Summary
TL1A is a member of the TNF superfamily and its expression is increased in the mucosa of inflammatory bowel disease (IBD) patients. Neutralizing anti-mouse TL1A Ab attenuates chronic colitis in two T cell driven murine models, suggesting that TL1A is a central modulator of gut mucosal inflammation in IBD. We showed previously that TL1A is induced by immune complexes (IC) via the FcγR signaling pathway. In this study, we report that multiple bacteria, including gram negative organisms (E. coli, E. coli Nissle 1917, S. typhimurium), gram positive organisms (L. monocytogenes, S. epidermidis), partial anaerobes (C. jejuni), and obligate anaerobes (B. thetaiotaomicron, B. breve, Clostridium A4) activate TL1A expression in human APC, including monocytes and monocyte-derived DC. Bacterially induced TL1A mRNA expression correlates with the detection of TL1A protein levels. TL1A induced by bacteria is mediated in part by the TLR signaling pathway and inhibited by downstream blockade of p38 MAPK and NF-κB activation. Microbial induction of TL1A production by human APC potentiated CD4+ T cell effector function by augmenting IFN-γ production. Our findings suggest a role for TL1A in pro-inflammatory APC-T cell interactions and implicate TL1A in host responses to enteric microorganisms.
Introduction
TL1A (TNFSF15) is a member of the TNF superfamily that binds to death domain receptor 3 (DR3, TNFRSF25), activates NF-κB, and costimulate IFN-γ production in T cells [1–3]. In particular, TL1A augments IFN-γ production by IL-12/IL-18 stimulated CD4+/CCR9+ T cells, which are enriched in the intestinal immune compartment [4, 5]. In addition to mediating Th1 response, the role of TL1A in Th2-mediated functions and disease pathology was demonstrated in a mouse model of allergic lung inflammation where TL1A augmented effector function in Th2-polarized CD4 cells and co-stimulated IL-13 production [6]. Furthermore, TL1A was recently shown to be important in promoting Th17 effector cell function in a Th17 mediated autoimmune disease mouse model [7]. Interestingly, a report using DR3 deficient mice showed that TL1A/DR3 signaling is dispensable for polarization of naïve CD4+ T cells into Th1, Th2, or Th17 effector cell subtypes. Instead, DR3 expression is required on T cells for immunopathology, local T cell accumulation, and cytokine production, suggesting that TL1A/DR3 signaling is important to costimulate Ag-induced expansion of primed T cells in the target organ of T cell-mediated autoimmune and inflammatory diseases [8].
Several studies implicate the TL1A/DR3 signaling pathway in mucosal inflammation. TL1A and DR3 expression is increased on T cells and macrophages in the mucosa of patients with IBD [2, 9], and in inflamed gut mucosa of two distinct murine models of ileal inflammation [10]. Neutralizing anti-mouse TL1A Ab attenuated inflammation in both the dextran sulfate sodium induced chronic colitis and a Gαi2−/− T cell transfer colitis model [11], suggesting a role for TL1A in the pathology of mucosal inflammation in IBD [12, 13]. Moreover, genome-wide association studies revealed a significant association of genetic variants of the TL1A gene with Crohn's disease (CD) in Japanese patients, in several European cohorts [14–17], in U.S. Jewish patients [18], and in pediatric IBD patients [19]. In fact, genetic association between TL1A and CD has been found in every ethnic and age groups that were studied.
Although the exact pathogenesis of IBD remains uncertain, it is proposed that IBD is caused by a dysregulated immune response to enteric microflora in genetically susceptible hosts (Reviewed in [20]). The importance of the microflora in the induction and maintenance of disease has been demonstrated in various murine models of colitis. For example, mice that are deficient in the cytokines IL-2 or IL-10 or rats containing the HLA-B27 transgene develop IBD in the presence of a normal gut microflora but not in a sterile germ-free environment [21–25]. Experimental colitis is attenuated when animals are treated with a wide range of antibiotics [26]. Moreover, T cells reactive to cecal bacterial antigens induce colitis in immunodeficient mice [27].
