Abstract
Mitochondrial DNA (mtDNA) mutations underlie a variety of human genetic disorders and are associated with the aging process. mtDNA polymorphisms are widely used in a variety of evolutionary applications. Although mtDNA mutation spectra are known to differ between distantly related model organisms, the extent to which mtDNA mutation processes vary between more closely related species and within species remains enigmatic. We analyzed mtDNA divergence in two sets of 250-generation Caenorhabditis briggsae mutation-accumulation (MA) lines, each derived from a different natural isolate progenitor: strain HK104 from Okayama, Japan, and strain PB800 from Ohio, United States. Both sets of C. briggsae MA lines accumulated numerous large heteroplasmic mtDNA deletions, whereas only one similar event was observed in a previous analysis of Caenorhabditis elegans MA line mtDNA. Homopolymer length change mutations were frequent in both sets of C. briggsae MA lines and occurred in both intergenic and protein-coding gene regions. The spectrum of C. briggsae mtDNA base substitution mutations differed from the spectrum previously observed in C. elegans. In C. briggsae, the HK104 MA lines experienced many different base substitution types, whereas the PB800 lines displayed only C:G → T:A transitions, although the difference was not significant. Over half of the mtDNA base substitutions detected in the C. briggsae MA lines were in a heteroplasmic state, whereas all those previously characterized in C. elegans MA line mtDNA were fixed changes, indicating a narrower mtDNA bottleneck in C. elegans as compared with C. briggsae. Our results show that C. briggsae mtDNA is highly susceptible to large deletions and that the mitochondrial mutation process varies between Caenorhabditis nematode species.
Keywords: bottleneck, heteroplasmy, mutation-accumulation line, nematode
Introduction
Mitochondrial DNA (mtDNA) is one of the most widely used molecules for studying diverse evolutionary processes. The mitochondrial genomes of most animal species are maternally inherited, encode the same set of genes, and mutate at rates ∼10 times higher than nuclear DNA (Lightowlers et al. 1997; Baer et al. 2007; Gissi et al. 2008)—these features have made mtDNA a favorite target for numerous phylogenetic studies of closely related species and within-species analyses at the population level. A segment of the cytochrome oxidase I mtDNA protein-coding gene was selected by the Consortium for the Barcode of Life to serve as the focal “barcode” DNA region for global species identification (Savolainen et al. 2005). mtDNA mutations also underlie many mitochondrial myopathies in humans and have been associated with numerous neurodegenerative diseases, including Alzheimer's disease and Parkinson's disease (Howell et al. 2005; Wallace 2005; Biskup and Moore 2006). Large heteroplasmic mitochondrial genome deletions are observed to accumulate to high levels in aging somatic tissue from a variety of animal species from humans to Caenorhabditis elegans (Holt et al. 1988; Melov et al. 1994; Arai et al. 2003) and are implicated as potential causative agents in the degenerative aging process (Barazzoni et al. 2000; Wei and Lee 2002; Biskup and Moore 2006). These large deletion events are usually associated with flanking direct repeat sequences. In nematodes, repeat-associated deletions have been attributed to intragenomic recombination events (Lunt and Hyman 1997). Electron transport chain dysfunction associated with deletions involving mtDNA protein-coding genes is thought to be a major underlying cause of the degenerative phenotypes associated with aging, mitochondrial myopathies, and other human disorders (Conley et al. 2007).
Between- and within-species variation in the rate and spectrum of mitochondrial genome mutations has the potential to confound evolutionary analyses where the mitochondrial mutation process is presumed to be constant (Baer et al. 2007). Furthermore, extensive mtDNA mutational variation in humans has the potential to render some individuals or lineages more susceptible to mitochondrial disease and earlier onset of degenerative aging-related phenotypes than others. Our knowledge on mitochondrial mutational variation has increased over the last decade with the analysis of mtDNA from model organism mutation-accumulation (MA) line experiments. In MA line studies, organisms are bottlenecked to one (for self-fertile species) or two (for obligate outcrossing species) individuals across many generations to allow for the essentially neutral accumulation of mutations over time. Results from C. elegans and Drosophila melanogaster MA experiments (Denver et al. 2000; Haag-Liautard et al. 2008) revealed highly similar per-generation mtDNA mutation rate estimates but different base substitution mutation spectra. Furthermore, between-species mtDNA mutation pattern variation became evident (Montooth and Rand 2008) with results from a Saccharomyces cerevisiae MA experiment (Lynch et al. 2008) and an analysis of mtDNA mutator mice (Stewart et al. 2008). Although these combined studies have provided an important step in understanding between-species mtDNA mutational variation, the model organisms used in these studies are all very distantly related from one another. The extent to which mtDNA mutation processes vary between more closely related species and within species remains enigmatic.
The mtDNA mutation process is a complicated one in that many mitochondrial genomes are present inside an individual mitochondrion and many mitochondria are present inside an individual cell (Upholt and Dawid 1977; Rand 2001). Thus, a mitochondrial mutation originates on a single mtDNA molecule and then must traverse a complex path of heteroplasmy (condition where more than one nucleotide sequence is present at a given site in the pool of mitochondrial genomes within an individual) to become a fixed difference between species or between individuals within a species. mtDNA heteroplasmy has proven a difficult challenge in a variety of contexts, from diagnosing the conditions under which mitochondrial disorder symptoms are manifested (Wong 2007) to its complicating effects on molecular evolutionary and ecological studies (White et al. 2008). In mammals, mtDNA sequence variants are observed to turn over rapidly between generations despite the high mtDNA copy number in the oocyte, leading to the interpretation that there is a very narrow “mtDNA bottleneck” in the female germ line (Jenuth et al. 1996). The extent to which the mtDNA bottleneck width varies in different species, however, remains unclear.
