Abstract
RanGTPase belongs to the Ras superfamily of small GTPases. It possesses a distinctive acidic C-terminal DEDDDL motif and predominantly localizes to the nucleus. RanGTPase is known to regulate nucleocytoplasmic trafficking as well as mitotic spindle and nuclear envelope formation. Ran-directed nucleocytoplasmic trafficking is an energy-dependent directional process that also depends on nuclear import or export signals. Ran-directed nucleocytoplasmic trafficking is also facilitated by several cellular components, including RanGTPase, karyopherins, NTF2 and nucleoporins. GTP-bound Ran is asymmetrically distributed in the nucleus, while GDP-bound Ran is predominantly cytoplasmic. Controlled by RanGEF and RanGAP, RanGTPase cycles between the GDP- and GTP-bound states enabling it to shuttle cargoes in an accurate spatial and temporal manner. RanGTPase plays a role in the nuclear import in such a way that GTP-bound Ran dissociates importin:cargo complex in the nucleus and recycles importin back to cytoplasm. Likewise, RanGTPase plays a role in the nuclear export in such a way that nuclear GTP-bound Ran triggers the aggregation of Ran:exportin:cargo trimeric complex which is then transported to cytoplasm while hydrolysis of RanGTP to RanGDP releases the export cargoes in cytoplasm. RanGTPase has been reported to be essential for cell viability and its over-expression is linked to tumorigenesis. Thus, RanGTPase plays a crucial role in regulating key cellular events and alterations in its expression may lead to cancer development and/or progression.
Keywords: Exportins, Importins, Nucleocytoplasmic Trafficking, RanGTPase, Ras, RBEL1
Introduction
In eukaryotes, cell membranes act as barriers to separate individual cells from the surrounding environment and likewise, intracellular membranes, including nuclear membrane, serve to compartmentalize intracellular components. To facilitate the exchange of molecules across membranes, transmembrane protein channels have evolved spanning the cell membranes. In the case of nuclear membrane, nuclear pore complexes have been identified that are embedded in the nuclear envelope and act as gates to selectively filter and transport proteins/RNAs between cytoplasm and the nucleus. For example, the p53 tumor suppressor is a transcription factor that constantly shuttles between the nucleus and cytoplasm and its nuclear influx and efflux are determined by its post-translational modifications such as phosphorylation (1) and ubiquitination (1, 2). By increasing the nuclear influx of p53 and/or blocking the nuclear efflux, cells can reversibly shift their growth control machinery from proliferation to growth arrest. Hence, cells maintain homeostasis and, more importantly, execute immediate responses through the control of nuclear import or export of proteins.
The nucleocytoplasmic trafficking is a well-regulated process that involves a number of major components, including RanGTPase, karyopherins (carrier proteins), NTF2 (Ran nuclear import factor) and nucleoporins (proteins of NPC). RanGTPase, in particular, acts as a key controller of the entire nucleocytoplasmic trafficking process. Here we discuss the current state of knowledge on the role of RanGTPase in controlling nucleocytoplasmic trafficking.
Overview of nucleocytoplasmic trafficking
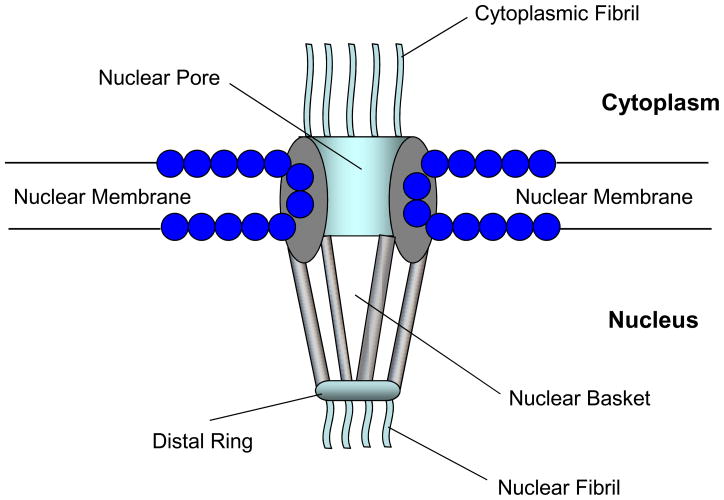
Nucleocytoplasmic trafficking is utilized by cells to shuttle materials between cytoplasm and the nucleus through the nuclear pore complex (NPC). NPC-an assembly of more than 30 different protein called nucleoporins (Nups)-is a basket-like symmetric, octagonal structure embedded in the nuclear envelope (3) (Figure 1). Its major function is to selectively filter proteins across nuclear membrane. High Resolution Electron Microscopy (HREM) has deciphered the structure of NPC and indicates that there are eight flexible cytoplasmic fibrils protruding from the NPC, while eight nuclear fibrils extending from the basket base towards the nucleoplasm (4), and that they possess FG repetitive motifs. The FG motifs serve to interact with carrier proteins, and this interaction is believed to mediate cargo translocation (3). For example, through a series of weak association and dissociation between karyopherins and the FG motifs, a translocation force is generated that is believed to propel the cargo (5). Although water, ions and molecules smaller than 40kD are able to freely diffuse across the NPC (6), macromolecules, such as mRNA, ribosomal proteins, and transcription factors have to depend on nucleocytoplasmic trafficking carrier proteins and the RanGTPase to facilitate their movement.
Figure 1.
A schematic showing structural components of the nuclear pore complex.