We have previously shown that TL1A is induced by IC and the FcγR signaling pathway [28]. Other stimuli of TL1A production are likely. In the present study, we investigated the hypothesis that interaction between enteric microorganisms and the TL1A signaling pathway participate in activation of the pro-inflammatory pathway. Several types of microbial organisms can induce TL1A expression in human monocytes and monocyte-derived DC. Microbial activation of TL1A was in part mediated by a TLR signaling pathway in APC and was dependent on downstream p38 MAPK and NF-κB activation. Furthermore, in autologous monocyte and monocyte-derived DC-T-cell co-cultures, microbial induction of TL1A production potentiated IFN-γ production by CD4+ T cells.
Results
Microbial organisms induce TL1A expression in human monocytes and monocyte-derived DC
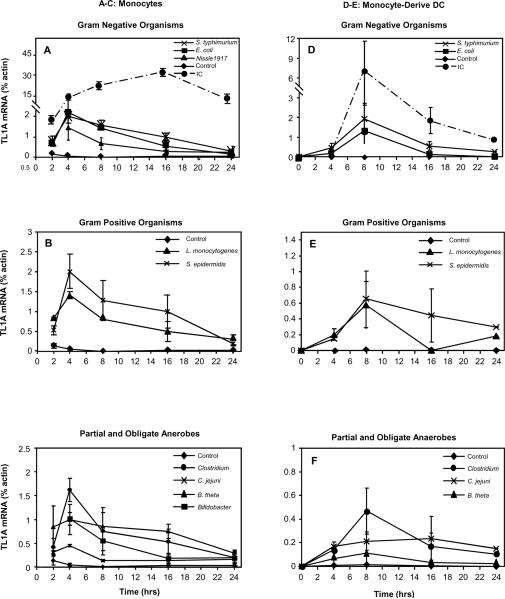
To investigate the hypothesis that microbes can induce TL1A expression, we stimulated CD14+ monocytes and monocyte-derived DC with gram negative organisms (E. coli, E. coli Nissle1917 and S. typhimurium), gram positive organisms (L. monocytogenes and S. epidermidis), a microaerophilic organism (C. jejuni), and obligate anaerobes (Bifidobacterium breve, B. thetaiotaomicron, Clostridium A4). Cells were stimulated for up to 24 h and the TL1A mRNA was quantified by real-time PCR. All the microbial organisms tested, including probiotic bacteria (E. coli Nissle 1917 and Bifidobacterium breve), were capable of stimulating TL1A mRNA expression (Fig. 1). In general, aerobic organisms were better inducers of TL1A expression than anaerobes (Fig. 1A–C and D–F). In monocytes, bacteria induced TL1A mRNA expression peaked at 4 h and decreased over 24 h (Fig. 1A–C); whereas in DC, bacteria stimulated TL1A expression peaked at 8 h and decreased over 24 h (Fig. 1D–F). The kinetics of microbial induction of TL1A expression was similar to FcγR induction of TL1A mRNA in DC (Fig. 1D–F)[28] but different from FcγR activation of TL1A in monocytes where its expression levels continued to increase for up to 16 h (Fig. 1A–C)[28].
Figure 1. Microbial organisms can induce TL1A mRNA in APCs.
Fresh monocytes from blood or monocyte-derived DC were stimulated by live microbial organisms or plate coated IC. TL1A transcripts at the indicated times were quantitated by real-time RT-PCR and expressed as percent β-actin level for monocytes (A–C) or monocyte-derived DC (D–F). Data shown are means ± SEM of at least three independent time-course experiments.
