within a genome that is attributable to copy number variation and structural variation. However, formal analysis of population genetics of duplicates is lacking. Two studies have formally examined the population genetics of copy-number variants one in flies (Emerson et al. 2008) and one in humans (Conrad et al. 2009). Both studies found purifying selection to be involved in patterns of copy-number variants, but this information cannot be considered to be exhaustive.
Recently, microarray-based methods have identified copy-number variants on a genome-wide scale in Drosophila melanogaster (Dopman and Hartl 2007; Emerson et al. 2008; Turner et al. 2008). Array-based approaches have suggested the existence of thousands of duplications and insertion/deletion (indel) mutations in natural populations. In spite of their success, arrays suffer from several limitations. First, the array must be designed specifically for each species of interest, making genome-wide surveys of the recently sequenced Drosophila species (Drosophila 12 Genomes Consortium 2007) currently infeasible. Second, such arrays are limited to the analysis of those portions of the genome to which reliable probes can be designed, resulting in subtle ascertainment biases (Emerson et al. 2008). Third, the inference of copy-number variation is indirect, relying on probe intensity rather than a direct observation of a rearranged sequence or sequence break point.
An attractive alternative to arrays is high-throughput sequencing of paired-ends. Tuzun et al. (2005) pioneered this approach, using Sanger sequencing to end sequence a fosmid library. By mapping the end-sequences back to the published human genome sequence, they were able to directly identify rearrangements (see also Kidd et al. 2008). High-throughput sequencing methods may also be applied to this problem, obtaining paired-ends from size-selected fragments of sheared genomic DNA (e.g., Korbel et al. 2007; Bentley et al. 2008; Doniger et al. 2008; Wang et al. 2008). However, paired-end data sets have more difficulty detecting structural variation in certain genomic regions, such as rearrangements flanked by transposable elements. The difficulty arises due to being unable to align reads from complex break points (which may contain many small indels as well as many single-nucleotide polymorphisms [SNPs] close to the break point) to the reference sequence. However, coverage can alleviate this issue by providing a complementary metric by which to detect rearrangements (Yoon et al. 2009). Insert size can also constrain the ability of the paired-end method to detect variants, but performing multiple library preparations of the same sample with different insert sizes can surmount this issue, though this can be expensive.
For the case of SNP detection, coverage is the key variable determining accuracy of SNP calls (Bentley et al. 2008; Ossowski et al. 2008; Smith et al. 2008; Wang et al. 2008). Here, we ask how much coverage is required to accurately detect rearrangement break points using paired-end sequencing. Korbel et al. (2007) reported a ∼58% confirmation rate for break points identified using the 454 sequencing platform. Other studies, utilizing either array or high-throughput sequencing methods have confirmed only a few selected break points (Urban et al. 2006; Wang et al. 2008). Our method was able to confirm a much higher percentage of structural variants and did not rely on any previous information to identify potential structural variants to be identified and validated.
We performed paired-end sequencing on three isofemale lines from a population sample of D. melanogaster collected from Victoria Falls, Zimbabwe, Africa (Haddrill et al. 2005), utilizing the Illumina platform. We were able to detect three categories of structural events. These categories are indels as well as two other categories that we have defined; Class 1 events, which are either tandem duplications or translocations and Class 2 events, which are either inversions or duplications with a change in orientation (fig. 2). Our experimental confirmation of the structural events, through polymerase chain reaction (PCR) and sequencing, showed that we were able to detect Class 1 and Class 2 events with great accuracy; we were able to confirm all of these structural events that were indicated by eight or more read-pairs. This method allowed us to validate the utility of the paired-end method to detect structural variants and analyze the amount of coverage that is required to accurately detect structural variants with paired-end data. As high-throughput sequencing methods become more commonly used, it is important to assess the accuracy of events detected with this method by experimental confirmation of events indicated by paired-end data.
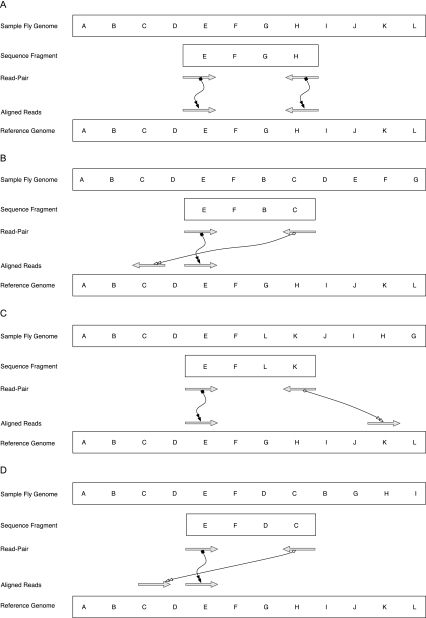
FIG. 2.—
Alignment of read-pairs to the reference genome: (A). Expected alignment of read-pairs to the reference genome. (B). Class 1 (tandem duplication). (C). Class 2 (inversion). (D). Class 2 (duplication with a change in orientation).
This study also found that most structural variants include genic, broadly defined as exonic and intronic, regions and that a number of functional categories, including cell adhesion, cytoskeletal structure, and receptor proteins are enriched in our set of structural variants. We were able to detect several whole-gene duplications, including a novel duplication of Or22a, which has previously been found to be polymorphic for a different copy-number variant (Aguade 2008; Turner et al. 2008). We also find copy-number variants for genes previously reported to be under positive selection, like Cyp6g1, which is involved in dichlorodiphenyltrichloroethane (DDT) resistance (Daborn et al. 2002) and is located in one of several hot spots of structural variation that were detected. Finally, a significant excess of rare alleles in our population sample strong suggests that duplications segregating in the population are likely to be deleterious and may ultimately be lost from the genome due to selection against them.
Materials and Methods
Fly Lines and DNA Extractions
This study examined a population sample of 21 isofemale lines of D. melanogaster collected from Victoria Falls, Zimbabwe, Africa (Haddrill et al. 2005). Genomic DNA for paired-end sequencing was extracted from 15 pooled adult females for each of three lines (Zw104, Zw106, and Zw109, lines 1, 2, and 3 hereafter) using the Qiagen DNeasy Blood and Tissue Kit protocol including the optional RNase treatment. Sample concentrations and purity were verified on a NanoDrop 1000 Spectrophotometer (Thermo Scientific) before sequencing. The Gentra Puregene Cell Kit protocol (Qiagen) was used to extract additional genomic DNA from 10 to 15 pooled adult individuals for each of the 21 Zimbabwean lines as well as the reference strain for PCR and sequencing validation (Adams et al. 2000).
Paired-End Sequencing
Five microgram of genomic DNA per line sequenced was sent to Prognosys Biosciences (La Jolla, CA) for further sample preparation and sequencing. Each sample was sequenced on a single lane of a flow cell with an Illumina Genome Analyzer 2 with the paired-end module attachment. Samples were prepared to produce a mean fragment size of ∼430 bp and 36 bp reads. Paired-end sequencing files have been submitted to the National Center for Biotechnology Information Short Read Archive (accession number: SRA009785.1).
Quality Control of Illumina Reads
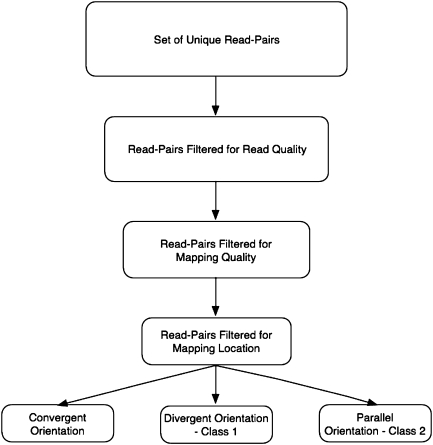
The set of read-pairs from Prognosys was first filtered to remove redundant copies of read-pairs that existed more than once in our data set. Then, read-pairs where either one or both sequences matched a transposable element, read-pairs where the sequences were reverse complements of each other, and read-pairs with sequences of all one nucleotide were filtered from the data set. This was done to help remove from the data set read-pairs that were of low quality or read-pairs that did not map uniquely to the genome. Additional filters were applied to reduce the data set to a subset of read-pairs that were unique and of high quality, mapping to the reference sequence with few mismatches and no gaps (fig. 1).
FIG. 1.—
Read-pairs included in the analysis were filtered in three ways: 1) Read Quality: Pairs where either read was of all one letter and pairs where the reads were reverse complements of each other were removed. 2) Mapping Quality: At an e value of 10−7 both sequences in the read-pair must match exactly once to the genome and must align over all 36 bp with no gaps and up to one mismatch per read. 3) Mapping Location: Both sequences in the read-pair must map to the same chromosome and not to chromosome U or to heterochromatin.
Alignment of Reads to the Reference
Reads were aligned individually to the D. melanogaster reference sequence (version 5.1 was downloaded locally from FlyBase [www.flybase.org] on March 2, 2008) using BlastN (Altschul et al. 1990) with an e value of 10−7. The e value represents the number of alignments for the given sequence that are expected in the database by chance and a lower e value means that a sequence is less likely to be in the database by chance. In addition to aligning the reads to the entire genome, reads were aligned to three other databases (in each case version 5.1 was downloaded locally from FlyBase [www.flybase.org] on 2 March 2008): a database containing only coding sequence, a database containing only intronic sequence, and a database containing only intergenic sequence. Alignments to these databases were used to determine the genomic context (genic vs. intergenic) of the reads and the specific gene to which the read aligned, if applicable. We categorized our reads in this way because this can be directly compared with the data reported in figure 1 of Emerson et al. (2008).
