Abstract
Colonization of the gastrointestinal tract (GIT) of human infants with a suitable microbial community is essential for numerous aspects of health, but the progression of events by which this microbiota becomes established is poorly understood. Here, we investigate two previously unexplored areas of microbiota development in infants: the deployment of functional capabilities at the community level and the population genetics of its most abundant genera. To assess the progression of the infant microbiota toward an adult-like state and to evaluate the contribution of maternal GIT bacteria to the infant gut, we compare the infant’s microbiota with that of the mother at 1 and 11 months after delivery. These comparisons reveal that the infant’s microbiota rapidly acquires and maintains the range of gene functions present in the mother, without replicating the phylogenetic composition of her microbiota. Microdiversity analyses for Bacteroides and Bifidobacterium, two of the main microbiota constituents, reveal that by 11 months, the phylotypes detected in the infant are distinct from those in the mother, although the maternal Bacteroides phylotypes were transiently present at 1 month of age. The configuration of genetic variants within these genera reveals populations far from equilibrium and likely to be undergoing rapid growth, consistent with recent population turnovers. Such compositional turnovers and the associated loss of maternal phylotypes should limit the potential for long-term coadaptation between specific bacterial and host genotypes.
Keywords: Bacteroides, Bifidobacterium, gut microbiota, community genomics, bacterial population genetics
Introduction
The gastrointestinal tract (GIT) is host to the most abundant and diverse bacterial community in humans, containing, in adults, on the order of 1014 bacterial cells compared with 1013 human cells for the entire body. This complex microbiota plays essential roles for gut maturation, food digestion, modulation of the immune system, and protection from pathogens (Savage 1977; Stark and Lee 1982; Cebra 1999; Mackie et al. 1999; Hooper et al. 2001; Favier et al. 2002; Fanaro et al. 2003; Schumann et al. 2005; Adlerberth et al. 2006; Ley, Peterson, and Gordon 2006; Ley, Turnbaugh, et al. 2006; O’Mahony et al. 2008). At birth, the GIT is germ free, but it rapidly enters an extensive and a complex process of colonization by a variety of microbes. Several studies have monitored this process at different stages by means of 16S ribosomal RNA (rRNA) analyses (Favier et al. 2002; Hopkins et al. 2005; Park et al. 2005; Penders et al. 2006; Palmer et al. 2007) and have shown that it is far from a strict microbial succession. Rather, GIT colonization is highly variable among individuals and is influenced by numerous factors, including mode of birth, feeding habits, health status, and sanitary conditions (Fanaro et al. 2003; Adlerberth et al. 2006; Penders et al. 2006; Palmer et al. 2007). However, 16S rRNA analyses can only appraise the phylogenetic composition of a sample and provide no direct information about its functional capabilities. An additional issue of fundamental importance remains unexplored: no studies have addressed the population dynamics of individual bacterial species as the infant gut is colonized. However, an appreciation of the amount, pattern, origin, and persistence of population-level genetic variation within an infant’s GIT is crucial for understanding the evolutionary forces at work during the process of colonization. In particular, a specific aspect of population dynamics—the degree to which an infant’s microbiota is vertically inherited from the maternal GIT—will determine the potential for natural selection to foster long-term bacterial adaptations to specific host genotypes.
Here, we address these issues by focusing on the GIT microbiota in a single mother–infant pair, revealing not only the coding capabilities and phylogenetic composition of these bacterial communities but also the microdiversity, population biology, and degree of maternal inheritance within their main constituents. The infant in our analysis was a healthy male, vaginally delivered at full term in an urban hospital in Tucson, AZ. He was exclusively breast-fed for 5 months, after which solid food was introduced to his diet, although he continued to ingest breast milk until 2 years of age. We analyzed the GIT microbiota of this infant and that of his mother at two time points during the first year of life, at 1 month (I-1m and M-1m) and 11 months (I-11m and M-11m) after birth. These two stages represent well-differentiated phases of microbiota development in the infant (Cooperstock and Zead 1983). By 1 month, the initial phase of rapid acquisition of microbes after birth is thought to be over and, in exclusively breast-fed infants, to have produced a breast milk–adapted microbiota that will remain until the introduction of solid foods (Cooperstock and Zead 1983; Mackie et al. 1999). By 11 months, the baby has been exposed to a complex diet for a significant period of time and should be reaching an adult-like microbiota composition (Stark and Lee 1982; Mackie et al. 1999; Favier et al. 2002; Fanaro et al. 2003; Park et al. 2005; Penders et al. 2006; Palmer et al. 2007). Our analyses revealed that these two stages contained bacterial communities that were largely distinct in phylogenetic composition but remarkably similar and adult like in their global functional capabilities. Overall, this sampling strategy indicated that the GIT microbiota remains very dynamic throughout the first year of life, with highly diverse and actively growing bacterial populations, and that maternal GIT phylotypes can be transmitted to the infant but need not persist in the long term.
Materials and Methods
Sample Collection
We obtained fecal samples from a healthy mother–infant pair at 1 and 11 months after delivery. Samples were collected at the University of Arizona, with informed written consent from the infant’s parents, using protocols approved by the institutional review boards of the Lawrence Berkeley National Laboratory and the University of Arizona.
Preparation of High–Molecular Weight DNA and Fosmid Library Construction
In order to capture extensive genomic fragments from the bacterial species in the GIT microbiota, we generated fosmid libraries from high–molecular weight bacterial DNA isolated directly from the fecal samples. Fosmid libraries enable the elucidation of both the coding capabilities and the phylogenetic positions of the members of a given bacterial community while providing the possibility of recovering long contiguous sequences from interesting clones. To obtain high–molecular weight bacterial DNA, fresh fecal samples were harvested and immediately processed. In short, 5 g of sample was suspended in 50 ml phosphate buffer saline (PBS) and subjected to a series of filtration steps to eliminate particulate matter and large eukaryotic cells. Sequential filtrations were performed with a Micro Filtration System (Millipore) using 100-, 20-, and 11-μm nylon filters and were followed by centrifugation at 250 × g to remove eukaryotic nuclei. Bacterial cells were then pelleted, washed, and resuspended in 5 ml PBS. Visual inspection under the microscope revealed a large variety of bacterial cell types and no trace of eukaryotic cells. Bacterial cells were embedded in agarose plugs and lysed with a variety of enzymes (lysozyme, N-acetylmuramidase, and achromopeptidase) capable of digesting cell walls from phylogenetically diverse bacteria plus proteinase K and detergent (Hayashi et al. 2002). Agarose plugs containing bacterial DNA were washed exhaustively and stored at 4°C in 50 mM ethylenediaminetetraacetic acid. For fosmid library construction, DNA was extracted from the agarose plugs and mechanically fragmented by hydroshearing. Forty-kilobase DNA fragments were separated by pulsed-field gel electrophoresis and cloned into pCC1Fos (Epicentre Corp.).