We have previously shown that IC induced both surface TL1A expression (44–53% of monocytes and 8.6–17.4% of DC) and soluble protein (average of 3 ng/mL/106 monocytes and 4 ng/mL/106 DC) [28]. To determine whether induction of TL1A mRNA by bacteria leads to protein expression, we quantified both membrane bound and secreted TL1A. Incubation in the presence of bacterial organisms led to increased membrane bound TL1A expression on both monocytes and DC (Fig. 2). Interestingly, Clostridium A4 was more efficient than other microbes in the induction of membrane bound TL1A in monocytes (Fig. 2A). In monocyte-derived DCs, Clostridium A4 was similarly efficient in the induction of membrane TL1A (Fig. 2C). In contrast to IC, the microbial organisms lead to modest levels of soluble TL1A protein in monocytes and DC (Fig. 2B, D). TNF-α converting enzyme (TACE) is a metalloprotease involved in the release of a number of cellular proteins including TNF. To determine whether the increase in membrane bound TL1A in Clostridium stimulated monocytes is due to lower induction of TACE, we assayed the TACE activity. We did not observe differences in TACE activity between monocytes stimulated by IC, E. coli, or Clostridium A4 (Fig. 3). Taken together, microbial organisms up-regulate TL1A mRNA and protein levels in APC, but the degree of activation is modest compared to FcγR induction of TL1A.
Figure 2. Expression of soluble and membrane TL1A by APCs.
TL1A expression on the surface of monocytes (A) and monocyte-derived DC (C) that were stained with TL1A specific monoclonal Ab (O4H08) and evaluated by flow cytometry. TL1A secretion was measured in media by ELISA for monocytes (B) and monocyte-derived DC (D). Data shown are means ± SEM of at least three independent experiments.
Figure 3. TACE activity in human monocytes.
Freshly isolated monocytes were incubated with plate-coated IC, E. coli, or Clostridium A4. The cellular extracts were collected and analyzed using an ELISA based assay for TACE activity. Data shown are normalized to monocytes alone; closed circles represent values for each individual experiment, bars represent the mean value for each condition.
TLR signaling partially reconstitutes TL1A expression in human monocytes
We previously found that TLR 1, 2, and 4 induced low levels of TL1A transcripts, but not detectable protein levels [28]. To determine if microbial antigens utilize other TLR to induce TL1A expression, we stimulated monocytes with TLR ligands 1 through 9. In addition, we also tested whether Muramyl dipeptide (MDP), a ligand for NOD2 (CARD15) [29], could induce TL1A expression. TL1A transcript was quantified at 4 h post stimulation because the bacteria dependent TL1A mRNA level peaked between 4–8 h (Fig. 1). Consistent with previous findings, LPS and Pam3CSK4 induced low levels of TL1A, while flagellin was an ineffective inducer of TL1A transcripts in human monocytes [28] (Fig. 4A). Of the additional TLR ligands that we tested, HKLM (TLR 2 agonist, 0.36% of β-actin) and ODN2006 (TLR 9 agonist, 0.75% of β-actin) were also inducers of TL1A expression (Fig. 4A). Stimulation of monocytes with various combinations of TLR agonists did not reconstitute 100% of bacteria dependent TL1A expression (Fig. 4A and data not shown). To test if live bacteria induce more TL1A mRNA than dead bacteria, monocytes were stimulated with either sonicated or heat lysed bacteria. We found that TL1A expression was significantly decreased when microbes were killed (Fig. 4B).
Figure 4. Induction of TL1A is partly mediated by TLR signaling pathway and dependent on live bacteria, NF-κB and p38 MAPK activity.