Because the initial analysis, a multitude of short-read aligners have become available, for example (Maq [Li, Ruan, et al. 2008]; SOAP [Li, Li, et al. 2008], and Mosaik [http://bioinformatics.bc.edu/marthlab/Mosaik]). We therefore compared the results of our read alignment using BlastN to results obtained with Mosaik 0.9.0891. The results are presented in the supplementary material (Supplementary Material online). Briefly, 99.9% of reads mapped uniquely by BlastN with our alignment criteria were also identified by Mosaik. Reads mapped using our BlastN criteria but not by Mosaik were low complexity, and many reads mapped by Mosaik but not BlastN were identified in our BlastN searches, but with alignment lengths <36 bp. Importantly, these results show that our BlastN analysis is conservative (e.g., we could have mapped more reads by relaxing requirements on alignment length) and has an extremely low false positive rate, by which we mean that we identified zero cases of BlastN calling a read unique that Mosaik identified as nonunique.
Categories of Structural Rearrangements
For read-pairs where both reads map uniquely to the same chromosome of a reference genome, there are three possible mapping orientations. The first orientation is convergent (-> <-) and is expected for the majority of read-pairs under the null hypothesis that rearrangements with respect to the reference are rare. However, differences in the mapping distance between these read-pairs from the expected distance will indicate indels. The second two mapping orientations indicate structural rearrangements. Read-pairs whose reads mapped in divergent orientation (<- ->) were hypothesized to be best explained by tandem duplications with no change in orientation (the “FB” junction in fig. 2). In these cases, the sequenced fragment was hypothesized to span one break point of the duplication. Under the assumption that the rearrangement is a tandem duplication, the paired-ends detect the sole novel junction of the rearrangement (fig. 2). However, it is also possible that reads in divergent orientation represent translocations and the identified break point is one of two novel break points. It is not possible to distinguish between the two types of events given the information in our data set. Read-pairs with reads in parallel orientation (-> ->) may also be due to two types of rearrangements—inversions or tandem duplications with a change in orientation (fig. 2). Again, it is not possible to distinguish between the two types of events given the available information. Here, we designate read-pairs in divergent orientation as Class 1 structural events and read-pairs in parallel orientation as Class 2 structural events.
Coverage
Two different calculations of coverage were computed for each line. Raw sequence coverage was calculated from the uniquely mapped reads. We also calculated, for each position in the genome, the average distance between convergently oriented read-pairs flanking that position. The average distance between these read-pairs at each point was then compared with the overall mean distance between convergently oriented read-pairs. Regions that indicated that the average distance between read-pairs were greater or less than the sample mean were hypothesized to be deletions or insertions in the fly line with respect to the reference sequence, respectively, and a two-tailed P value was calculated based on the empirical distribution of the distance statistics. For this calculation, we used only reads that mapped within 1,000 bp of each other when aligned to the reference. The choice of 1,000 bp was arbitrary, but the distribution of P values was largely unaffected by the use of larger cutoffs, 99.8% of convergently oriented read-pairs mapped within 1,000 bp of each other (data not shown). We have designated this second coverage statistic “indel coverage” as it indicates the number of convergently oriented read-pairs that provide information about a given region of the genome.
Identification of and Experimental Confirmation of Structural Events
All structural variants identified in this study are variants in one or more of the sequenced fly lines with respect to the published reference sequence. We first determined how many uniquely mapping read-pairs indicated each event. We chose to only attempt to experimentally confirm events that were indicated by two or more read-pairs, in order to minimize the chance of false positives, even though these were only a small subset of the read-pairs in these categories. We did this for three reasons. First, it should clearly reduce the false positive rate. Second, this makes our confirmation rate comparable with previous studies, such as Korbel et al. (2007), who suggested that events indicated by a single pair of reads may represent artifacts of the sample preparation, and therefore only confirmed events suggested by at least two data points; also see Doniger et al. (2008). Finally, it is more representative of the types of events that will be detected in future data sets. Our data were collected in May 2008, soon after the introduction of the paired-end module for the Illumina platform. As protocols improve and sequencing platforms evolve, data collected now will naturally yield much higher coverage, making it reasonable to target higher coverage events for confirmation.
Particular rearrangements, Class 1 events, Class 2 events, and Indels, were hypothesized to occur in 1, 2, or all 3 African lines based on the Illumina sequencing data. To verify that our filtering pipeline excluded reads that were located in repetitive areas of the genome and to assist in the design of unique primers, a repeat masked version of the genome was also generated using RepeatMasker 3.2 (www.repeatmasker.org, Smit et al. 2004), and putative structural events to be experimentally confirmed were compared with this file to see if they occurred in regions of low complexity.
Primers were designed using Primer 3 (http://primer3.sourceforge.net/, downloaded and run on the command line) so that each pair of primers aligned uniquely to the genome and was in the same relative orientation as the read-pairs that suggested each event. Thus, for example, a hypothesized Class 1 event will be validated using a pair of primers that point away from each other in the reference. This design will result in the amplification of a short stretch of sequence only if the indicated structural event is real. This is also the confirmation strategy used by Emerson et al. (2008); the difference here is that paired-ends give a direct prediction of the break point, whereas the break point must be inferred statistically when using arrays to detect structural variants.
A total of 1,222 putative indels were detected with a P value of less than 0.001. We used RepeatMasker 3.2 (www.repeatmasker.org; Smit et al. 2004) to remove from the set events that were in repetitive areas of the genome. This reduced our set of Indels to 794 from which we chose an initial sample of 96 events. The average size of predicted events in our sample was between 71.8 and 166.0 bp for insertions and between 30.7 and 106.9 bp for deletions. Our chosen set included 90 insertions and 6 deletions. The maximum size of deletions we could expect to detect with this experiment was constrained to medium-sized deletions < ∼570 bp for our indel coverage statistic because we used only read-pairs that mapped within 1 Kb of each other, 99.8% of the total number of convergently oriented read-pairs, and our sequence fragments were on average 392 bp. Similarly the size of insertions we could expect to detect with this method was constrained to ∼358 bp.
PCR was performed for each pair of primers in all three of the Illumina sequenced lines as well as the reference strain. All PCR products were of the expected size and were sent to Agencourt Bioscience (Beverly, MA) for Sanger sequencing in both directions. The resulting sequences were aligned to the reference using BlastN, and the nature of the structural event was checked by eye. Events were considered confirmed if the sequencing data verified the event as predicted by the paired-end data. Sequences confirming the paired-end data have been submitted to GenBank (accession numbers: GU014579–GU014692).
Coverage between Read-Pairs of Confirmed Class 1 Events
For the set of confirmed Class 1 events, for each line, we calculated the average coverage for the region of the genome that lies between the locations where the two sets of read-pairs align in the reference. We considered a line to have a structural event if the event was indicated either by the paired-end data, the PCR/sequencing confirmation or both. We used a Welch two-sample t-test to compare the average coverage of fly lines that possessed each event to the average coverage of fly lines that did not.
Identification of Unique Sequence in the Reference Genome
The efficacy of assembly of short reads to a reference sequence depends on the ability to accurately map reads back to the genome. This ability depends to a large extent on the “uniqueness” of the region from which the read is generated. We identified nonoverlapping windows that contained 500 kb of sequence in which each 36-mer mapped uniquely to the genome. To do this, we generated all possible 36 bp sequences from the reference sequence and mapped them back to the reference using blat (Kent 2002). For any 36-mer that mapped to more than one location with one or fewer mismatches and an alignment length of 36 with no gaps, the positions of all matches were recorded, resulting in a genome sequence for which each base pair is labeled either “unique” or “repetitive.” A window is defined as containing 500,000 nucleotides from the unique class.
Location of Structural Events
We first identified the subset of Class 1 events that were separated by a distance of 32 kb or less. We chose this distance because it corresponds to the largest distance between a set of two or more read-pairs indicating a single Class 1 event that was confirmed in our PCR and sequencing confirmation. Although we did not validate any Class 1 events indicated by only one read-pair in this size category, we have several reasons for believing that the majority of these events are real. The high rate of confirmation of events indicated by only two read-pairs in this size range gives us confidence that the majority of these events are real, and the total number of events we detect, regardless of the number of read-pairs indicating the event, is also similar to the number of duplications detected by Emerson et al. (2008), Dopman and Hartl (2007), and Turner et al. (2008). A Kolmogorov–Smirnov test was performed to compare the distribution of distances between events on each chromosome with an exponential distribution, in order to test a model in which events arise according to a Poisson process. We then calculated the mean number of events per unique 500 Kb, using nonoverlapping windows, for each chromosome, which is the maximum-likelihood estimator of the rate from a Poisson distribution, as well as the 95% confidence interval for the rate. An analysis of variance (ANOVA) was performed to determine if there was an effect of chromosome on the rate. Finally, we mapped the number of Class 1 events along the repeat masked genome to identify any potential duplication hot spots.
Gene Functional Analysis with DAVID
For the set of Class 1 events that were less than 32 kb, we examined both gene-term enrichment and gene annotation group enrichment using the Functional Annotation Clustering tool, which is part of the DAVID tool set (http://david.abcc.ncifcrf.gov/, Dennis et al. 2003; Huang et al. 2009). We examined read-pairs whose reads mapped to genic (exonic + intronic) regions. Annotation groups are created by clustering terms based upon similar annotation. Functional annotation clustering takes into account composition of the background genome, and thus the enrichment score is not influenced by gene family size (Huang et al. 2009). We used the DAVID analysis tools default settings to select the annotation categories that were examined in this analysis as well as the stringency levels utilized by the analysis tools. In our analysis of annotation categories, we examined categories where the enrichment score was ≥1.3, corresponding to a nonlog scale of 0.05. Within these categories, we considered those terms with a P value of ≤0.05 as enriched. DAVID also performs multiple test correction and we considered terms with a Benjamini corrected P value of ≤0.05 as “significantly” enriched. We repeated this analysis for the set of Class 1 structural events that were confirmed via sequencing.