Although most naturally occurring gene function–disrupting mtDNA deletions that have been studied occur as nonheritable and sporadic somatic events (Chinnery et al. 2004), Caenorhabditis briggsae natural populations are known to suffer varying levels of a large, heteroplasmic, and heritable deletion of the NADH dehydrogenase subunit 5 (ND5) protein-coding gene (Howe and Denver 2008)—the deletion is associated with 21-bp direct repeats, one copy in the ND5 gene itself and the second ∼880 bp upstream in a pseudogene region named ΨND5-2. Howe and Denver (2008) showed that different C. briggsae natural isolates harbor varying levels of this ΨND5-2/ND5 boundary deletion, ranging from 0% to ∼50% of the total mtDNA pool within an individual L1 larval-stage nematode, and that there was a significant negative correlation between isolate-specific ND5 deletion level and lifetime fecundity, suggesting that the deletion has a negative impact on C. briggsae fitness. Furthermore, certain C. briggsae natural isolates were discovered to have fixed ΨND5-2 direct repeat substitutions that result in imperfect matches to the downstream ND5 repeat and are associated with reduced ND5 deletion levels. The unusual ND5 deletion dynamics observed in C. briggsae populations suggests that mtDNA mutation processes might differ between this species and its congener C. elegans in which no mtDNA pseudogenes or associated deletion events have been observed in mitogenomic analyses of natural isolates (Denver et al. 2003).
Here, we characterize the mtDNA mutational spectrum of two sets of C. briggsae MA lines—one set derived from natural isolate HK104 from Okayama, Japan, and the second set derived from natural isolate PB800 from Ohio, United States—that were previously shown to experience significantly higher per-generation rates of fitness decline and higher rates of mutation at nuclear microsatellite loci as compared with C. elegans MA lines (Baer et al. 2005; Phillips et al. 2009). We observed substantial mtDNA mutational differences between C. briggsae and its congener C. elegans, providing evidence that mtDNA mutation processes can vary between related animal species.
Materials and Methods
Polymerase Chain Reaction Amplification and DNA Sequencing
Caenorhabditis briggsae MA line mitochondrial genomes were amplified from total genomic DNA preparations (many thousands of nematodes included in each lysis) as four overlapping polymerase chain reaction (PCR) products, ranging in size from ∼2,800 to ∼4,900 bp, using the Expand Long Range dNTPack, containing a proofreading DNA polymerase blend (Roche). Six of the long PCR primers used were first described in a methods paper for long PCR of parasitic nematode mitochondrial genomes (Hu et al. 2002). Two C. briggsae sequence-specific primers were designed by D.K.H. for this and a previous study (Howe and Denver 2008). The primer sequences and product sizes used for long PCRs are provided in supplementary table S1 (Supplementary Material online). A total of 13,900 bp was analyzed in each HK104 MA line; 13,894 bp was analyzed in each PB800 MA line. Entire mitochondrial genomes were sequenced with the exception of the “AT” region (prohibitively difficult to sequence) for both sets. PCR products were visualized on 1.5% agarose gels to ensure amplification success and screen for large heteroplasmic deletion events. Prior to DNA sequencing, all PCR products were purified using solid phase reversible immobilization (Elkin et al. 2001). PCR and internal primers were used in direct sequencing reactions. DNA sequencing was carried out on an ABI3730 capillary sequencer at the Oregon State University Center for Genome Research and Biocomputing. All mutations were confirmed on both strands of DNA. MEGA 4.0 was used for DNA sequence alignments and to visually scrutinize chromatogram trace files (Tamura et al. 2007). Caenorhabditis briggsae MA line mtDNA sequences were submitted to GenBank under accession numbers (GU452324-GU452381).
Detection and Treatment of Heteroplasmy
To distinguish between fixed MA line mtDNA changes and those where sequence differences are in a heteroplasmic state (either in progenitor or in MA line), we carefully scrutinized chromatogram data (for base substitution polymorphisms and homopolymer mutations) and the results of PCR amplification on agarose gels (for large deletion events). Large deletions observed in the MA lines were marked as heteroplasmic if there was evidence of the original intact progenitor mtDNA on the agarose gel. We also used primers internal to the deleted regions in PCR reactions to probe for the presence of intact mitochondrial genome copies wherever possible—in all cases analyzed, wild-type copies were detected. For the ΨND5-2/ND5 deletion region, a previous analysis of this deletion in C. briggsae natural isolates showed that a simple standard PCR band scoring method for estimating relative levels of deletion-bearing and intact mitochondrial genomes (applied here—see supplementary fig. S1, Supplementary Material online) yielded values that were highly comparable with and correlated positively and significantly (Spearman rank correlation = 0.74, P < 10−15) with values derived from a more sensitive quantitative PCR approach to estimating deletion levels (Howe and Denver 2008). For homopolymer loci changes at smaller (6–10 bp) runs, mutations were defined as fixed if a clean chromatogram resulted from the sequencing reaction that clearly showed uniform evidence of the homopolymer mutation—that is, no “stutter” sequence after the homopolymer in the chromatogram. Homopolymer changes were defined as heteroplasmic if there was strong evidence in the peaks associated with the homopolymer in the chromatogram data, with the presence of stutter sequence after the homopolymer sequence presumably reflecting the presence of other homopolymer length variants in the MA line mitochondrial genome pool. For the two longest homopolymer repeats analyzed here, (A:T)14 in HK104 and (A:T)12 in PB800, stutter peaks were present in all data analyzed, though the predominant repeat length present in each MA line and progenitor could be discerned from the data (supplementary fig. S2, Supplementary Material online).
MA line base substitutions were marked as heteroplasmic if there was any evidence of the original progenitor base present in the chromatogram data (supplementary fig. S3, Supplementary Material online). For heteroplasmic base substitution mutations, we estimated the frequencies of the mutant bases in the total mtDNA pool using the comparative peak height approach applied by a previous analysis of D. melanogaster MA line mtDNA divergence (Haag-Liautard et al. 2008). We searched for sites within 20 bp of the heteroplasmic base substitution site that were the same nucleotide as the mutant base; likewise, we also searched for nearby sites that were the same nucleotide as the progenitor (wild type) site. The peak heights of those two sites were measured and compared with the corresponding heights of mutant and wild-type peaks present at the heteroplasmy site. We used equation 1 of Haag-Liautard et al. (2008) to then estimate the frequencies of the mutant bases. The correction approach was carried out using data from both strands of DNA in all cases.