Sequencing and Analysis of Fosmid Ends
In order to characterize the phylogenetic composition and gene content of the obtained libraries, inserts from 1,536 randomly selected fosmids from each library were sequenced at both ends. All sequencing reactions were performed by the Sanger method using BigDye Terminators in ABI 3730 sequencers at the DOE Joint Genome Institute. Only high-quality sequences (≥Q20), as assessed by Phred (Ewing and Green 1998; Ewing et al. 1998), were retained for analysis after being subjected to LUCY (Chou and Holmes 2001) for quality trimming and vector contamination removal. Assembly was attempted with Phrap (http://www.phrap.org), but due to the low percentage of reads assembling into contigs, all subsequent analyses were performed on unassembled collections of sequences for each sample. All sequences were deposited in GenBank (EF990944–EF998850) and IMG/M (Markowitz et al. 2008). Ab initio gene calling was performed in IMG/M using GeneMark (Besemer et al. 2001), and the identified protein-coding genes were further analyzed in IMG/M to provide estimates of the phylogenetic composition and the functional repertoire of each sample. Taxonomic affiliation was registered only for genes with best hits having >90% identity to IMG/M reference isolate genomes as determined by BlastP. Genes were assigned to COGs based on RPS-Blast (Reverse Position–Specific Blast) applying default IMG/M parameters (E value < 10−2). In addition, two fosmid clones that had matching end sequences with best BlastP hits to Bacteroides were selected for complete sequencing. These fosmids were fragmented by hydroshearing, subcloned into pUC18, sequenced, base called in Phred, partially assembled with Phrap, and annotated with NCBI’s open reading frame finder (GenBank GU362641 and GU362642).
Statistical Comparison of the Functional Profiles of GIT Samples
The abundances of different gene functions in each sample were compared by means of D scores for each individual COG as well as D ranks for COG pathways and categories, as described in IMG/M (Markowitz et al. 2008). D ranks were also used to evaluate differences among samples in terms of the abundances of COG groups containing genes potentially involved in the utilization of human milk oligosaccharides (HMOs) or plant carbohydrates. D scores are derived under a binomial assumption and can be translated into P values at different levels of significance. The D rank represents a normalization ranking of each pairwise comparison and is calculated by adding the D scores of all COGs assigned to a particular functional pathway, category or other defined group, and normalizing by the square root of the number of these categories, including those with no genes assigned. COGs with zero counts are assigned a zero value in the computation of D ranks, which avoids overestimating the statistical significance of the differences in gene counts for sparsely populated functional categories. In addition, a principal component analysis (PCA) of the overall COG category profiles was undertaken to establish the relative similarity of samples to one another and to other human gut samples included in IMG (Gill et al. 2006). The PCA was performed in the R software environment (Ihaka and Gentleman 1996) after normalizing the gene count in each COG category by the total gene count in the corresponding metagenome and using the R function prcomp (x, center=T, scale=T) to center the values around zero and scale to unit variance.
Genus-Specific 16S rRNA Phylogenies
We investigated the extent of variation within Bacteroides and Bifidobacterium, two of the main constituent genera of the GIT microbiota in the infant and maternal microbial communities. 16S rRNA was polymerase chain reaction (PCR) amplified from the agarose-embedded DNA (above) using primers and reaction conditions determined for each genus. The primer pairs employed were forward Bac32F (5′-AACGCTAGCTACAGGCTT-3′) and reverse Bac708R (5′-CAATCGGAGTTCTTCGTG-3′) for Bacteroides (Bernhard and Field 2000) and forward lm26 (5′-GATTCTGGCTCAGGATGAACG-3′) and reverse lm3 (5′-CGGGTGCTICCCACTTTCATG-3′) for Bifidobacterium (Kaufmann et al. 1997), yielding PCR products of 677 and 1,418 bp, respectively. PCR products were cloned into pCR4-TOPO (Invitrogen), and 50 randomly selected clones from each library were sequenced from both ends. Sequence reads were vector trimmed, assembled, quality checked, and chimera checked using the software package Genelib (Kirton E, unpublished data). The resulting sequences were aligned in ARB (Ludwig et al. 2004) to a backbone alignment from the Greengenes 16S rRNA database (http://greengenes.lbl.gov) (DeSantis et al. 2006). Alignments were refined manually taking into account rRNA secondary structures. Neighbor-joining trees were produced to identify those sequences in the Greengenes backbone alignment that were most similar to the reads from our samples. These sequences were exported for phylogenetic analysis along with our reads and sequences representing the extent of diversity within each genus analyzed. Prior to phylogenetic reconstruction, alignments were pruned of redundant sequences and poorly aligned regions in Gblocks (Castresana 2000), yielding 39- and 48-sequence alignments for Bacteroides and Bifidobacterium, respectively. Maximum likelihood phylogenetic analysis with 1,000 bootstrap replicates was performed using Garli (http://www.bio.utexas.edu/faculty/antisense/garli/Garli.html) (Zwickl 2006) with the best-fitting sequence evolution model (general time reversible with invariant sites and gamma distribution for site rate variation) and parameters determined in Modeltest v. 3.7 (Posada and Crandall 2001).
Population Genetic Analyses
Genealogical relationships among the different haplotypes recovered within the main constituent genera of the GIT microbiota were estimated by statistical parsimony (Templeton et al. 1992), as implemented in the computer program TCS (Clement et al. 2000). Gene diversity (H) was computed as
where is the estimated frequency of allele i in a sample of n alleles (Nei 1987). Expected numbers of total haplotypes E(k) were computed according to Ewens (1972) as
where θ = H/(1 − H). Expected numbers of haplotypes present singly (singletons) E(k1) were computed according to Maruyama and Fuerst (1984) as
Data Deposition
GenBank (EF990944–EF998850) and (GU362547–GU362642).
Results
Fosmid Libraries and End Sequencing
To characterize and compare the microbial communities present in the infant and the mother at 1 and 11 months after delivery, we generated four fosmid libraries from bacterial DNA isolated directly from fecal samples (50,000 clones each; 8 Gb total). Inserts from 1,536 randomly selected fosmids from each library were sequenced at both ends, yielding a total of 12,288 sequence reads and more than 8.4 Mb of high-quality sequence (Q20) as assessed by Phred (Ewing and Green 1998; Ewing et al. 1998). After further quality trimming and vector contamination removal, the sequences of each sample were independently assembled using phrap (http://www.phrap.org). The majority of sequences (62% on average) could not be incorporated into assemblies, and most of the assembled contigs contained only two reads. But because bacterial genes are compact and intronless, ab initio gene calling in IMG/M (Markowitz et al. 2008) was able to detect more than 7,500 genes in the unassembled reads of the four samples (table 1).
Table 1.
Characterization of Mother and Infant Samples Based on Fosmid-End Sequencing
| M-1m | M-11m | I-1m | I-11m | |
| Analyzed reads | 1,981 | 2,056 | 1,778 | 2,092 |
| Analyzed bases | 1,317,583 | 1,400,487 | 1,033,600 | 1,422,416 |
| Total genes | 1,916 | 2,041 | 1,522 | 2,077 |
| Protein-coding genes | 1,891 | 2,003 | 1,507 | 2,045 |
| Functionally assigned genesa | 869 | 1,218 | 894 | 1,152 |
| COG-assigned genes | 719 | 1,067 | 767 | 1,030 |
| Taxonomically assigned genes | 1,039 | 1,160 | 1,128 | 1,434 |
| Identified genera | 22 | 20 | 9 | 16 |
Functional assignment was performed within IMG/M (Markowitz et al. 2008) with basis on COG (Tatusov et al. 1997), Pfam (Bateman et al. 2004), TIGRfam (Selengut et al. 2007), and KEGG (Kanehisa et al. 2004) classifications.