Population Frequency Analysis
For each Class 1 structural event that was confirmed by sequencing, we determined if the event was present in the other 18 Zimbabwean lines via PCR. The presence of a band of the expected size was considered evidence for the event in the line, and the total number of lines that showed each event was determined. Although there is no ascertainment bias in the site frequency spectrum for the three lines that were paired-end sequenced, an ascertainment bias is introduced by surveying only events confirmed in one or more of these three lines in our population sample (Marth et al. 2004). We calculated Watterson’s θ (Watterson 1975) where S = the number of Class 1 events.
The expectation of the folded site frequency spectrum was obtained using Hudson’s “ms” program (Hudson 2002) and custom C++ code (Thornton 2003). Our ascertainment scheme was based on discovering events in a sample of three African isofemale lines plus the reference strain, for a panel depth of four. Hudson’s program was used to simulate 100,000 replicates of n = 22 chromosomes, each with 100 segregating sites and no recombination (for neutral models, the expected site frequency spectrum is independent of the recombination rate). For each site, our program looked at the genotype of the first individual (the “reference” individual) at that site (either 0 or 1) and then tallied the number of occurrences of the other allele in the next three individuals. If that count was >0 and ≤ 3, the site was kept, and the frequency of the alternate allele counted in the 21 nonreference individuals. From this list of frequencies, the expected folded site frequency spectrum was calculated, conditional on our ascertainment scheme. We performed simulations both under the standard neutral model of a Wright-Fisher population and also under the demographic model inferred by Li and Stephan (2006). For the latter model, the reference individual was drawn from the non-African population, and the remaining 21 individuals are drawn from the simulated African population.
We performed χ2 tests to compare the number of observed events at frequencies 1, 2, and ≥3 with the number expected under the infinite-sites model where the ancestral state in unknown, the number expected under the neutral model conditional on our ascertainment scheme and the number expected under the demographic model inferred by Li and Stephan (2006).
Results
Paired-End Sequencing Data
Between 3,118,233 and 4,866,329 pairs of reads were generated for each fly line, of which between 50% and 72% of read-pairs were left after removing redundant copies of read-pairs that appeared more than once in our set of reads (table 1). Reads were aligned to the reference genome using BlastN and we then applied a series of additional filters to reduce our data set to reads that were high quality and mapped to uniquely to the genome (fig. 1). A total of 3,804,391 read-pairs passed all the applied quality filters and were included in the further analysis (table 1).
Table 1.
Total Number of Read-Pairs Sequenced and the Numbers Included in the Analysis
| Line 1 | Line 2 | Line 3 | Total | |
| No. read-pairs | 3118233 | 4311872 | 4866329 | 12296434 |
| No. unique read-pairs | 1560110 | 3103166 | 3302690 | 7965966 |
| No. read-pairs passing all filters | 714919 | 1534112 | 1555360 | 3804391 |
| No. in convergent orientation | 713889 | 1532189 | 1553171 | 3799249 |
| No. in divergent orientation | 402 | 725 | 823 | 1950 |
| No. in parallel orientation | 628 | 1198 | 1366 | 3192 |
The observed mean distance between convergently oriented read-pairs with a maximum distance between pairs of 1,000 bp was 383.28 bp with a standard deviation of 50.69 bp. The median distance was 392 bp for read-pairs. We calculated raw sequence coverage for the three lines to be ∼0.8×. In addition, we calculated an indel coverage statistic (for details, see Materials and Methods). Indel coverage was 2.39×, 4.56×, and 5.05× for lines 1, 2, and three, respectively.
Analysis and Confirmation of Structural Events
The filtered data set contained 1,950 Class 1 read-pairs. We further removed from the data set additional read-pairs which mapped to the same location but with up to one difference per read, leaving only one read-pair at each location in the data set. This left 1,754 read-pairs, which indicated 1,489 Class 1 events. Of these events, 1,410 were indicated by a single uniquely mapping read-pair (table 2). Seventy-nine events were indicated by two or more read-pairs (table 2).
Table 2.
Summary of Structural Rearrangements Detected by At Least Two Read-Pairs
| Indicated by | Single read-pair |
Multiple read-pairs |
||
| Class 1 | Class 2 | Class 1 | Class 2 | |
| No. events chromosome arm | 1410 | 2808 | 79 | 16 |
| X | 230 | 439 | 10 | 3 |
| 2L | 218 | 472 | 16 | 3 |
| 2R | 236 | 426 | 22 | 2 |
| 3L | 281 | 587 | 15 | 1 |
| 3R | 441 | 881 | 14 | 7 |
| 4 | 4 | 3 | 2 | 0 |
| No. discovered per line | ||||
| Line 1 | 276 | 510 | 30 | 5 |
| Line 2 | 544 | 1086 | 44 | 11 |
| Line 3 | 590 | 1202 | 39 | 9 |
| Estimated sample frequency | ||||
| 1 | — | — | 48 | 8 |
| 2 | — | — | 28 | 7 |
| 3 | — | — | 3 | 1 |
The data set also contained 3,192 Class 2 read-pairs. The data set was reduced as above to remove additional read-pairs mapping in the same location leaving 2,808 Class 2 events that were indicated by a single read-pair. An additional 16 events were indicated by two or more read-pairs (table 2). The majority of structural events were found in one line only, and only four events were predicted in all three lines (table 2). In contrast with the set Class 1 read-pairs, which included events indicated by up to 23 read-pairs (table 3), for the set of Class 2 events no more than three read-pairs ever indicated the same event (table 4).
Table 3.
PCR and Sequencing Confirmation of Class 1 Events
| PCR confirmation |
Sequencing confirmation |
|||||
| No. paired-ends (coverage) | No. Class 1 | No. confirmed | %Confirmed | No. Class 1 | No. confirmed | %Confirmed |
| 2 | 23 | 21 | 91.30 | 21 | 21 | 100.00 |
| 3 | 14 | 13 | 92.86 | 13 | 12 | 92.31 |
| 4 | 18 | 18 | 100.00 | 18 | 18 | 100.00 |
| 5 | 9 | 9 | 100.00 | 9 | 8 | 88.89 |
| 6 | 3 | 3 | 100.00 | 3 | 2 | 66.67 |
| 7 | 5 | 5 | 100.00 | 5 | 4 | 80.00 |
| 8 | 2 | 2 | 100.00 | 2 | 2 | 100.00 |
| 13 | 1 | 1 | 100.00 | 1 | 1 | 100.00 |
| 17 | 2 | 2 | 100.00 | 2 | 2 | 100.00 |
| 23 | 1 | 1 | 100.00 | 1 | 1 | 100.00 |
| Total | 78 | 75 | 96.15 | 75 | 71 | 94.67 |
Table 4.
PCR and Sequencing Confirmation of Class 2 Events
| PCR confirmation |
Sequencing confirmation |
|||||
| No. paired-ends (coverage) | No. Class 2 | No. confirmed | %Confirmed | No. Class 2 | No. confirmed | %Confirmed |
| 2 | 13 | 9 | 69.00 | 9 | 5 | 56.00 |
| 3 | 3 | 3 | 100.00 | 3 | 1 | 33.00 |
| Total | 16 | 12 | 75.00 | 12 | 6 | 50.00 |
We chose to attempt to experimentally confirm structural events that were indicated by two or more read-pairs (for details, see Materials and Methods). Primers were designed for 78 of the 79 Class 1 events indicated by ≥2 read-pairs; we were not able to design reliable pair of primers for one event due to its location in a low-complexity region of chromosome 4. Primers were also designed for all 16 Class 2 events.
We were able to amplify bands of the expected size for 75 out of 78 predicted Class 1 events for an overall PCR confirmation rate of 96% (table 3, fig. 3). For 69 of these events, we were able to amplify bands in all the lines in which the event was predicted. Of the 16 Class 2 events predicted, 12 were confirmed by PCR (table 4). In 11 cases, we were able to amplify a band in all the lines in which the event was predicted.
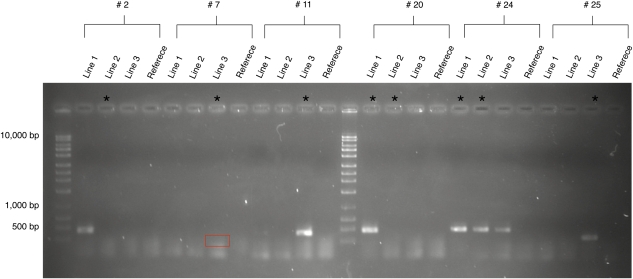
FIG. 3.—
PCR confirmation of six Class 1 structural events. Asterisks indicate the lines in which the event was predicted by the paired-end data. Class 1 #2 is an example of an event that was confirmed in a different line than the one in which it was predicted. Class 1 #20 shows an event that was confirmed in one of the two lines in which it was predicted. Class 1 #24 shows an event that was predicted in two lines and confirmed in all three lines. The box in Class 1 event #7 highlights a faint band.
All PCR products were Sanger sequenced and subsequent BlastN searches confirmed 71 of 75 predicted Class 1 events, resulting in a sequencing confirmation rate of 94.7% (table 3). Of the 71 Class 1 events that were confirmed via sequencing the majority of events were small with a mean of 3,264.6 bp and a median size of 2,504 bp. Likewise, 6 of 12 Class 2 events were confirmed by sequencing (table 4).
For the four Class 1 and six Class 2 events that were not confirmed via sequencing nine events were not confirmed due to nonspecific PCR amplification. One event was not confirmed because the sequence amplified was of poor quality and was generated when we attempted to amplify bands using an annealing temperature lower than the optimal temperature following failure to amplify any bands at the predicted optimal temperature. Importantly, 100% of the sequences that we successfully obtained showed evidence of a rearrangement when compared with the reference sequence.