(A) Human CD14+ monocytes were incubated with plate coated IC, E. coli, PAM3CSK4, HKLM, Poly (I:C), LPS, flagellin, FSL, imiquimod, ssRNA, ODN2006, or MDP for 4 h. TL1A mRNA was analyzed by RT-PCR and expressed as percent β-actin. (B) Human CD14+ monocytes were incubated with live and dead E. coli (lysed by either heat or sonication) for 4 h. TL1A mRNA was analyzed by RT-PCR and expressed as percent β-actin. (C) Activated NF-κB is detected using an ELISA based assay. IC and E. coli induced NF-κB activity in monocyte nuclear extracts. There was a reduction NF-κB activity when MG-132, a NF-κB inhibitor, was added. WT, but not mutant NF-κB consensus binding oligonucleotide, reduced the detection of activated NF-κB. (D) In vitro kinase assay were performed and assayed for the amount of phosphorylated ATF-2 using Western immunoblot. IC and E. coli increased ATF-2 phosphorylation. The addition of p38 MAPK inhibitor, SB-203580, reduced phosphorylation of ATF-2. Data shown are means ± SEM of at least three independent experiments. *, p<0.05.
TL1A expression is dependent on p38 MAPK and NF-κB activation
Because NF-κB and MAPK are downstream mediators of TLR signaling pathways, we investigated their role in TL1A expression. We incubated monocytes and DC that were stimulated with E. coli in the presence of either a p38 MAPK inhibitors (SB-203580 [30] and SB-239063 [31]), NF-κB inhibitors (MG132 [32, 33], QNZ [34], IKK-NBD [35]), or NF-κB negative control inhibitor (IKK-Control). To assess NF-κB activity, we isolated nuclear extracts from stimulated monocytes and showed that both IC and E. coli induced NF-κB activity and that the NF-κB activity could be inhibited by MG-132 (Fig. 4C). The specificity of activated NF-κB detection is illustrated by the fact that WT, but not mutant consensus binding site oligonucleotide, prevented the detection of activated NF-κB (Fig. 4C). We then showed that the NF-κB inhibitors, but not its negative control (IKK-Control), significantly reduced E. coli induced TL1A expression at the RNA and protein levels in both monocytes (Fig. 5A–C) and monocyte-derived DC (data not shown).
Figure 5. p38 MAPK inhibitor and NF-κB inhibitor reduced microbial activation of TL1A expression.
Monocytes were incubated with either p38 MAPK inhibitors (SB-203580 or SB-239063), NF-κB inhibitors (MG132, QNZ, or IKK-NBD), or negative control for NF-κB inhibitor (IKK-Control) in combination with E. coli. (A) TL1A mRNA was analyzed by real-time RT-PCR and expressed as relative expression. (B) Cells were stained for surface TL1A expression and analyzed by flow cytometry. (C) TL1A in supernatants was quantified by ELISA. Data shown are means ± SEM of at least three independent experiments. *, p<0.05. **, p<0.001.
To assess p38 MAPK activity, we performed an in vitro kinase assay to measure the amount of phosphorylated ATF-2 using stimulated monocyte total cell extracts. We showed that both IC and E. coli can increase the activity of p38 MAPK by enhanced ATF-2 phosphorylation (Fig. 4D). The addition of p38 MAPK inhibitory compound (SB-203580) suppressed the phosphorylation of ATF-2 (Fig. 4D). We then showed that p38 MAPK inhibitors (SB-203580 [30] and SB-239063 [31]) significantly reduced E. coli induced TL1A mRNA in both monocytes (Fig. 5A–C) and monocyte-derived DC (data not shown). These results suggested that p38 MAPK and NF-κB activity were important signaling components to induce TL1A expression.
E. coli enhances IFN-γ production by CD4+ T cells through induction of TL1A expression
Using an IL-12/IL-18 primed CD4+ T cell co-culture system, we previously demonstrated that FcγR signaling of monocytes enhanced IFN-γ production by CD4+ T cells partially through induction of TL1A expression [28]. To determine whether microbial induction of TL1A expression in monocytes yields a similar functional consequence, we used the same system. Because the induction of IFN-γ by bacteria between different donors can vary by up to 10 fold (10 – 50 ng/mL), we normalized the IFN-γ value by E. coli or IC stimulation. Normalized IFN-γ value is shown in Figure 6, and absolute values are documented in parentheses below.
