Abstract
Rice is one of the most important crops, feeding more than half of the world population. There are two cultivated species, the African rice Oryza glaberrima and the Asian rice O. sativa. Although the African species is gradually replaced by O. sativa in most of African rice agrosystems, this species represents an important reservoir of genes of agronomical interest. Their exploitation for the development of modern African rice varieties requires a good understanding of the genetic relationships between the two cultivated species. We took advantage of the recent availability of the sequence of the chromosome 3 short arm of O. glaberrima to estimate the date of radiation between O. glaberrima and O. sativa lineages, using all the long terminal repeat (LTR)-retrotransposons as paleogenomic markers. We first demonstrated that in two distinct lineages, LTR-retrotransposons mutate at the same rate. Based on LTR-retrotransposons shared by both species in orthologous position, we then estimated that O. glaberrima and O. sativa progenitors diverged 1.2 Ma. This constitutes one of the first studies using such a large sample of transposable elements to reconstruct the phylogeny of species. Given the number of genome sequencing projects, there is no doubt that such approach will allow to resolve phylogenetic incongruities. The application of this method to other plant genomes will also facilitate further understanding of evolution of LTR-retrotransposons and eventually of the whole genome in divergent plant lineages.
Keywords: LTR-retrotransposons, mutation rate, paleogenomic markers, phylogeny, genome evolution
Introduction
Rice is one of the most important crops, representing the primary source of food for more than half of the world population. There are two cultivated rice species, Asian rice Oryza sativa and African rice O. glaberrima. Based on both archaeological and molecular data, O. sativa is thought to have originated from two independent domestications in Asia from the wild species O. rufipogon. The subspecies O. sativa ssp. japonica originated in the yellow river basin; whereas, the subspecies O. sativa ssp. indica originated from the southern foothills of the Himalayan chain (Second 1982; Vitte et al. 2004). These two subspecies were thus domesticated from two distinct gene pools of the wild relative O. rufipogon that were genetically isolated by the Himalayas. African rice, on the other hand, was domesticated from the wild relative O. barthii 2,000 to 3,000 years ago in the Niger river delta (Viguier 1939; Porteres 1962, 1976). However, it has been gradually replaced by the Asian species O. sativa since its introduction in the 15th century by the Portuguese (Porteres 1962; Linares 2002). Several cultivars of O. glaberrima are still grown in traditional African rice agrosystems. Moreover, the species is known to exhibit various agronomically favorable traits, such as a good tolerance to various biotic and abiotic stress. It therefore represents an important reservoir of genes of interest for the varietal improvement of modern African rice (Jones et al. 1997). As an example, a new germplasm (New Rice for Africa [NERICA]) has been developed in the late 90’s through interspecific cross between O. sativa and O. glaberrima constituting a promising variety for upland rice agrosystems (Jones et al. 1997). In this context, the understanding of the evolution and phylogenetic relationships between the cultivated rice species is essential in order to fully exploit the genetic diversity of the available gene pools. Recently, a number of studies clarified the phylogenetic relationship between cultivated and wild rice belonging to the AA genome type but the time of radiation between O. sativa and O. Glaberrima progenitors (i.e., the two gene pools at the origin of both domesticated species) remains unclear (Zhu and Ge 2005; Duan et al. 2007; Xu et al. 2007; Zou et al. 2008).
Transposable elements are mobile DNA sequences able to move from one location to another in their host genome. They are divided into two classes according to their mode of transposition. Most of class II elements or transposons, transpose through a cut and paste mechanism, that is, the element is excised and integrated elsewhere in the genome. Class I elements, the retrotransposons transpose through a “copy and paste” mechanism, that is, after their transcription, the RNA is reverse transcribed and integrated into the genome, leading to the duplication of the original copy. Long terminal repeat (LTR)-retrotransposons are elements of class I which are ubiquitous in plants. It has been shown that these elements are the main cause of genome size increase in the genus Oryza (Piegu et al. 2006), in cotton (Hawkins et al. 2006) or in maize (SanMiguel et al. 1998). There is a very low probability that a transposable element insertion occurs twice independently at an exact orthologous position in two distinct genomes. The presence of a LTR-retrotransposon at the same position in several genomes indicates that the insertion occurred before the radiation of the species as LTR-retrotransposons do not excise after insertion. Therefore, the presence of an element at a given locus in a lineage and its absence in the genome of another one indicates that the insertion is posterior to the radiation. In addition, the age of a LTR-retrotransposon insertion can be easily estimated by using the divergence observed between the two LTRs of the element (SanMiguel et al. 1998). In this study, we used the LTR-retrotransposons found in O. sativa ssp. japonica, O. sativa ssp. indica, and O. glaberrima as paleogenomic markers to estimate the age of the radiation between these three species progenitors.