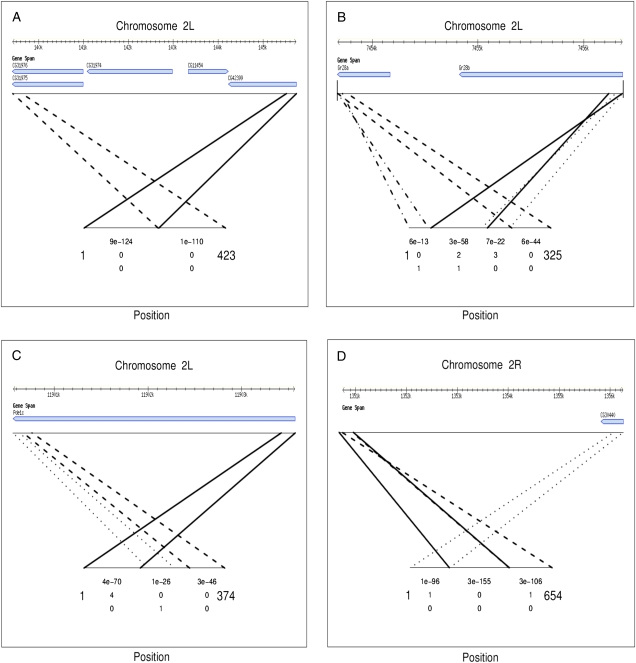
Sixty-five of the 71 confirmed Class 1 events showed evidence of a simple break point sequence when aligned to the reference sequence; the five prime end of the sequenced fragment aligns to one region of the genome and the three prime end aligns to region upstream of the location where the five prime end aligns (panel A in fig. 4). This is consistent with either tandem duplication or a translocation event. However, there were six events with additional structural differences nearby (panels B through D in fig. 4). Five of these six events include genic regions.
FIG. 4.—
Alignment of confirmed Class 1 sequences to the reference genome. The reference sequence is represented at the top of each panel with information from FlyBase (www.flybase.com) regarding genes located in the region in question. The sample sequence is shown at the bottom. e values, the number of mismatches and the number of gaps (top to bottom) for each aligning portion of the sample sequence are shown underneath the corresponding sequence. (A). A typical example confirming a Class 1 event. (B). A Class 1 event that includes multiple duplications and inversions within the coding regions of the gustatory receptor genes Gr28a and Gr28b. (C). A Class 1 event with a small, nearby insertion. (D). A Class 1 event with an additional inverted duplication.
Two of the confirmed duplications contained transposable element sequence. These events were not removed by our initial filtering because the read-pairs indicating the duplications did not contain transposable element (TE) sequence; however, during our sequencing confirmation, we recovered sequence that matched the TE database. We compared these sequences with the list of known D. melanogaster TEs, corresponding to the version of the reference sequence to which we aligned our reads, to determine the specific TEs involved as well as the family of TEs to which they belonged. One is a long terminal repeat retroposon element which appears to be a duplication of the diver2 element located just upstream of CG3107. In the second instance, both regions of the duplication match multiple families of TEs.
Confirmed Class 2 sequences showed a consistent alignment pattern: part of the sequence matched one region of the genome and the other part of the sequence matched a different region though in an inverted orientation. This indicates that there is a structural event that includes a change in orientation, but the available data cannot be used to discriminate inversions from nontandem duplications or some other more complex event.
Coverage in Class 1 Presence versus Absence Lines
For the set of confirmed Class 1 events, the average coverage between the locations where Class 1 read-pairs align in the reference for strains where a Class 1 event is present was 0.8618, whereas the coverage for strains where the event is absent was 0.5402. We found there to be a significant difference between the mean coverage for these two categories (Welch two-sample t-test, P = 3.55 × 10−9).
Confirmation of Indels
For 88 of 96 predicted indels, a band was amplified in the line in which the event was predicted. Four additional pairs of primers amplified a band in a line other than the predicted line but not in the predicted line. From the resulting sequences, we found that in 32 of 96 cases an indel of greater than 3 bp was found in the line for which the indel was predicted. A total of 14.6% (14/96) of sequenced potential indel events were found to have indels whose actual size was within 10% of the predicted size range for that indel.
Genomic Context of Structural Events
Confirmed Class 1 events mapped to genic regions in 78.9% of cases, as indicated by their alignment to sequences in either our intronic or exonic sequences database (table 5). Confirmed Class 2 events mapped to genic regions in 66.7% of cases (table 5). Because each read from a read-pair is mapped individually a single pair can have portions that are both genic and intergenic. A list of genes involved in these structural events is given in supplementary table 1 (Supplementary Material online).
Table 5.
Number of Class 1 and Class 2 Events and the Percentage of Events Located in Genic and Intergenic Regions
| Class 1 |
Class 2 |
|||
| All | Confirmed | All | Confirmed | |
| No. Reads | 1489 | 71 | 2824 | 6 |
| % Genic | 78.2 | 78.9 | 78.2 | 66.7 |
| % Intergenic | 64.1 | 54.9 | 64.0 | 66.7 |
NOTE.—Because the two reads of each read-pair are aligned separately a single read-pair might match both genic and intergenic regions.
Location of Structural Events
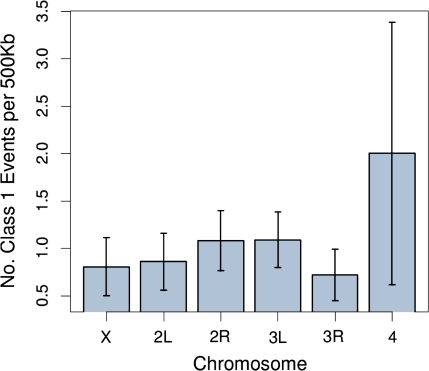
We now shift focus to looking at genome-wide patterns of structural variation. To attempt to guard against including false positives in the analysis, we conditioned the mapping distance between reads to be ≤32 kb, which is the maximum distance between reads in our set of 71 confirmed Class 1 events. This reduced our set of 1,489 Class 1 structural events to 201 events that were considered for this and further analyses. The comparison of the distribution of distances between these 201 Class 1 events on each chromosome to an exponential distribution suggested that the distribution of events followed a Poisson distribution (Kolmogorov–Smirnov test P > 0.05 for each chromosome arm after Bonferroni correction). We calculated the mean number of Class 1 events per 500 Kb of unique sequence (for definition of unique sequence, see Materials and Methods). The mean is the maximum-likelihood estimate of the rate of a Poisson process, and the corresponding 95% confidence interval of that rate for each chromosome is shown in figure 5. An ANOVA found no effect of chromosome on the rate of Class 1 events per 500 Kb.
FIG. 5.—
Mean number of Class 1 events per 500 Kb and 95% confidence limits assuming that events arise according to a Poisson process (for details, see text).
Hot spots of Putative Structural Events
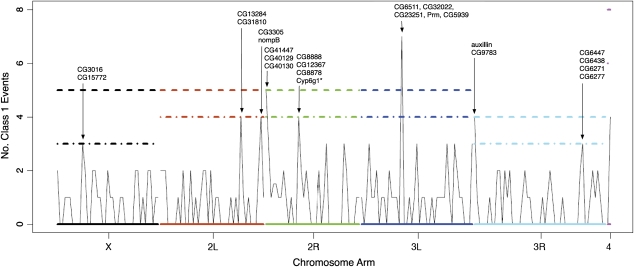
To identify potential hot spots of structural variation, we examined the locations of Class 1 read-pairs of 32 kb or less from the entire set of mapped pairs, using a nonoverlapping, sliding window approach. Plotting the number of Class 1 events across the unique portions of the chromosome revealed that while Class 1 events appear to be generally uniform within chromosomes there are a few peaks which indicate regions with elevated numbers of Class 1 events (fig. 6). Several of these peaks have P values ≤ 0.01, and one peak on chromosome arm 3L has P < 0.001, and includes reads mapping to an exon of Prm and to the exons and introns of several other genes of unknown function in the region. Some of these read-pairs map to an intron of the hairy locus, and others are adjacent to chorion protein genes. The reads mapping near the chorion protein genes completely surround these genes and therefore suggest complete duplications, assuming that Class 1 reads represent tandem duplications. This hot spot of duplication was also identified in a microarray study by Turner et al. (2008). Other hot spots include reads that map to exons of auxillin, which is involved in protein kinase activity and adenosine triphosphate binding, nompB which is involved in flagellum assembly and sound perception, and Cyp6g1 which has been identified as conferring resistance to DDT (fig. 6).
FIG. 6.—
The number of Class 1 read-pairs of 32 kb or less in nonoverlapping 500 Kb windows. This includes confirmed Class 1 events detected by two or more read-pairs and Class 1 events indicated by only one read-pair. The dashed line represents the a P value of ≤ 0.001, given the chromosome mean and assuming a Poisson process (for details, see text). The dot-dashed line represents P ≤ 0.01. The P values are not corrected for multiple tests. The genes listed above the peaks in the figure are genes located in that window identified by read-pairs in our data set. Genes marked with an asterisk are from our confirmed set.
Genes between Read-Pairs
If we assume that Class 1 reads represent tandem duplications, which we consider to be likely given our finding of higher sequence coverage in regions between Class 1 read-pairs, then the read-pair identifies the sole novel junction of the duplication (fig. 2). Thus, the portion of the reference sequence between where the two reads map is an estimate of both the size of the duplication and indicates which genes are duplicated. One example of this is our Class 1 event 4 that is indicated by two read-pairs and was confirmed by sequencing. In this instance, the read-pairs map to intergenic regions flanking Or22a. If we assume that this event is a tandem duplication, then these data suggest a whole-gene duplication of Or22a.