Nucleocytoplasmic trafficking is a directional, signal peptide and energy-dependent process (Figure 2). This is a rapid process, and in the presence of RanGTPase and soluble carrier proteins, it can reach its maximum rate of a few hundred macromolecules per second per NPC (3, 7). The soluble carrier proteins are collectively named as karyopherins and can be subcategorized into 2 subgroups, based on their functions, as “importins” for nuclear import and “exportins” for nuclear export. Both types of karyopherins possess 3 functional domains, including (i) Ran-binding domain at the N-terminus, (ii) nucleoporin-binding domain in the middle and (iii) cargo-binding domain at the C-terminus (8).
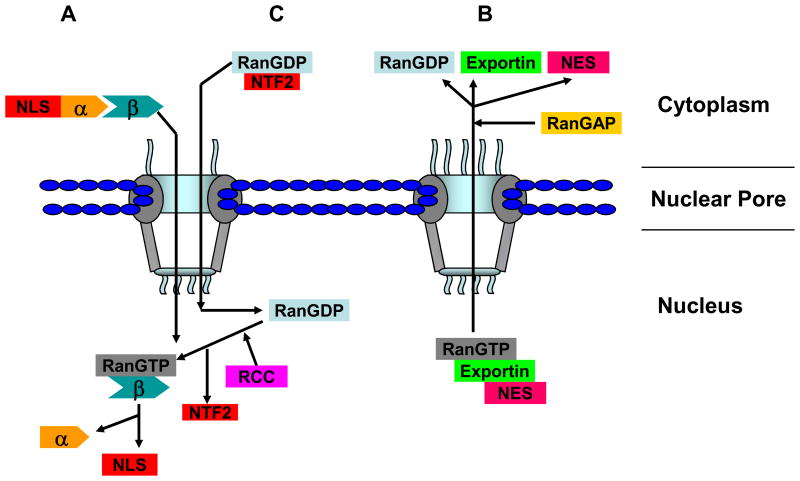
Figure 2. A schematic illustration depicting the events associated with nucleocytoplasmic trafficking.
Nuclear import is shown on the left (A) while nuclear export is shown on the right (B) through the nuclear pore complex (NPC). As shown in (A), a NLS (nuclear localization signal)-containing nuclear import cargo (NLS) binds with importin α and β (α and β) in cytoplasm and this complex is transported into the nucleus through NPC. When in the nucleus, the nuclear RanGTP interacts with this complex dissociating α from the complex to release the NLS cargo in the nucleus, meanwhile capturing importin β for recycling. As shown in (B), RanGTP triggers the formation of RanGTP:exportin:NES complex which is then exported to cytoplasm. After the complex reaches cytoplasm, RanGAP activates the GTP hydrolysis facilitating the release of exportin from Ran. (C) Recycling of Ran from cytoplasm to nucleus. RanGDP interacts with NTF2 (nuclear transporter factor 2) and reaches the nucleus through NPC. After RCC1 (RanGEF) activates Ran by inducing GDP to GTP exchange, RanGTP dissociates from NTF2. Arrows show the direction of the movement, NLS: nuclear localization signal-containing cargo and NES: nuclear export signal-containing cargo. This illustration is modified from Sorokin AV, et al., (69).
To date, 10 members in the human importin family have been identified of which importin α and β are the 2 most extensively studied members. In the human nuclear export family, there are about 7 members with Crm1 (transporter of NES-containing cargo), exportin-t (transporter of t-RNA) and CAS (transporter of importin α) as the most extensively studied members (8). Interestingly, importin 13 in humans (8), and Msn5p in yeast (9) are believed to be involved in not only nuclear import of specific groups of proteins, but also nuclear export of different groups of proteins.
In general, importins and exportins recognize the localization signal peptide on the cargo protein to execute their functions (Figure 2). Therefore, importins and exportins have to form a complex with proteins that possess a nuclear localization signal (NLS) and a nuclear export signal (NES) respectively, prior to shuttling. Although lacking an absolute consensus sequence, NLSs are usually a cluster of continuous basic amino acids (mainly lysine and arginine residues) called monopartite NLS. Some proteins however, harbor two clusters of such sequences which accordingly is called bipartite NLS (10, 11). In the case of NESs, the clusters usually harbor leucine-rich sequence and other hydrophobic amino acids (12). Based on the computational alignment studies, the consensus for about 75% of NES can be formulated as: φ-X(2-3)-φ-X(2-3)-φX-φ (φ=any hydrophobic amino acid residue representing I, L, V, F, M; X=any amino acid) (13, 14).
After the carrier proteins form complex with the localization peptide on the cargoes, the complex is then docked onto NPC through interactions with nucleoporins and finally translocation ensues. Although this is the classical and widely accepted model of nucleocytoplasmic trafficking, there remains a possibility that alternative nucleocytoplasmic trafficking pathways may exist. For example, the nuclear import of the glucocorticoid receptor and the SV40 large T antigen requires an association of their NLSs with Heat Shock Protein 70 (HSP70). It has been suggested that HSP70 may behave as a transporter of specific sets of cargo, rather than acting as a general transporter (15).
Discovery of RanGTPase and its characteristics
RanGTPase, also known as TC4, was first discovered through screening of a teratocarcinoma (TC) cDNA library using oligonucleotide probes corresponding to the GTPase core domain of H-ras (16). In that screen, 4 clones were found to be homologous, but not identical, to Ras sequence and all 4 possessed the putative core guanine nucleotide binding domain of a functional GTPase. Further studies revealed that the Ran/TC4 clone did not resemble the typical GTPase of the Ras superfamily at the carboxyl terminus. Ran/TC4 lacked a consensus CAAX motif which was a characteristic signature of membrane-associated Ras for post-translational modification of protein prenylation (17, 18). In addition, Ran/TC4 was found to uniquely localize to the nucleus. In view of these unique features, TC4 was named as Ras-related nuclear protein now known as RanGTPase.
