Abstract
Despite striking differences in cognition and behavior between humans and our closest primate relatives, several studies have found little evidence for adaptive change in protein-coding regions of genes expressed primarily in the brain. Instead, changes in gene expression may underlie many cognitive and behavioral differences. Here, we used digital gene expression: tag profiling (here called Tag-Seq, also called DGE:tag profiling) to assess changes in global transcript abundance in the frontal cortex of the brains of 3 humans, 3 chimpanzees, and 3 rhesus macaques. A substantial fraction of transcripts we identified as differentially transcribed among species were not assayed in previous studies based on microarrays. Differentially expressed tags within coding regions are enriched for gene functions involved in synaptic transmission, transport, oxidative phosphorylation, and lipid metabolism. Importantly, because Tag-Seq technology provides strand-specific information about all polyadenlyated transcripts, we were able to assay expression in noncoding intragenic regions, including both sense and antisense noncoding transcripts (relative to nearby genes). We find that many noncoding transcripts are conserved in both location and expression level between species, suggesting a possible functional role. Lastly, we examined the overlap between differential gene expression and signatures of positive selection within putative promoter regions, a sign that these differences represent adaptations during human evolution. Comparative approaches may provide important insights into genes responsible for differences in cognitive functions between humans and nonhuman primates, as well as highlighting new candidate genes for studies investigating neurological disorders.
Keywords: gene expression, transcriptional evolution, Tag-Seq, noncoding RNA
Introduction
Some of the most striking differences between humans and our closest relatives are related to changes in the brain. During human evolution, alterations in cranial morphology and neural patterning and function (Carroll 2003; Thompson et al. 2003; Jobling et al. 2004) have allowed for large alterations in cognitive phenotypes and human social behaviors relative to other primates (Tomasello and Call 1997). Due to the paucity of functional differences in protein-coding regions of the genome between humans and chimpanzee (Chimpanzee Sequencing and Analysis, Chimpanzee Sequencing and Analysis Consortium 2005), it has been hypothesized that many of these phenotypic changes may have been driven by changes in transcriptional regulation rather than protein function per se.
Previous studies investigating large-scale changes in gene expression in primates in multiple tissues employed microarray technologies (Caceres et al. 2003; Gu J and Gu X 2003; Karaman et al. 2003; Khaitovich et al. 2004, 2005, 2006a; Uddin et al. 2004; Gilad et al. 2005, 2006; Blekhman et al. 2008; Somel et al. 2009). A subset of these studies identified numerous transcripts that are differentially expressed between chimpanzee and human neocortex (Enard et al. 2002; Caceres et al. 2003; Khaitovich et al. 2004, 2005; Uddin et al. 2004; Somel et al. 2009). These studies showed that there are many differences in protein-coding expression in the cortex between humans and other primate species and that many of the changes may be related to neural function and metabolism, suggesting that changes in transcriptional regulation have, indeed, played an important role in the evolution of human neural phenotypes. The advent of high-throughput sequencing technologies provides substantial improvements in our ability to assay the abundance of protein-coding transcripts in comparative studies. Additionally, we can now explore genome-wide strand-specific changes in expression of previously uncharacterized noncoding RNAs (ncRNAs) conserved between primate species, something not possible with the previously used microarray-based platforms.
Gene expression assays using deep sequencing may improve the resolution of those differences, include new protein-coding regions, and assist in answering outstanding questions about the evolution of noncoding transcripts. Technologies such as Tag-Seq offer several advantages including the ability to accurately measure a larger dynamic range of transcript abundances, genome-wide coverage, avoiding probe hybridization effects due to inter- and intraspecific sequence variation, and the ability to assay polyadenylated RNA transcripts not previously characterized (Lister et al. 2008; Marioni et al. 2008; Mortazavi et al. 2008; t Hoen et al. 2008; Morrissy et al. 2009). Digital gene expression: tag profiling (here called Tag-Seq, also called DGE:tag profiling), specifically, uses a restriction enzyme (NlaIII) with a common recognition site (CATG) to create “tags” from all polyadenylated transcripts, which are sequenced in an known orientation relative to the poly-A tail giving strand-specific information (for details, see Materials and Methods).
The ability to assay polyadenylated RNA transcripts in an unbiased manner, from multiple functional categories, offers an especially exciting insight into new mechanisms by which changes in transcription may influence organismal phenotypes. Recently, a number of studies have documented transcription throughout most of the genome, with different classes of ncRNA having different levels of conservation between human and mouse (Pang et al. 2006). For example, microRNAs (miRNA) and small nucleolar RNAs (snoRNAs) are well conserved between human and mouse, although this may be due to how they are defined (Ambros et al. 2003). In contrast, many longer ncRNAs are not well conserved at the sequence level. These ncRNA show an average of <70% identity between human and murine sequence, comparable with the conservation seen within introns (Pang et al. 2006). It is necessary to examine species more recently diverged with human than mouse to get a clear understanding of the tempo of change in the sequence of these ncRNA molecules over evolutionary time, and which of those ncRNAs are unique to humans. Even when the amount of sequence conservation is known, very little is known about conservation in expression levels between species. Understanding the extent to which expression levels are conserved will give us even more insight into functional constraints.
Here, we describe analyses of differential transcript expression between primate cortexes using Tag-Seq, a sequencing-based assay of expression (t Hoen et al. 2008; Morrissy et al. 2009). We were able to quantify differences for 12,990 genes for which orthology could be assigned in all three primate species and were expressed in at least one individual in all three species in our samples. We were also able to analyze expressed tags that map outside of annotated coding regions and were expressed in all three species. Analyses of these data support three basic findings. First, a number of the noncoding tags are conserved among all three species in sequence and genomic position (by synteny), and a subset of these are also conserved in expression level. These patterns of conservation suggest a functional role for a specific subset of the ncRNAs. Second, ∼15% of genes are differentially regulated among human and chimpanzee frontal cortex, and enrichments of functional categories for the protein-coding transcripts reveal that many differentially regulated genes are related to neuronal signaling and energy metabolism, especially aerobic energy metabolism. Third, a subset of coding transcripts come from genes showing both significant differences in expression and a signature of positive selection on adjacent, putatively regulatory, regions. This overlap provides a way to identify candidate mutations responsible for gene expression differences between species and to enlarge the set of candidate genes containing mutations that underlie the origin of uniquely human cognitive traits.
Materials and Methods
Sample Preparation and Sequencing
The frontal cortex samples used in this study were from 3 humans, 3 chimpanzees, and 3 macaques individuals. All samples were obtained through opportunistic sampling; thus, no primates were sacrificed for the purposes of this research. Human samples were obtained from BioChain. The nonhuman primate samples are from the Southwest Foundation for Biomedical Research and the New England Primate Center (supplementary table 6, Supplementary Material online). Postmortem tissue samples were collected within 12 h of time of death. All samples are frontal cortex from adult males (except one female macaque). Total RNA was isolated with an RNeasy kit (Qiagen) including a DNaseI treatment step, and the quality of the total RNA verified by Experion (BioRad) analysis. Only total RNA samples with high quality 18S and 28S ribosomal bands with no obvious contamination and good 28:18S rRNA ratios were used.