Mutation Rate Calculations
Mutation rates for large deletion events and at homopolymer loci were calculated with the equation μ = m/(LT), where μ is the mutation rate (per nucleotide site per generation), m is the number of observed mutations, L is the number of MA lines, and T is the time in generations, as previously described and applied to C. elegans MA line analyses (Denver et al. 2000). The probability of ultimate fixation of a neutral mutation is its current frequency in the population. Thus, for base substitution mutations, we simply used the summed observed frequencies of the mutations to estimate m and used equation 2 of Haag-Liautard et al. (2008) to calculate the mutation rate. The standard errors for individual mutation rates were calculated as [μ/(LnT)]1/2, as previously described (Denver et al. 2000). Approximate 95% confidence intervals were estimated using the quantiles approach applied to the standard error estimates. Tests of between-species and between-strain differences in mutational spectra were done by Fisher’s Exact test. Although this method will be biased if base composition differs between groups, the slight differences in base composition between species (%A/T = 75.6% in C. elegans and 75.1% in C. briggsae) and the even smaller difference between strains of C. briggsae (<0.1%) mean that the test is essentially unbiased as applied to our data.
Data Deposition
DNA sequences generated for this study were submitted to GenBank under accession numbers GU452324-GU452381.
Results
Experimental Overview
To investigate mtDNA mutational variation in Caenorhabditis, we sequenced nearly complete mitochondrial genomes from a set of 250-generation C. briggsae MA lines: 21 MA lines derived from natural isolate HK104 (Okayama, Japan) and 18 MA lines derived from natural isolate PB800 (Dayton, OH). Results were compared with a previous mtDNA analysis of 214-generation C. elegans N2 (laboratory strain) MA lines (Denver et al. 2000). In the HK104 MA lines, 13,900 bp of sequence was collected; in the PB800 MA lines, 13,894 bp was collected (entire mitochondrial genomes except the AT region for both sets). Although these two isolates have very different geographic origins, a previous analysis of natural mtDNA variation in C. briggsae (Howe and Denver 2008) showed that they are closely related to one another at the mtDNA level (0.4% pairwise mtDNA sequence divergence). mtDNA was PCR amplified in each MA line (products ranging in size from ∼2,800 to ∼4,900 bp) and then directly sequenced using PCR and internal primers (see Materials and Methods)—the same strategy employed for a previous analysis of C. briggsae natural isolate mtDNA variation (Howe and Denver 2008). This strategy was also highly similar to the previous analysis of C. elegans MA line mtDNA mutation (Denver et al. 2000), though in the C. elegans study more overlapping individual PCR products of smaller size (19 amplicons, 821 bp average size) were generated for the analysis. When referring to specific mtDNA nucleotide pair sites in this paper, the coding strand nucleotide is first denoted, followed by a colon and then the nucleotide on the noncoding strand.
Sequence differences between progenitor and MA line mtDNA were identified in DNA sequence alignments and by evaluating DNA sequence chromatogram data. Heteroplasmy was found to be a significant variable in our analysis, requiring the establishment of careful criteria for subcategorizing different mutation types. mtDNA mutations originating along a specific MA line lineage might still be in a heteroplasmic state at the particular generation analyzed or be a fixed difference (no heteroplasmy). There is also the possibility that preexisting heteroplasmies in the progenitor strain, or their origination in the expansion phase leading up the establishment of MA lines, might differentially segregate in different MA lines. Our approaches to distinguishing fixed-difference mutations from heteroplasmic changes for different mutation types are described in the Materials and Methods.
Large Deletion Mutations
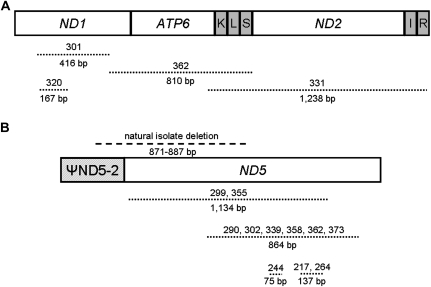
We identified large mtDNA deletion mutations in the C. briggsae MA lines through visual analysis of PCR amplicons on agarose gels and subsequent direct DNA sequencing analysis of PCR amplicons. Where possible, we confirmed that the deletions were in a heteroplasmic state, though it is likely that all of them are heteroplasmic because in nematodes such deletions have never been observed to accumulate to >60% levels (Liau et al. 2007; Howe and Denver 2008). Although the two C. briggsae natural genotypes used as the MA line progenitors showed only 0.4% pairwise sequence divergence across all mtDNA sequences, the MA line progenitors differed dramatically in terms of starting ΨND5-2/ND5 boundary deletion levels: the inbred HK104 progenitor produced a PCR result suggesting high (∼30–50%) ΨND5-2/ND5 deletion levels, whereas analysis of the inbred PB800 progenitor revealed much lower (∼0–5%) deletion levels (Howe and Denver 2008)—see supplementary fig. S1 (Supplementary Material online). Consistent with this observation, PB800 encodes a ΨND5-2 direct repeat allele with substitutions rendering it an imperfect match (19/21 bp match) to the downstream ND5 repeat, whereas the HK104 ΨND5-2 direct repeat allele is a perfect match to the downstream ND5 repeat (Howe and Denver 2008). PCR and DNA sequencing analysis of this region in the MA lines (see Materials and Methods) revealed that 16/21 HK104 MA lines continued to harbor high levels of the same specific ΨND5-2/ND5 boundary deletion observed in the HK104 progenitor and other C. briggsae natural isolates. Two of the HK104 MA lines bearing high ΨND5-2/ND5 deletion levels (MA217 and MA264) were also observed to bear an additional identical deletion (137 bp) downstream in the truncated ND5 coding sequence (fig. 1, supplementary table S2, Supplementary Material online). Two HK104 MA lines (MA262 and MA263) were observed to bear primarily intact “wild-type” mitochondrial genomes in the entire ΨND5-2/ND5 region. HK104 line MA244 mtDNA contained intact DNA in the ΨND5-2/ND5 boundary region but also had a 75-bp deletion downstream in the ND5 coding region (fig. 1). Finally, two HK104 MA lines (MA290 and MA299) were found to bear separate large mitochondrial genome deletions entirely in the ND5 gene that overlapped the ΨND5-2/ND5 boundary deletion observed in C. briggsae natural isolates and the HK104 progenitor strain.