Similar Distribution of Gene Functions in the GIT Microbiota of Mother and Infant
The functional repertoire within the different GIT samples was appraised within IMG/M with basis on different classifications, including COG (Tatusov et al. 1997), Pfam (Bateman et al. 2004), TIGRfam (Selengut et al. 2007), and KEGG (Kanehisa et al. 2004). Combined, all classifications allowed for functional categorization of 45–60% of the genes detected within each sample, with 38–52% of total genes assigned to COG clusters (table 1). The overall distribution of COG functional classes was remarkably similar in all four libraries (fig. 1A). Carbohydrate and amino acid transport and metabolism were the most common COG categories overall—categories G and E—and together represented 21–27% of the COG assignments per sample. In contrast, lipid transport and metabolism (COG category I) accounted for only 2–3% of these assignments.
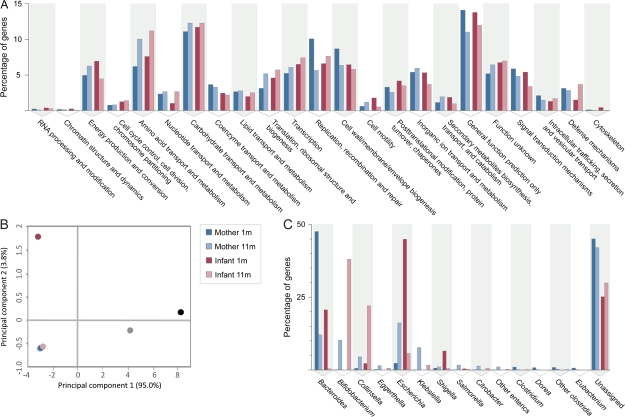
FIG. 1.—
Phylogenetic composition and general functional capabilities across GIT microbiota samples. (A) Distribution of COG functional categories in the GIT microbiota of mother and infant at 1 and 11 months after delivery. Distributions of COG functional classes are remarkably similar; observed differences across samples do not reach significance (D rank analysis; P > 0.05). (B) PCA of overall COG category profiles in the mother–infant GIT microbiota. Two additional samples (black and gray circles) from adult humans (Gill et al. 2006) are included for comparative purposes. Note the overlapping positions of the two maternal microbiota samples and the infant sample taken at 11 months. The large separation of I-1m from these three samples on the second PC axis stems from the fact that this is the only sample containing the COG category Extracellular Structures (W); the first PC axis separates the four samples in our study from those of Gill et al. (2006) due to the higher numbers of individual COGs represented within each category in these larger samples. (C) Bacterial diversity present in the gastrointestinal microbiota of mother and infant at 1 and 11 months after delivery. Taxonomic affiliation was assigned to the high-scoring (>90% identity) best BlastP matches of protein-coding genes in fosmid-end sequence reads against IMG/M reference isolate genomes. Most identified genera belong to only four phyla: Bacteroidetes (Bacteroides), Actinobacteria (Bifidobacterium, Collinsella, and Eggerthella), Proteobacteria (Escherichia, Klebsiella, Shigella, Salmonella, Citrobacter, and other enterics), and Firmicutes (Clostridium, Dorea, other clostridia, and Eubacterium).
To evaluate any potential significance of function abundance differences among samples, we computed D scores for each individual COG as well as D ranks for COG pathways and categories as defined in IMG/M (Markowitz et al. 2008). Pairwise comparisons among samples did not identify significant differences in the distribution of individual COGs, pathways or categories (all P > 0.05). In contrast, a PCA of the COG category profiles grouped closely the M-1m, M-11m, and I-11m samples to the exclusion of I-1m (fig. 1B). Inspection of the contribution of the different COG categories to the principal components indicates that it is the presence of the COG category W (Extracellular Structures) in I-1m that separates this sample on the second principal component (PC) axis, whereas the large distance between our samples and the larger adult human gut samples from Gill et al. (2006) (black and gray circles) on the first axis is due to the higher numbers of individual COGs represented within each category in the latter. The Extracellular Structures category is only represented in I-1m by four sequences; these sequences match COG5295, which codes for autotransporter adhesins, secreted bacterial proteins that may exhibit diverse virulence functions. Best hits to reference genomes in IMG indicated that the four I-1m COG5295 sequences were likely encoded by Escherichia coli.
To further evaluate potential differences in functionality between infant and maternal samples, we investigated the distribution of those COGs in the carbohydrate transport and metabolism category (G) that were previously found to be overrepresented in human gut microbiomes (Kurokawa et al. 2007). The human gut overrepresented G COGs include 32 COGs (group I) encoding proteins that could transport or metabolize the different sugars found in human milk, mostly lactose and other oligosaccharides containing glucose, galactose, N-acetylglucosamine, fucose, or sialic acid residues (table S1, Supplementary Material online). In addition, the human gut overrepresented G COGs also include 13 COGs (group II) encoding proteins specialized for the transport and metabolism of carbohydrates of plant origin (table S2, Supplementary Material online). To search for specific differences among our samples potentially related to diet, we specifically analyzed the distribution of human gut overrepresented G COGs belonging to these two groups.
We recovered in at least one of our samples 74% of the human gut overrepresented G COGs, including 13 COGs encoding glycosyl hydrolases and 5 for different components of sugar transporters. The two infant samples (I-1m and I-11m) contained very similar numbers of group I COGs, 12 and 11, respectively, although only 5 of these were common to both (COG0676, COG1129, COG1263, COG1486, and COG2017). Notably, the two maternal samples (M-1m and M-11m) contained 18 group I COGs each, and 15 of these were in common. In terms of the overall number of reads assigned to group I COGs in each of the four samples (table S1, Supplementary Material online), there were no significant differences as evaluated by IMG/M D rank scores (P > 0.05), but some differences were apparent in terms of the presence/absence of specific group I COGs among samples. Six COGs were recovered exclusively from infants and not from the maternal samples, including COG0676, encoding uncharacterized enzymes related to aldose 1-epimerase, and COG1486, encoding α-galactosidases/6-phospho-β-glucosidases. In addition to these two COGs common to both infants, COG1299 (fructose-specific IIC component of the phosphotransferase system) and COG4668 (galactose-1-phosphate uridyltransferase) were unique to I-1m, whereas COG3414 (galactitol-specific IIB component of the phosphotransferase system) and COG3669 (α-L-fucosidase) were unique to I-11m. On the other hand, the maternal samples also contained several group I COGs not recovered in the infant samples, including COGs encoding enzymes for the metabolism of galactose (COG0153, COG1874, and COG3345), fructose (COG0205, COG1762, and COG2893), and fucose (COG0738 and COG2407) (table S1, Supplementary Material online).
Among group I COGs, we recovered in our samples three out of four glycosidase-encoding COGs found in a HMO utilization cluster recently discovered in Bifidobacterium longum subsp. infantis (Sela et al. 2008). Of these, COG3669 (α-L-fucosidase) was unique to I-11m, whereas COG3250 (β-galactosidase/β-glucuronidase) and COG3525 (β-hexosaminidase) were present in I-1m and both of the maternal samples but not in I-11m (table S1, Supplementary Material online). The fourth HMO cluster COG (COG4409, sialidase) was not recovered from any of the samples in our study. Best hits to reference genomes in IMG indicated that COG3250 and COG3525 were likely encoded by Bacteroides in the mother and the infant at 1 month but by E. coli and an organism belonging to the Firmicutes in the 11-month maternal sample, whereas COG3669 in I-11m was represented in reads of likely bifidobacterial origin.