One to two micrograms of total RNA were used as starting material for the creation of the Tag-Seq mRNA library. Library construction was performed with the Tag-Seq profiling for NlaIII Sample Prep Kit (Illumina). Briefly, Tag-Seq isolation involves binding polyadenylated mRNA to beads, which is then made into double-stranded cDNA. The cDNA is then digested with the NlaIII restriction enzyme and a 5′ adapter primer is added. A second digest with MmeI cleaves the tag sequences from the beads, and adapters are then added to the 3′ end of the tag. Cluster generation and tag sequencing were performed in the Duke Institute for Genome Sciences & Policy Sequencing Core Facility. Approximately 32 million tag sequences 18 bases in length were generated on a single-flow cell. Each individual sample was run in one lane of a flow cell on an Illumina GA2. Verified data will be deposited into gene expression omnibus, and the fastq sequence files into the short read archive.
Sequence Quality Filters and Orthology Assignment
Because the 5′ of each tag corresponds to a cut site for NlaIII, the corresponding 4 bp site (CATG) was appended to the beginning of each tag. Tags were then aligned to the species-appropriate University of California, Santa Cruz (UCSC) genome assembly: build36 (hg18) for human, build 2 version 1(panTro2) for chimpanzee, and Mmul_051212 (rheMac2) for macaque (Karolchik et al. 2003). We employed the maq sequence alignment software (Li et al. 2008) to align sequences and to filter low-quality sequence and sequences with adapter contamination. For sequences aligning to multiple locations, maq randomly assigns tags to one of those locations. We removed tags that align to more than four positions to reduce artifacts but to allow for annotations of genes in recent segmental duplications, although we could not reliably assign a signal to a specific duplicated sequence. We also discarded alignments to certain regions of the human genome for which the sequence is underrepresented in the sequence assembly as compared with the actual genome. These primarily consist of satellite sequences and rRNA genes. Finally, of these usable alignments, we also mapped those that are unique to only one location in the reference genome. All tags sequenced from multiple sites within the same RefSeq defined mRNA (Pruitt et al. 2007) on the sense strand (relative to the direction of transcription) were added together to create a cumulative count for each transcript.
Orthologous protein-coding regions between all three species were defined by using alignments of human RefSeq mRNAs to the macaque and chimpanzee genomes in the UCSC Genome Browser. RefSeq RNAs were aligned against the chimpanzee genome using blat; those with an alignment of less than 15% were discarded. When a single RNA aligned in multiple places, the alignment having the highest base identity was identified. Only alignments having a base identity level within 0.1% of the best and at least 96% base identity with the genomic sequence were kept. We used this approach because there are very few annotated transcripts for the macaque genome and we wanted a standard filter for all three species. Coordinates for genes in each species were extracted from the UCSC Genome Browser. We discarded genes for which good orthology assignment in the three species does not exist, that is, if the corresponding RefSeq mRNA did not align to a nonrandom portion (must be part of the primary assembled sequence) of each of the three genomes. Coordinates of predicted ncRNAs were downloaded from the UCSC Genome Browser RNAGene track (http://genome.ucsc.edu/). We utilized this database as it uses the same build and coordinates for the human genome as our other analyses. The ncRNAs listed in this database are a combination of experimentally tested and computationally predicted ncRNAs. We removed predicted pseudogenes and transcripts that are not transcribed by PolII (e.g., rRNAs and tRNAs that would not have been sequenced based on our method of library preparation).
Normalization of Tag Counts and Assessment of Significant Expression Differences
To normalize for variation in the distribution of tags between libraries as well as to compensate for variable numbers of tags generated for each sample, we employed the program edgeR (http://bioconductor.org/packages/2.5/bioc/html/edgeR.html; Robinson and Smyth 2007, 2008). This program is designed to test differences in SAGE and Tag-Seq data, specifically, when there are small numbers of replicates or individuals being tested and when transcripts with low-expression levels are present. To normalize counts between libraries, first, the data are fit to a negative binomial distribution and then a quantile-adjusted conditional maximum likelihood estimation is employed to moderate overdispersion. Significance values for differences in expression levels were determined using a modified exact test, similar to Fisher’s exact test. P values were adjusted using a false discovery rate (FDR) = 5% (Storey and Tibshirani 2003; supplementary table 7, Supplementary Material online). It is important to note that Tag-Seq data appear to be very robust to the method of normalization. Even the most basic normalization method, dividing by the total number of reads in that library, has an extremely high correlation with the method described above (Spearman correlation of R2 = 0.9994).
Correlations and Microarray Comparison
Correlations within and between species were performed on data sets of all sequenced tags for a given individual. In order to compare our data with other platforms, we also examined expression using a microarray platform. RNA was isolated from AG16409 human fibroblast cells from the Coriell Institute (Camden, New Jersey) using an RNeasy kit (Qiagen). This same RNA preparation was used for a Tag-Seq library and sequenced on the Illumina GA2 (as described above), as well as for an Affymetrix Human Genome U133 Plus 2.0 microarray, which was run at the Duke Microarray Facility. The raw image was visually inspected for overall quality of the array. The array was normalized using the MAS5.0 algorithm implemented in the affy R package (Gautier et al. 2004) available from www.bioconductor.org. Only unique probe sets (_at probe sets) were considered and when a Unigene identifier was mapped to multiple probe sets, the average of their signal intensities was assigned to this Unigene entry (Pedotti et al. 2008). We uniquely mapped this expression subset to the Tag-Seq expression data by matching gene symbols. Entries with ambiguous mapping were removed from the final data set. In total, 10,381 gene IDs were present in both the Affymetrix array and the Tag-Seq data set. A Spearman correlation was calculated to assess the correlation between the two platforms.
Assessing Branch-Specific Changes
We analyzed possible human and chimp branch-specific expression changes utilizing the expression data from macaques. Significant differences in mean species expression (P < 0.05) were determined by pairwise comparisons between all three species using edgeR (Robinson and Smyth 2007, 2008) as described above. To assess the direction of these changes, we first looked at the subset of genes where the mean macaque expression is intermediate to the human and chimpanzee values (μH > μM > μC or μH < μM < μC). Cases in which the macaque expression values are intermediate to the human and chimpanzee may be more consistent with a scenario in which the macaque represents an ancestral expression level (Blekhman et al. 2008). Alternatively, we also used a pattern-matching approach to assess whether one species’ expression was different from the other two, rather than assuming that the macaque level of expression provides an estimate of the ancestral expression profile. Genes were determined to be significantly differentially expressed using a modified Fisher’s exact test with the FDR set at 5% (Robinson and Smyth 2007, 2008). Genes significantly differentially expressed (P < 0.05) in humans relative to both chimp and macaque were labeled dH and likewise dC when chimp was differentially expressed. We also examined instances where all three species had significantly different levels of expression (P < 0.05) for all pairwise comparisons.
Categorical Enrichment
To determine functional category enrichment for the differentially expressed genes, we employed the PANTHER (HMM Library Version 6.0; Mi et al. 2005) and GO (The Gene Ontology Consortium, 2000) gene ontology databases. Our background set of genes were those genes measured in our tissue samples. PANTHER and GO category enrichment scores were computed using the top 5% of the hypergeometric probability distribution. Python code used to perform all enrichments is available at: http://www.duke.edu/∼ofedrigo/Olivier_Fedrigo/PythonScripts.html.