Gene Functional Analysis
In the set of 201 Class 1 events less than 32 kb apart (fig. 6), there were four annotation categories with enrichment scores ≥1.3 which represents the top 5% of enriched categories. Within these categories, we found 34 terms that were significantly enriched of which four remained significantly enriched following Benjamini correction for multiple tests (table 6). These terms included proteins involved in the cytoskeletal structure, proteins involved in cell adhesion, and receptor proteins that combine with neurotransmitters or other signaling molecules to cause a change in cell function.
Table 6.
Functional Term Analysis for Class 1 Structural Events of ≤32 kb
| Functional annotation clustering—all | ||||
| Cluster 1 | Enrichment score: 3.28 | Count | P value | Benjamini |
| Spectrin repeat | 5 | 7.50 × 10−05 | 2.70 × 10−01 | |
| SPEC | 5 | 7.60 × 10−05 | 4.00 × 10−02 | |
| Cytoskeleton | 10 | 2.60 × 10−02 | 9.50 × 10−01 | |
| Cluster 2 | Enrichment score: 1.82 | |||
| Integrin complex | 7 | 6.50 × 10−09 | 4.60 × 10−06 | |
| Receptor complex | 7 | 6.50 × 10−06 | 2.30 × 10−03 | |
| Actin cytoskeleton | 8 | 1.20 × 10−04 | 2.90 × 10−02 | |
| Cytoskeletal protein binding | 11 | 5.90 × 10−04 | 7.10 × 10−01 | |
| Actin binding | 7 | 4.70 × 10−03 | 9.60 × 10−01 | |
| Mesoderm development | 7 | 1.20 × 10−02 | 1.00 | |
| Integral to plasma membrane | 8 | 1.40 × 10−02 | 9.20 × 10−01 | |
| Intrinsic to plasma membrane | 8 | 1.50 × 10−02 | 8.80 × 10−01 | |
| Cytoskeleton | 10 | 2.60 × 10−02 | 9.50 × 10−01 | |
| Plasma membrane part | 10 | 2.70 × 10−02 | 9.40 × 10−01 | |
| Cluster 3 | Enrichment score: 1.6 | |||
| Vitamin C | 3 | 1.50 × 10−02 | 7.70 × 10−01 | |
| Dioxygenase | 3 | 1.90 × 10−02 | 7.40 × 10−01 | |
| Cluster 4 | Enrichment score: 1.55 | |||
| Tetratricopeptide repeat | 6 | 2.70 × 10−03 | 5.20 × 10−01 | |
| Myoblast development | 4 | 7.10 × 10−03 | 1.00 | |
| Myoblast maturation | 4 | 7.10 × 10−03 | 1.00 | |
| Myoblast differentiation | 4 | 7.70 × 10−03 | 1.00 | |
| Tetratricopeptide-like helical | 6 | 8.40 × 10−03 | 1.00 | |
| Cell maturation | 4 | 1.10 × 10−02 | 1.00 | |
| Muscle cell differentiation | 4 | 1.40 × 10−02 | 1.00 | |
| Tetratricopeptide region | 5 | 1.50 × 10−02 | 1.00 | |
| Developmental maturation | 4 | 1.50 × 10−02 | 1.00 | |
| Plasma membrane fusion | 3 | 2.70 × 10−02 | 1.00 | |
| Syncytium formation by plasma membrane fusion | 3 | 2.70 × 10−02 | 1.00 | |
| Syncytium formation | 3 | 2.70 × 10−02 | 1.00 | |
| Myoblast fusion | 3 | 2.70 × 10−02 | 1.00 | |
| Myotube differentiation | 3 | 2.70 × 10−02 | 1.00 | |
| Skeletal muscle fiber development | 4 | 3.10 × 10−02 | 1.00 | |
| Muscle fiber development | 4 | 3.60 × 10−02 | 1.00 | |
| Membrane fusion | 3 | 4.20 × 10−02 | 1.00 | |
| Ankyrin | 5 | 4.40 × 10−02 | 1.00 | |
| ANK | 5 | 4.40 × 10−02 | 1.00 | |
| Functional annotation clustering—confirmed | ||||
| Cluster 1 | Enrichment score: 1.83 | |||
| Iron | 5 | 9.90 × 10−04 | 4.30 × 10−01 | |
| Metal binding | 8 | 1.30 × 10−03 | 3.10 × 10−01 | |
| Vitamin C | 3 | 1.50 × 10−03 | 2.50 × 10−01 | |
| Dioxygenase | 3 | 1.90 × 10−03 | 2.40 × 10−01 | |
| Oxidoreductase | 6 | 4.30 × 10−03 | 3.80 × 10−01 | |
| Monooxygenase | 3 | 2.20 × 10−02 | 8.40 × 10−01 | |
| Oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen | 3 | 4.50 × 10−02 | 1.00 |
NOTE.—Both P values and Benjamini corrected P values are given.
For the set of 71 confirmed Class 1 events, only one annotation category with an enrichment score ≥1.3 was found. In this set, there were seven terms that were significantly enriched although none of them survived multiple test correction. This may be due to the small size of the data set. In the set of proteins involved in metal binding, Vitamin C cofactors and proteins involved in oxygen reduction reactions were enriched. Many of the other functional terms that were reported in the whole-genome data set were also present, but they were not significantly enriched in this set nor were they in an enrichment category of ≥1.3.
Population Frequencies of Duplications
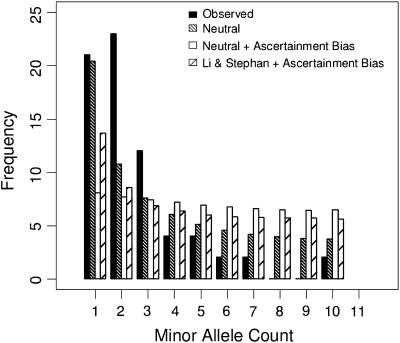
Next, we then tested each of the additional 18 fly lines in our population sample for the presence of our 71 confirmed Class 1 events. We used pooled genomic DNA from 15 females per line so that we would be very likely to detect duplicates that may still be segregating within our isofemale lines. We considered the presence of a band of the appropriate size to be confirmation for the presence of the Class 1 event since, due to our primer design strategy only the presence of a duplication would result in any amplification (for details, see Materials and Methods). For the set of confirmed Class 1 events, we found that the majority of events are present in only a few of the 21 fly lines in our sample population. Only one of the Class 1 events confirmed in our study is fixed in the population, and several events are present at intermediate frequencies (fig. 7).
FIG. 7.—
The folded site frequency spectrum for the 70 confirmed segregating Class 1 events.
In order to test for a departure of the observed site frequency spectrum from a neutral model, we must consider that Drosophila populations are not at demographic equilibrium, and there is considerable differentiation between African and non-African population samples (e.g., Begun and Aquadro 1992; Haddrill et al. 2005; Li and Stephan 2006; Thornton and Andolfatto 2006). As our sample from which we detected copy-number variants consists of three African lines and the non-African reference strain, we use the demographic model of Li and Stephan (2006) as one of our null models. This model has been proposed as a good fit to SNP data from African and European D. melanogaster. The observed number of events at frequencies 1, 2, and ≥3 in the population differed from the expected number of events at the same frequencies under the infinite-sites model (χ2 test; P = 0.00111, degrees of freedom [df] = 2); the infinite-sites model conditional on our ascertainment scheme (χ2 test; P = 5.30 × 10−15, df = 2); the demographic model inferred by Li and Stephan (2006) conditional on our ascertainment scheme (χ2 test; P = 4.88× 10−9, df = 2). We also repeated the analysis ignoring singletons because our detection strategy may miss most singletons in the population. The observed number of events at frequencies 2, 3, and ≥4 in the population differed from the expected number of events under all three models (infinite-sites model: χ2 test; P = 2.09 × 10−05, df = 2; infinite sites conditional on our ascertainment scheme: χ2 test; P = 1.56 × 10−11, df = 2; demographic model conditional on our ascertainment scheme χ2 test; P = 1.54 × 10−09, df = 2).
Discussion
We have identified polymorphic structural variants in a natural population and found evidence that many of these variants are deleterious. A number of hot spots of Class 1 events, many of which contained genes, were also detected and these regions may be of interest to future studies. Certainly, examination of these regions in other Drosophila species would be informative with regard to the evolutionary forces that shape levels of variation across the genome. Similar to the results of Emerson et al. (2008) and contrasting with the results of Dopman and Hartl (2007), we find most structural events to involve genic regions. Also, these events are enriched for a variety of functional categories including receptor proteins and cytoskeletal structural proteins.
Our experimental confirmation of structural events indicated by at least two read-pairs demonstrates that the paired-end data can be assumed to accurately portray structural variants in a genome of interest provided that there is sufficient coverage of the sample genome. Given that we were able to confirm all the structural events in our data set that were indicated by eight or more read-pairs and the majority of events indicated by two or more read-pairs, we estimate that a genome sequenced to 8× coverage would accurately identify the majority of simple structural events in that genome. We also point out that this level of coverage is relatively modest by current standards. Future studies can further focus on the characteristics and forces acting upon structural variants segregating in populations by further using this technique.