Figure 6. E. coli activated TL1A expression enhances IFN-γ production by CD4+ T cells.
CD4+ T cells were incubated with IL-12 and IL-18 were either cultured alone, co-cultured in the presence or absence of blocking TL1A Ab (04H08) or isotype control Ab with autologous monocytes (A, C) or monocyte-derived DC (B) that had been preincubated with bacteria or IC. After 48 h, media were collected and analyzed for IFN-γ production by ELISA. Closed circles represent values for each individual experiment, bars represent the mean value for each condition. *, p<0.05. **, p<0.001.
CD4+ T cells primed with IL-12/IL-18 express low levels of IFN-γ by itself (average of 0.91 ng/mL/106 CD4+ T cells), with E. coli stimulation (average of 2.78 ng/ml/106 CD4+ T cells) or with unstimulated monocytes (average of 2.56 ng/mL/106 CD4+ T cells). We first confirmed our previous findings that there was enhanced IFN-γ production by IL-12/IL-18 primed CD4+ T cells co-cultured with IC treated monocytes compared to untreated monocytes (Fig. 6A)[28]. Furthermore, addition of blocking TL1A Ab inhibited IFN-γ production and the maximal inhibitory efficiency was 50% (Fig. 6A)[28] indicating that TL1A induced by FcγR signaling of monocytes could enhance IFN-γ production by CD4+ T cells. When IL-12/IL-18 primed CD4+ T cells were co-cultured with autologous monocytes that had been stimulated by E. coli, there was an increase in IFN-γ production compared to the co-culture with unstimulated monocytes (average of 19.1 vs. 2.56 ng/mL/106 CD4+ T cells, p<0.0001) (Fig. 6A). To determine whether this additional increase of IFN-γ production was in part due to microbe dependent TL1A expression, we used blocking TL1A Ab. We showed that anti-TL1A Ab, but not its isotype control, significantly and dose dependently reduced IFN-γ levels by an average of 25% (19.1 vs. 14.23 ng/mL/106 T cells, p<0.0001 and Fig. 6A).
We next used monocyte-derived DC to determine whether TL1A induced by FcγR signaling or bacteria stimulation of other APC could similarly enhance IFN-γ production in CD4+ T cells. Primed CD4+ T cells express low levels of IFN-γ when co-cultured with unstimulated DCs (average of 3.88 ng/mL/106 CD4+ T cells). Similar to co-culture with stimulated monocytes, IC- and bacteria- treated DC increased IFN-γ production by IL-12/IL-18 primed CD4+ T cells (average of 21.08 and 18.7 ng/mL/106 CD4+ T cells, respectively) (Fig. 6B). We also demonstrated that neutralizing TL1A Ab, but not its isotype controls, can dose dependently inhibit IFN-γ production by CD4+ T cells when co-cultured with either IC (21%, 21.08 vs. 16.54 ng/mL/106 CD4+ T cells, p<0.0001 and Fig. 6B) or E. coli (12%, 18.7 vs. 16.1 ng/mL/106 CD4+ T cells, p<0.002 and Fig. 6B).
We performed co-culture experiments on anaerobic bacteria (B. thetaiotaomicron, Clostridium A4) because these microorganisms induce TL1A less efficiently. There was an increase in IFN-γ production in co-culture with monocytes stimulated with B. thetaiotaomicron (7.3 ng/mL/106 CD4+ T cells) and Clostridium A4 (9.2 ng/mL/106 CD4+ T cells) compared to unstimulated monocytes (2.56 ng/mL/106 CD4+ T cells). However, neutralizing-TL1A Ab did not reduce IFN-γ levels (Fig. 6C). We additionally performed co-culture experiments using probiotic microorganisms E. coli Nissle 1917 and Bifidobacterium breve and found that monocytes stimulated with these bacteria increased IFN-γ production in CD4+ T cells (9.4 and 7.8 ng/mL/106 CD4+ T cells for E. coli Nissle 1917 and Bifidobacterium breve, respectively) compared to unstimulated monocytes (2.56 ng/mL/106 CD4+ T cells). We found neutralizing TL1A Ab reduced IFN-γ levels in co-culture with E. coli Nissle 1917 but not with Bifidobacterium breve stimulated monocytes (Fig. 6C). Taken together, these data showed that microbe dependent TL1A expression in APC could enhance IFN-γ production by CD4+ T cells.