Permutation Tests on Correlations of Intergenic Tag and Downstream Gene Expression
Significance of correlations between expression of intergenic tags and an adjacent gene was determined by performing random permutation tests on human only, and human–chimpanzee conserved tag location between these two species. Tests were done for the following genomic compartments: 5′ flanking sense strand, 5′ flanking antisense strand, 3′ flanking sense strand, and 3′ flanking antisense strand. Initially, intergenic compartments were defined as the 5 MB flanking protein-coding regions. Further analyses were performed for tags transcribed within a 1 kb, 5 kb, 10 kb, or 20 kb window upstream of the nearest protein-coding gene for both of the 5′ flanking compartments or downstream of the nearest gene for both of the 3′ flanking compartments. Single-tail permutation tests were done as we had specific hypotheses about the sign of the correlation based on previously published data for intergenic sense (Khaitovich et al. 2006a) and antisense (Kapranov et al. 2007a; Mazo et al. 2007; He et al. 2008) correlations with expression of nearby genes.
Scores for Positive Selection
P values for signatures of positive selection in regulatory regions were taken from Haygood et al. (2007), in which 5 kb 5′ upstream of genes were assayed, and (Pollard et al. 2006a) in which human accelerated regions were analyzed. For the Pollard data set, we assigned noncoding regions to the closest gene (Haygood R. et al. submitted. Strong contrasts between adaptive coding and noncoding changes during human evolution. Proc Natl Acad Sci U S A.) using the UCSC known genes list (Hsu et al. 2006). When several regions were assigned to one gene, we combined their P values using the Simes’ method (Simes 1986). We uniquely mapped each of the positive selection studies to the Tag-Seq data set using their gene symbols and RefSeqs. Ambiguous matches were discarded from the final data set. Spearman rank correlations were performed for adjusted P values for the Tag-Seq data and the P values from Pollard et al. (2006a) and Haygood et al. (2007).
Results
Tag-Seq Expression Measurements in Frontal Cortex from Three Primate Species
Tag-Seq libraries were constructed for frontal cortex tissue from 3 humans, 3 chimpanzees, and 3 macaques individuals. Of the 6 to 8 million tags generated for each of the libraries, ∼70–73% of the tags mapped uniquely to the species-specific genome (supplementary table 1, Supplementary Material online). For each individual, approximately 3 million tags (ca. 60% of total mapped tags) originated from the sense strand of protein-coding transcriptional regions, as defined by human RefSeq mRNAs (Pruitt et al. 2007) and aligned to each species’ genome assembly. The remaining tags are located in intronic or intergenic regions. From the pool of tags that mapped to the sense strand of RefSeq transcripts, we were able to analyze the expression levels for 12,990 genes expressed in the frontal cortex of all three species. This number represents a significant increase over previous microarray studies that were limited to transcripts conserved, at the sequence level, between species (e.g., Somel et al. [2009] measured expression in 7,958 genes between human and chimpanzee and 3,075 in all three species).
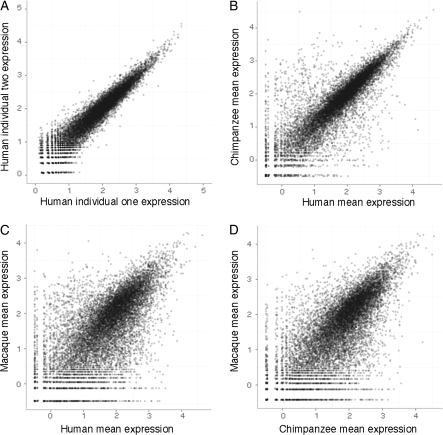
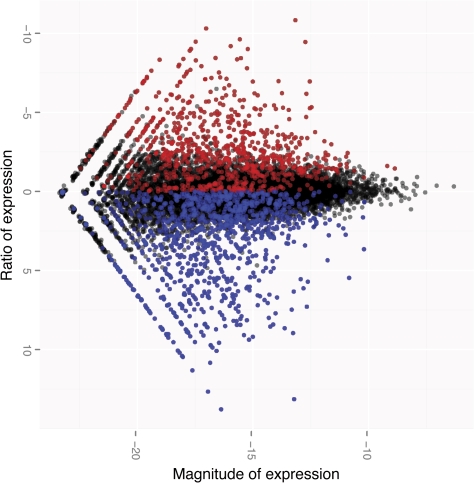
In order to first assess the consistency of gene expression measurements using this platform, we checked correlations between normalized Tag-Seq libraries within species (see Materials and Methods). The correlations are very strong between both a technical replicate (Spearman R2 = 0.96) of the same biological sample as well as between individuals within the same species (R2 = 0.92; fig. 1). We then compared expression profiles across species: as expected the correlations are reduced over increased evolutionary distances between species (e.g., the human–chimpanzee correlation [R2 = 0.75] is higher than that between human–macaque [R2 = 0.47] and chimpanzee–macaque [R2 = 0.53]). To understand how Tag-Seq compares with microarray assays, we compared expression data generated from the same human sample assayed by both Tag-Seq and by an Affymetrix Human Genome U133 Plus 2.0 microarray (supplementary fig. 1, Supplementary Material online). The correlations show a similar pattern (with a Spearman correlation of 0.54) to previous reports comparing results of microarrays and other DGE data (t Hoen et al. 2008; Morrissy et al. 2009) or RNA-Seq data (Wang et al. 2009), where genes at intermediate expression levels are more correlated between platforms than very high- or low-expressed genes. In addition, the Tag-Seq data show an approximately equal amount of higher expressing genes in humans versus the nonhuman primate levels of expression (fig. 2), in contrast to previous reports (Enard et al. 2002; Caceres et al. 2003; Gu J and Gu X 2003).
FIG. 1.—
Correlations in gene expression between Tag-Seq libraries. Each data point is slightly transparent in order to assist in visualizing the density of data points. (A). Biological replicates between two human individuals. (B). Human–chimpanzee comparison. (C). Human–macaque comparison. (D) Chimpanzee–macaque comparison. Spearman correlation R2 values are 0.92, 0.75, 0.47, and 0.53, respectively.
FIG. 2.—
MA plot of the human and chimpanzee brain expression data. The magnitude (x axis) and ratio (y axis) of expression are plotted here for all genes measured. Genes with higher expression levels in humans are the negative ratios here (left of graph). Each data point is slightly transparent in order to assist in visualizing the density of data points. Significantly differentially expressed genes (P < 0.05) are colored by higher expression in humans (red) or chimpanzees (blue).