Paired-End Sequencing
Our confirmation rate for Class 1 events was 100% for events indicated by 8 or more read-pairs, and the rate of confirmation for Class 1 events indicated by fewer read-pairs remained high. This gives us an estimate of the number of read-pairs and therefore the coverage that would be required across the genome to be able to accurately detect all simple structural events (i.e., those not possessing complex break points) present in that genome with paired-end sequencing. However, this does not mean that we will detect all structural variants at 100% accuracy at 8× coverage. Break points with many differences from the reference sequence may be at a coverage that is less than the genome average due to the difficulty of aligning reads to highly polymorphic areas of the genome. In these cases, alternative strategies, such as read depth, can be implemented to augment the number of variants found (Yoon et al. 2009). In the case of the Class 2 events, a much smaller proportion of events were confirmed experimentally; however, the greatest number of read-pairs indicating an event in this category was three, whereas Class 1 events were indicated by up to 23 read-pairs. Therefore, coverage is the main factor determining the accuracy of detecting structural variation, which is not surprising given published results on the effect of coverage on SNP calling (Bentley et al. 2008; Ossowski et al. 2008; Wang et al. 2008). Despite the low coverage of this study (∼0.8× raw sequence coverage and between 2.39× and 5.05× indel coverage per lane), we still detected a lot of variation; our results compare well with previous microarray studies designed to detect copy-number variants.
Finally, the heterogeneity in the number of read-pairs indicating Class 1 versus Class 2 events may be ascribed to two different factors. First, many of our Class 2 events suggest a very large inversion, on the order of a significant portion of a single chromosome. Events of this type are rare and a large number of events of this type being found in a single fly strain would be highly unlikely. Instead Class 2 events may be more likely to be artifacts of the sequencing process, which further illustrates the importance of having multiple read-pairs indicating each event. Inversions may also be harder to detect with paired-end sequences in general due to the likelihood of increased sequence complexity at the break points of the event. For example, the In(2L)t proximal break point in D. melanogaster and D. simulans shows a large number of indel polymorphisms at the break point (Andolfatto and Kreitman 2000) and similar results are found for the In(3R)P break point (Matzkin et al. 2005), suggesting that reads from such a rearrangement break point would be difficult to align to the reference with current-generation aligners.
Properties of Identified Structural Events
Of the 71 Class 1 and 6 Class 2 events confirmed by PCR and sequencing, 65 Class 1 events and all the Class 2 structural events were straightforward rearrangements with simple break points. However, there were six Class 1 events that showed additional structural variation to what was indicated with the paired-end data (fig. 4).
The majority of Class 1 structural rearrangements found in this study involve genic regions (78%), whereas a smaller fraction (64%) involve intergenic regions. This pattern holds for the subset of Class 1 events ≤32 kb; 86% genic and 73% intergenic. Due to the spatial relationship between paired reads, a single pair can contain both genic and nongenic sequence. Our results are more similar to the findings of Emerson et al. (2008), who also found that the majority of events they detected were genic and contrast with the findings of Dopman and Hartl (2007), who determined that tandem repeats were elevated in regions containing noncoding and intergenic regions but not coding regions. However, the reason for the discrepancy between Dopman and Hartl (2008) and the current study, and Emerson et al. (2008) is unclear.
Of our confirmed break point sequences, only two contained any transposable element sequence. In Drosophlia, the population frequencies of TEs at a given position in the genome are quite low and few mRNAs include TE sequences (Charlesworth and Langley 1989; Lipatov et al. 2005). Furthermore, there is evidence in Drosophila for ectopic exchange between TEs at different positions (Charlesworth and Langley 1989), and the repair of such events may lead to structural variants of the types detected in this work. Fiston-Lavier et al. (2007) proposed a model of duplication dependent strand annealing that explains how segmental duplications arise in D. melanogaster via TEs and suggest that there should be an increase in the density of duplications in TE-rich regions of the genome. They also showed that TEs are associated with segmental duplications in heterochromatic regions (Fiston-Lavier et al. 2007), whereas we examined mostly read-pairs that mapped to the euchromatic portions of the genome. Other mechanisms of producing new genes, such as illegitimate recombination, have been recently suggested in Drosophila (Arguello et al. 2006), and some of the break points we identified may represent chimeric gene structures.
There are several reasons that may explain why we see few examples of TE sequence in rearrangement break points. First, the association of TE sequence with segmental duplication reported by Fiston-Lavier et al. (2007) is largely due to an effect in heterochromatic sequence, whereas our events are mostly found in euchromatin. Second, a novel TE insertion would be larger than the insert size of our paired-end library and, therefore, would not be sequenced. Third, during our filtration of read-pairs we excluded from our set any pair where either one or other read matched a transposable element. We did this because we only wanted read-pairs that would align uniquely to the genome in order to be confident that the inferences made from our paired-end data were correct. Also, in the cases of structural events that were confirmed through PCR and sequencing, the sequences we amplified were generally only ∼400 to ∼1,000 bp in length. This means that not only did we initially screen out sequences matching transposable elements but also we may not have captured enough sequence from these new duplications to detect TEs near to the identified break points. Fourth, the analysis performed by Fiston-Lavier et al. (2007) was on the published reference sequence. It is quite likely that most of the duplication events in the reference are fixed (e.g., in the largest resequencing study of duplicate genes done thus far Thornton and Long [2005] were able to amplify over 90% of duplicated alleles, which is a similar success rate to resequencing studies of single-copy regions in D. melanogaster [Haddrill et al. 2005], indicating that closely related duplications of coding sequence in the reference sequence are all fixed in population samples), and it is possible that the break points of polymorphic structural variants may differ from those of fixed events, particularly if Emerson et al.'s (2008) inference of a large number of partial duplications of DNA is correct—many copy-number variants segregating in Drosophila populations may be pseudogenes or deleterious mutations destined to be lost from the species.
The majority of break points detected in this study are “clean” break points, where the sequence near the break point shows few differences from the reference, which suggests that these break points are the result of illegitimate recombination events. We detected a few complex break points that showed evidence of significant differences from the reference sequence, which included duplications, deletions, and/or inversions at the location of the break point; these are likely to have originated through nonhomologous recombination mechanisms at sites of microhomology (Hastings et al. 2009). However, Hastings et al. (2009) suggests that most copy-number changes are the result of mechanisms that act on microhomology. Again these discrepancies may be the result of differences between the mechanisms that produce variants that are fixed and mechanisms that produce variants that will be lost.
Class 1 and Class 2 Structural Variants
Our method indicates a total of 1,489 Class 1 and 2,824 Class 2 structural events in our three sample lines, which is similar to the number of duplications detected by Emerson et al. (2008), Dopman and Hartl (2007), and Turner et al. (2008). Of these there are 201 Class 1 events that are ≤32 kb, corresponding to the size of the largest confirmed Class 1 event.
We believe that, whereas we did not validate structural events indicated by only one read-pair, the majority of the events identified in our data set are real. First, we have a high rate of confirmation for structural events indicated by two or more read-pairs. Our data also compare well with previous microarray experiments (Emerson et al. 2008); not only in the number of detected duplications but also specific genes, gene enrichment categories, and the percentage of events that involve exons versus noncoding DNA. Finally, certain gene families, such as the larval cuticle proteins that are known to be polymorphic for copy number in Drosophila and it is thought that some of this variation is of recent origin (Charles et al. 1997), also appear in our set of Class 1 events.
Although read depth can be used to augment the detection of structural rearrangements in the genome, we chose not to do so extensively here because our sequence coverage was ∼0.8×, whereas previous experiments that have successfully detected variation using read depth have done so with coverage ∼30× (Yoon et al. 2009). However, our finding of a significant difference between coverage in regions between read-pairs in strains where a Class 1 event was present versus strains where a Class 1 event was absent further supports the results of our paired-end data and indicates that these events are tandem duplications. We do believe that read depth could and should be applied as a complementary strategy to detecting structural variation in further paired-end sequencing projects.
Structural variants that were detected in this study also compare well with the sizes of structural variants found by a number of previous studies. The average size of the 201 Class 1 structural variants of less then 32 kb in this study was 4,200.55 bp with a median size of 2,331 bp. Both Dopman and Hartl (2008) and Emerson et al. (2008) found that the majority of events detected by their studies were in this range. Emerson et al. (2008) calculated a mean duplication size of 367 bp with a median size of 1,117 bp. Dopman and Hartl (2008) found that regions smaller than single genes are most likely to have copy-number variation and that the median size for structural variants is about 3 kb with a maximum duplication size of 12 kb. Additionally, the average size of a recent gene duplication in the reference sequence, from start to stop codon, is 1.5 kb (data from Thornton and Long 2002), which underestimates the true size as it does not consider duplication of adjacent noncoding DNA. The congruence of results across studies (and technologies) is reassuring, as it remains an open empirical question how well duplication sizes are inferred from either arrays or paired-end sequencing.
Indels
Our attempt to detect indels in this experiment was less successful than our detection of other structural events. Of the 96 events we examined, about 14% were found to be indels within 10% of the expected size range when Sanger sequenced. However, we later discovered that, as a result of the sample preparation, there was more variation in the sizes of fragments than we originally assumed (Chee M, personal communication; Prognosys Biosciences). This resulted in greater variability in the distance between read-pairs and detracted significantly from our ability to make accurate predictions about indels. However, our difficulties with indel detection does not preclude this technique from being used to detect this type of event in future studies. Preparation of sample libraries has been refined to a point where the variance in the size of the fragments generated is much smaller (Mark Chee, personal communication; Prognosys Biosciences) and additional coverage will also improve the accuracy at which indels can be called.
Population Genetics of Structural Events
The population frequency analysis showed that most Class 1 duplications are rare in the sample, with an excess of rare alleles compared with the prediction of the standard neutral model, and the neutral model taking into account our ascertainment bias and the demographic expansion model of Li and Stephan (2006). This excess of rare variants is consistent with the analysis of Emerson et al. (2008), who also observed an excess of rare alleles, suggesting that segregating duplications may often be deleterious. This suggests that natural selection is acting against these events and that many segregating duplications may ultimately be lost from the population.