Even though the 1-month-old infant had no exposure to any food other than breast milk, I-1m contained representatives of four group II COGs, encoding proteins specialized for the transport and metabolism of carbohydrates of plant origin, within the range of group II COG types found in the partially weaned 11-month infant (7) and the two maternal samples (3 and 6). Moreover, as was the case for group I COGs, there were no significant differences in the overall number of reads assigned to group II COGs in each sample (P > 0.05 for all IMG/M D rank scores). The four group II COGs in I-1m were involved in the transport or metabolism of xylose (COG2115 and COG3507), cellobiose (COG1455), and melibiose (COG2211). There was no COG absent from I-1m and present in all others, and only two COGs (COG1440, IIB component of the cellobiose-specific phosphotransferase system, and COG2160, L-arabinose isomerase) were absent from I-1m and present in two other samples (table S2, Supplementary Material online). Even though COG1440 was not represented in I-1m, this sample did contain COGs encoding the IIA (COG1447) and IIC (COG1455) components of the cellobiose-specific phosphotransferase system, such that this function is likely present in the 1-month-old infant gut.
Different Phylogenetic Compositions across GIT Microbiota Samples
Phylogenetic composition was established through BlastP queries of the identified protein-coding genes. Requiring a BlastP identity threshold to assign genes to a specific taxonomic group resulted in the identification and classification of 72% and 56% of genes in infant and mother samples, respectively. The distribution of these high-scoring BlastP hits revealed that, in spite of the functional similarity already noted, there were large differences in phylogenetic membership among libraries, indicating turnover in the GIT microbiota over time in both infant and mother. Although the total numbers of phyla represented were similarly low in all samples (four to five: Bacteroidetes, Actinobacteria, Proteobacteria, and Firmicutes, plus an occasional Fusobacteria), the presence and frequencies of different genera within these phyla varied substantially. In the 1-month-old infant, only 9 genera were recovered, but by 11 months, the number of genera increased and approached that found in the maternal samples (16 genera in I-11m vs. 22 in M-1m and 20 in M-11m). The microbiota in the 1-month-old infant consisted primarily of Escherichia (45%) and Bacteroides (21%), but by 11 months, these genera had been drastically reduced to 6% and 0.4%, whereas the previously absent Bifidobacterium (38%) and the related actinobacterium Collinsella (22%) dominated the community. During the same time period, the maternal microbiota also exhibited a substantial reduction in Bacteroides (from 48% at 1 month to 12% at 11 months) and the appearance of Bifidobacterium (10%), although in this case, these changes were accompanied by a large increase in enteric genera (3–29%) (fig. 1C).
Microdiversity within Bacteroides and Bifidobacterium
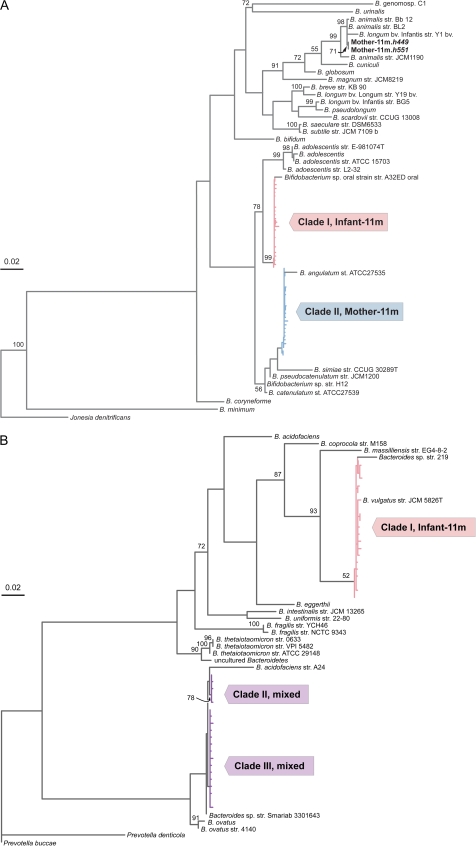
In that Bacteroides and Bifidobacterium are main constituents of the gastrointestinal microbiota and play basic roles in the metabolism of carbohydrates in the gut, we investigated the extent of variation within these genera in the infant and maternal microbial communities. To this end, we employed 16S ribosomal DNA (rDNA) primers specific to Bacteroides (Bernhard and Field 2000) or Bifidobacterium (Kaufmann et al. 1997) and measured the species (97% sequence identity) and phylotype (99% sequence identity) diversity in each of these genera. The Bacteroides-specific primers amplified the 16S rDNA gene in all samples, but Bifidobacterium primers produced amplicons only in the samples recovered at 11 months after childbirth, in accordance with the results obtained by fosmid-end sequencing (fig. 1C). A maximum likelihood phylogeny of the 48 nonidentical Bifidobacterium sequences (i.e., haplotypes) recovered from the 11-month samples resolved the main Bifidobacterium haplotypes into distinct clades according to their source of origin (fig. 2A). The 24 Bifidobacterium sequences from the infant at 11 months after delivery (I-11m) form a single phylotype, most closely related to an oral Bifidobacterium strain and to Bi. adolescentis, whereas the vast majority of haplotypes from the mother at this sampling point (M-11m) form a second phylotype related to Bi. angulatum (two other M-11m sequences reside within the Bi. animalis clade). The presence of distinct phylogenetic lineages in mother and infant suggests that each independently acquired bifidobacteria from external sources during the period between 1 and 11 months after birth.
FIG. 2.—
Phylogeny of GIT haplotypes in relation to diversity within the corresponding genus. Phylogenies of the (A) Bifidobacterium and (B) Bacteroides 16S rRNA haplotypes with representative samples from each genus were derived by maximum likelihood with 1,000 bootstrap replicates under a general time-reversible model with invariant sites and gamma-distributed site rate variation. Scale bar represents 0.02 nucleotide substitutions per site. “Mixed” clades contain haplotypes recovered from both the mother and the infant.
The phylogeny of the 39 Bacteroides 16S haplotypes obtained across the four libraries portrays a very different pattern of acquisition and transmission (fig. 2B). In contrast to the Bifidobacterium tree topology, two of the three Bacteroides clades (II and III) are intermixed with sequences from both maternal samples (M-1m and M-11m) and the 1-month infant sample (I-1m), whereas clade I consists only of sequences recovered from the 11-month infant (I-11m). The 19 sequences in clade I form a single phylotype related to Bacteroides vulgatus. Clades II and III represent distinct but closely related phylotypes (98.5% sequence identity), containing 5 and 15 haplotypes, respectively. Thus, the maternal GIT harbored the same two Bacteroides phylotypes over a period of 10 months, and these phylotypes were transmitted from the mother to the infant before 1 month of age. Nonetheless, 10 months later, a new phylotype belonging to a different Bacteroides species had replaced the original maternal phylotypes in the infant GIT.
Fosmid Sequencing Confirms that Mother and 1-Month-Old Infant Contain the Same Bacteroides Strain
To test if the mother and the 1-month-old infant contained identical strains, we culled our collection of fosmid-end sequences and recovered two Bacteroides clones, one from the mother and one from the infant, with extended regions of 100% identity. From these, we generated draft shotgun sequences that provided large contigs with 26 kb of overlapping sequence. Alignment of the 26-kb regions from each host did not reveal a single nucleotide difference, confirming that mother and infant shared the same strain of Bacteroides.