The full genomes of O. sativa ssp. japonica and of O. sativa ssp. indica are currently available (Yu et al. 2002; International Rice Genome Sequencing Project 2005). Wild rice species represent a reservoir of genetic diversity and of genes of interest used for crop improvement. A large amount of genomic resources (such as BAC end sequences or BES) has been generated by the Oryza Map Alignment Project concerning 12 wild rice species (http://www.omap.org/, Ammiraju et al. 2006). This project also generated the complete sequence of the short arm of chromosome 3 for different species, including O. glaberrima. The availability of these genomic resources enabled us to perform a wide paleogenomic analysis on LTR-retrotransposons with the objective of dating the radiation between the Asian and the African rice species.
Materials and Methods
Identification of LTR-Retrotransposon in Orthologous Position
The LTR-retrotransposons inserted into the chromosome 3 short arm of O. sativa japonica have been identified with LTR-finder (http://tlife.fudan.edu.cn/ltr_finder/, Xu and Wang 2008). For each element of O. sativa japonica, we extracted 500 bp flanking the insertion. The chromosome 3 short arm of O. glaberrima is 17.1 Mbp long, and it has been recently sequenced through a BAC to BAC sequencing strategy based on a physical map of the genome. This sequence was kindly provided by Rod Wing and Steve Rounsley. The 500 bp extracted from the genome of O. sativa was used to perform a Blast search (Altschul et al. 1990) against the sequence of the chromosome 3 short arm of O. glaberrima. When it yielded more than one hit (indicating that the flanking sequence is repeated) or if there was no hit (indicating the absence of the flanking sequence in the genome), the insertion was considered as noninformative. When a single hit was obtained, a region of 25 kb spanning the flanking sequence was extracted from the genomic sequence of O. glaberrima. This region was compared with its ortholog in O. sativa using a dot plot analysis (Sonnhammer and Durbin 1995). When the element and its flanking sequence were found in both genomes, then it was considered to have been inserted before the speciation. The genomic sequence of O. sativa indica has been obtained by a whole-genome shotgun sequencing (Yu et al. 2002). We performed a similar Blast search using the nonassembled indica contigs, using the method proposed by Vitte et al. (2004). For a given element of O. sativa japonica, we eventually obtained information about its presence in orthologous position in O. sativa indica and O. glaberrima.
Using the same method (described in the previous paragraph), a reverse analysis was performed using the LTR-retrotransposons identified by LTR-finder in the sequence of the short arm sequence of O. glaberrima as query.
Computation of Sequence Divergence
For each complete element identified through LTR-finder (i.e., harboring both LTRs and at least a part of the internal region), both LTRs were aligned using ClustalX (Thompson et al. 1997). The alignment was corrected by hand using SEAVIEW software (Galtier et al. 1996), and the sequence divergence (dintra) was computed with ClustalW (Larkin et al. 2007). The observed divergence was translated into an insertion date using a substitution rate of 1.3 × 10−8 mutation/site/year (Ma and Bennetzen 2004).
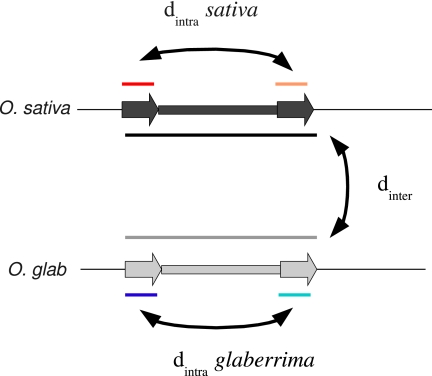
For each element identified in orthologous position, the sequences of O. sativa and O. glaberrima were aligned using ClustalX. The alignment was corrected by hand using SEAVIEW software, and the sequence divergence (dinter) was computed with ClustalW (fig. 1).