Gene Enrichment
We found a number of functional terms in our whole-genome data set of structural variants ≤32 kb that were enriched relative to the background of the D. melanogaster genome. A functional term analysis on our subset of genes that were confirmed by sequencing produced similar results including most of the same functional term categories but at a lower enrichment score. This is probably due to a combination of the small size of the data set; DAVID enrichment analyses generally have higher power for larger gene lists and the low-sequence coverage for our paired-end sequenced lines. Genes involved in signal reception are significantly increased even following Benjamini correction as a genes involved in cytoskeletal structure and cell adhesion. Other genes, like Cyp6g1, which is involved in DDT resistance and is known to be under positive selection (Daborn et al. 2002), which was identified in both Dopman and Hartl (2008) and Emerson et al. (2008) is also identified in our overall data set though the associated functional terms do not survive Benjamini correction. Many of the genes identified as enriched by DAVID belong to multigene families. Although DAVID takes into account the composition of the genome to which the sample is being compared and thus will not artificially show enrichment of large gene families, it is also likely that gene family size and duplication rate are related and that the enrichment we observe in large gene families is due to an increase in duplication rates in those families. To be able to actually discriminate between mutation and selection polymorphism and divergence data would be required so that appropriate tests could be carried out (e.g., McDonald and Kreitman 1991).
An analysis of just gene ontology (GO) terms revealed the same set of significantly enriched terms as the functional annotation clustering but limited to GO terms. Comparing this analysis with the GO term analysis done by Dopman and Hartl (2008), we find no similarities between their set of overrepresented GO categories and our enriched terms.
Whole-Gene Duplications
Turner et al. (2008) and Aguade (2008) described a structural variant of the Or22a and Or22b genes. They found that portions of these genes are deleted at high frequency in some populations, and the deletion is not present in other populations. Although we do not detect this variant, we do detect a novel duplication of Or22a in our African sample, and the break point for which was confirmed by PCR and sequencing. This is a novel duplication of Or22a and differs from the rearrangement described by Turner et al. (2008) and Aguade (2008), which is a fusion of Or22a and Or22b that presumably arose from a large deletion. This duplication is rare in Africa, being present in 2 of the 21 lines in our Zimbabwe sample. Thus, the olfactory receptor gene family appears to segregate at least two different structural variants species-wide.
We also detected whole-gene duplications of chorion proteins, as described above. Other read-pairs indicate a number of whole-gene duplications including Cyp28d1 and Cyp28d2 which are important in heme binding and electron carrier activity, dro2 and dro3 which are involved in defense responses to fungus, and Es2 which is involved in nervous system development to name a few examples. These genes are all members of larger gene families that have presumably undergone duplication events in their respective pasts. Further studies, which sequence genomes of interest at deeper coverage, should be able to detect more of these events directly.
Current Limitations Paired-End Sequencing
Paired-end sequencing also has limitations that are reflected in this study. In our data set, there were three main limitations; distinguishing tandem duplications from translocations, detecting rearrangements caused by TEs, and detecting rearrangements where the sample sequence may have many differences from the reference. Luckily, many of these issues can be resolved by sequencing at higher coverage. A higher level of coverage would increase the probability that both novel junctions of a translocation event would be detected, and thus this type of event could be clearly distinguished from tandem duplication (Faddah et al. 2009). Likewise, higher coverage would allow for the accurate calling of SNPs in repetitive regions that could be used to distinguish rearrangements associated with repetitive regions like transposable elements. Finally, higher coverage would increase the number of reads that map to regions of the genome that are quite different between the sample genome and the reference sequence. These structural variants may be the result of nonhomologous end-joining mechanisms, and the reads would be difficult to align to a reference. Previous studies have identified that certain types of rearrangements like inversions (Andolfatto and Kreitman 2000) and transposed duplications (Yang et al. 2008) have many indel polymorphisms near the break point. However, it is also difficult to predict here the number of false negatives because we do not have enough coverage in this study to be certain that we have detected all the events we could detect with this method. To do this, we would need to be certain that we had reached a level of coverage where we would detect no new events with additional coverage.
Multiple library preparation to produce sequence fragments of various sizes for each sample genome of interest would also increase the number and type of events that could be detected with this method. Sample preparation is also key to generating paired-end data from which accurate inferences can be drawn. This study, which uses data produced very early in the adoption of paired-end Illumina technology, did not generate as many read-pairs per lane of sequencing as is now available.
Conclusions
This study has highlighted a number of interesting evolutionary results with respect to structural variation in a natural population. First, we have detected evidence of natural selection acting upon segregating duplications within a natural population. Additionally, we have found that the majority of structural variation that we see involves genic sequence, and we detect many previously-described copy-number variants and some that are novel, such as a duplication of Or22a. Genome-wide, there appears to be regional variation in levels of structural polymorphism, and functional term analysis revealed that a variety of biological processes are enriched in our data set. Future work, at higher coverage and in larger samples, will allow quantitative analysis of the evolutionary dynamics of structural polymorphism, as well as more detailed analyses of enrichment of functional classes among structural variants.
We have also shown that paired-end sequencing accurately detects structural variation, when read-pairs are aligned to a reference sequence. Detection of rearrangements is very accurate at modest coverage, and the majority of our sequenced break points have a simple structure. The paired-end technique, combined with the limited analysis of coverage to detect structural variants, indicates that this method can quickly and accurately detect structural variants in a genome of interest. Furthermore, our results compare very well with previous microarray studies (Dopman and Hartl 2007; Emerson et al. 2008). This independent examination of structural variation using a different technique not only affirms the validity of the technique but also supports the conclusions of previous work in the area.
Supplementary Material
Supplementary table 1 and supplementary raw data are available at Genome Biology and Evolution online (http://www.oxfordjournals.org/our_journals/gbe/).
Acknowledgments
We would like to thank Tony Long and other members of the Long laboratory for useful comments on the manuscript as well as three anonymous reviewers. This research was supported through start-up funds from University of California Irvine and National Institutes of Health grant GM085183 to K.R.T.
References
- Adams MD, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Aguade M. Nucleotide and copy-number polymorphism at the odorant receptor genes Or22a and Or22b in Drosophila melanogaster. Mol Biol Evol. 2008;26:61–70. doi: 10.1093/molbev/msn227. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J of Mol Biol. 1990;3:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Andolfatto P, Kreitman M. Molecular variation at the In(2L)t proximal breakpoint site in natural populations of Drosophila melanogaster and D. simulans. Genetics. 2000;154:1681–1691. doi: 10.1093/genetics/154.4.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arguello JR, Chen Y, Yang S, Wang W, Long M. Origination of an X-linked testes chimeric gene by illegitimate recombination in Drosophila. PLoS Genet. 2006;20:e77. doi: 10.1371/journal.pgen.0020077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun DJ, Aquadro CF. African and North-American populations of Drosophila melanogaster are very different at the DNA level. Nature. 1992;365:548–550. doi: 10.1038/365548a0. [DOI] [PubMed] [Google Scholar]
- Bentley DRS, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;53:59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betran E, Bai Y, Motiwale M. Fast protein evolution and germ line expression of a Drosophila parental gene and its young retroposed paralog. Mol Biol Evol. 2006;23:2191–2202. doi: 10.1093/molbev/msl090. [DOI] [PubMed] [Google Scholar]
- Betran E, Long M. Dntf-2r, a young Drosophila retroposed gene with specific male expression under positive Darwinian selection. Genetics. 2003;164:977–988. doi: 10.1093/genetics/164.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betran E, Thornton K, Long M. Retroposed new genes out of the X in Drosophila. Genome Res. 2002;12:1854–1859. doi: 10.1101/gr.604902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles JP, Chihara C, Nejad S, Riddiford LM. A cluster of cuticle protein genes of Drosophila melanogaster at 65A: sequence, structure and evolution. Genetics. 1997;147:1213–1224. doi: 10.1093/genetics/147.3.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Langley CH. The population genetics of Drosophila transposable elements. Annu Rev Genet. 1989;23:251–287. doi: 10.1146/annurev.ge.23.120189.001343. [DOI] [PubMed] [Google Scholar]
- Chen W-K, Swartz JD, Rush LJ, Alvarex CE. Mapping DNA structural variation in dogs. Genome Res. 2009;19:500–509. doi: 10.1101/gr.083741.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad DF, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2009 doi: 10.1038/nature08516. doi: 10:1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daborn PJ, Yen JL, Bogwitz MR, et al. A single P450 allele associated with insecticide resistance in Drosophila. Science. 2002;297:2253–2256. doi: 10.1126/science.1074170. [DOI] [PubMed] [Google Scholar]
- Dennis G, et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4 doi: 10.1186. [PubMed] [Google Scholar]