Bacteroides and Bifidobacterium Population Genetics
The population biology and transmission patterns in our mother–infant system were addressed by analyzing the distribution, frequency, and relationships of haplotypes within the Bifidobacterium and Bacteroides clades (fig. 2A and B). Genealogical relationships among haplotypes were estimated by statistical parsimony (Templeton et al. 1992; Clement et al. 2000); this approach is required for population-level analyses because, unlike methods aimed at reconstructing lineage phylogenies, it integrates information about haplotype frequencies, allows for the ancestral haplotype to be represented in the sample, and does not force a bifurcating network topology.
In the case of Bacteroides, the maternal 1-month sample contained 12 haplotypes, the most common of which were represented by 28 (M-1m.h1, clade III) and 4 (M-1m.h12, clade II) identical sequences. All of the haplotypes in this sample were also recovered at nearly identical frequencies 10 months later in the maternal sample taken at 11 months after delivery (table 2). In addition, two novel haplotypes were represented by individual sequences in the 11-month maternal sample. In contrast, only three of the maternal haplotypes were recovered in the 1-month-old infant. These haplotypes included the two most prevalent and one of the singletons and were recovered in proportions similar to those observed in the mother (31, 8, and 1 sequences; table 2). Aside from harboring three haplotypes detected in the mother, the 1-month-old infant also contained seven unique singletons. Although the specific haplotypes differed between mother and infant, the total number of haplotypes, the proportion of singletons, and the overall gene diversity were similarly high in both individuals (table 2). Figure 3 presents the genealogy of all the Bacteroides haplotypes recovered from the mother and the 1-month-old infant (corresponding to clades II and III in fig. 2B). Irrespective of their sample of origin, all of the singletons and other low-frequency alleles were derived from the most common haplotypes by one or two mutational steps. These changes are unlikely to represent sequencing errors, as average Phred quality scores (Ewing and Green 1998; Ewing et al. 1998) for our quality-trimmed and assembled 16S contigs are approximately Q60, such that the average error is <10−6. Moreover, the fact that we recover very similar haplotype distributions in M-1m and M-11m, with nearly all the singletons being shared between the two, strongly indicates that the variants detected in the Bacteroides populations are real and not significantly affected by errors in sequencing.
Table 2.
Persistence of Haplotypes of Bacteroides and Bifidobacterium in Mother and Infant Samples
| M-1m | M-11m | I-1m | I-11m | |
| Bacteroides haplotypesa | ||||
| M-1m.h1 (clade III) | 28 (0.65) | 27 (0.60) | 31 (0.66) | 0 |
| M-1m.h12 (clade II) | 4 (0.09) | 5 (0.11) | 8 (0.17) | 0 |
| M-1m.h4 (clade III) | 2 (0.05) | 2 (0.04) | 0 | 0 |
| M-1m.h30 (clade II) | 1 (0.02) | 1 (0.02) | 1 (0.02) | 0 |
| M-1m.h23 (clade II) | 1 (0.02) | 1 (0.02) | 0 | 0 |
| M-1m.h5 (clade III) | 1 (0.02) | 1 (0.02) | 0 | 0 |
| M-1m.h8 (clade III) | 1 (0.02) | 1 (0.02) | 0 | 0 |
| M-1m.h10 (clade III) | 1 (0.02) | 1 (0.02) | 0 | 0 |
| M-1m.h13 (clade III) | 1 (0.02) | 1 (0.02) | 0 | 0 |
| M-1m.h19 (clade III) | 1 (0.02) | 1 (0.02) | 0 | 0 |
| M-1m.h20 (clade III) | 1 (0.02) | 1 (0.02) | 0 | 0 |
| M-1m.h25 (clade III) | 1 (0.02) | 1 (0.02) | 0 | 0 |
| I-11m.h441 (clade I) | 0 | 0 | 0 | 14 (0.30) |
| I-11m.h440 (clade I) | 0 | 0 | 0 | 11 (0.24) |
| I-11m.h445 (clade I) | 0 | 0 | 0 | 4 (0.09) |
| I-11m.h433 (clade I) | 0 | 0 | 0 | 2 (0.04) |
| Singletons not present in M-1m | 0 | 2 | 7 | 15 |
| Total singletonsb | 9 [1] | 11 [2] | 8 [1] | 15 [5] |
| Total haplotypesb | 12 [5] | 14 [6] | 10 [5] | 19 [13] |
| Gene diversity (H) and θc | 0.57 [1.35] | 0.63 [1.73] | 0.54 [1.19] | 0.85 [5.76] |
| Bifidobacterium haplotypesa | ||||
| M-11m.h450 | 0 | 8 (0.21) | 0 | 0 |
| M-11m.h552 | 0 | 4 (0.11) | 0 | 0 |
| M-11m.h740 | 0 | 3 (0.08) | 0 | 0 |
| M-11m.h549 | 0 | 2 (0.05) | 0 | 0 |
| M-11m.h637 | 0 | 2 (0.05) | 0 | 0 |
| I-11m.h478 | 0 | 0 | 0 | 13 (0.29) |
| I-11m.h658 | 0 | 0 | 0 | 2 (0.04) |
| I-11m.h475 | 0 | 0 | 0 | 2 (0.04) |
| I-11m.h576 | 0 | 0 | 0 | 2 (0.04) |
| Total singletonsb | 0 | 19 [12] | 0 | 26 [9] |
| Total haplotypesb | 0 | 24 [21] | 0 | 30 [18] |
| Gene diversity (H) and θc | NA | 0.95 [18.00] | NA | 0.92 [11.22] |
NOTE.—NA, not applicable.
Only haplotypes recovered multiple times are shown individually. Clade designations are from figure 2B. Haplotype frequencies are shown in parentheses.
Expected numbers of singletons or total haplotypes in a steady-state population are bracketed.
θ values are bracketed.
FIG. 3.—
Genealogy of Bacteroides haplotypes. (A) Haplotypes forming clade II in figure 2B, including haplotypes isolated from the mother at 1 month after delivery (M-1m; dark blue circles) as well as new haplotypes not present in the M-1m sample but detected in the infant at 1 month (I-1m; red) or the mother at 11 months after delivery (M-11m; light blue). (B) Haplotypes forming clade III in figure 2B, following the same color scheme described above. Haplotypes are numbered with the prefix “h.” Area of circles denotes haplotype frequency in the M-1m, M-11m, and I-1m samples combined; the smallest circles represent singletons and the large central circles the likely ancestors from which other haplotypes derive. Unmodified lines connecting haplotypes indicate single mutational steps, and tick marks on the lines indicate additional steps. Note that most derived haplotypes differ from the ancestor by a single mutational step.
In the three new populations present at 11 months (i.e., the mother and infant Bifidobacterium phylotypes and the new Bacteroides phylotype in the infant), we also detected very large numbers of haplotypes, many of which were singletons derived by one or a few mutations from the most common haplotype (table 2, fig. S1, Supplementary Material online), although in the case of the longer Bifidobacterium contigs, some of these changes might represent sequencing errors because most bases were covered by a single sequencing pass. The large numbers of singletons and total haplotypes present in all the GIT populations analyzed suggest that these populations are generally far from genetic equilibrium. Populations at equilibrium have characteristic haplotype frequency configurations that can be predicted based on the overall level of gene diversity (Ewens 1972; Nei and Li 1976; Maruyama and Fuerst 1984). For each of the Bacteroides and Bifidobacterium populations in our samples, we computed gene diversity indices based on the observed haplotype frequencies and derived the numbers of singletons and total haplotypes that would be expected in populations at equilibrium containing such level of gene diversity. In all cases, the predicted equilibrium frequencies of singletons and of total haplotypes were lower than those observed (table 2). Potential biases in the data introduced by undersampling are not likely to have caused the observed deviations from equilibrium frequencies; Good’s (1953) nonparametric coverage estimator indicates that we likely recovered 70–80% and 40–50% of the haplotypes present in the Bacteroides and Bifidobacterium populations, respectively, and, as missed haplotypes would likely be those present at low frequencies, the numbers of haplotypes and singletons we detected are probably underestimated.