FIG. 1.—
Large heteroplasmic deletion mutations. The schematics show the boundaries of large heteroplasmic deletion events observed in the Caenorhabditis briggsae MA line mtDNA. (A) Shows the ND1/ATP6/ND2 region and (B) shows the ΨND5-2/ND5 region. Protein-coding genes are indicated by white boxes, transfer RNA (tRNA) genes by gray boxes, and the ΨND5-2 pseudogene is shown as a hashed box. Lines with small dashes show deleted regions observed only in MA line genomes; the line with wider dashes shows the ΨND5-2/ND5 boundary deletion observed to predominate in the HK104 MA lines and some C. briggsae natural isolates. The numbers above the lines indicate the MA lines in which the deletion was observed—HK104 MA lines are numbered in the 200s and PB800 MA lines in the 300s. The numbers below the lines show the size (in base pair) of the deletions. Specific details on deletion sizes and associated direct repeat motifs are provided in supplementary table S2 (Supplementary Material online).
None of the PB800 MA lines analyzed accumulated high levels of the ΨND5-2/ND5 boundary region deletion. The observation that the PB800 MA lines do not accumulate these ΨND5-2/ND5 deletions across 250 generations of bottlenecking is consistent with the hypothesis (Howe and Denver 2008) that its ΨND5-2 direct repeat genotype (imperfect repeat relative to downstream repeat in ND5) renders this locus more resistant to repeat-mediated deletion events as compared with ΨND5-2 direct repeat sequences that are identical to downstream ND5 repeats. Although none of the PB800 MA lines were found to harbor the particular ΨND5-2/ND5 boundary deletion observed in the HK104 MA lines and C. briggsae natural isolates, 5/18 PB800 MA lines analyzed accumulated high levels of a nearby direct repeat–associated 864-bp deletion entirely within the ND5 gene boundaries (fig. 1, supplementary table S2, Supplementary Material online). The high frequency of this deletion among the PB800 MA lines suggests that it might have originated during the MA line expansion phase; however, this particular deletion type was also observed in one HK104 MA line, suggesting that this might be a deletion mutational hot spot. Another large ND5 deletion (1,134 bp) was detected in a single PB800 MA line; this same deletion was also observed in a single HK104 MA line.
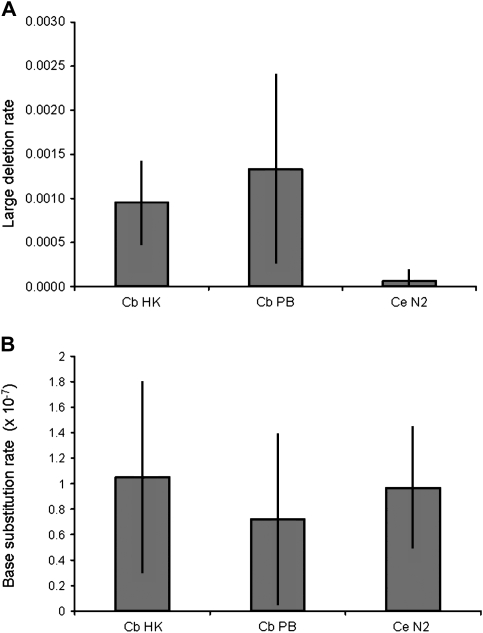
We also detected large mtDNA deletions outside of the ΨND5-2/ND5 region in the C. briggsae MA lines, none of which were observed in C. briggsae natural isolates (Howe and Denver 2008). In the HK104 MA lines, a relatively small (10 bp) deletion was observed in the ND6 gene (supplementary table S2, Supplementary Material online). In the PB800 MA lines, four different large deletions were found to accumulate in the ND1/ATP6/ND2 region of the mitochondrial genome (fig. 1). No deletions were found in this region in the HK104 MA lines. Two of the large gene deletions in this region were not associated with flanking direct repeats (supplementary table S2, Supplementary Material online). We calculated per-generation mutation rates specific for large heteroplasmic mtDNA deletions (see Materials and Methods) and discovered >10-fold higher average rate estimates for both sets of C. briggsae MA lines relative to C. elegans under the conservative assumption that all deletions shared by more than one line resulted from single events during the MA line expansion phase (fig. 2A). No significant large deletion mutation rate differences were observed between the two C. briggsae strains (P = 0.8, Fisher’s Exact test). Although a significant difference in large deletion mutation rate was detected between the C. briggsae (HK104 and PB800 data combined) and C. elegans MA lines (P < 0.001, Fisher's Exact test), this finding must be interpreted with caution because large-amplicon PCRs were used in the current study, whereas the previous C. elegans study (Denver et al. 2000) relied on smaller PCR amplicons and as a consequence might have missed some large heteroplasmic deletions (see Discussion).
FIG. 2.—
Caenorhabditis mtDNA mutation rates. The per-generation, per-genome rates of large mitochondrial genome deletions are shown in (A) for Caenorhabditis briggsae HK104 (Cb HK) and PB800 (Cb PB) MA lines as well as Caenorhabditis elegans N2 (Ce N2) MA lines (Denver et al. 2000). For these estimates, identical deletions observed in more than one MA line of a common ancestral genotype are presumed to have originated from a single event. The per-generation, per–base pair rates of base substitution are shown in (B) for the same sets of MA lines. For both estimates, substitutions observed in more than one MA line of a common ancestral genotype are presumed to have originated from a single event. Error bars show approximate 95% confidence intervals (see Materials and Methods).