- Doniger SW, et al. A catalog of neutral and deleterious polymorphism in yeast. PLoS Genet. 2008;4:e1000183. doi: 10.1371/journal.pgen.1000183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dopman EB, Hartl DL. A portrait of copy-number polymorphism in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2007;104:19920–19925. doi: 10.1073/pnas.0709888104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Emerson J, Cardoso-Moreira M, Borevitz JO, Long M. Natural selection shapes genome wide patterns of copy-number polymorphism in Drosophila melanogaster. Science. 2008;320:1629–1631. doi: 10.1126/science.1158078. [DOI] [PubMed] [Google Scholar]
- Faddah DA, et al. Systematic identification of balanced transposition polymorphisms in Saccharomyces cerevisiae. PLoS Genet. 2009;5:e1000502. doi: 10.1371/journal.pgen.1000502. doi: 10.1371/journal.pgen.1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadista J, Nygaard M, Holm L-E, Thomsen B, Bendixen C. A snapshot of CNVs in the pig genome. 2008;3:e3916. doi: 10.1371/journal.pone.0003916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C, Long M. A new retroposed gene in Drosophila heterochromatin detected by microarray-based genomic hybridization. J Mol Evol. 2007;64:272–283. doi: 10.1007/s00239-006-0169-9. [DOI] [PubMed] [Google Scholar]
- Fiston-Lavier A-S, Axolabehere D, Quesneville H. A model of segmental duplication formation in Drosophila melanogaster. Genome Res. 2007;17:1458–1470. doi: 10.1101/gr.6208307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graubert TA, et al. A high-resolution map of segmental DNA copy number variation in the mouse genome. PLoS Genet. 2007;3(1):e3. doi: 10.1371/journal.pgen.0030003. doi: 10.1371/journal.pgen.0030003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Lupski JR, Rosenberg SM, Grzegorz I. Mechanisms of change in gene copy number. Nature Reviews Genetics. 2009;10:551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddrill PR, Thornton KR, Charlesworth B, Andolfatto P. Multilocus patterns of nucleotide variability and the demographic and selection history of Drosophila melanogaster populations. Genome Res. 2005;15:790–799. doi: 10.1101/gr.3541005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4 doi: 10.1038/nprot.2008.211. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hudson RR. Generating samples under a Wright-Fisher neutral model. Bioinformatics. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- Jones CD, Custer AW, Begun DJ. Origin and evolution of a chimeric fusion gene in Drosophila subobscura, D. madeirensis and D. guanche. Genetics. 2005;170:207–219. doi: 10.1534/genetics.104.037283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern AD, Begun DJ. Current deletion and gene presence/absence polymorphism: telomere dynamics dominate evolution at the tip of 3L in Drosophila melanogaster and D. simulans. Genetics. 2008;179:1021–1102. doi: 10.1534/genetics.107.078345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd JM, et al. Haplotype sorting using human fosmid clone end-sequence pairs. Genome Res. 2008;18:2016–2023. doi: 10.1101/gr.081786.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov FA, Kondrashov AS. Role of selection in fixation of gene duplications. J Theor Biol. 2006;239:141–151. doi: 10.1016/j.jtbi.2005.08.033. [DOI] [PubMed] [Google Scholar]
- Korbel JO, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange BW, Langley CH, Stephan W. Molecular evolution of Drosophila metallothionein genes. Genetics. 1990;126:921–932. doi: 10.1093/genetics/126.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Stephan W. Inferring the demographic history and rate of adaptive substitution in Drosophila. PLoS Genet. 2006;2(10):e166. doi: 10.1371/journal.pgen.0020166. doi: 10.1371/journal.pgen.0020166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Li Y, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24:713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- Lipatov M, Lenkov K, Petrov DA, Bergman CM. Paucity of chimeric gene-transposable element transcripts in the Drosophila melanogaster genome. BMC Biol. 2005;3:24. doi: 10.1186/1741-7007-3-24. doi: 10.1 186/1741-7007-3-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M, Betran E, Thornton K, Wang W. The origin of new genes: glimpses from the young and old. Nat Rev Genet. 2003;4:865–875. doi: 10.1038/nrg1204. [DOI] [PubMed] [Google Scholar]
- Long MY, Langley CH. Natural-selection and the origin of jingwei, a chimeric processed functional gene in Drosophila. Science. 1993;260:91–95. doi: 10.1126/science.7682012. [DOI] [PubMed] [Google Scholar]
- Lootens S, Burnett J, Friedman TB. An intraspecific gene duplication polymorphism of the urate oxidase gene of Drosophila virilis: a genetic and molecular analysis. Mol Biol Evol. 1993;10:635–646. doi: 10.1093/oxfordjournals.molbev.a040028. [DOI] [PubMed] [Google Scholar]
- Loppin B, Lepetit D, Dorus S, Couble P, Karr TL. Origin and neofunctionalization of a Drosophila paternal effect gene essential for zygote viability. Curr Biol. 2005;15:87–93. doi: 10.1016/j.cub.2004.12.071. [DOI] [PubMed] [Google Scholar]
- Maroni G, Wise J, Young JE, Otto E. Metallothionein gene duplications and metal tolerance in natural populations of Drosophila melanogaster. Genetics. 1987;117:739–744. doi: 10.1093/genetics/117.4.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marth G, Czabarka A, Murvai J, Sherry ST. The allele frequency spectrum in genome-wide human variation data reveals signals of differential demographic history in three large world populations. Genetics. 2004;166:351–372. doi: 10.1534/genetics.166.1.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzkin LM, Merritt TJS, Zhu CT, Eanes WF. The structure and population genetics of the breakpoints associated with the cosmopolitan chromosomal inversion In(3R)Payne in Drosophila melanogaster. Genetics. 2005;170(3):1143–1152. doi: 10.1534/genetics.104.038810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maydan JS, et al. Efficient high-resolution deletion discovery in Caenorhabditis elegans by array comparative genomic hybridization. Genome Res. 2007;17(3):337–347. doi: 10.1101/gr.5690307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- Ossowski S, et al. Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res. 2008;18:2024–2033. doi: 10.1101/gr.080200.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp AJ, et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat Genet. 2006;38:1038–1042. doi: 10.1038/ng1862. [DOI] [PubMed] [Google Scholar]
- Shih H-J, Jones CD. Patterns of amino acid evolution in the Drosophila ananassae chimeric gene, siren, parallel those of other Adh-derived chimeras. Genetics. 2008;180:1261–1263. doi: 10.1534/genetics.108.090068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P. Repeat Masker Open 3.2 [Internet] 2004. Technical report. [cited 2010 Jan 27]. Available from: http://www.repeatmasker.org. [Google Scholar]
- Smith DR, et al. Rapid whole-genome mutational profiling using next-generation sequencing technologies. Genome Res. 2008;18:1638–1642. doi: 10.1101/gr.077776.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stambuk BU, Dunn B, Alves SL, Jr., Duval EH, Sherlock G. Industrial fuel ethanol yeasts contain adaptive copy number changes in genes involved in vitamin B1 and B6 biosynthesis. Genome Res. 2009;19(12):2271–2278. doi: 10.1101/gr.094276.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano T, Kusakabe S, Koga A, Mukai T. Polymorphism for the number of tandemly multiplicated glycerol-3-phosphate dehydrogenase genes in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1989;86:5000–5004. doi: 10.1073/pnas.86.13.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton K. Libsequence, a C++ class library for evolutionary genetic analysis. Bioinformatics. 2003;19(17):2325–2327. doi: 10.1093/bioinformatics/btg316. [DOI] [PubMed] [Google Scholar]
- Thornton K, Andolfatto P. Approximate Bayesian inference reveals evidence for a recent strong bottleneck in non-African populations of Drosophila melanogaster. Genetics. 2006;172:1607–1619. doi: 10.1534/genetics.105.048223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton K, Long M. Rapid divergence of gene duplicates on the Drosophila melanogaster X chromosome. Mol Biol Evol. 2002;19:918–925. doi: 10.1093/oxfordjournals.molbev.a004149. [DOI] [PubMed] [Google Scholar]
- Thornton K, Long M. Excess of amino acid substitutions relative to polymorphism between X-linked duplications in D. melanogaster. Mol Biol Evol. 2005;22:273–284. doi: 10.1093/molbev/msi015. [DOI] [PubMed] [Google Scholar]
- Turner TL, Levine MT, Eckert ML, Begun DJ. Genomic analysis of adaptive differentiation in Drosophila melanogaster. Genetics. 2008;179:455–473. doi: 10.1534/genetics.107.083659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuzun E, et al. Fine-scale structural variation of the human genome. Nat Genet. 2005;37:727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- Urban AE, et al. High-resolution mapping of DNA copy alterations in human chromosome 22 using high-density tiling oligonucleotide arrays. Proc Natl Acad Sci U S A. 2006;103 doi: 10.1073/pnas.0511340103. doi: 10.1073/pnas.0511340103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Brunet FG, Nevo E, Long M. Origin of sphinx, a young chimeric RNA gene in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2002;99:4448–4453. doi: 10.1073/pnas.072066399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Yu H, Long M. Duplication-degeneration as a mechanism of gene fission and the origin of new genes in Drosophila species. Nat Genet. 2004;5:523–537. doi: 10.1038/ng1338. [DOI] [PubMed] [Google Scholar]
- Wang W, Zhang JM, Alvarez C, Llopart A, Long M. The origin of the jingwei gene and the complex modular structure of its parental gene, yellow emperor, in Drosophila melanogaster. Mol Biol Evol. 2000;17:1294–1301. doi: 10.1093/oxfordjournals.molbev.a026413. [DOI] [PubMed] [Google Scholar]
- Watterson GA. On the number of segregating sites in genetical models without recombination. Theor Popul Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- Yang S, Arguello JR, Li X, Ding Y, Zhou Q, Chen Y, Zhang Y, Zhao R, Brunet F, Peng L, Long M, Wang W. Repetitive element-mediated recombination as a mechinism for new gene origination in Drosophila. PLoS Genet. 2008;4(1):e3. doi: 10.1371/journal.pgen.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S, Xuan Z, Makarov V, Ye K, Sebat J. Sensitive and accurate detection of copy number variants using read depth of coverage. Genome Res. 2009;19:1586–1592. doi: 10.1101/gr.092981.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease and evolution. Annu Rev Genomics Hum Genet. 2009;10:451–481. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Dean AM, Brunet F, Long M. Evolving protein functional diversity in new genes of Drosophila. Proc Natl Acad Sci U S A. 2004;101:16246–16250. doi: 10.1073/pnas.0407066101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, et al. On the origin of new genes in Drosophila. Genome Res. 2008;18:1446–1455. doi: 10.1101/gr.076588.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.