Discussion
One of the widely held hypotheses in the pathogenesis of IBD is that both CD and ulcerative colitis (UC) are a result of an overly aggressive host response to enteric microbes. Signaling via TNF family members is a key costimulatory mechanism modulating T cell responses to TCR and MHC-peptide engagement. Of the TNF members, TL1A has emerged as an important mediator of gut inflammation [2, 3, 5, 9, 10, 13, 15]. In this study, we investigate the interaction between microbes and the TL1A signaling pathway in APCs and its functional consequences.
In general, aerobic organisms induce higher TL1A expression than anaerobic organisms (Fig. 1). Reduced TL1A induction by anaerobic microbes may be due to species-specific differences that result in decreased expression of TL1A. For example, the anaerobic proteome profile may be less efficient in inducing pathways that lead to TL1A expression. Alternatively, these differences may reflect the toxic effect of oxygen on anaerobic bacteria because anaerobes are not alive under aerobic culturing conditions and dead bacteria induce less TL1A transcripts compared to live bacteria (Fig. 4B). Even though anaerobic bacteria are less efficient in the induction of TL1A in APCs, they are capable of initiating colitis. This is likely due to the production of other pro-inflammatory cytokines including IL-6 and TNF-α (data not shown). In addition, anaerobic bacteria such as B. thetaiotaomicron, B. breve, and Clostridium A4 were able to enhance IFN-γ production in CD4+ T cells (Fig. 6C).
Compared to other bacteria, Clostridium A4 induced more membrane bound than secreted TL1A protein, suggesting that the processing of TL1A into the secreted form is not as efficient when monocytes are stimulated by this organism. (Fig 2A). This is not observed in Clostridium A4 stimulated DC (Fig 2C). Clostridium A4 has a flagellin gene that is highly similar to the FlaX flagellin, to which patients with CD have seroreactivity [36, 37]. We tested the hypothesis that TACE may mediate the release of membrane TL1A to its secreted form [28]. We did not observe any differences in TACE activity (Fig. 3) to account for the disconnect between the levels of membrane and secreted TL1A in Clostridium A4 stimulated monocytes. Other investigations are currently ongoing to better understand the processing to TL1A to its secreted form.
Bacteria induced TL1A mRNA peaked by 4 h and decreased over the next 24 h while IC induced a gradual transcript increase over 16 h in monocytes (Fig. 1A–C)[28]. This indicates that both bacteria and IC (through FcγR pathway) can initiate TL1A expression. Yet, continued activation of TL1A expression requires FcγR signaling, implying a role of opsonized pathogens in the sustained amplification of TL1A and the pro-inflammatory pathway. However, the sustained expression of TL1A by FcγR is not observed in monocyte-derived DC (Fig. 1D–F)[28] and suggests that the mechanism involved in the kinetics of TL1A expression in monocytes may be different than that in DC.
Despite the finding that microbial organisms induce less TL1A protein than IC, the amount of bacteria induced TL1A in APC is physiologically relevant. CCR9+ CD4+ T cells represent a small subset of peripheral blood T cells involved in small intestinal immune and inflammatory responses [38]. T cells expressing CCR9 are enriched in the lamina propria and intraepithelial lymphoid compartment of the small intestine [4, 39] as well as in the peripheral blood of CD patients [40]. Importantly, small bowel associated CCR9+ CD4+ T cells are extremely sensitive to TL1A stimulation since even 100 pg of TL1A could enhance IFN-γ production by up to 10 fold [5]. This is the range of TL1A that is secreted by bacteria stimulated monocytes (approximately 100 pg/mL) and DC (approximately 300 pg/mL) (Fig. 2B, D). The significance of microbial induction of TL1A production by APC was further supported by the finding that neutralizing TL1A Ab partially but significantly inhibit APC mediated induction of IFN-γ production by T cells (Fig. 6).