Homopolymer Mutations
Homopolymeric nucleotide runs occur frequently in nematode mitochondrial genomes both in intergenic space and in coding regions. These sequence motifs are known to be mutational hot spots for insertion–deletion mutations as a consequence of slip-strand mispairing during replication (Denver et al. 2000, 2004; Fan and Chu 2007). Only smaller (6–10 bp) homopolymer loci analyzed for mutation here yielded clean chromatogram data where the presence of predominantly fixed changes (clean sequences data continue after homopolymer) could be discriminated from those still in a heteroplasmic state (stutter sequences clearly present after homopolymer). Although the two largest homopolymer loci analyzed—(A:T)14 in HK104 progenitor and (A:T)12 in PB800 progenitor (same intergenic mtDNA locus)—consistently yielded stutter peaks in all MA line chromatograms, progenitor–MA line differences in the size of the predominant homopolymer run length present in the mtDNA pool could be discerned from the data (supplementary fig. S2, Supplementary Material online).
In HK104, 10 different specific homopolymer loci were observed to suffer mutation, 6 of which were in protein-coding gene sequence (table 1). Four HK104 MA lines were observed to have experienced a shift in the predominant homopolymer run length at the intergenic (A:T)14 present in the progenitor. Aside from this locus, nine other HK104 mtDNA homopolymer mutations at smaller repeats showed clear signs of heteroplasmy (stutter peaks after repeat); for the remaining four HK104 homopolymer mutations, no evidence of heteroplasmy was detected in either the progenitor or the MA line. In PB800, mutations were detected at four homopolymer loci in the mitochondrial genome, three of which were in protein-coding gene sequence. The large intergenic (A:T)12 run in the PB800 progenitor was observed to be predominantly (A:T)11 in 8/18 PB800 MA lines, (A:T)13 in 3/18 lines, and (A:T)14 in 1/18 lines. Single–base pair length change mutations were observed at each of three smaller homopolymer runs in coding sequence in the PB800 MA lines, two of which showed clear signs of heteroplasmy (table 1). In the HK104 MA lines, 11/17 homopolymer length changes were single–base pair insertions and 6/17 were single–base pair deletions. In the PB800 MA lines, 4/12 homopolymer length changes were single–base pair insertions, 7/12 were single–base pair deletions, and one 2-bp insertion was observed. In the previous analysis of C. elegans MA line mtDNA (Denver et al. 2000), seven (A:T)n and (T:A)n homopolymer mutations were detected across both intergenic and coding sequences: 2/7 were single–base pair insertions and 5/7 were single–base pair deletions. Although greater numbers of homopolymer loci changes were detected in the C. briggsae MA lines relative to C. elegans, the results are not directly comparable as the largest homopolymer analyzed in C. elegans was a (A:T)11, whereas a (A:T)14 was available for analysis in the C. briggsae HK104 MA lines and a (A:T)12 was present in the PB800 MA lines. Homopolymers generally experience increasing mutation rates with increasing run lengths (Denver et al. 2004). Thus, inherent size differences in the homopolymers analyzed in the different MA line sets, coupled with uncertainties regarding the heteroplasmy status of many homopolymer changes before and after MA, preclude meaningful quantitative between- and within-species comparative analyses of homopolymer mutation processes here.
Table 1.
Homopolymer Mutations
| Lines | Position | Mutation | Gene |
| HK104 MA lines | |||
| 254, 261 | 131 | (T:A)9 → (T:A)10a | ND6 |
| 232, 261 | 199 | (T:A)8 → (T:A)7 | ND6 |
| 293 | 199 | (T:A)8 → (T:A)9 | ND6 |
| 264 | 1482 | (A:T)7 → (A:T)6a | ssu rRNA |
| 287 | 2051 | (T:A)7 → (T:A)8a | ND1 |
| 206, 290 | 3250 | (A:T)14a → (A:T)15a | IG |
| 217, 236 | 3250 | (A:T)14a → (A:T)13a | IG |
| 299 | 4669 | (T:A)8 → (T:A)9a | ΨND5-1 |
| 206, 288 | 7986 | (T:A)9 → (T:A)10a | IG |
| 244 | 12697 | (T:A)8 → (T:A)9a | ND5 |
| 206 | 13581 | (T:A)7 → (T:A)6a | ND5 |
| 244 | 13832 | (T:A)8 → (T:A)9 | ND5 |
| PB800 MA lines | |||
| 339 | 207 | (T:A)6 → (T:A)7 | ND6 |
| 8 linesb | 3244 | (A:T)12a → (A:T)11a | IG |
| 306, 355, 388 | 3244 | (A:T)12a → (A:T)13a | IG |
| 366 | 3244 | (A:T)12a → (A:T)14a | IG |
| 306 | 6189 | (T:A)7 → (T:A)6a | COIII |
| 355 | 7014 | (T:A)6 → (T:A)5a | ND4 |
NOTE.—Cytochrome oxidase III, COIII.
Indicates heteroplasmic loci.
The eight lines with predominant single–base pair deletions at this locus are MA302, 308, 316, 339, 358, 373, 374, and 380. Positions denoted are with respect to the progenitor mtDNA sequence. IG indicates intergenic.
Base Substitution Mutations
We identified 19 mtDNA base substitution differences in the C. briggsae MA lines relative to their respective progenitors (table 2)—9 in the HK104 lines and 10 in the PB800 lines. Fourteen of the base substitutions were observed in protein-coding gene sequence, and all but one resulted in a nonsynonymous change (including nonsense mutations), consistent with the expectation of neutral MA and previous observations in C. elegans and D. melanogaster MA line mtDNA (Denver et al. 2000; Haag-Liautard et al. 2008). In the HK104 MA lines, multiple different specific base substitution types were observed, 4/9 were in a heteroplasmic state, all substitutions were at different sites in the mitochondrial genome, and all occurred in different specific MA lines. Thus, each of these nine differences most likely constitutes de novo mutation events. The most prevalent base substitution type in the HK104 MA lines was G:C → T:A transversions (3/9). By contrast, 10/10 base substitution polymorphisms observed in the PB800 MA lines were C:G → T:A transitions. Furthermore, the 10 PB800 substitution differences were observed across only 5 nucleotide sites. At position 7549, two PB800 MA lines were observed to carry fixed substitutions and two other lines were observed to carry heteroplasmic substitutions. Although no clear evidence of a preexisting T:A heteroplasmic variant was discernable from the PB800 progenitor chromatogram data (supplementary fig. S3, Supplementary Material online), the fact that this substitution was discernible in 4/18 MA lines suggests that it might have originated as a single event in the expansion phase of the MA line experiment. Position 10460 in PB800 was a similar case: 3/18 MA lines analyzed had a heteroplasmic T:A at the position, whereas the progenitor and all other PB800 MA lines only showed evidence of the progenitor C:G at that position. One conclusion is that there were five de novo base substitution events in the PB800 MA lines under the conservative assumption that those observed in >1 line resulted from single mutations in the MA line expansion phase. The possibility cannot be ruled out, however, that positions 7549 and 10460 are mutational hot spots.