Since the discovery of RanGTPase, tremendous effort has been spent to elucidate the role of Ran in the nucleocytoplasmic trafficking process. For example, Gerace et al., (19) pioneered the in-vitro approaches by incubating fluorescence-labeled SV40 NLS peptides with digitonin-permeabilized cells, to identify the essential effectors in nuclear import. This method utilizes a low concentration of the non-ionic detergent digitonin that permeabilizes the cell membrane and it allows the removal of cytosolic proteins while keeping the nuclear envelope intact. By adding fluorescence-labeled NLS containing cargo with NTP, importins, RanGTPase and NTF2, nuclear import can be reconstituted. These studies laid the groundwork for additional studies investigating the fundamental roles of Ran in nucleocytoplasmic trafficking.
It is now widely held that almost all eukaryotic cells rely on RanGTPase, as it is believed to play a number of important roles including nucleocytoplasmic trafficking, mitotic spindle and nuclear envelope assembly (20). Furthermore, the expression of RanGTPase is not limited to mammalian cells as its homologues have also been found in Arabidopsis, rodent, drosophila, c. elegans and yeast. For example, Fyt1/spi1 in fission yeast showing 80% homology to human RanGTPase is believed to be yeast equivalent of human RanGTPase (21). In budding yeast, by contrast, there are 2 isoforms of RanGTPase called GSP1/CNR1 and GSP2/CNR2 (22, 23). The expression of GSP1 is about 10-fold higher than that of GSP2 and is essential (not GSP2) for cell viability (23). Surprisingly, the lethality due to GSP1 deficiency can be rescued by over-expressing GSP2 that indicates overlapping functions of GSP1 and GSP2 in budding yeast (23, 24). Also, four putative isoforms of Ran have been found in plant Arabidopsis (25). Based on the evidence from multiple RanGTPase models in the budding yeast and Arabidopsis plant, it is possible that there are other proteins, possibly another GTPase that could complement the role of RanGTPase in mammalian cells. The alternative RanGTPase hypothesis can be further supported by the fact that for nuclear transport of a number of proteins such as β-catenin and calmodulin, the mechanism remains to be elucidated and their nuclear shuttling appears to occur independent of RanGTPase (12, 26).
Theoretically, it is feasible to screen for an alternative GTPase that is cable of substituting the RanGTPase function by utilizing the GSP1-null cells rescue approach followed by GTP overlay assay. For example, a human cDNA expression library (deficient in RanGTPase cDNA) can be introduced in GSP1-null mutant and the clone(s) capable of rescuing the GSP1-null lethal phenotype can be identified. Subsequently, all positive clones in the GSP1 rescue assay can also be subjected to GTP overlay assay to check their GTP-binding ability and further investigations to elucidate their function as GTPase(s). Any clone that is positive in both assays and encodes a known or a novel GTPase is a likely candidate to substitute Ran function.
In our recent studies, we have reported the identification of a novel gene that we have named RBEL1 (Rab-like protein 1) (27). RBEL1 gene has four alternatively spliced transcripts that encode four isoforms named RBEL1A, B, C and D (27, 70). RBEL1 proteins share significant sequence homology with the Ras superfamily proteins at their N-terminus but with the highest degree similarity with Ran and Rab proteins (27, 70). All RBEL1 proteins are capable of binding to GTP and GDP albeit with some difference. For example, RBEL1A and RBEL1B are predominantly GTP-bound whereas RBEL1C and RBEL1D bind to both GTP and GDP (70). All four RBEL1 variants (A, B, C, D) are found to reside in both cytosol and the nucleus although there are some differences. For instance, RBEL1A is found to be predominantly cytoplasmic while RBEL1B is mostly nuclear; RBEL1C and RBEL1D, on other hand, are evenly distributed between cytoplasm and nucleus. Our unpublished data also indicate that RBEL1A protein interacts with some nuclear proteins and appears to shuttle between the nucleus and cytoplasm. Future studies are needed to determine whether the RBEL1 family GTPases have Ran-like function in nucleocytoplasmic trafficking.
RanGTPase and its regulators
RanGTPase and its associated proteins are constantly shuttling into and out of the nucleus. It has been demonstrated by fluorescent microscopy that more than 105 copies of Ran are exported out of a nucleus per second (28, 29). This rapid trafficking has been demonstrated to require energy, e.g., ATP (30, 31) and how this is accomplished remains to be elucidated. One hypothesis suggests that ATP rather than acting as an energy source could be converted into GTP (32). Interestingly however, recent in vitro studies have demonstrated that reversal of the concentration gradient of RanGTP could reverse the directionality of the nucleocytoplasmic trafficking, i.e. cytoplasmic RanGTP becomes higher than nuclear RanGTP (33, 34). These findings seem to suggest that the RanGTP/RanGDP gradient may govern the directionality of the transport.
RanGTPase functions as a molecular switch cycling between the GTP-bound form and the GDP-bound form, which is regarded as the active and inactive states respectively (20). Interestingly, GTP-bound Ran is asymmetrically distributed in the nucleus while GDP-bound Ran is asymmetrically distributed in the cytoplasm (20). This steep and asymmetrical pattern is tightly regulated by two GTPase regulatory proteins namely Guanine Exchange Factor (GEF) and GTPase Activating Protein (GAP). RanGEF is able to activate Ran function by triggering the exchange of GDP to GTP in RanGTPase, whereas RanGAP is cable of terminating Ran function by catalyzing the hydrolysis of GTP to GDP in RanGTPase (35) (Figure 2). Recent data have shown that the distinctive Ran DEDDDL acidic motif is essential for mediating its interaction with RanGAP and other proteins such as RanBP1 (Ran-binding protein 1) or RCC1 (regulator of chromosome condensation 1) (36, 37).