Patterns of Tag Conservation Outside of Coding Regions
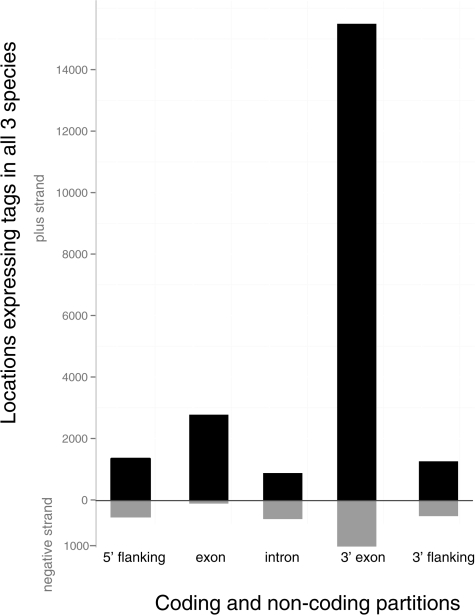
Conservation of noncoding regulatory regions has been important in locating noncoding functional regulatory elements (Ahituv et al. 2005; Siepel et al. 2005; Pennacchio et al. 2006; Visel et al. 2008), and so we took a similar comparative approach to look for conserved genomic partitions across these three species. In this study, we assayed only polyadenlyated RNAs expressed in these species, however, polyA-RNAs may also contain a large amount of nonannotated functional transcripts. We examined the distribution of tags both inside and outside of RefSeq transcript regions. For each individual assayed, approximately 2 million mapped tags (ca. 40% of total per sample) are located outside of traditional coding transcripts; defined as not within exons or the untranslated regions or not on the sense strand relative to that gene’s transcription (fig. 3). It is important to note here that the Tag-Seq library preparation is a 3′ biased method, where tags usually come from the location of the 3′-most restriction enzyme cutsite in a transcript (for details, see Materials and Methods). To mask out very rare tags, we filtered tags to include only those observed five or more times in each of the three species. In order to explore the conservation of both protein-coding and noncoding transcripts further, we partitioned all uniquely mapped tags, conserved between all three species, into five groups: 5′ flanking to a coding region, non 3′ exonic, intronic, 3′ exonic, and 3′ flanking, based on their relationship to known transcripts. Because of the strand-specific nature of the Tag-Seq assay, the tags in each of these five groups were further characterized as being transcribed in the sense or antisense direction relative to the direction of transcription of the nearest gene. As expected, the large majority of tag locations conserved between all three species are located on the sense strand in the 3′-most exon of coding transcripts (fig. 3). The second largest partition contains sense-strand tags from other exons. These tags come from transcripts where the 3′-most exons do not contain an NlaIII cutsite or from tags arising from other NlaIII cutsites within the transcript. Nonexonic partitions have fewer locations where tags are being transcribed, with the fewest in the 5′ exons of the gene, on the antisense strand (fig. 3). There are also a number of tags that show conservation in the noncoding compartments of the genome, and so we explored those regions further.
FIG. 3.—
Distribution of tag locations across genomic compartments. Histogram of the number of locations (not the number of tags sequenced from each location) that have conserved locations across an individual of all three species. Tag locations were considered conserved if all species had five or more tags sequenced from the same exact location. Note that a given transcript may have more than one tag site sequenced from it. Compartments where the tags were sequenced on the sense strand (relative to the direction of transcription for each gene) are in black. Antisense transcriptional conserved regions are shown in gray.
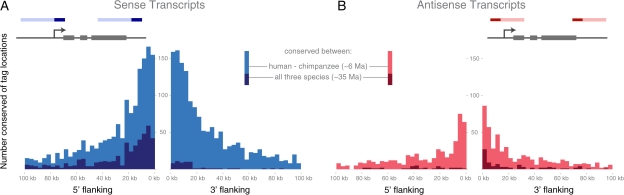
For all the intergenic compartments, there is a highly significant enrichment of tags nearby protein-coding regions (Wilcox test, P << 0.0001). Outside of the sense strand coding partitions between humans and chimpanzees, the 5′ and 3′ flanking regions on the sense strand show the most conservation (here defined as the same sequence in a syntenic location between species; fig. 4A and B). The 5′ flanking sense tags are the most conserved between all three species (fig. 4A). Looking at tags within 5 Mb of protein-coding transcripts between just human and chimpanzee, the distribution of conserved tag locations on the sense strand drops off ∼20 kb from the 5′ and 3′ ends of protein-coding regions, with the median of the distribution occurring within 40 or 60 kb of coding regions, respectively (fig. 4A). In contrast, the number of conserved tag sites is lower on the antisense strand, and loss of conservation distally occurs much more quickly, with most tags coming from locations within 5–10 kb of a protein-coding region. This pattern shows that although conserved tags are mostly located near to protein-coding regions, some are distributed up to tens of kilobases away from genes. The rapid loss of conservation in some compartments (most notably in the 3′ flanking, sense strand) going out to macaque is striking and illustrates the importance of examining species that are related more closely than mouse is to human.
FIG. 4.—
The distribution of conserved intragenic tags located near genes. The x axis shows the distance from a RefSeq region either upstream (left) or downstream (right) of a gene. Each count on the y axis is the number of locations to which five or more tags map and are conserved in sequence between the following two or three species. (A). Sense tags conserved between humans, chimpanzees, and macaques. (B). Antisense tags conserved between humans, chimpanzees, and macaques. The panels are colored based on conserved transcription. For tags sequenced from the sense strand conservation is indicated between humans and chimpanzees (blue) or between humans, chimpanzees, and macaques (dark blue). For tags from the antisense strand, relative to the proximal gene’s direction of transcription, conservation is labeled between humans and chimpanzees (red) or between humans, chimpanzees, and macaques (dark red). The schematic in the upper corners of each histogram are to illustrate where the tags are coming from relative to the coding regions (gray boxes, the grey arrow indicates the transcriptional start site). The sense (dark blue) or antisense (dark red) tags are coming from RNA transcripts of unknown length (here illustrated in blue [sense] or red [antisense]).
Where we found tags to be conserved in location between species, we also examined the correlation between tag expression levels between humans and chimpanzee or between all three species from these partitions (supplementary table 2, Supplementary Material online). Between humans and chimpanzees, the highest correlations were seen in the 3′ exonic partitions as well as the category of other exons (r = 0.86–0.87). The noncoding tags, however, also show high degrees of correlation (r = 0.67–0.84), with the lowest correlations between expression coming from 5′ antisense exon and intron sense-strand tags. This indicates that the expression of noncoding tags is evolving slightly faster than the expression of tags within exons and that there is some variation in that rate between compartments. The comparisons between all three species show that even though fewer tags are conserved over this evolutionary distance, when they are conserved in sequence, the expression levels are conserved as well.
In order to examine any functional annotations of the noncoding transcripts, we also examined whether any of our intergenic tags fall within the genomic coordinates of annotated human ncRNAs according the UCSC Genome Browser. After removing transcripts that are not polyadenylated or that were had low prediction values, we were able to examine 559 predicted ncRNAs. By overlapping our tag coordinates with these coordinates, we found 24 tags expressed in our samples that correspond to annotated ncRNAs. These are comprised predominately of snoRNAs, with some RNaseP, RNAs, and miRNAs. This distribution of functional types is similar to that seen in the UCSC database. Of these 24 ncRNAs, 11 are expressed only in our human brain samples, 10 are expressed in only the human and chimpanzee samples, 1 in the humans and macaques, and 2 are expressed in brain tissue in all 3 species. Therefore, we do see a similar pattern of conservation to that seen with the other noncoding tags in our study. We expect that as additional ncRNAs are annotated as a result of ENCODE and other projects, many more of the transcript tags will be found to correspond to functional ncRNAs.