FIG. 1.—
Calculation of dintra et dinter. The arrows represent LTRs sequences. dintra is calculated using both LTRs of a given element, for each species. dinter is calculated using the two whole sequences of LTR-retrotransposons found in orthologous position in Oryza sativa and O. glaberrima.
Computation of the O. sativa/O. glaberrima Molecular Clock Ratio
The ratio of the divergence rate of the LTR-retrotransposons between the O. sativa and the O. glaberrima lineages was computed, taking into account the intraelement divergence for each species (dintraS and dintraG for O. sativa and O. glaberrima, respectively), the interelement divergence (dinter), the sequence divergence between the two LTRs before radiation dorg, the respective divergence rate of O sativa and O. glaberrima HS and HG, and the time since speciation. The explanation of the computation method is detailed in supplementary figure S1 (Supplementary Material online).
Statistical Analysis
All statistical analysis was performed using the R software (R Development Core Team 2005). The test of Shapiro–Wilk was used to test the normality of the distribution of the divergence rate ratio in the case of orthologous insertions. The conformity of mean has been tested with t-test for one variable measurement, and the correlation between dintraS and dintraG was tested with Pearson coefficient test.
Results
56 LTR-Retrotransposons Are Found in Orthologous Position between O. sativa and O. glaberrima
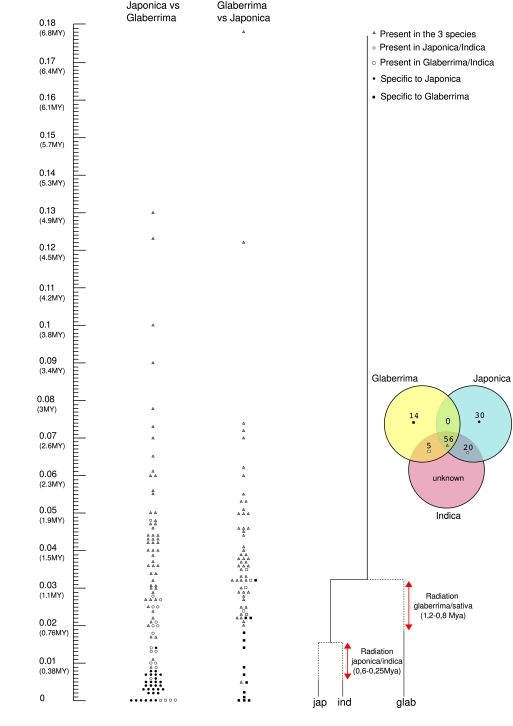
From this study, we identified 167 and 91 LTR-retrotransposons on the short arm of the chromosome 3 of O. sativa japonica and O. glaberrima, respectively, using LTR-finder (Xu and Wang 2008). Due to the small size of the shotgun reads, the sequence of the chromosome 3 short arm of indica's was not submitted to LTR-finder search. Only 106 and 74 LTR-retrotransposons in O. sativa japonica and O. glaberrima, respectively, were subjected to further analyses because clear orthologous relationships of the insertion site were established between the three genomes of O. sativa japonica, O. glaberrima, and O. sativa indica (supplementary table S1, Supplementary Material online). The graphical representation of these elements is provided in a venn diagram where the number of elements is given per species (fig. 2). Figure 2 shows that 56 LTR-retrotransposon insertions are common to the three genomes, 20 in O. sativa japonica and O. sativa indica (but not in O. glaberrima), 5 in O. glaberrima and O. sativa indica (but not in O. sativa japonica), 30 in O. sativa japonica only, and 14 in O. glaberrima only.
FIG. 2.—
Gaphical representation of the LTR-retrotransposons found in Oryza sativa and O. glaberrima in function of their dintra. The scale corresponds to the divergence observed, translated in a date provided in million years (Myr, see Materials and methods). The left part corresponds to the elements identified in O. sativa. The right part corresponds to the elements found in O. glaberrima. The venn diagram shows the number of LTR-retrotransposons shared between the three species.