RCC1 is the only known RanGEF and it enhances the release of GDP in RanGTPase by 105 fold (38). In addition, Ran-binding protein 3 (RanBP3) has been found to assist RCC1 in nucleotide exchange (39). X-ray crystallographic analysis and modeling have suggested RCC1 to insert a β-wedge structure into RanGTPase and thereby help to release the GDP nucleotide (40). Given the fact that RCC1 is exclusively expressed in the nucleus and the fact that GTP is more abundant than GDP in cells (41), RCC1 could readily catalyze the formation of active GTP-bound Ran in the nucleus. After RanGTPase has been charged with GTP, it becomes activated and serves as an initiator for the formation of the nuclear export complex and importin recycling complex (42). Subsequently, GTP-bound Ran and the carriers could facilitate the translocation of both complexes. Unlike nuclear RCC1, RanGAP is predominantly expressed in cytoplasm. With the assistance from RanBP1 and RanBP2/Nup358, RanGAP catalyzes the enzymatic hydrolysis of GTP to GDP in Ran after the nuclear RanGTP:exportins:cargo complex is translocated to cytoplasm (8). GTP hydrolysis induces conformational alternation in Ran triggering the release of cargo from Ran as well as the dissociation of cargo from exportins in cytoplasm.
Classical nuclear import: RanGTPase, importin α, importin β, and NLS
Nuclear import is the movement of cargo from cytoplasm to nucleus facilitated by importins and RanGTPase. There are various known nuclear importers and cargo combinations, but each combination utilizes similar import steps. Nuclear import has been shown to be a multi-step event involving (i) recognition of NLS by importins, (ii) docking of cargo-importins complex with FG repeats on nucleoporins in NPC, (iii) translocation of the complex through the NPC, (iv) cargo release and finally (v) recycling of importins to cytoplasm by RanGTPase (43).
In general, import cargo that bears a classical basic NLS is able to form a complex directly with importin β (44), or form a complex mediated by an adaptor protein, called importin α. One end of importin α interacts with NLS while the other end interacts with importin β through a NLS like-importin β binding (IBB) domain (45, 46). An in vitro study has shown that nuclear import could be reconstituted by incubating Ran, importin α, importin β and NLS-containing cargo in permeabilized cells without Ran GTP hydrolysis (32). After nuclear import is concluded, free nuclear RanGTP recognizes importin β and associates with its Ran-binding domain. The association process triggers a conformational change in importin β and subsequently, relieves the interaction between importin β and importin α:cargo complex by reducing the affinity to IBB domain. The free IBB domain then releases the cargo from importin α through a self-inhibitory mechanism (43). On the other hand, nuclear RanGTP:importin β complex appears to be transported to cytoplasm for recycling purposes, whereas free importin α binds to exportin CAS and RanGTP for recycling purposes (46).
Once the complex traverses to cytoplasm, RanGAP activates Ran GTP hydrolysis to trigger the release of importin β from RanGDP. The free importin β can initiate another nuclear import event, while free RanGDP is recycled back into the nucleus by forming a complex with the specific nuclear importer of Ran, nuclear transporter factor 2 (NTF2) (also known as p10 or pp15) (47). It has been shown that NTF2 forms a homodimer, which creates two reactive hydrophobic pockets to bind with RanGDP while the hydrophobic ends of NTF2 dimer are able to interact with FXFG motifs from nucleoporins to induce the nuclear translocation (48, 49, 50). Once this complex reaches the nucleus, nuclear RCC1 (RanGEF) activates the exchange of GDP to GTP in Ran and also it induces the release of NTF2 from Ran. Then, the free GTP-bound Ran is ready for another round of nuclear export while the free NTF2 is re-transported back to cytoplasm. To summarize, RanGTPase plays a role in the nuclear import in such a way that GTP-bound Ran dissociates importin:cargo complex in the nucleus and recycles importin back to cytoplasm.
Classical nuclear export: RanGTPase, exportin 1 (Crm1) and NES
Nuclear export is the movement of cargo from nucleus to cytoplasm facilitated by exportins and RanGTPase. There are about 7 exportins found in humans and all show preferential cargo specificity (51–53). Surprisingly, all exportins follow a general nuclear export principle that involves (i) recognition of export cargo by exportins:RanGTP complex, (ii) docking of cargo-exportin-RanGTP complex with FG repeats on nucleoporins in NPC, (iii) translocation of the complex through the NPC, (iv) induction of GTPase hydrolysis by RanGAP and dissociation of cargo, exportin and RanGDP, and (v) recycling of exportin and RanGDP to nucleoplasm. Here, a classical nuclear export of NES-containing cargo facilitated by Crm1 and RanGTPase will be discussed as an example.