It well established that some intergenic RNAs are capable of regulating the expression of nearby genes (Lapidot and Pilpel 2006; Kapranov et al. 2007a; Mazo et al. 2007; Prasanth and Spector 2007). Therefore, we also examined the correlations between conserved (tag locations conserved between human and chimpanzee) intergenic tag expression and the expression of the nearest protein-coding gene for the following genomic compartments using the data from humans alone: 5′ flanking sense strand, 5′ flanking antisense strand, 3′ flanking sense strand, and 3′ flanking antisense strand. No correlation was found when looking at a large window of 5 Mb flanking the protein-coding region. However, an analysis of smaller size windows of 1 kb, 5 kb, 10 kb, and 20 kb flanking the nearest downstream (for both of the 5′ flanking compartments) or upstream (for both of the 3′ flanking compartments) gene revealed weak but significant correlations in two compartments. First, there is a positive correlation between tag expression in the 5′ flanking sense-strand tags located <1 kb upstream of a protein-coding region and the protein-coding transcript downstream (Spearman correlation = 0.285, n = 22, P = 0.044, one-tailed permutation test). Second, the 5′ flanking antisense transcripts compartment revealed a significant negative correlation for tags <10 kb upstream of the gene (Spearman r = −0.193, n = 119, P = 0.028, one-tailed permutation test), where higher expression of 5′ flanking antisense transcripts correlates with reduced expression of the downstream gene. Although these correlations do not survive a strict correction for multiple testing, the patterns are suggestive and consistent with results of previous studies (Khaitovich et al. 2006a; Kapranov et al. 2007a; Mazo et al. 2007; He et al. 2008).
Patterns and Enrichments of Tag Expression in Protein-Coding Regions
In genic regions, we found 1,872 genes differentially expressed between human and chimpanzee frontal cortex (modified Fisher’s exact test, corrected for a FDR of 5%; for details, see Materials and Methods). To get a better understanding of higher order patterns of expression differences, we performed categorical enrichment analyses using both the GO (The Gene Ontology Consortium 2000) and PANTHER (Mi et al. 2005) ontology databases. The enrichments were performed using the largest absolute differences in expression (as opposed to looking only at upregulated changes) between the mean human and mean chimpanzee expression levels (table 1 and supplementary table 3, Supplementary Material online). A few clear patterns emerge from these enrichment analyses. Many of the biological process categories that show large differences in expression concern synaptic transmission and transport within the cell, as might be expected based on tissue being analyzed. More interestingly, there are also a number of categories related to aerobic energy metabolism and the nuclear-encoded genes that function inside of the mitochondrial electron transport chain. The third group of categories are involved in cellular repair and apoptosis. Lastly, categories involved in lipid metabolic processes appear multiple times in enrichments using GO.
Table 1.
Categorical Enrichments for Differentially Expressed Genes between the Human and Chimpanzee Individuals
 |
NOTE.—The results for the biological process domain of both the GO and PANTHER ontologies are shown. Categorical enrichments are for the top 5% of a hypergeometric probability distribution. The right-hand columns show the number of genes in the top 5%, as well as the total number of genes evaluated. Categories that evaluated less than 10 genes total are not shown. Categories are further colored according to hierarchically related ontology terms: nucleic acid metabolism (green), electron transport (yellow), neuronal activity (blue), transport, extra- and intracellular protein traffic (pink), and lipid metabolism (purple).
Next, we explored the polarity of changes underlying differences in expression between human and chimpanzee. Investigating the polarity of expression differences between human and chimpanzees requires information from an outgroup species; therefore, in order to examine lineage-specific changes in expression, we compared expression between human, chimpanzees, and macaques. Our first approach was to look at the subset of genes where the mean macaque level of expression is intermediate to the mean human and chimpanzee levels of expression. In this case, there are 1,001 genes where the human is highest and 1,371 where the human is lowest; a trend also seen in other tissues in a previous studies (Gilad et al. 2006; Blekhman et al. 2008). The second approach was to look for genes where the human level of expression was significantly different from both the chimpanzee and macaque, but the chimpanzee and macaque were not significantly different from each other. Using this criterion, we found that 309 genes are specifically and significantly different only on the human branch. In contrast, there are 1,326 genes for which all three species are significantly differentially expressed (where each pairwise P value between species is <0.05). Categorical overrepresentations for the subset of human branch-specific changes in expression also highlight aerobic energy metabolism and transport, as well as multiple categories related to RNA interference and protein targeting (supplementary table 4, Supplementary Material online).
The Intersection of Expression Differences and Positive Selection in Regulatory Regions
Of the 1,872 genes that we find to be significantly differentially expressed when comparing human and chimpanzee expression levels, it is possible that many of these changes in expression levels are due to neutral evolutionary processes. There is notable lack of positive selection in the coding regions of genes involved in neurogenesis and neural function (Clark et al. 2003; Bustamante et al. 2005; Kosiol et al. 2008). However, some important changes in gene expression in human brain evolution may be due to positive selection on specific transcriptional regulatory elements, leading to functionally important changes in transcriptional levels. To explore this possibility, we tested for a correlation between evidence of directional selection in regulatory regions and differential expression. Regulatory regions showing evidence of selection in humans were obtained from Haygood et al. (2007) and Pollard et al. (2006a). Due to the complexities of gene regulation, the null hypothesis would be that there is little or no correlation between positive selection in what is typically just a subset of the total regulatory region and expression of the corresponding gene in just one tissue, at just one developmental stage. Consistent with that hypothesis, with expression data from only one tissue, we see no correlation between positive selection and changes in expression level across all the genes assayed (for Haygood et al. [2007]: r = −0.0040; for Pollard et al. [2006a]: r = 0.0047). Yet, for the few genes where there is a correlation, this may be suggestive of changes in expression due to selective pressures and regulatory regions where follow-up functional analyses would be valuable. Of genes measured for expression in this study, 4,331 genes overlap with Haygood et al. (2007) and 2,328 with Pollard et al. (2006a). Of these, 97 from Haygood et al. (2007) and 35 from Pollard et al. (2006a) (132 loci total) have significant P values (P < 0.05) for both differential expression as well as signatures of selection (supplementary table 5, Supplementary Material online). An enrichment analysis, albeit based on this small number of genes, shows enrichments for genes involved in electron transport (electron transport chain: P = 0.0198; mitochondrial electron transport, NADH to ubiquinone: P = 0.0020) and transport (P = 0.0274).
Discussion
The Utility of Sequencing-Based Expression Assays for Comparative Analyses
Tag-Seq, and other sequencing-based methods of quantifying transcript expression, provides a powerful alternative to hybridization-based assays of gene expression, particularly for cross-species comparisons. Tag-Seq has a wider dynamic range than microarrays or SAGE (t Hoen et al. 2008; Morrissy et al. 2009), it can also measure expression from unannotated and noncoding transcripts, and it provides strand-specific information for each of the tags. In comparison with another common sequencing-based protocol, RNA-Seq, Tag-Seq can provide accurate quantification of tags at much lower coverage (Morrissy et al. 2009); however, RNA-Seq has the added benefit of providing information about transcript structure over the entire length of the transcript. However, the strand-specific information provided by Tag-Seq also allows us to explore the possible functional importance of antisense transcripts by looking at their evolutionary conservation. With this additional type of information, sequencing-based assays of expression will make comparative analyses more comprehensive and accurate over larger evolutionary distances.