Table 2.
Base Substitution Mutations
| Lines | Position | Mutation | Frequency | Gene | Effect |
| HK104 MA lines | |||||
| 206 | 834 | T:A → C:Ga | 0.93 | tRNA-Trp | |
| 262 | 1549 | G:C → A:Ta | 0.53 | ssu rRNA | |
| 202 | 5217 | G:C → T:Aa | 0.71 | Cyt b | Gly → Val |
| 244 | 5604 | G:C → T:A | 1.00 | Cyt b | Ser → Met |
| 298 | 6881 | G:C → A:Ta | 0.51 | ND4 | Val → Ile |
| 293 | 8610 | G:C → T:A | 1.00 | COI | Ser → Met |
| 288 | 9671 | T:A → C:G | 1.00 | tRNA-Cys | |
| 258 | 11849 | T:A → A:T | 1.00 | ND3 | Met → Lys |
| 205 | 12160 | C:G → T:A | 1.00 | ψND5-2 | |
| PB800 MA lines | |||||
| 308 | 1390 | C:G → T:A | 1.00 | ssu rRNA | |
| 347 | 2598 | C:G → T:A | 1.00 | ND1 | Silent |
| 347 | 3414 | C:G → T:Aa | 0.85 | tRNA-Ser | |
| 316, 373 | 7549 | C:G → T:A | 1.00, 1.00 | ND4 | Leu → Phe |
| 358, 362 | 7549 | C:G → T:Aa | 0.82, 0.55 | ND4 | Leu → Phe |
| 339, 358, 380 | 10460 | C:G → T:Aa | 0.79, 0.60, 0.62 | COII | Gln → STOP |
NOTE.—Methods for estimating mutation frequencies at heteroplasmic sites are described in the Materials and Methods. Effect shows the impact of mutations on protein-coding function. Cytochrome oxidase I, COI.
Indicates heteroplasmic mutations.
Estimation of the per-generation, per–base pair base substitution mutation rate (μbs) for C. briggsae mtDNA based on our data comes with a number of complicating factors. First, for the PB800 MA lines, we are unable to pinpoint the precise origins of certain base substitution events. For both sets of lines, many mutations were in a heteroplasmic state and detected using direct DNA sequencing alone; thus, we were unable to effectively identify and account for low-frequency variants (Hancock et al. 2005; Theves et al. 2006). Our analyses were also based on bulk DNA prepared from many thousands of nematodes—it is possible that there is variation among individual MA line nematodes and/or among somatic cells within an individual nematode for which we are unable to account. μbs Values can be estimated for the C. briggsae MA line mtDNA, with the preceding caveats in mind and under the assumption that all base substitution differences shared by more than one MA line of a common progenitor genotype resulted from a single mutational event in the expansion phase prior to MA line bottlenecking. Under these conditions, we estimate μbs = 1.1 × 10−7 (standard error of mean [SEM] = 3.8 × 10−8) for HK104 mtDNA and μbs = 7.2 × 10−8 (SEM = 3.4 × 10−8) for PB800 mtDNA (see Materials and Methods). These values are very close to the μbs estimate for C. elegans mtDNA (Denver et al. 2000), 9.7 × 10−8 (SEM = 2.7 × 10−8). Approximate 95% confidence intervals overlap all three estimates (fig. 2B), and application of Fisher’s Exact test revealed no significant μbs differences (P > 0.8 for both between- and within-species comparisons). We thus conclude that the rates of base substitution events are likely highly similar between C. elegans and C. briggsae and between the two C. briggsae natural isolate genotypes analyzed.
Despite the similarities in μbs estimates between species and between C. briggsae strains, marked differences were observed among all three sets of MA lines in terms of the spectrum of base substitution types. We tested for significance in the base substitution spectrum, under the same assumptions and caveats applied to the μbs analysis and including all heteroplasmic changes, in terms of the numbers of observed nonstrand-specific base substitution types (six categories). We found a significant difference between C. briggsae (HK104 and PB800 data combined) and C. elegans (P = 0.012, Fisher's Exact test), though the difference between the two C. briggsae strains was just outside of the conventional cutoff for statistical significance (P = 0.082, Fisher's Exact test). We thus conclude that the spectrum of mtDNA base substitution types differs between Caenorhabditis species. The spectrum might also vary within C. briggsae; a larger scale of mtDNA mutational analysis will be required to effectively address this possibility.