We showed that TLR 1, 2, 4, and 9 could induce low levels of TL1A mRNA in human monocytes (Fig. 4A). However, activating TLR 1–9 either individually or in combination (Fig. 4A) did not reconstitute 100% of the bacteria dependent TL1A expression, which suggested additional mechanisms mediating live bacteria induction of higher TL1A expression. Our observation that TL1A expression was significantly decreased when microbes were killed either by heat or sonication (Fig. 4B) supports this notion. The more efficient induction of TL1A by live bacteria over dead bacteria or combination of TLR ligands may be due to differences in efficiency in the uptake of bacteria by APCs, bacteria processing, or in the generation of ROS. These hypotheses are being investigated.
We found that p38 MAPK and NF-κB are involved in the signaling pathway leading to TL1A expression. We used multiple agents using non-overlapping mechanisms to inhibit p38 MAPK and NF-κB to minimize non specific effects of pharmacologic compounds. For example, because both MG 132 (inhibits NF-κB activation by preventing IκB degradation) and IKK-NBD (inhibits NF-κB activation by blocking an IKB complex regulatory protein) reduced bacteria dependent TL1A expression (Fig. 5 and data not shown), then it is more likely that NF-κB is involved in TL1A expression. In addition, negative control NF-κB inhibitor (IKK-Control) did not reduce TL1A expression (Fig 5). The specificity of the inhibitors is further illustrated by the fact that WT, but not mutant consensus binding site oligonucleotide, reduced NF-κB activity in the nuclear extracts (Fig. 4C) and that p38 MAPK inhibitors reduced the amount of phosphorylated ATF-2 (Fig. 4D).
We and others showed that low levels of IL-12 and IL-18 are needed for TL1A to augment IFN-γ production [2, 3, 10, 28]. We used the conditions that weakly activate T cells because in titration experiments, these conditions yielded the highest augmentation of IFN-γ production by TL1A in synergy with IL-12/IL-18. This is also consistent with our previous findings that TL1A is most potent for augmentation of IFN-γ production under low rather than high intensity T-cell stimulation conditions [2, 10].
We showed that microbe induced TL1A in APC enhanced the production of IFN-γ by CD4+ T cells. The enhanced IFN-γ release could be partially blocked with an anti-TL1A Ab with certain but not all bacteria (Fig. 6). The reduced IFN-γ level with neutralizing TL1A Ab is correlated with the amount of secreted TL1A by stimulated APC. For example, anti-TL1A Ab reduced IFN-γ levels in co-culture with monocytes stimulated by E. coli K19 and Nissle 1917 (Fig. 6) because the amount of secreted TL1A by monocytes is significantly higher (Fig. 2B). In contrast, there was no significant reduction of IFN-γ with addition of neutralizing TL1A Ab in co-culture with monocytes stimulated with Bifidobacter, Clostridium, or Bacteriodes (Fig. 6) because there is significantly less secreted TL1A (Fig. 2B). These findings indicate that the enhanced IFN-γ secretion is partially TL1A mediated.
The reduction of IFN-γ with a blocking TL1A Ab in monocyte co-cultures was generally greater than with DC (Fig. 6) despite a similar amount of TL1A expressed in the co-culture (data not shown). This may be due to a higher basal IFN-γ level when CD4+ T cells were co-cultured with unstimulated DC (3.88 ng/mL) than with unstimulated monocytes (2.56 ng/mL) and thus, the degree of IFN-γ reduction is blunted in DC. Alternatively, this may reflect a greater activation of CD4+ lymphocytes by other pro-inflammatory cytokines (e.g. IL-6) or molecules (e.g. CD80, CD86), therefore, the contribution by the TL1A/DR3 pathway may be proportionally less in co-culture with DC. The reduction of IFN-γ (up to 25%, Fig. 6) by blocking the TL1A/DR3 pathway induced by E. coli is significant because there are other pro-inflammatory cytokines (e.g. IL-6, LIGHT) and molecules (e.g. CD80, CD86) present in the co-culture and that the maximal inhibition of IFN-γ in the co-culture experiment by neutralizing TL1A Ab was previously shown to be at most 50% [28].