Noncoding Transcriptional Units Conserved Over Evolutionary Time
It is clear that transcription is not only confined to protein-coding regions of the genome but also includes many of the noncoding regions as well (Kapranov et al. 2002, 2007b; Okazaki et al. 2002; Bertone et al. 2004; Carninci et al. 2005). It is not yet clear precisely what fraction of these noncoding transcripts play a functional role (Kapranov et al. 2007a; Prasanth and Spector 2007; Mattick 2009). One approach to understanding if a particular transcript is functional is to ask whether it is evolutionarily conserved in sequence, position, and expression level. For example, many functional miRNAs and snoRNAs are conserved over long stretches of evolutionary time, with 80–90% sequence identity between human and mouse for these classes of RNA (Pang et al. 2006). Although a conserved location of noncoding transcription could be due to transcriptional noise (Struhl 2007), it has been shown that some categories of noncoding transcripts are functional as well (Guttman et al. 2009), even those located nearby transcribed genes (He et al. 2008; Preker et al. 2008). There is even one example of human-specific changes in the sequence of a novel ncRNA that may be associated with changes in neural migration during human brain development (Pollard et al. 2006b).
Specifically comparing between humans and chimpanzees, there is a high correlation of both tag sequence and expression levels in noncoding tags for many partitions (although somewhat less strong than those between tags within exons) (supplementary table 2, Supplementary Material online). These correlations are very consistent with those seen using human tiling microarrays (Khaitovich et al. 2006a). That study used exonic and intergenic probes in the 1% of the genome surveyed in the ENCODE pilot project regions (Birney et al. 2007) and found similar, but slightly higher, amount of conservation between intergenic probes expressed in both human and chimpanzee. Differences seen between this study and Khaitovich et al. (2006a) could be due to differences in the measurement platforms, biological noise, or the much larger number of genes considered here. Importantly, however, both studies find evidence for functional constraints in regions of noncoding transcription between humans and chimpanzees based on conservation of expression.
The correlation of conserved regions decreases if we look at intronic or intergenic tags conserved between human, chimpanzee, and macaque (fig. 4). These three species last shared a common ancestor ∼25 MYA, an intermediate divergence time compared with human–chimpanzee (5–7 MYA; Kumar and Hedges 1998; Glazko and Nei 2003) and human–mouse (ca. 90 MYA; Waterston et al. 2002). Between all three species, the sense tags, and specifically, the 5′ flanking sense tags that are the most conserved. The number of 3′ flanking tags on both strands are decreased, with conservation concentrated directly adjacent to exonic regions. Conserved sense-strand tags within intronic regions could be due to incomplete or alternative splicing, transcription along the stretch of an open region of chromatin around actively transcribed genes, or functional RNAs coming from these regions. That both 5′ and 3′ sense flanking region tags also have a number of conserved regions of expression may alternatively point to previously unannotated 5′ and 3′ untranslated regions of the nearby genes (fig. 4A). Other studies have also found an enrichment of expressed intergenic regions nearby to genes (Bertone et al. 2004; Khaitovich et al. 2006a) using tiling arrays, and a positive correlation between conserved upstream sense transcription and the expression of downstream genes (Khaitovich et al. 2006a).
These intergenic tags may also be due to the transcription of short polyadenylated RNAs on both the sense and antisense strand concentrated around promoter regions, some of which may play a role in gene regulation (Kapranov et al. 2007b; Core et al. 2008; He et al. 2008; Preker et al. 2008; Seila et al. 2008). We did find some tags that overlap with annotated snoRNAs and miRNAs, and it is likely that that number will increase dramatically as additional ncRNA functional types are annotated for the human genome. It is likely that some of the tags near genes are due to truncated transcription of short RNAs around actively transcribed genes, which may promote a constitutively open chromatin conformation could be driving the high numbers of tags located directly upstream to genic regions. However, the distribution of conserved tags as one moves away from the protein-coding regions is much larger than a typical region of open chromatin for an active promoter of several hundred base pairs, based on direct sequencing data (Core et al. 2008; He et al. 2008; Preker et al. 2008; Seila et al. 2008) as well as DNase hypersensitivity assays of open chromatin regions near genes (Boyle et al. 2008).
The conservation of antisense tags is also intriguing. The reduced amount of conservation for these tag locations relative to noncoding sense tags across all three species may mean that many of these RNAs are the result of transcriptional noise (fig. 4B). Yet, the amount of conservation between chimpanzee and human and the fact that expression from these regions are relatively correlated between species, may mean that some of the antisense transcripts are functional and could provide interesting candidate RNAs for future functional studies. Another line of evidence that these conserved antisense transcripts are functional comes from the negative correlation of the 5′ flanking antisense transcripts and the expression of the downstream gene. This correlation suggests that antisense transcripts may play a regulatory role for nearby genes. There is substantial experimental evidence for this phenomenon in human cell culture assays (Lapidot and Pilpel 2006; Kapranov et al. 2007a; Mazo et al. 2007), and antisense transcription is pervasive throughout the genome near protein-coding regions (He et al. 2008). Focusing on expressed antisense RNAs that are conserved over evolutionary time may provide insights about the pervasiveness of antisense intergenic RNA that regulate nearby protein-coding genes, and the selective pressures under which they evolve.
Sequencing technologies that allow for longer reads, such as RNA-Seq, as well as strand-specific sequence information will greatly assist in understanding the conservation of structure, and possibly function, of these RNAs. It is also likely that different categories of genomic location as well as of the type of ncRNA produced will evolve at different rates. For example, long intergenic transcripts performing a regulatory function may be much more highly conserved than shorter transcripts due to transcription of open chromatin near promoters. Looking more broadly over multiple lineages, it will be interesting to see which structural and functional classes of RNA are differentially conserved within and between lineages and which are subject to stabilizing, neutral, or even positive, selective pressures.
Changes in Gene Expression between Human and Chimpanzee in the Frontal Cortex
Investigations of expression differences of brain gene expression between humans and nonhuman primates have been done for several brain regions using microarrays (Enard et al. 2002; Caceres et al. 2003; Khaitovich et al. 2004, 2005; Uddin et al. 2004; Somel et al. 2009). These studies found significant differences in gene expression between humans and other primate species in a number of different brain regions; although, it is important to note that studies looking at multiple brain regions noted that there are very few expression differences between neocortical regions themselves (e.g., frontal cortex vs. temporal cortex) within a species (Khaitovich et al. 2004; Uddin et al. 2004; Roth et al. 2006; Johnson et al. 2009). A few previous reports noted elevated gene expression levels in the human brain as opposed to other nonhuman primate species (Enard et al. 2002; Caceres et al. 2003), although other studies did not find this asymmetry of expression in the brain (Uddin et al. 2004) or other tissues (Blekhman et al. 2008). A first exploration of our data was to see if expression is generally higher in one species. In agreement with these later studies, we also did not find a difference in the number of those genes that are more highly expressed in chimpanzees versus humans (fig. 2). From a biological perspective, this would imply that it is not a global change in the amount of transcript but rather expression differences in specific gene pathways, which has led to phenotypic changes between human and chimpanzee brains.
The results from our categorical enrichment analyses are generally concordant with results from previous studies of gene expression differences in human and nonhuman primate brain. An enrichment analysis of the brain expression data from Khaitovich et al. (2005) showed enrichments for similar ontology categories such as transport and metabolism (Khaitovich et al. 2006b). Uddin et al. (2004) also saw an upregulation of genes related to neuronal function as well as components of the electron transport chain (table 1, and discussed below). This signal also remains when we look at enrichments on the human branch, when differentially expressed from both chimpanzee and macaque. The interpretation of categories involved in cellular repair and apoptosis is less clear; although it is known that DNA repair is active in neurons as they are especially sensitive to reactive oxygen species produced by mitochondrial activity (reviewed in Bohr et al. 2007). Lastly, our enrichments also show categories linked to lipid, and to a lesser extent protein, metabolism. Differential expression of these categories, possibly related to the dramatic changes in the human diet, as compared with nonhuman primates (reviewed in Leonard et al. 2007) has also been observed in liver, heart, and kidney expression as well (Blekhman et al. 2008).