Discussion
The goal of our study was not to generate an exhaustive enumeration of the bacterial species in the human GIT but to compare the main compositional and functional features of the microbiota at different stages of development. Given the ease of obtaining human fecal samples, compared with the alternative invasive procedures to sample the contents of the GIT, we chose to analyze the bacterial composition of fecal material to characterize the gastrointestinal microbiota of mother and infant, as performed in most previous studies in humans (Zoetendal et al. 1998; Gewolb et al. 1999; Favier et al. 2002; Hayashi et al. 2002; Fanaro et al. 2003; Hopkins et al. 2005; Park et al. 2005; Adlerberth et al. 2006; Gill et al. 2006; Ley, Turnbaugh, et al. 2006; Penders et al. 2006; Kurokawa et al. 2007; Palmer et al. 2007; Turnbaugh et al. 2009). Feces, which comprise 40% bacterial cells, are thought to be most representative of the microbiota in the colonic lumen, although, due to normal mucus excretion, epithelial turnover, and peristaltic movements, they are also likely to contain lumen- and mucosa-associated bacteria originating in other areas of the GIT (Moore and Holdeman 1975; Eckburg et al. 2005). Thus, although feces cannot fully represent the complexity of the GIT microbial ecosystem, they probably recover a substantial proportion of the bacterial species in this environment (Eckburg et al. 2005), and their analysis should be adequate to investigate differences between individuals or sampling times (Turnbaugh et al. 2009).
Our community composition and microdiversity analysis based on fecal samples indicate that between 1 and 11 months, the infant GIT microbiota increased substantially in complexity while undergoing a significant turnover of taxa at several phylogenetic levels. In terms of taxonomic composition, we detected Escherichia and Bacteroides as the most abundant organisms in the 1-month-old infant, whereas Bifidobacterium only appeared and grew to dominate the infant GIT microbiota at some later stage between 1 and 11 months. Although Bifidobacterium is thought to dominate the early GIT microbiota in most breast-fed babies (Stark and Lee 1982; Mackie et al. 1999; Favier et al. 2002; Hopkins et al. 2005; Penders et al. 2006), a preponderance of Escherichia and Bacteroides has also been commonly observed, especially in cases where Bifidobacterium is not present (Stark and Lee 1982; Mackie et al. 1999). Remarkably, the mother followed a compositional shift at the genus level between 1 and 11 months that was similar to that of the infant: in both cases, Bifidobacterium appeared and rose to a substantial proportion of the GIT community, whereas Bacteroides decreased importantly (fig. 1C). However, it is important to note that the mother in our study followed a course of prescribed antibiotics a few days before delivery, and the perinatal administration of antibiotics, currently a frequent practice in industrialized countries, likely altered the composition of the maternal microbiota (Spaetgens et al. 2002). In particular, human GIT Bifidobacterium species are known to be highly sensitive to antibiotic treatment (Gewolb et al. 1999; Delgado et al. 2005), which might account for their absence in our study at 1 month after delivery.
Although our sequencing depth was not high and we did not repeat samplings for given time points, it is unlikely that undersampling could explain the large shifts in relative taxa abundances and the observed changes in dominant taxa between samples. Moreover, the general features of the pattern of changes in community complexity in the mother–infant pair concur with those reported in previous analyses based on 16S rDNA or on culture-dependent techniques, with a limited number of taxa in the 1-month-old infant, a large increase during the first year of life (Stark and Lee 1982; Mackie et al. 1999; Favier et al. 2002; Fanaro et al. 2003; Hopkins et al. 2005; Park et al. 2005; Penders et al. 2006; Palmer et al. 2007), and a stable number of taxa in the mother across the same time period (Zoetendal et al. 1998; Ley, Turnbaugh, et al. 2006).
Even though mother and infant incurred similar genus-level compositional shifts between 1 and 11 months (fig. 1C), microdiversity analyses revealed that the populations of Bifidobacterium that had been acquired by each individual by 11 months belonged to different species and that the Bacteroides populations of maternal origin present in the 1-month-old infant had been replaced at this point by a novel one. These results indicate that genera that are shared by mother and infant may have undergone species-level turnover or represent independent acquisitions from external sources, which might include food, other individuals, or other components of the environment rather than vertical inheritance. Two recent analyses compared the overall composition of the GIT microbiota between mothers and infants or adult daughters with varying conclusions regarding the potential role of vertical transmission (Palmer et al. 2007; Turnbaugh et al. 2009). However, these analyses did not report whether the most prevalent genera in their mother–child samples were represented by identical species.
At an even finer diversity scale, our analyses provide the first insights into the population biology of GIT bacteria. In the case of Bacteroides, identities between 16S and fosmid sequences recovered from mother and 1-month infant samples presented clear evidence for an early transmission of the maternal strain. In addition to the recognition of identical sequences, the high similarity between the frequencies of the most abundant haplotypes in the maternal and the I-1m samples confirmed the likelihood of a transmission event (table 2), and further analysis of the frequency distribution of different 16S haplotypes across these samples provided additional information regarding the process by which Bacteroides was transmitted among hosts. The fact that the two most abundant maternal haplotypes, but only one of the low-frequency haplotypes, were identified in the infant suggests that colonization involved a small maternal inoculum, one in which most low-frequency alleles were not represented. Despite this likely bottleneck, the fact that I-1m contained seven new haplotypes, each derived from those inherited from the mother by a single nucleotide change (fig. 3), suggests that colonization of the infant gut by a small inoculum was followed by the quick appearance of novel genetic variants. This process is expected when a population expands rapidly after a brief bottleneck (Maruyama and Fuerst 1984), as would probably occur after the maternal Bacteroides strain entered the GIT of the young infant. Alternatively, the seven singletons found uniquely in I-1m might have been present in the mother at very low frequencies and be strongly selected for after transmission to the infant. Although this scenario is possible, it does not seem likely, as it would imply that the seven singletons were able to outcompete most of the haplotypes detected in both maternal samples, which would presumably also have been transmitted in the maternal inoculum, and without substantially altering the frequencies of the two major haplotypes. Similarly, it does not seem parsimonious that seven haplotypes, each differing from the most abundant maternal haplotype by a single nucleotide, would have been acquired from the environment through an independent inoculation.