Discussion
This study shows that the mitochondrial mutation process can vary in a variety of ways between animal species of the same genus. Both sets of C. briggsae MA lines were observed to accumulate large heteroplasmic deletions at >10-fold higher average rates than C. elegans. The ΨND5-2/ND5 region is prone to deletions in both the HK104 and the PB800 MA lines, and one deletion type (the ΨND5-2/ND5 boundary deletion) persists in multiple genetically diverse C. briggsae natural isolates at varying levels in nature (Howe and Denver 2008). The ND1/ATP6/ND2 region, however, was only observed to accumulate large deletions in the PB800 MA lines. A previous analysis of C. elegans MA line mtDNA (Denver et al. 2000) revealed the presence of only one large heteroplasmic deletion out of 74 MA line mitochondrial genomes analyzed at 214 generations; the C. elegans deletion involved the cytochrome b region rather than the ΨND5-2/ND5 and ND1/ATP6/ND2 regions observed here to be deletion hot spots in C. briggsae. No large heteroplasmic deletions were observed in D. melanogaster MA line mitochondrial genomes (Haag-Liautard et al. 2008). The previous C. elegans MA line mtDNA analysis (Denver et al. 2000), however, used an approach whereby 19 small PCR products were used to probe for mutations around the mitochondrial genome rather than 4 large overlapping amplicons as applied here to C. briggsae. Thus, it is possible that the C. elegans large deletion rate is an underestimate because some large deletion events, involving sequences outside the PCR primer boundaries, were missed. Although this possibility cannot be ruled out, a very large deletion (416 bp) was detected in the C. elegans study and 5/10 large mtDNA deletion types observed here in the C. briggsae MA lines were smaller than the mean amplicon size in the C. elegans study of Denver et al. (2000). These observations, combined with the fact that C. briggsae nematodes are known to experience large mtDNA deletions in natural populations (Howe and Denver 2008), suggest that C. briggsae mtDNA is inherently more susceptible to large deletion events than is C. elegans mtDNA. It is also noteworthy that the C. briggsae mitochondrial genomes analyzed here harbor two pseudogene regions (ΨND5-1 and ΨND5-2) that are not present in C. elegans (Howe and Denver 2008). These combined observations suggest that there are differences between C. briggsae and its congener C. elegans in terms of their susceptibilities to large mtDNA insertion events, as well. Although we cannot pinpoint the underlying reasons why C. briggsae is much more prone to large mtDNA deletions than C. elegans, we speculate that between-species variation in the frequency of intra/intergenomic recombination processes, known to occur in nematodes (Lunt and Hyman 1997), might be responsible for both the high deletion rate and the presence of ND5-derived mtDNA pseudogenes in C. briggsae.
The pattern of mtDNA base substitution also differed between C. briggsae and C. elegans MA lines. The previous study of C. elegans MA line mtDNA mutation processes revealed 16 base substitution events, all of which were in a fixed state (Denver et al. 2000). The majority of base substitutions observed in the C. elegans MA lines were T:A → C:G transitions (8/16), which deviates from patterns observed in both C. briggsae MA line sets analyzed here, though especially in the PB800 MA lines where C:G → T:A transitions were the only type of base substitution observed. Although the small numbers of observed base substitution mutation events specific to a set of MA lines likely preclude a meaningful statistical analysis of within-species variation in mtDNA base substitution mutation processes, the observed pattern of variation between HK104 and PB800 (table 2) suggests that there might be large differences among different C. briggsae natural isolates. Multiple MA line base substitution polymorphisms were observed to be in a heteroplasmic state in both sets of 250-generation C. briggsae MA lines, whereas all base substitutions previously identified in 214-generation C. elegans MA lines were fixed differences, possibly reflecting a wider mtDNA bottleneck in C. briggsae relative to C. elegans.
Although our analysis approach successfully revealed some aspects of the heteroplasmic nature of the mtDNA mutation process, many features of nematode mtDNA heteroplasmy remain mysterious—in particular, the abundance and persistence patterns of low-frequency variants and the linkage relationships between different variants in the population of mitochondrial genomes within an individual. We are optimistic, however, that the application of new high-throughput DNA sequencing technologies to larger numbers of MA lines will provide an avenue for better understanding the enigma of heteroplasmy and natural variation in mitochondrial mutation processes.
Supplementary Material
Supplementary tables S1 and S2 and figures S1, S2, and S3 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We thank Mark Dasenko at the Oregon State University Center for Genome Research and Biocomputing for sequencing support and Frank Shaw at Hamline University for statistical advice. This work was supported by National Institutes of Health grant (GM087678) to C.F.B. and D.R.D.
References
- Arai T, et al. Age-related mitochondrial DNA deletion in human heart: its relationship with cardiovascular diseases. Aging Clin Exp Res. 2003;15:1–5. doi: 10.1007/BF03324472. [DOI] [PubMed] [Google Scholar]
- Baer CF, Miyamoto MM, Denver DR. Mutation rate variation in multicellular eukaryotes: causes and consequences. Nat Rev Genet. 2007;8:619–631. doi: 10.1038/nrg2158. [DOI] [PubMed] [Google Scholar]
- Baer CF, et al. Comparative evolutionary genetics of spontaneous mutations affecting fitness in rhabditid nematodes. Proc Natl Acad Sci U S A. 2005;102:5785–5790. doi: 10.1073/pnas.0406056102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barazzoni R, Short KR, Nair KS. Effects of aging on mitochondrial DNA copy number and cytochrome c oxidase gene expression in rat skeletal muscle, liver, and heart. J Biol Chem. 2000;275:3343–3347. doi: 10.1074/jbc.275.5.3343. [DOI] [PubMed] [Google Scholar]
- Biskup S, Moore DJ. Detrimental deletions: mitochondria, aging and Parkinson’s disease. Bioessays. 2006;28:963–967. doi: 10.1002/bies.20471. [DOI] [PubMed] [Google Scholar]