Intersection with Signals of Positive Selection
Differential expression is only one step in understanding the genetic changes underlying adaptive phenotypic changes. A separate line of evidence indicative of adaptive pressures lies in signatures of selection in DNA sequences. We looked for correlations between the Tag-Seq expression data and signatures of selection in putative regulatory regions from two previously published scans for selection (Haygood et al. 2007) and (Pollard et al. 2006a) with evidence of selection on the human branch. It is important to note that we would not expect a good global correlation between signatures of positive selection and differential expression; we are only measuring expression in one tissue at one developmental stage. Nonetheless, for genes where there is an overlap, this may be indicative of an adaptive change in expression. Here, with expression data from only one tissue, we see no correlation for all the genes assayed. However, the few genes (135 total here) that lay in the intersection between differential expression and signatures of positive selection may provide important candidate genes for more detailed functional analyses (bsupplementary table 5, Supplementary Material online). For instance, SNX19, a nexin with a phospholipid-binding motif involved in intracellular transport (Worby and Dixon 2002), is shared between the two selection studies (Pollard et al. [2006a]: P = 4.8 × 10−05, Haygood et al. [2007]: P = 2.3 × 10−03), and also shows differential expression in this study (P = 1.3 × 10−08).
Shifts in both phenotype and diet during human evolution (Aiello and Wheeler 1995) may have necessitated shifts in the regulation of core metabolic pathways. Blekhman et al. (2008) analyzed signatures of selection based on patterns of expression differences found an enrichment for genes involved in metabolic pathways, as we do here. In contrast, the enrichments for transport and electron transport that we observe may be due to different assays for selection than in Blekhman et al. (2008) but are more likely being driven by the unique cellular components and energy requirements of the brain. The strong and consistent signal of change in aerobic energy metabolism and protein transport within and outside of the cell are intriguing as there is also evidence of positive selection acting on protein-coding regions of the electron transport genes in the mitochondrial genome in anthropoid primates (Wu et al. 1997, 2000; Wildman et al. 2002; Goldberg et al. 2003). As the brain increased in size and complexity through certain lineages of primate evolution, so too did its energy requirements. Specifically in the lineage leading to humans, genes involved in aerobic energy metabolism may have been under positive selection at both protein-coding and regulatory loci during human evolutionary history.
Conclusion
Sequence-based assays of expression have substantial promise for comparative genomics. This is especially true for comparative primate genomics, where genomic resources exist, but samples are difficult to gather. Comparative Tag-Seq studies, along with other related technologies, can illuminate the relatively unexplored area of ncRNA conservation over shorter time scales. Our results show conservation in both location and expression levels between these primate species, possibly suggesting a functional role for these transcripts. Within coding regions, we found the most expression differences between human, chimpanzee, and macaques in genes involved in neuronal signaling and transport, as well as essential metabolic categories, with the strongest signal including those involved in aerobic energy metabolism. Lastly, the overlap between differentially expressed genes and those showing a signature of positive selection in putative promoter regions is enriched for genes involved in transport and aerobic energy metabolism. Future functional studies will be necessary to test if signatures of adaptive change in regulatory regions are the drivers of the differences in expression.
Supplementary Material
Supplementary tables 1–7 and supplementary figure 1 are available at Genome Biology and Evolution online (http://www.oxfordjournals.org/our_journals/gbe/).
Acknowledgments
We would like to thank the Primate Genomics Initiative at Duke University for assistance in obtaining samples and the Duke University Institute for Genome Sciences & Policy Sequencing Facility for sequencing the libraries. We thank Jera Pecotte and Mary Jo Aivaliotis for the biological materials obtained from the Southwest National Primate Research Center. We thank Elizabeth Curran for biological materials from the New England Regional Primate Research Center. We would also like to thank Sayan Mukherjee for discussions concerning data normalization. Lastly, we would like to thank David Garfield, Jenny Tung, Lomax Boyd, Lisa Warner, and all the members of the Wray laboratory for helpful discussion and comments. This project was funded by National Science Foundation grant NSF-BCS-08-27552 (HOMINID) and the Institute for Genome Sciences & Policy at Duke University.
References
- Ahituv N, et al. Mapping cis-regulatory domains in the human genome using multi-species conservation of synteny. Hum Mol Genet. 2005;14:3057–3063. doi: 10.1093/hmg/ddi338. [DOI] [PubMed] [Google Scholar]
- Aiello LC, Wheeler P. The expensive-tissue hypothesis—the brain and the digestive-system in human and primate evolution. Curr Anthropol. 1995;36:199–221. [Google Scholar]
- Ambros V, et al. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertone P, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the encode pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blekhman R, et al. Gene regulation in primates evolves under tissue-specific selection pressures. PLoS Genet. 2008;4:e1000271. doi: 10.1371/journal.pgen.1000271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohr VA, et al. Genome instability and DNA repair in brain, ageing and neurological disease. Neuroscience. 2007;145:1183–1186. doi: 10.1016/j.neuroscience.2007.03.015. [DOI] [PubMed] [Google Scholar]
- Boyle AP, et al. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante CD, et al. Natural selection on protein-coding genes in the human genome. Nature. 2005;437:1153–1157. doi: 10.1038/nature04240. [DOI] [PubMed] [Google Scholar]
- Caceres M, et al. Elevated gene expression levels distinguish human from non-human primate brains. Proc Natl Acad Sci U S A. 2003;100:13030–13035. doi: 10.1073/pnas.2135499100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Carroll SB. Genetics and the making of homo sapiens. Nature. 2003;422:849–857. doi: 10.1038/nature01495. [DOI] [PubMed] [Google Scholar]
- Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Clark AG, et al. Inferring nonneutral evolution from human-chimp-mouse orthologous gene trios. Science. 2003;302:1960–1963. doi: 10.1126/science.1088821. [DOI] [PubMed] [Google Scholar]
- Core LJ, et al. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enard W, et al. Intra- and interspecific variation in primate gene expression patterns. Science. 2002;296:340–343. doi: 10.1126/science.1068996. [DOI] [PubMed] [Google Scholar]
- Gautier L, et al. Affy—analysis of affymetrix genechip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- Gilad Y, et al. Multi-species microarrays reveal the effect of sequence divergence on gene expression profiles. Genome Res. 2005;15:674–680. doi: 10.1101/gr.3335705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad Y, et al. Expression profiling in primates reveals a rapid evolution of human transcription factors. Nature. 2006;440:242–245. doi: 10.1038/nature04559. [DOI] [PubMed] [Google Scholar]
- Glazko GV, Nei M. Estimation of divergence times for major lineages of primate species. Mol Biol Evol. 2003;20:424–434. doi: 10.1093/molbev/msg050. [DOI] [PubMed] [Google Scholar]
- Goldberg A, et al. Adaptive evolution of cytochrome c oxidase subunit VIII in anthropoid primates. Proc Natl Acad Sci U S A. 2003;100:5873–5878. doi: 10.1073/pnas.0931463100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Gu X. Induced gene expression in human brain after the split from chimpanzee. Trends Genet. 2003;19:63–65. doi: 10.1016/s0168-9525(02)00040-9. [DOI] [PubMed] [Google Scholar]
- Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haygood R, et al. Promoter regions of many neural- and nutrition-related genes have experienced positive selection during human evolution. Nat Genet. 2007;39:1140–1144. doi: 10.1038/ng2104. [DOI] [PubMed] [Google Scholar]
- He YP, et al. The antisense transcriptomes of human cells. Science. 2008;322:1855–1857. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu F, et al. The UCSC known genes. Bioinformatics. 2006;22:1036–1046. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- Jobling MA, et al. Human evolutionary genetics: origins, people, and disease. New York: Garland Science; 2004. [Google Scholar]
- Johnson MB, et al. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron. 2009;62:494–509. doi: 10.1016/j.neuron.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, et al. Large-scale transcriptional activity in chromosomes 21 and 22. Science. 2002;296:916–919. doi: 10.1126/science.1068597. [DOI] [PubMed] [Google Scholar]
- Kapranov P, et al. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007a;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- Kapranov P, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007b;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- Karaman MW, et al. Comparative analysis of gene-expression patterns in human and African great ape cultured fibroblasts. Genome Res. 2003;13:1619–1630. doi: 10.1101/gr.1289803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, et al. The UCSC genome browser database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich P, et al. Regional patterns of gene expression in human and chimpanzee brains. Genome Res. 2004;14:1462–1473. doi: 10.1101/gr.2538704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich P, et al. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science. 2005;309:1850–1854. doi: 10.1126/science.1108296. [DOI] [PubMed] [Google Scholar]
- Khaitovich P, et al. Functionality of intergenic transcription: an evolutionary comparison. PLoS Genet. 2006a;2:e171. doi: 10.1371/journal.pgen.0020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich P, et al. Positive selection on gene expression in the human brain. Curr Biol. 2006b;16:R356–R358. doi: 10.1016/j.cub.2006.03.082. [DOI] [PubMed] [Google Scholar]
- Kosiol C, et al. Patterns of positive selection in six mammalian genomes. PLoS Genet. 2008;4(8):e1000144. doi: 10.1371/journal.pgen.1000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Hedges SB. A molecular timescale for vertebrate evolution. Nature. 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- Lapidot M, Pilpel Y. Genome-wide natural antisense transcription: coupling its regulation to its different regulatory mechanisms. EMBO Rep. 2006;7:1216–1222. doi: 10.1038/sj.embor.7400857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard WR, et al. Effects of brain evolution on human nutrition and metabolism. Annu Rev Nutr. 2007;27:311–327. doi: 10.1146/annurev.nutr.27.061406.093659. [DOI] [PubMed] [Google Scholar]
- Li H, et al. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni JC, et al. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet. 2009;5:e1000459. doi: 10.1371/journal.pgen.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazo A, et al. Transcriptional interference: an unexpected layer of complexity in gene regulation. J Cell Sci. 2007;120:2755–2761. doi: 10.1242/jcs.007633. [DOI] [PubMed] [Google Scholar]
- Mi HY, et al. The panther database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 2005;33:D284–D288. doi: 10.1093/nar/gki078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissy AS, et al. Next generation tag sequencing for cancer gene expression profiling. Genome Res. 2009;19:1825–1835. doi: 10.1101/gr.094482.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, et al. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Okazaki Y, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- Pang KC, et al. Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends Genet. 2006;22:1–5. doi: 10.1016/j.tig.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Pedotti P, et al. Can subtle changes in gene expression be consistently detected with different microarray platforms? BMC Genomics. 2008;9:124. doi: 10.1186/1471-2164-9-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio LA, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- Pollard KS, et al. Forces shaping the fastest evolving regions in the human genome. PLoS Genet. 2006a;2:1599–1611. doi: 10.1371/journal.pgen.0020168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006b;443:167–172. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- Prasanth KV, Spector DL. Eukaryotic regulatory RNAs: an answer to the 'genome complexity' conundrum. Genes Dev. 2007;21:11–42. doi: 10.1101/gad.1484207. [DOI] [PubMed] [Google Scholar]
- Preker P, et al. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- Pruitt KD, et al. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Smyth GK. Moderated statistical tests for assessing differences in tag abundance. Bioinformatics. 2007;23:2881–2887. doi: 10.1093/bioinformatics/btm453. [DOI] [PubMed] [Google Scholar]
- Robinson MD, Smyth GK. Small-sample estimation of negative binomial dispersion, with applications to sage data. Biostatistics. 2008;9:321–332. doi: 10.1093/biostatistics/kxm030. [DOI] [PubMed] [Google Scholar]
- Roth RB, et al. Gene expression analyses reveal molecular relationships among 20 regions of the human CNS. Neurogenetics. 2006;7:67–80. doi: 10.1007/s10048-006-0032-6. [DOI] [PubMed] [Google Scholar]
- Seila AC, et al. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simes RJ. An improved Bonferroni procedure for multiple tests of significance. Biometrika. 1986;73:751–754. [Google Scholar]
- Somel M, et al. Transcriptional neoteny in the human brain. Proc Natl Acad Sci U S A. 2009;106:5743–5748. doi: 10.1073/pnas.0900544106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol. 2007;14:103–105. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- t Hoen PAC, et al. Deep sequencing-based expression analysis shows major advances in robustness, resolution and inter-lab portability over five microarray platforms. Nucleic Acids Res. 2008;36:e141. doi: 10.1093/nar/gkn705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JL, et al. Patterns of growth and development in the genus homo. New York: Cambridge University Press; 2003. [Google Scholar]
- Tomasello M, Call J. Primate cognition. New York: Oxford University Press; 1997. [Google Scholar]
- Uddin M, et al. Sister grouping of chimpanzees and humans as revealed by genome-wide phylogenetic analysis of brain gene expression profiles. Proc Natl Acad Sci U S A. 2004;101:2957–2962. doi: 10.1073/pnas.0308725100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, et al. Ultraconservation identifies a small subset of extremely constrained developmental enhancers. Nat Genet. 2008;40:158–160. doi: 10.1038/ng.2007.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, et al. RNA-seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Wildman DE, et al. Episodic positive selection in ape cytochrome c oxidase subunit IV. Mol Biol Evol. 2002;19:1812–1815. doi: 10.1093/oxfordjournals.molbev.a004005. [DOI] [PubMed] [Google Scholar]
- Worby CA, Dixon JE. Sorting out the cellular functions of sorting nexins. Nat Rev Mol Cell Biol. 2002;3:919–931. doi: 10.1038/nrm974. [DOI] [PubMed] [Google Scholar]
- Wu W, et al. Molecular evolution of cytochrome c oxidase subunit IV: evidence for positive selection in simian primates. J Mol Evol. 1997;44:477–491. doi: 10.1007/pl00006172. [DOI] [PubMed] [Google Scholar]
- Wu W, et al. Molecular evolution of cytochrome c oxidase subunit I in primates: is there coevolution between mitochondrial and nuclear genomes? Mol Phylogenet Evol. 2000;17:294–304. doi: 10.1006/mpev.2000.0833. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.