Dating the Speciation of Cultivated Rice
LTR-Retrotransposons Evolve at a Similar Rate in O. sativa and O. glaberrima Lineages
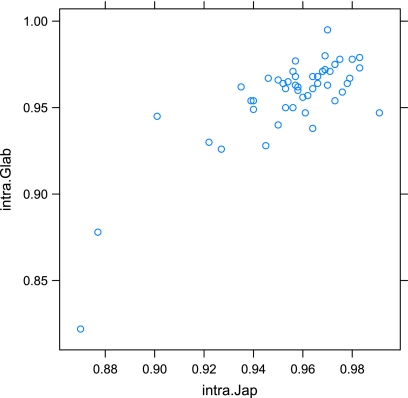
From a total of 56 elements conserved in orthologous position between O. sativa japonica and O. glaberrima, only 46 were complete in both species, that is, harboring at least the two LTRs and a part of the internal region necessary for the computation of the divergence rate ratio HS/HG. For these 46 elements, we estimated the observed divergence between their LTRs in each species (dintra.). We also computed the divergence between the two complete elements between each species (dinter, fig. 1). In the case of O. sativa indica, the computation of dintra and dinter was not possible due to the short size of the shotgun reads. The HS/HG ratio (supplementary table S1, Supplementary Material online) fits a normal distribution (W = 0.9769, P value = 0.4988) and the mean of the distribution (m = 1.15) is not significantly different from 1 (t = 1.0001, degrees of freedom = 45, P value = 0.3226). These results show that there is no significant difference in the divergence rate of the LTR-retrotransposons between the O. sativa and the O. glaberrima lineages. This is further confirmed by the clear correlation between the dintraS and dintraG values (r2 = 0.8, P value < 0.001, see fig. 3). We therefore used the same molecular clock for both species (i.e., the one proposed by Ma and Bennetzen 2004) to estimate the radiation date between both species.
FIG. 3.—
Correlation between dintraS and dintraG. The correlation coefficient r2 is equal to 0.8 (P value < 0.0001).
Dating the Divergence of O. sativa Subspecies and O. glaberrima Progenitors using Orthologous LTR-Retrotransposons
Figure 2 shows the temporal distribution of the LTR-retrotransposon insertions for both O. sativa and O. glaberrima lineages, based on the sequence divergence of their LTRs (dintra). We used this data to tentatively date the radiation between the two species progenitors, which should have occurred after the last common insertion and before the first polymorphic insertion. Concerning the subspeciation between O. sativa japonica and indica progenitors, the oldest polymorphic LTR-retrotransposon insertion corresponds to an element exhibiting a sequence divergence between its two LTRs of 1.7%, whereas the most recent common insertion exhibits a 0.6% divergence. Using the molecular clock of 1.3 × 10−8 substitution/site/year, we estimated that the two subspecies progenitors diverged between 0.6 and 0.25 Ma. Using the same method, we estimated that O. sativa and O. glaberrima progenitors diverged between 1.2 and 0.8 Ma (fig. 2).
Introgressions may cause a bias in the estimation of the radiation date, resulting in the presence of old transposable element (TE) insertions in both genomes. Many rice species and more particularly O. sativa and O. glaberrima have been in contact very recently (few hundred years ago). We postulated that introgressed elements shouldn’t have enough time to diverge significantly and should harbor a very low sequence divergence (more or less equal to 0%) between both genomes. Our study revealed four elements perfectly conserved (i.e., harboring 0% of sequence divergence between O. sativa japonica and indica [fig. 2]). The variety 93-11 is not a pure indica type because it has been introgressed by japonica (Huang et al. 2008). We therefore considered the four elements perfectly conserved to have been introgressed from one subspecies to the other. These have therefore not been taken into account for the dating of the subspeciation japonica/indica. Interestingly, one element found in orthologous position in the three genomes harbors a dintra of 0.5% in O. glaberrima (triangle harboring less than 0.5% of sequence divergence in figure 2, O. glaberrima vs. O. sativa japonica), whereas it harbors a dintra of 3% in O. sativa japonica and the ratio HS/HG = 6.55. This element clearly evolved slower in O. glaberrima than in O. sativa and therefore has not been taken into account for the dating of the radiation of both species.