In a growing population, new mutants accumulate and produce an excess of low-frequency haplotypes relative to the theoretical expectation for a population that has achieved a steady state; based on an observed level of gene diversity, it is possible to compute the total number of haplotypes and the number of singletons expected in a theoretical equilibrium population (Ewens 1972; Nei and Li 1976; Maruyama and Fuerst 1984). Such calculations confirmed that the Bacteroides population in the 1-month-old infant contained excesses of singletons and of total haplotypes (table 2), as is characteristic of a population expanding after a bottleneck, and supporting the hypothesis of colonization by a small inoculum followed by in situ diversification by mutation in a rapidly growing population. In fact, low-frequency haplotypes were also present in excess in each of the Bifidobacterium and Bacteroides populations analyzed from every sample (table 2). It is not surprising to detect indications of rapid growth in the populations that were novel at 11 months (i.e., Bifidobacterium in the mother, and both Bifidobacterium and the new Bacteroides strain in the infant) because these could have been recently founded. The lack of equilibrium in genetic diversity is more difficult to interpret for the older maternal Bacteroides population that was already present at the 1-month sampling, especially given the likely alterations in population dynamics caused by the administration of antibiotics before delivery. However, other 16S analyses of adult GIT samples have also detected the existence of very shallow fans of diversity for different genera (Eckburg et al. 2005; Ley, Peterson, and Gordon 2006), suggesting that bacterial populations in the gut may commonly exist far from genetic equilibrium and may be due to frequent oscillations in population size or to selection for rare variants. Experimental inoculation and subsequent monitoring of strains in model animals should enable a better understanding of bacterial population dynamics in the GIT.
In regard to coding capabilities, characterization of the COG functional classes present in the infant GIT at 1 and 11 months after birth identified no broad differences between these two stages or between the infant and the mother (D rank analysis; P > 0.05) in the face of remarkable change in phylogenetic composition (fig. 1A and C). Analysis of COG functional classes can distinguish bacterial communities inhabiting different environments (Tringe et al. 2005; Dinsdale et al. 2008). This suggests that, in spite of dietary and other differences, the infant and adult GIT offer largely similar environments for their bacterial inhabitants. In both, genes related to carbohydrate and amino acid transport and metabolism were the most abundant, whereas those involved in lipid transport and metabolism were rare, in accordance with previous metagenomic analyses of the human gut microbiota (Gill et al. 2006; Kurokawa et al. 2007; Turnbaugh et al. 2009). Carbohydrates and peptides, rather than fats, are therefore likely to represent the main sources of nutrients for GIT bacteria in the exclusively breast-fed 1-month-old infant as well as in the 11-month-old infant and the adult.
Furthermore, analyses targeted to functional groupings most likely to sense dietary differences between the two infant stages and the mother also failed to detect significant variation among samples. We hypothesized that 1) COGs encoding proteins able to transport or metabolize the different oligosaccharides that are abundant in human milk (group I) could be most frequent in the exclusively breast-feeding stage (I-1m) than in the adult and at the partially weaned stage when solid foods have already been introduced to the infant diet (I-11m), whereas 2) COGs encoding proteins specialized for the transport and metabolism of plant carbohydrates (group II) could be most frequent in the adult and partially weaned stages. However, the proportions of sequences belonging to these two COG groups were not different between the different samples by D rank analysis, and the number of group I COGs represented was actually higher in the maternal than in the infant samples. Previous analyses also indicated that similar families of glycosyl hydrolases were present in the infant and adult gut microbiota, including enzymes that degrade carbohydrates of plant origin (Kurokawa et al. 2007). This suggests that the global makeup of the unweaned infant microbiota is not particularly adapted to a breast milk diet but rather that this bacterial community exploits the oligosaccharides of breast milk with a repertoire of enzymes similar to that present in the older infant and the mother.
It is necessary to note that although pairwise comparisons of the abundance of different functions and functional groupings between samples did not identify significant differences, a PCA of overall COG category profiles suggested that relative similarity was greatest for the two maternal microbiota samples and the infant sample taken at 11 months to the exclusion of I-1m (fig. 1B). This result reflects the fact that one of the COG categories (W—Extracellular Structures) is uniquely represented in the 1-month infant sample. However, the few W COG category sequences recovered from I-1m are orthologs of E. coli secreted adhesins often associated with intestinal and nonintestinal infections (Restieri et al. 2007), and their presence is not likely to represent a functional adaptation specific to the gut environment of the 1-month-old infant.
In conclusion, our analyses present a dynamic GIT microbiota, capable of accommodating a significant turnover of taxa at different phylogenetic levels while maintaining a remarkable constancy in total functional capabilities. Furthermore, this study has generated the first insight into the bacterial population-level processes that shape the coevolution between the GIT microbiota and the human host. However, 16S rRNA analyses have repeatedly shown that there is a large degree of interindividual variability among infants in the development of the microbiota (Favier et al. 2002; Adlerberth et al. 2006; Penders et al. 2006; Palmer et al. 2007), and, therefore, numerous studies of this sort will be required to evaluate the generality of the trends detected here; large-scale studies of microbiota development in cohorts of infants are now enabled by the increasing facility of high-throughput sequencing. If typical, the dynamic patterns detected in this study would have significant evolutionary and medical implications. On one hand, our analyses suggest a lack of long-term maintenance of maternal phylotypes in the infant, although it is very difficult to rule out a low-level presence of a strain in a given environment, and further in-depth sequencing in other mother–infant pairs will be necessary to address this issue. If confirmed, the phylotype turnover suggested here would limit a tight coadaptation between specific strains and host genotypes and the adaptive diversification of bacterial lineages within host family lines (Ley, Peterson, and Gordon 2006); rather, an intermittent pattern of interactions between different bacterial strains and host genotypes would likely result in a diffuse process of coevolution with little local adaptation and moderate functional differentiation among strains (Futuyma and Slatkin 1983; Thompson 1994). On the other hand, the observed turnovers of genera, species, and strains offer heartening news for medical approaches aimed at modulating the composition of the gut microbiota because they suggest that the phylogenetic composition of this community is malleable and should be responsive to probiotic intervention. Finally, the observation that similar functional repertoires can be attained in the GIT microbiota in the face of substantial differences in phylogenetic composition supports the notion that diseased states of this community may be best identified by atypical distributions of functional gene categories, as recently shown for obesity (Turnbaugh et al. 2009).
Supplementary Material
Supplementary tables S1–S2 and figure S1 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We thank Falk Warnecke for introducing us to the ARB package, Edward Kirton for processing sequencing data through the Genelib pipeline, Kostas Mavrommatis and Ernest Szeto for assistance with IMG, Juan José Abellán for statistical advice, and Becky Nankivell and Shubhangi Kadhe for help in the preparation of the figures. This work was supported by the National Institutes of Health (grant number R01 DK66288 to M.P.F.). This work has been performed under the auspices of the US Department of Energy’s Office of Science, Biological and Environmental Research Program and by the University of California, Lawrence Berkeley National Laboratory under contract number DE-AC03-76SF00098.