As mentioned earlier, RanGTP is predominantly localized in the nucleus. It has been found that RanGTP is required for Crm1-mediated nuclear export (50). Compared with RanGDP, this nuclear RanGTP is highly cooperative with Crm1 (8). The association of RanGTP and Crm1 induces the association of NES-containing cargo with Crm1 (8). By contrast, dissociation of Ran from Crm1 triggers the dissociation of cargo from Crm1 (8). After cargo:Crm1:RanGTP complex is formed, the complex interacts with nucleoporins and transverses through the nuclear pore complex (NPC) to the cytoplasm. Once the trimeric complex passes through the NPC, the cytoplasmic RanGAP triggers the hydrolysis of GTP to GDP in Ran, which concomitantly dissociates Ran from Crm1 and releases cargo in the cytoplasm leaving the free Crm1 and the RanGDP to recycle back into the nucleus. For its transport, the cytoplasmic RanGDP interacts with NTF2, and the RanGDP:NTF2 complex is transported into the nucleus through the interaction with FG motifs in the nucleoporins. Once the complex arrives at the nucleoplasm, RanGDP is then recharged with GTP by RCC1 (RanGEF). The re-charged RanGTP is able to perform yet another round of nucleocytoplasmic trafficking. To summarize, RanGTPase plays a role in the nuclear export in such a way that nuclear GTP-bound Ran triggers the aggregation of Ran:exportin:cargo trimeric complex which is then transported to cytoplasm while hydrolysis of RanGTP to RanGDP releases the export cargoes in cytoplasm.
Additional function of RanGTPase
RanGTPase has also been reported to play an important role in mitotic spindle formation. RanGTPase has been found to stabilize the nucleation of microtubules and the formation of spindle fibers on chromatins through the assistance of chromatin associated RCC1. The underlying mechanism is believed to be that RCC1 catalyzes the formation of RanGTP and it establishes a higher concentration gradient of RanGTP centered on chromatins (54). Then, the RanGTP activates the microtubule initiation factor TPX2 by releasing inhibitory importin α (55) and importin β (56). Once TPX2 is released, it initiates spindle formation on chromatin. Furthermore, additional evidence has shown that the activated TPX2 is able to bind and activate Aurora A kinase, which is an important kinase in the formation centrosome and spindle fibers (57). Therefore, RanGTPase orchestrates not only the nucleocytoplasmic trafficking, but also play a role in the mitotic spindle formation.
RanGTPase and its link to cancer
It has been suggested that RanGTPase is a potential candidate linked to carcinogenesis, specifically its role in cellular transformation (58) and mitosis (59). A number of studies have reported that RanGTPase is extensively upregulated in a wide spectrum of cancer cells representing prostate (60), breast (60), colon (60), kidney (61), ovary (62), sarcoma (59), ALK-positive anaplastic large cell lymphoma (59), and nasopharyngeal carcinoma (63). Interestingly, Xia et al., (60) have reported that knockdown of RanGTPase expression in cancer cells by RNA interference approach significantly induces p53/Bax-independent apoptosis and disrupts mitotic spindle formation (64). On the other hand, normal cells such as human fibroblasts WS-1, HEK293 human embryonic kidney cells and Human Umbilical Vein Endothelial (HUVEC) cells do not show considerable alternations in cell cycle and mitotic spindle formation after RanGTPase knockdown. These findings would suggest that only low levels of RanGTPase appear to be required for normal cell viability, while excessive RanGTPase appears to deregulate normal cell function thereby predisposing to tumorigenesis.
RanGTPase has also been reported to be upregulated in androgen-dependent (65) as well as androgen-independent (66) prostate cancers. In some androgen-independent prostate cancers, androgen receptors are activated in the absence of ligand (androgen) binding and it is possible that androgen receptors are translocated into the nucleus by an unknown nuclear trafficking mechanism. Given the fact that Ran is over-expressed in cancer cells and it can strongly interact with androgen receptor through the acidic DEDDDL domain (67, 68), a possibility exists that Ran may trigger deregulation of androgen receptor nuclear import and hence, the development of androgen-independent state. Further studies are however, needed to explore these possibilities and to also firmly establish the role, if any, of RanGTPase in cancer development and/or progression.
Conclusion
Ran plays a key role in controlling nucleocytoplasmic trafficking, a process in which many accessory proteins involved in nuclear import and export pathways have also been implicated. It is clear that RanGTPase is crucial for both the nuclear import and export processes. In nuclear import, Ran dissociates importin β:cargo complex in the nucleus and recycles importin β to cytoplasm. In nuclear export, Ran triggers the formation of the export complex and RanGTPase hydrolysis releases the export-cargo in cytoplasm. Even though there is a broader understanding of nucleocytoplasmic trafficking, numerous fundamental questions remain unanswered. For example, which process utilizes energy to shuttle cargo against the concentration gradient? What is the exact mechanism involving the interactions between transporter proteins and nucleoporins to achieve translocation? How does the translocation take place for proteins devoid of NLS or NES? What triggers RanGTPase-independent translocation of proteins such as β–catenin and calmodulin? Although these are challenging questions, further investigations seeking to address these issues are expected to provide valuable insights into the precise mechanisms by which this key protein modulate nucleocytoplasmic trafficking. Likewise, investigations are also needed to firmly establish a link between the role of RanGTPase and cancer development and progression.
Acknowledgments
This work was supported in part by grants from the NIH (CA113868 and CA128096 to YH). KL is supported by a predoctoral fellowship from the U.S. Department of Defense (Grant BC083017).
Footnotes
Conflicts of Interest
No potential conflicts of interest to disclose.