- Chinnery PF, et al. Risk of developing a mitochondrial DNA deletion disorder. Lancet. 2004;364:592–596. doi: 10.1016/S0140-6736(04)16851-7. [DOI] [PubMed] [Google Scholar]
- Conley KE, Marcinek DJ, Villarin J. Mitochondrial dysfunction and age. Curr Opin Clin Nutr Metab Care. 2007;10:688–692. doi: 10.1097/MCO.0b013e3282f0dbfb. [DOI] [PubMed] [Google Scholar]
- Denver DR, et al. Abundance, distribution, and mutation rates of homopolymeric nucleotide runs in the genome of Caenorhabditis elegans. J Mol Evol. 2004;58:584–595. doi: 10.1007/s00239-004-2580-4. [DOI] [PubMed] [Google Scholar]
- Denver DR, Morris K, Lynch M, Vassilieva LL, Thomas WK. High direct estimate of the mutation rate in the mitochondrial genome of Caenorhabditis elegans. Science. 2000;289:2342–2344. doi: 10.1126/science.289.5488.2342. [DOI] [PubMed] [Google Scholar]
- Denver DR, Morris K, Thomas WK. Phylogenetics in Caenorhabditis elegans: an analysis of divergence and outcrossing. Mol Biol Evol. 2003;20:393–400. doi: 10.1093/molbev/msg044. [DOI] [PubMed] [Google Scholar]
- Elkin CJ, et al. High-throughput plasmid purification for capillary sequencing. Genome Res. 2001;11:1269–1274. doi: 10.1101/gr.167801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H, Chu JY. A brief review of short tandem repeat mutation. Genomics Proteomics Bioinformatics. 2007;5:7–14. doi: 10.1016/S1672-0229(07)60009-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gissi C, Iannelli F, Pesole G. Evolution of the mitochondrial genome of Metazoa as exemplified by comparison of congeneric species. Heredity. 2008;101:301–320. doi: 10.1038/hdy.2008.62. [DOI] [PubMed] [Google Scholar]
- Haag-Liautard C, et al. Direct estimation of the mitochondrial DNA mutation rate in Drosophila melanogaster. PLoS Biol. 2008;6:e204. doi: 10.1371/journal.pbio.0060204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock DK, Tully LA, Levin BC. A Standard Reference Material to determine the sensitivity of techniques for detecting low-frequency mutations, SNPs, and heteroplasmies in mitochondrial DNA. Genomics. 2005;86:446–461. doi: 10.1016/j.ygeno.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Holt IJ, Harding AE, Morgan-Hughes JA. Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature. 1988;331:717–719. doi: 10.1038/331717a0. [DOI] [PubMed] [Google Scholar]
- Howe DK, Denver DR. Muller's Ratchet and compensatory mutation in Caenorhabditis briggsae mitochondrial genome evolution. BMC Evol Biol. 2008;8:62. doi: 10.1186/1471-2148-8-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell N, Elson JL, Chinnery PF, Turnbull DM. mtDNA mutations and common neurodegenerative disorders. Trends Genet. 2005;21:583–586. doi: 10.1016/j.tig.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Hu M, Chilton NB, Gasser RB. Long PCR-based amplification of the entire mitochondrial genome from single parasitic nematodes. Mol Cell Probes. 2002;16:261–267. doi: 10.1006/mcpr.2002.0422. [DOI] [PubMed] [Google Scholar]
- Jenuth JP, Peterson AC, Fu K, Shoubridge EA. Random genetic drift in the female germline explains the rapid segregation of mammalian mitochondrial DNA. Nat Genet. 1996;14:146–151. doi: 10.1038/ng1096-146. [DOI] [PubMed] [Google Scholar]
- Liau WS, Gonzalez-Serricchio AS, Deshommes C, Chin K, LaMunyon CW. A persistent mitochondrial deletion reduces fitness and sperm performance in heteroplasmic populations of C. elegans. BMC Genet. 2007;8:8. doi: 10.1186/1471-2156-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightowlers RN, Chinnery PF, Turnbull DM, Howell N. Mammalian mitochondrial genetics: heredity, heteroplasmy and disease. Trends Genet. 1997;13:450–455. doi: 10.1016/s0168-9525(97)01266-3. [DOI] [PubMed] [Google Scholar]
- Lunt DH, Hyman BC. Animal mitochondrial DNA recombination. Nature. 1997;387:247. doi: 10.1038/387247a0. [DOI] [PubMed] [Google Scholar]
- Lynch M, et al. A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc Natl Acad Sci U S A. 2008;105:9272–9277. doi: 10.1073/pnas.0803466105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melov S, Hertz GZ, Stormo GD, Johnson TE. Detection of deletions in the mitochondrial genome of Caenorhabditis elegans. Nucleic Acids Res. 1994;22:1075–1078. doi: 10.1093/nar/22.6.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montooth KL, Rand DM. The spectrum of mitochondrial mutation differs across species. PLoS Biol. 2008;6:e213. doi: 10.1371/journal.pbio.0060213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips N, Salomon M, Custer A, Ostrow D, Baer CF. Spontaneous mutational and standing genetic (co)variation at dinucleotide microsatellites in Caenorhabditis briggsae and Caenorhabditis elegans. Mol Biol Evol. 2009;26:659–669. doi: 10.1093/molbev/msn287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand DM. The units of selection on mitochondrial DNA. Annu Rev Ecol Syst. 2001;32:415–448. [Google Scholar]
- Savolainen V, Cowan RS, Vogler AP, Roderick GK, Lane R. Towards writing the encyclopedia of life: an introduction to DNA barcoding. Philos Trans R Soc Lond B Biol Sci. 2005;360:1805–1811. doi: 10.1098/rstb.2005.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JB, et al. Strong purifying selection in transmission of mammalian mitochondrial DNA. PLoS Biol. 2008;6:e10. doi: 10.1371/journal.pbio.0060010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Theves C, et al. Detection and quantification of the age-related point mutation A189G in the human mitochondrial DNA. J Forensic Sci. 2006;51:865–873. doi: 10.1111/j.1556-4029.2006.00163.x. [DOI] [PubMed] [Google Scholar]
- Upholt WB, Dawid IB. Mapping of mitochondrial DNA of individual sheep and goats: rapid evolution in the D loop region. Cell. 1977;11:571–583. doi: 10.1016/0092-8674(77)90075-7. [DOI] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei YH, Lee HC. Oxidative stress, mitochondrial DNA mutation, and impairment of antioxidant enzymes in aging. Exp Biol Med (Maywood). 2002;227:671–682. doi: 10.1177/153537020222700901. [DOI] [PubMed] [Google Scholar]
- White DJ, Wolff JN, Pierson M, Gemmell NJ. Revealing the hidden complexities of mtDNA inheritance. Mol Ecol. 2008;17:4925–4942. doi: 10.1111/j.1365-294X.2008.03982.x. [DOI] [PubMed] [Google Scholar]
- Wong LJ. Diagnostic challenges of mitochondrial DNA disorders. Mitochondrion. 2007;7:45–52. doi: 10.1016/j.mito.2006.11.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.