References
- Adlerberth I, et al. Reduced enterobacterial and increased staphylococcal colonization of the infantile bowel: an effect of hygienic lifestyle? Pediatr Res. 2006;59:96–101. doi: 10.1203/01.pdr.0000191137.12774.b2. [DOI] [PubMed] [Google Scholar]
- Bateman A, et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhard AE, Field KG. Identification of nonpoint sources of fecal pollution in coastal waters by using host-specific 16S ribosomal DNA genetic markers from fecal anaerobes. Appl Environ Microbiol. 2000;66:1587–1594. doi: 10.1128/aem.66.4.1587-1594.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besemer J, Lomsadze A, Borodovsky M. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001;29:2607–2618. doi: 10.1093/nar/29.12.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Cebra JJ. Influences of microbiota on intestinal immune system development. Am J Clin Nutr. 1999;69:1046S–1051S. doi: 10.1093/ajcn/69.5.1046s. [DOI] [PubMed] [Google Scholar]
- Chou HH, Holmes MH. DNA sequence quality trimming and vector removal. Bioinformatics. 2001;17:1093–1104. doi: 10.1093/bioinformatics/17.12.1093. [DOI] [PubMed] [Google Scholar]
- Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Mol Ecol. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- Cooperstock MS, Zedd AJ. Intestinal flora of infants. In: Hentges DJ, editor. Human intestinal microflora in health and disease. New York: Academic Press; 1983. pp. 79–99. [Google Scholar]
- Delgado S, Florez AB, Mayo B. Antibiotic susceptibility of Lactobacillus and Bifidobacterium species from the human gastrointestinal tract. Curr Microbiol. 2005;50:202–207. doi: 10.1007/s00284-004-4431-3. [DOI] [PubMed] [Google Scholar]
- DeSantis TZ, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinsdale EA, et al. Functional metagenomic profiling of nine biomes. Nature. 2008;452:629–632. doi: 10.1038/nature06810. [DOI] [PubMed] [Google Scholar]
- Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewens WJ. The sampling theory of selectively neutral alleles. Theor Popul Biol. 1972;3:87–112. doi: 10.1016/0040-5809(72)90035-4. [DOI] [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Fanaro S, Chierici R, Guerrini P, Vigi V. Intestinal microflora in early infancy: composition and development. Acta Paediatr Suppl. 2003;91:48–55. doi: 10.1111/j.1651-2227.2003.tb00646.x. [DOI] [PubMed] [Google Scholar]
- Favier CF, Vaughan EE, De Vos WM, Akkermans AD. Molecular monitoring of succession of bacterial communities in human neonates. Appl Environ Microbiol. 2002;68:219–226. doi: 10.1128/AEM.68.1.219-226.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futuyma DJ, Slatkin M. Coevolution. Sunderland (MA): Sinauer Associates; 1983. [Google Scholar]
- Gewolb IH, Schwalbe RS, Taciak VL, Harrison TS, Panigrahi P. Stool microflora in extremely low birthweight infants. Arch Dis Child Fetal Neonatal Ed. 1999;80:F167–F173. doi: 10.1136/fn.80.3.f167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SR, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good IL. The population frequencies of species and the estimation of population parameters. Biometrika. 1953;40:237–264. [Google Scholar]
- Hayashi H, Sakamoto M, Benno Y. Fecal microbial diversity in a strict vegetarian as determined by molecular analysis and cultivation. Microbiol Immunol. 2002;46:819–831. doi: 10.1111/j.1348-0421.2002.tb02769.x. [DOI] [PubMed] [Google Scholar]
- Hooper LV, et al. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 2001;291:881–884. doi: 10.1126/science.291.5505.881. [DOI] [PubMed] [Google Scholar]
- Hopkins MJ, Macfarlane GT, Furrie E, Fite A, Macfarlane S. Characterisation of intestinal bacteria in infant stools using real-time PCR and northern hybridisation analyses. FEMS Microbiol Ecol. 2005;54:77–85. doi: 10.1016/j.femsec.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Ihaka R, Gentleman R. R: a language for data analysis and graphics. J Comput Graph Stat. 1996;5:299–314. [Google Scholar]
- Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann P, Pfefferkorn A, Teuber M, Meile L. Identification and quantification of Bifidobacterium species isolated from food with genus-specific 16S rRNA-targeted probes by colony hybridization and PCR. Appl Environ Microbiol. 1997;63:1268–1273. doi: 10.1128/aem.63.4.1268-1273.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa K, et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007;14:169–181. doi: 10.1093/dnares/dsm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- Ludwig W, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackie RI, Sghir A, Gaskins HR. Developmental microbial ecology of the neonatal gastrointestinal tract. Am J Clin Nutr. 1999;69:1035S–1045S. doi: 10.1093/ajcn/69.5.1035s. [DOI] [PubMed] [Google Scholar]
- Markowitz VM, et al. IMG/M: a data management and analysis system for metagenomes. Nucleic Acids Res. 2008;36:D534–D538. doi: 10.1093/nar/gkm869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T, Fuerst PA. Population bottlenecks and nonequilibrium models in population genetics. I. Allele numbers when populations evolve from zero variability. Genetics. 1984;108:745–763. doi: 10.1093/genetics/108.3.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore WE, Holdeman LV. Discussion of current bacteriological investigations of the relationships between intestinal flora, diet, and colon cancer. Cancer Res. 1975;35:3418–3420. [Google Scholar]
- Nei M. Molecular evolutionary genetics. New York: Columbia University Press; 1987. [Google Scholar]
- Nei M, Li WH. The transient distribution of allele frequencies under mutation pressure. Genet Res. 1976;28:205–214. doi: 10.1017/s0016672300016906. [DOI] [PubMed] [Google Scholar]
- O’Mahony C, et al. Commensal-induced regulatory T cells mediate protection against pathogen-stimulated NF-kB activation. PLoS Pathog. 2008;4:e1000112. doi: 10.1371/journal.ppat.1000112. doi:1000110.1001371/journal.ppat.1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HK, et al. Molecular analysis of colonized bacteria in a human newborn infant gut. J Microbiol. 2005;43:345–353. [PubMed] [Google Scholar]
- Penders J, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118:511–521. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. Selecting the best-fit model of nucleotide substitution. Syst Biol. 2001;50:580–601. [PubMed] [Google Scholar]
- Restieri C, Garriss G, Locas MC, Dozois CM. Autotransporter-encoding sequences are phylogenetically distributed among Escherichia coli clinical isolates and reference strains. Appl Environ Microbiol. 2007;73:1553–1562. doi: 10.1128/AEM.01542-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- Schumann A, et al. Neonatal antibiotic treatment alters gastrointestinal tract development gene expression and intestinal barrier transcriptome. Physiol Genomics. 2005;23:235–245. doi: 10.1152/physiolgenomics.00057.2005. [DOI] [PubMed] [Google Scholar]
- Sela DA, et al. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc Natl Acad Sci U S A. 2008;105:18964–18969. doi: 10.1073/pnas.0809584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selengut JD, et al. TIGRFAMs and Genome Properties: tools for the assignment of molecular function and biological process in prokaryotic genomes. Nucleic Acids Res. 2007;35:D260–D264. doi: 10.1093/nar/gkl1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaetgens R, et al. Perinatal antibiotic usage and changes in colonization and resistance rates of group B streptococcus and other pathogens. Obstet Gynecol. 2002;100:525–533. doi: 10.1016/s0029-7844(02)02068-9. [DOI] [PubMed] [Google Scholar]
- Stark PL, Lee A. The microbial ecology of the large bowel of breast-fed and formula-fed infants during the first year of life. J Med Microbiol. 1982;15:189–203. doi: 10.1099/00222615-15-2-189. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- Templeton AR, Crandall KA, Sing CF. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics. 1992;132:619–633. doi: 10.1093/genetics/132.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JN. The coevolutionary process. Chicago (IL): University of Chicago Press; 1994. [Google Scholar]
- Tringe SG, et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoetendal EG, Akkermans AD, De Vos WM. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl Environ Microbiol. 1998;64:3854–3859. doi: 10.1128/aem.64.10.3854-3859.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwickl DJ. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. Austin (TX): The University of Texas at Austin; 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.