Discussion
LTR-Retrotransposons Evolution
Our results show that the divergence rate of LTR-retrotransposons is similar between two distinct lineages of the genus Oryza. This is the first time that such a study is conducted based on a full chromosome sequence that allows the establishment of clear orthologous relationships between TE insertions of two distinct species. TEs are known to be involved in gene regulation through several mechanisms that involve epigenetic modifications (Slotkin and Martienssen 2007), domestication of their promoters (Feschotte 2008), or genomic recombination mediated by repeated sequences (Raskina et al. 2008). Therefore, the understanding of their dynamics constitutes an important question in the field of molecular evolution. It is well known that LTR-retrotransposons are highly dynamic genomic components and that their transposition is rapidly counterbalanced by deletions leading to their elimination (Vitte and Panaud 2003; Ma et al. 2004; Vitte et al. 2007). Moreover, mutations are responsible for generating transposable elements family diversity. However, little is known about the variation in the dynamics of this process among plant lineages. Previous studies tried to estimate the mutation rate of LTR-retrotransposons in rice (Vitte and Panaud 2003; Ma and Bennetzen 2004) but the use of the molecular clock is still subject to debate. Studies have demonstrated that the mutation rate of genes (Smith and Donoghue 2008) and, more particularly, mitochondrial genes (Cho et al. 2004) is subject to variation between distant flowering plants, especially when species harbor different life-history traits (Smith and Donoghue 2008). No evidence is available that demonstrates that all LTR-retrotransposons evolve at the same rate in two distinct species or at least in a given genome. Here, we show that all but one LTR-retrotransposon that we found in orthologous position in O. sativa japonica and O. glaberrima have evolved at the same rate. Since cultivated and wild rice species display similar life-history trait (such as generation time, development time, and reproduction), we strongly conclude that, at least for this genus, the rate that was estimated by Ma and Bennetzen (2004) (1.3 × 10−8 mutations/site/years) could be applied with confidence for further paleogenomic studies. This approach could be extended to other Oryza species as the genomic sequences of full chromosomes become available for more species.
O. sativa and O. glaberrima Progenitors Diverged 1.2 Ma
Based on interspecific crosses and cytogenetic analysis (Morinaga et al. 1964), 10 genome types, 6 diploids (AA, BB, CC, EE, FF, and GG), and 4 tetraploids (BBCC, CCDD, HHJJ, and HHKK) have been identified (Khush 1997). Cultivated rice belongs to the AA genome type or O. sativa complex that is composed of eight species (Ge et al. 1999; Vaughan et al. 2005). A number of studies based on genes and/or transposable elements (Duan et al. 2007; Xu et al. 2007; Zou et al. 2008) clarified phylogenetic relationship within the AA genome type and, more recently, using introns sequences of four nuclear single-copy genes, the radiation of the A-genome type has been estimated to 2 Myr (Zhu and Ge 2005). The phylogenetic relationships that we inferred for the three studied species are congruent with the ones obtained by these other works and thus validate this approach. Moreover, the authors propose that the progenitors of both subspecies O. sativa japonica and indica diverged 0.4 Ma (Zhu and Ge 2005). These results are quite different from the ones obtained by Wang et al. (2008). Based on the comparison of the whole genome of japonica and indica, 15,916 LTR-retrotransposons have been identified, among which more than 3,100 are shared by both genomes. Using the intraelements divergence, Wang et al. (2008) estimated that the subspeciation occurred 0.6 Ma that corresponds to the estimation that we obtained. Zhu and Ge (2005) also concluded that O. sativa progenitor diverged 0.7 Ma from O. glaberrima progenitor. This estimation is also more recent than what we obtained, that is, 1.2 Ma. However, the study lead by Zhu and Ge (2005) was performed on a small sample of four nuclear genes for which they apply the mutation rate estimated for the Adh1 gene. This might introduce some bias in their estimation and could explain that we obtained a different estimation of the radiation date.