References
- 1.Bode AM, Dong Z. Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer. 2004;10:793–805. doi: 10.1038/nrc1455. [DOI] [PubMed] [Google Scholar]
- 2.Brooks CL, Gu W. p53 ubiquitination: Mdm2 and beyond. Mol Cell. 2006;21:307–15. doi: 10.1016/j.molcel.2006.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kubitscheck U, Grunwald D, Hoekstra A, et al. Nuclear transport of single molecules: dwell times at the nuclear pore complex. J Cell Biol. 2005;168:233–43. doi: 10.1083/jcb.200411005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang Q, Rout MP, Akey CW. Three-dimensional architecture of the isolated yeast nuclear pore complex: functional and evolutionary implications. Mol Cell. 1998;1:223–34. doi: 10.1016/s1097-2765(00)80023-4. [DOI] [PubMed] [Google Scholar]
- 5.Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- 6.Macara IG. Transport into and out of the nucleus. Microbiol Mol Biol Rev. 2001;65:570–94. doi: 10.1128/MMBR.65.4.570-594.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang W, Gelles J, Musser SM. Imaging of single-molecule translocation through nuclear pore complexes. Proc Natl Acad Sci USA. 2004;101:12887–92. doi: 10.1073/pnas.0403675101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pemberton LF, Paschal BM. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic. 2005;6:187–98. doi: 10.1111/j.1600-0854.2005.00270.x. [DOI] [PubMed] [Google Scholar]
- 9.Yoshida K, Blobel G. The karyopherin Kap142p/Msn5p mediates nuclear import and nuclear export of different cargo proteins. J Cell Biol. 2001;152:729–40. doi: 10.1083/jcb.152.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dingwall C, Laskey RA. Nuclear targeting sequences--a consensus? Trends Biochem Sci. 1991;16:478–81. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 11.Boulikas T. Nuclear localization signals (NLS) Crit Rev Eukaryot Gene Expr. 1993;3:193–227. [PubMed] [Google Scholar]
- 12.Stochaj U, Rother KL. Nucleocytoplasmic trafficking of proteins: with or without Ran? BioEssays. 1999;21:579–598. [Google Scholar]
- 13.la Cour T, Kiemer L, Mølgaard A, Gupta R, Skriver K, Brunak S. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng Des Sel. 2004;17:527–36. doi: 10.1093/protein/gzh062. [DOI] [PubMed] [Google Scholar]
- 14.Kutay U, Guttinger S. Leucine-rich nuclear-export signals: born to be weak. Trends Cell Biol. 2005;15:121–4. doi: 10.1016/j.tcb.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 15.Yang J, DeFranco DB. Differential roles of heat shock protein 70 in the in vitro nuclear import of glucocorticoid receptor and simian virus 40 large tumor antigen. Mol Cell Biol. 1994;14:5088–98. doi: 10.1128/mcb.14.8.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drivas GT, Shih A, Coutavas E, Rush MG, D’Eustachio P. Characterization of four novel ras-like genes expressed in a human teratocarcinoma cell line. Mol Cell Biol. 1990;10:1793–8. doi: 10.1128/mcb.10.4.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bokoch GM, Der CJ. Emerging concepts in the Ras superfamily of GTP-binding proteins. FASEB J. 1993;7:750–759. doi: 10.1096/fasebj.7.9.8330683. [DOI] [PubMed] [Google Scholar]
- 18.Sprang SR. G protein mechanisms: insights from structural analysis. Annu Rev Biochem. 1997;66:639–678. doi: 10.1146/annurev.biochem.66.1.639. [DOI] [PubMed] [Google Scholar]
- 19.Adam SA, Sterne-Marr R, Gerace L. Nuclear protein import using digitonin-permeabilized cells. Methods Enzymol. 1992;219:97–110. doi: 10.1016/0076-6879(92)19013-v. [DOI] [PubMed] [Google Scholar]
- 20.Joseph J. Ran at a glance. J Cell Sci. 2006;119:3481–4. doi: 10.1242/jcs.03071. [DOI] [PubMed] [Google Scholar]
- 21.Ren M, Drivas G, D’Eustachio P, Rush MG. Ran/TC4: a small nuclear GTP-binding protein that regulates DNA synthesis. J Cell Biol. 1993;120:313–23. doi: 10.1083/jcb.120.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sazer S. The search for the primary function of the Ran GTPase continues. Trends Cell Biol. 1996;6:81–85. doi: 10.1016/0962-8924(96)80992-5. [DOI] [PubMed] [Google Scholar]
- 23.Belhumeur P, Lee A, Tam R, DiPaolo T, Fortin N, Clark MW. GSP1 and GSP2, genetic suppressors of the prp20-1 mutant in Saccharomyces cerevisiae: GTP-binding proteins involved in the maintenance of nuclear organization. Mol Cell Biol. 1993;13:2152–61. doi: 10.1128/mcb.13.4.2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Villalva C, Trempat P, Greenland C, et al. Isolation of differentially expressed genes in NPM-ALK-positive anaplastic large cell lymphoma. Br J Haematol. 2002;118:791–8. doi: 10.1046/j.1365-2141.2002.03671.x. [DOI] [PubMed] [Google Scholar]
- 25.Vernoud V, Horton AC, Yang Z, Nielsen E. Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol. 2003;131:1191–208. doi: 10.1104/pp.013052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fagotto F, Gluck U, Gumbiner BM. Nuclear localization signal-independent and importin/karyopherin-independent nuclear import of beta-catenin. Curr Biol. 1998;8:181–90. doi: 10.1016/s0960-9822(98)70082-x. [DOI] [PubMed] [Google Scholar]