However, it is known that concerted evolution, hybridization, introgression, as well as rapid diversification, and allele sorting are responsible for phylogenetic incongruences (MasonGamer and Kellogg 1996; Zhu and Ge 2005; Doyle et al. 2008). We demonstrated that orthologous transposable elements have evolved at the same rate in both lineages, therefore, we propose that the use of a large sample of transposable elements constitutes a good alternative to the use of genes to date the radiation between lineages. Introgressions can also occur for transposable elements in the same manner as for genes. Recent studies have shown that 15% of the rice genome has undergone intersubspecies nonreciprocal recombinations between O. sativa japonica and O. sativa indica, based on comparative genomics studies of the varieties Nipponbare and 93-11 the genome of which is available (Wang et al. 2008). Moreover, O. sativa and O. glaberrima can cross naturally and give some fertile progenies, despite the presence of some fertility barriers such as the S1 locus which is known to be responsible of gametes abortion in O. sativa × O. glaberrima hybrids (Sano 1990). However, the development of new rice germplasm derived from such interspecific cross, that is, the NERICA rice, demonstrates that interspecific introgressions are possible (Jones et al. 1997). Because O. sativa was introduced in Africa several hundred years ago, one might expect that some introgression may have occurred between these two gene pools in the recent past. Using our approach, recent introgressions may induce a bias in the estimation because it may lead to the presence of old TE insertions in both genomes. As we demonstrated in a previous work (Vitte et al. 2004), very few introgressions between 93-11 (indica type) and Nipponbare (japonica type) occurred after domestication (i.e., after the last 10,000 years) and they don't affect the estimation of the radiation date. In our case, we found only four elements introgressed between O. sativa japonica and O. sativa indica and none between O. sativa japonica and O. glaberrima and therefore did not cause any bias in our analyses. Moreover, in an analysis of distribution of interspecies LTR divergences, these elements would be outliers and easily distinguished.
Transposable element sequences can also be under selective pressure, a process referred to as domestication, leading to the functionalization of either their regulatory or functional domains (Feschotte 2008). Several cases of domestication have been described (Feschotte 2008) but most of the examples concerned only some part of the element, notably the transposase-encoding sequence (Muehlbauer et al. 2006; Casola et al. 2007). However, given the number of transposable element families, domestication still remains an exception rather than a rule. On the other hand, it has also been demonstrated that LTR-retrotransposons genes are submitted to a strong purifying selection (Matsuoka and Tsunewaki 1999; Navarro-Quezada and Schoen 2002) but notably for families that have undergone recent transpositional activity (Baucom et al. 2009). However, except for the regions involved in the regulation of their transcription, LTRs are not supposed to be submitted to purifying or positive selection. At least, we consider that LTRs are subject to less selection than other functional sequences and that it cannot introduce a significant bias in our study. Given the high number of LTR-retrotransposons found in plant genomes, one could anticipate that a large part can be exploited as good markers for paleogenomic surveys such as the one presented here.
Conclusions
The method based on the use of transposable elements as diagnostic sequences has been proposed and applied by several authors (SanMiguel et al. 1998; Vitte et al. 2004; Zhu and Ge 2005) but this work constitutes one of the first study to use such a large sample of LTR-retrotransposons for which clear orthologous relationships can be established. The phylogeny that we obtained with our method is perfectly congruent with the ones based on genes. We demonstrated that LTR-retrotransposons evolve at the same rate in both species and therefore were able to estimate that the radiation between the African and the Asian rice occurred 1.2 Ma. With the availability of more genome sequences for several plant genera, such approach will allow to clarify the phylogenetic relationships among closely related taxa and thus date the timing of radiation events. There is no doubt that such approach will also provide the opportunity to better understand the dynamics of transposable elements and therefore genome evolution among divergent lineages.
Supplementary Material
Supplementary figure S1 and supplementary table S1 are available at Genome Biology and Evolution online (http://www.oxfordjournals.org/our_journals/gbe/).
Acknowledgments
The authors thank Rod Wing and Steve Rounsley for providing us with the complete sequence of chromosome 3 short arm of O. glaberrima. This work was supported by a PhD grant from the French ministry of education and from the CNRS.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ammiraju JSS, Luo M, Goicoechea JL, Wang W, Kudrna D, et al. The Oryza bacterial artificial chromosome library resource: construction and analysis of 12 deep-coverage large-insert BAC libraries that represent the 10 genome types of the genus Oryza. Genome Res. 2006;16(1):140–147. doi: 10.1101/gr.3766306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baucom RS, Estill JC, Leebens-Mack J, Bennetzen JL. Natural selection on gene function drives the evolution of LTR retrotransposon families in the rice genome. Genome Res. 2009;19(2):243–254. doi: 10.1101/gr.083360.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casola C, Lawing AM, Betran E, Feshotte C. PIF-like transposons are common in drosophila and have been repeatedly domesticated to generate new host genes. Mol Biol Evol. 2007;24(8):1872–1888. doi: 10.1093/molbev/msm116. [DOI] [PubMed] [Google Scholar]
- Cho Y, Mower JP, Qiu YL, Palmer JD. Mitochondrial substitution rates are extraordinarily elevated and variable in a genus of flowering plants. Proc Natl Acad Sci U S A. 2004;101(51):17741–17746. doi: 10.1073/pnas.0408302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Flagel LE, Paterson AH, Rapp RA, Soltis DE, et al. Evolutionary genetics of genome merger and doubling in plants. Annu Rev Genet. 2008;42:443–461. doi: 10.1146/annurev.genet.42.110807.091524. [DOI] [PubMed] [Google Scholar]
- Duan S, Lu B, Li Z, Tong J, Kong J, et al. Phylogenetic analysis of AA-genome Oryza species (Poaceae) based on chloroplast, mitochondrial, and nuclear DNA sequences. Biochem Genet. 2007;45:113–129. doi: 10.1007/s10528-006-9062-x. [DOI] [PubMed] [Google Scholar]
- Feschotte C. Transposable element and the evolution of regulatory networks. Nat Rev Genet. 2008;9(5):397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N, Gouy M, Gautier C. Seaview and phylo_win, two graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci. 1996;12(6):543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- Ge S, Sang T, Lu BR, Hong DY. Phylogeny of rice genomes with emphasis on origins of allotetraploid species. Proc Natl Acad Sci U S A. 1999;96(25):14400–14405. doi: 10.1073/pnas.96.25.14400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins JS, Kim H, Nason RA, Wendel JF. Differential lineage-specific amplification of transposable elements is responsible for genome size variation in Gossypium. Genome Res. 2006;16(10):1252–1261. doi: 10.1101/gr.5282906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Lu G, Zhao Q, Liu X, Han B. Genome-wide analysis of transposon insertion polymorphisms reveals intraspecific variation in cultivated rice. Plant Physiol. 2008;148:25–40. doi: 10.1104/pp.108.121491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project. The map-base sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- Jones P, Dingkuhn M, Aluko G-K, Mand S. Interspecific Oryza sativa L. x O. Glaberrima Steud. progenies in upland rice improvement. Euphytica. 1997;92:237–246. [Google Scholar]
- Khush GS. Origin, dispersal, cultivation and variation of rice. Plant Mol Biol. 1997;35:25–34. [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NAP, Chenna R, McGettigan PA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Linares O. African rice (Oryza glaberrima): history and future potential. Proc Natl Acad Sci U S A. 2002;99(25):16360–16365. doi: 10.1073/pnas.252604599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Bennetzen JL. Rapid recent growth and divergence of rice nuclear genomes. Proc Natl Acad Sci U S A. 2004;101(34):12404–12410. doi: 10.1073/pnas.0403715101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Devos KM, Bennetzen JF. Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res. 2004;14:860–869. doi: 10.1101/gr.1466204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MasonGamer RJ, Kellogg EA. Testing for phylogenetic conflict among molecular data sets in the tribe Triticeae (Gramineae) Syst Biol. 1996;45(4):524–545. [Google Scholar]
- Matsuoka Y, Tsunewaki K. Evolutionary dynamics of Ty1-copia group retrotransposons in grass shown by reverse transcriptase domain analysis. Mol Biol Evol. 1999;16(2):208–217. doi: 10.1093/oxfordjournals.molbev.a026103. [DOI] [PubMed] [Google Scholar]
- Morinaga T, Tsunoda S, Takahashi N. Rice genetics and cytogenetics. Amsterdam (The Netherlands): Elsevier; 1964. [Google Scholar]
- Muehlbauer GJ, Bhau BS, Syed NH, Heinen S, Cho S, et al. A hAT superfamily transposase recruited by the cereal grass genome. Mol Genet Genomics. 2006;275(6):553–563. doi: 10.1007/s00438-006-0098-8. [DOI] [PubMed] [Google Scholar]