- 27.Montalbano J, Jin W, Sheikh MS, Huang Y. RBEL1 is a novel gene that encodes a nucleocytoplasmic Ras superfamily GTP-binding protein and is overexpressed in breast cancer. J Biol Chem. 2007;282:37640–9. doi: 10.1074/jbc.M704760200. [DOI] [PubMed] [Google Scholar]
- 28.Smith AE, Slepchenko BM, Schaff JC, Loew LM, Macara IG. Systems analysis of Ran transport. Science. 2002;295:488–91. doi: 10.1126/science.1064732. [DOI] [PubMed] [Google Scholar]
- 29.Görlich D, Seewald MJ, Ribbeck K. Characterization of Ran-driven cargo transport and the RanGTPase system by kinetic measurements and computer simulation. EMBO J. 2003;22:1088–100. doi: 10.1093/emboj/cdg113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moore MS, Blobel G. The two steps of nuclear import, targeting to the nuclear envelope and translocation through the nuclear pore, require different cytosolic factors. Cell. 1992;69:939–50. doi: 10.1016/0092-8674(92)90613-h. [DOI] [PubMed] [Google Scholar]
- 31.Silver PA. How proteins enter the nucleus. Cell. 1991;64:489–97. doi: 10.1016/0092-8674(91)90233-o. [DOI] [PubMed] [Google Scholar]
- 32.Schwoebel ED, Talcott B, Cushman I, Moore MS. Ran-dependent signal-mediated nuclear import does not require GTP hydrolysis by Ran. J Biol Chem. 1998;273:35170–5. doi: 10.1074/jbc.273.52.35170. [DOI] [PubMed] [Google Scholar]
- 33.Izaurralde E, Kutay U, von Kobbe C, Mattaj IW, Görlich D. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 1997;16:6535–47. doi: 10.1093/emboj/16.21.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nachury MV, Weis K. The direction of transport through the nuclear pore can be inverted. Proc Natl Acad Sci U S A. 1999;96:9622–7. doi: 10.1073/pnas.96.17.9622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–77. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 36.Richards SA, Lounsbury KM, Macara IG. The C terminus of the nuclear RAN/TC4 GTPase stabilizes the GDP-bound state and mediates interactions with RCC1, RAN-GAP, and HTF9A/RANBP1. J Biol Chem. 1995;270:14405–11. doi: 10.1074/jbc.270.24.14405. [DOI] [PubMed] [Google Scholar]
- 37.Ren M, Villamarin A, Shih A, et al. Separate domains of the Ran GTPase interact with different factors to regulate nuclear protein import and RNA processing. Mol Cell Biol. 1995;15:2117–24. doi: 10.1128/mcb.15.4.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Renault L, Nassar N, Vetter I, et al. The 1.7 A crystal structure of the regulator of chromosome condensation (RCC1) reveals a seven-bladed propeller. Nature. 1998;392:97–101. doi: 10.1038/32204. [DOI] [PubMed] [Google Scholar]
- 39.Nemergut ME, Lindsay ME, Brownawell AM, Macara IG. Ran-binding protein 3 links Crm1 to the Ran guanine nucleotide exchange factor. J Biol Chem. 2002;277:17385–8. doi: 10.1074/jbc.C100620200. [DOI] [PubMed] [Google Scholar]
- 40.Renault L, Kuhlmann J, Henkel A, Wittinghofer A. Structural basis for guanine nucleotide exchange on Ran by the regulator of chromosome condensation (RCC1) Cell. 2001;105:245–55. doi: 10.1016/s0092-8674(01)00315-4. [DOI] [PubMed] [Google Scholar]
- 41.Bischoff FR, Krebber H, Smirnova E, Dong W, Ponstingl H. Co-activation of RanGTPase and inhibition of GTP dissociation by Ran-GTP binding protein RanBP1. EMBO J. 1995;14:705–15. doi: 10.1002/j.1460-2075.1995.tb07049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pemberton LF, Paschal BM. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic. 2005;6:187–98. doi: 10.1111/j.1600-0854.2005.00270.x. [DOI] [PubMed] [Google Scholar]
- 43.Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- 44.Lange A, Mills RE, Lange CJ, et al. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J Biol Chem. 2007;282:5101–5. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cingolani G, Petosa C, Weis K, Müller CW. Structure of importin-beta bound to the IBB domain of importin-alpha. Nature. 1999;399:221–9. doi: 10.1038/20367. [DOI] [PubMed] [Google Scholar]
- 46.Moroianu J. Nuclear import and export pathways. J Cell Biochem. 1999;(Suppl 32–33):76–83. doi: 10.1002/(sici)1097-4644(1999)75:32+<76::aid-jcb10>3.3.co;2-h. [DOI] [PubMed] [Google Scholar]
- 47.Ribbeck K, Lipowsky G, Kent HM, Stewart M, Görlich D. NTF2 mediates nuclear import of Ran. EMBO J. 1998;17:6587–98. doi: 10.1093/emboj/17.22.6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bayliss R, Leung SW, Baker RP, et al. Structural basis for the interaction between NTF2 and nucleoporin FxFG repeats. EMBO J. 2002;21:2843–53. doi: 10.1093/emboj/cdf305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stewart M, Kent HM, McCoy AJ. structural basis for molecular recognition between nuclear transport factor 2 (NTF2) and the GDP-bound form of the Ras-family GTPase Ran. J Mol Biol. 1998;277:635–46. doi: 10.1006/jmbi.1997.1602. [DOI] [PubMed] [Google Scholar]