- Navarro-Quezada A, Schoen DJ. Sequence evolution and copy number of Ty1-copia retrotransposons in diverse plant genomes. Proc Natl Acad Sci U S A. 2002;99(1):268–273. doi: 10.1073/pnas.012422299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piegu B, Guyot R, Picault N, Roulin A, Saniyal A, et al. Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res. 2006;16(10):1262–1269. doi: 10.1101/gr.5290206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porteres R. Primary agriculture birthplace in Africa (Berceaux agricoles primaires sur le continent africain) J Afr Hist. 1962;3(2):195–210. [Google Scholar]
- Porteres R. In: The origin of African plant domestication. Harlan J-R, de Wet J-M, Stemler ABL, editors. The Hague (The Netherlands): Mouton; 1976. pp. 409–452. [Google Scholar]
- R Development Core Team. R: a language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing; 2005. Available from: http://www.R-project.org. ISBN 3-900051-07-0. [Google Scholar]
- Raskina O, Barber JC, Nevo E, Belyayev A. Repetitive DNA and chromosomal rearrangements: speciation-related events in plant genomes. Cytogenet Genome Res. 2008;120(3–4):351–357. doi: 10.1159/000121084. [DOI] [PubMed] [Google Scholar]
- SanMiguel P, Gaut BS, Tikhonov A, Nakajima Y, Bennetzen JL. The paleontology of intergene retrotransposons of maize. Nat Genet. 1998;20(1):43–45. doi: 10.1038/1695. [DOI] [PubMed] [Google Scholar]
- Sano Y. The genic nature of gamete eliminator in rice. Genetics. 1990;125(1):183–191. doi: 10.1093/genetics/125.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Second G. Origin of the genetic diversity of cultivated rice (Oryza spp.): study of the polymorphism scored at 40 isozyme loci. Jpn J Genet. 1982;57:25–57. [Google Scholar]
- Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8(4):272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- Smith SA, Donoghue MJ. Rates of molecular evolution are linked to life history in flowering plants. Science. 2008;322(5898):86–89. doi: 10.1126/science.1163197. [DOI] [PubMed] [Google Scholar]
- Sonnhammer EL, Durbin R. A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene. 1995;167(3):GC1–G10. doi: 10.1016/0378-1119(95)00714-8. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The clustal_x windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25(24):4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan DA, Kadowaki KI, Tomooka N. On the phylogeny and biogeography of the genus Oryza. Breed Sci. 2005;55:113–122. [Google Scholar]
- Viguier P. Indigenous rice culture in Soudan (La riziculture indigène au Soudan français) In: Larose, editor. Paris (France): Larose; 1939. pp. 152–158. [Google Scholar]
- Vitte C, Ishii T, Lamy T, Brar D, Panaud O. Genomic paleontology provides evidence for two distinct origins of Asian rice (Oryza sativa L.) Mol Genet Genomics. 2004;272:504–511. doi: 10.1007/s00438-004-1069-6. [DOI] [PubMed] [Google Scholar]
- Vitte C, Panaud O. Formation of solo-LTRs through unequal homologous recombination counterbalances amplifications of LTR retrotransposons in rice Oryza sativa L. Mol Biol Evol. 2003;20(4):528–540. doi: 10.1093/molbev/msg055. [DOI] [PubMed] [Google Scholar]
- Vitte C, Panaud O, Quesneville H. LTR retrotransposons in rice (Oryza sativa, L.): recent burst amplifications followed by rapid DNA loss. BMC Genomics. 2007;8:218. doi: 10.1186/1471-2164-8-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Xu Z, Yu H. LTR retrotransposons reveal recent extensive inter-subspecies nonreciprocal recombination in Asian cultivated rice. BMC Genomics. 2008;27(9):565. doi: 10.1186/1471-2164-9-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu JH, Cheng C, Tsuchimoto S, Ohtsubo H, Ohtsubo E. Phylogenetic analysis of Oryza rufipogon strains and their relations to Oryza sativa strains by insertion polymorphism of rice SINEs. Genes Genet Syst. 2007;82:217–229. doi: 10.1266/ggs.82.217. [DOI] [PubMed] [Google Scholar]
- Xu Z, Wang H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 2008;35(Web Server issue):W265–W268. doi: 10.1093/nar/gkm286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK, Li S, et al. A draft sequence of the rice genome (Oryza sativa ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Ge S. Phylogenetic relationships among A-genome species of the genus Oryza revealed by intron sequences of four nuclear genes. New Phytol. 2005;167:249–265. doi: 10.1111/j.1469-8137.2005.01406.x. [DOI] [PubMed] [Google Scholar]
- Zou XH, Zhang F, Zhang JG, Zang LL, Tang L, et al. Analysis of 142 genes resolves the rapid diversification of the rice genus. Genome Biol. 2008;9(3):R49. doi: 10.1186/gb-2008-9-3-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.