- 50.Bullock TL, Clarkson WD, Kent HM, Stewart M. The 1.6 angstroms resolution crystal structure of nuclear transport factor 2 (NTF2) J Mol Biol. 1996;260:422–31. doi: 10.1006/jmbi.1996.0411. [DOI] [PubMed] [Google Scholar]
- 51.Bayliss R, Leung SW, Baker RP, et al. Structural basis for the interaction between NTF2 and nucleoporin FxFG repeats. EMBO J. 2002;21:2843–53. doi: 10.1093/emboj/cdf305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stewart M, Kent HM, McCoy AJ. Structural basis for molecular recognition between nuclear transport factor 2 (NTF2) and the GDP-bound form of the Ras-family GTPase Ran. J Mol Biol. 1998;277:635–46. doi: 10.1006/jmbi.1997.1602. [DOI] [PubMed] [Google Scholar]
- 53.Bullock TL, Clarkson WD, Kent HM, Stewart M. The 1.6 angstroms resolution crystal structure of nuclear transport factor 2 (NTF2) J Mol Biol. 1996;260:422–31. doi: 10.1006/jmbi.1996.0411. [DOI] [PubMed] [Google Scholar]
- 54.Caudron M, Bunt G, Bastiaens P, Karsenti E. Spatial coordination of spindle assembly by chromosome-mediated signaling gradients. Science. 2005;309:1373–6. doi: 10.1126/science.1115964. [DOI] [PubMed] [Google Scholar]
- 55.Schatz CA, Santarella R, Hoenger A, et al. Importin alpha-regulated nucleation of microtubules by TPX2. EMBO J. 2003;22:2060–70. doi: 10.1093/emboj/cdg195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dasso M. Running on Ran: nuclear transport and the mitotic spindle. Cell. 2001;104:32–14. doi: 10.1016/s0092-8674(01)00218-5. [DOI] [PubMed] [Google Scholar]
- 57.Barr AR, Gergely F. Aurora-A: the maker and breaker of spindle poles. J Cell Sci. 2007;120:2987–96. doi: 10.1242/jcs.013136. [DOI] [PubMed] [Google Scholar]
- 58.Rensen WM, Mangiacasale R, Ciciarello M, Lavia P. The GTPase Ran: regulation of cell life and potential roles in cell transformation. Front Biosci. 2008;1:4097–121. doi: 10.2741/2996. [DOI] [PubMed] [Google Scholar]
- 59.Sanderson HS, Clarke PR. Cell biology: Ran, mitosis and the cancer connection. Curr Biol. 2006;16:R466–8. doi: 10.1016/j.cub.2006.05.032. [DOI] [PubMed] [Google Scholar]
- 60.Xia F, Lee CW, Altieri DC. Tumor cell dependence on Ran-GTP-directed mitosis. Cancer Res. 2008;68:1826–33. doi: 10.1158/0008-5472.CAN-07-5279. [DOI] [PubMed] [Google Scholar]
- 61.Abe H, Kamai T, Shirataki H, Oyama T, Arai K, Yoshida K. High expression of Ran GTPase is associated with local invasion and metastasis of human clear cell renal cell carcinoma. Int J Cancer. 2008;122:2391–7. doi: 10.1002/ijc.23400. [DOI] [PubMed] [Google Scholar]
- 62.Ouellet V, Guyot MC, Le Page C, et al. Tissue array analysis of expression microarray candidates identifies markers associated with tumor grade and outcome in serous epithelial ovarian cancer. Int J Cancer. 2006;119:599–607. doi: 10.1002/ijc.21902. [DOI] [PubMed] [Google Scholar]
- 63.Li H, Ren CP, Tan XJ, et al. Identification of genes related to nasopharyngeal carcinoma with the help of pathway-based networks. Acta Biochim Biophys Sin (Shanghai) 2006;38:900–10. doi: 10.1111/j.1745-7270.2006.00235.x. [DOI] [PubMed] [Google Scholar]
- 64.Kurisetty VV, Johnston PG, Johnston N, et al. RAN GTPase is an effector of the invasive/metastatic phenotype induced by osteopontin. Oncogene. 2008;27:7139–49. doi: 10.1038/onc.2008.325. [DOI] [PubMed] [Google Scholar]
- 65.Urbanucci A, Waltering KK, Suikki HE, Helenius MA, Visakorpi T. Androgen regulation of the androgen receptor coregulators. BMC Cancer. 2008;8:219. doi: 10.1186/1471-2407-8-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chang HC, Chen SC, Chen J, Hsieh JT. In vitro gene expression changes of androgen receptor coactivators after hormone deprivation in an androgen-dependent prostate cancer cell line. J Formos Med Assoc. 2005;104:652–8. [PubMed] [Google Scholar]
- 67.Hsiao PW, Lin DL, Nakao R, Chang C. The linkage of Kennedy’s neuron disease to ARA24, the first identified androgen receptor polyglutamine region-associated coactivator. J Biol Chem. 1999;274:20229–34. doi: 10.1074/jbc.274.29.20229. [DOI] [PubMed] [Google Scholar]
- 68.Sampson ER, Yeh SY, Chang HC, et al. Identification and characterization of androgen receptor associated coregulators in prostate cancer cells. J Biol Regul Homeost Agents. 2001;15:123–9. [PubMed] [Google Scholar]
- 69.Sorokin AV, Kim ER, Ovchinnikov LP. Nucleocytoplasmic transport of proteins. Biochemistry (Mosc) 2007;72:1439–57. doi: 10.1134/s0006297907130032. [DOI] [PubMed] [Google Scholar]
- 70.Montalbano J, Lui K, Sheikh MS, Huang Y. Identification and characterization RBEL1 subfamily of GTPases in the Ras supergene family involved in cell growth regulation. J Bio Chem. 2009;284:18129–42. doi: 10.1074/jbc.M109.009597. [DOI] [PMC free article] [PubMed] [Google Scholar]