Even though microbial organisms induce TL1A in peripheral monocytes and DC, the potential for extrapolation of this pathway in the mucosal immune compartment remains to be determined. It is noted that intestinal lamina propria macrophages lack CD14 [41]. However, a network of mucosal DC has been identified within the murine intestine that express CD14 and FcγR [42], and the bacterial induction of the TL1A/DR3 pathway may be relevant in this population.
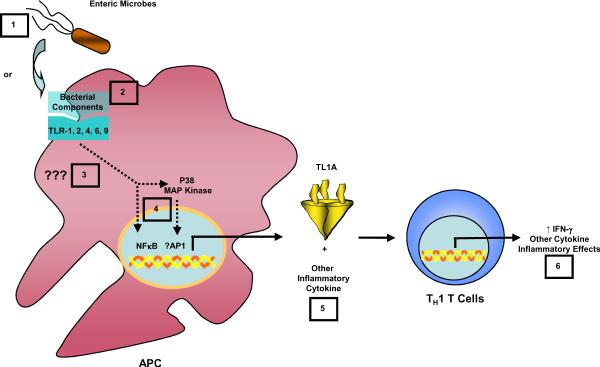
In summary, our findings demonstrate for the first time that TL1A could be induced by multiple types of bacteria in human APC. Bacterial induction of TL1A involved at least TLR 1, 2, 4, 6, and 9, and the activation of both p38 MAPK and NF-κB. Furthermore, bacteria activated TL1A on APC augmented IFN-γ production in CD4+ T cells, demonstrating the importance of microbial-host interaction in the induction of an IBD associated gene, TL1A, and its role in enhancing the Th1 immune response, in addition to its other known role in T cell responses including Th2 and Th17. Figure 7 summarizes our current hypothesis. Our findings prompt further investigation of the TL1A/DR3 pathway as a candidate target for intervention in the treatment of IBD.
Figure 7. Proposed model of microbial induction of TL1A in gut inflammation.
Either bacteria (1) or ligands for TLR 1, 2, 4, 6, and 9 receptors (2) are sensed by APC and activate a signaling pathway (3) that leads to the activation of the transcription factor NF-κB and p38 MAPK (4) to mediate the production of TL1A by APCs (5). The production of TL1A by APC then signals to CD4+ T cells to enhance IFN-γ expression and possibly other cytokines that augment Th1 immune response (6) in addition to its other known effect on Th2 and Th17 response.
Materials and Methods
Peripheral blood cell isolation and culture
Blood was obtained after informed consent in accordance with procedures established by the Cedars-Sinai Institutional Review Board. PBMC were isolated as previously described [28]. Monocyte preparations were >90% pure as determined by esterase stain (Sigma-Aldrich Inc, St. Louis, MO). Monocytes were cultured as described previously [28]. CD4+ T cells were isolated as described previously [28] and cultured for 12 h in the presence of human IL-12 (final concentration, 0.25ng/mL; PeproTech) and human IL-18 (final concentration, 6.25ng/mL; MBL) in RPMI complete medium prior to co-culture with autologous monocytes. CD4+ T lymphocytes were >90% pure as determined by CD4 stain. These stimulatory conditions (both duration and concentration of IL-12 and -18 used) were previously determined in titration experiments that demonstrated the highest augmentation of CD4+ T cell activation effects (fold induction of IFN-γ) when co-cultured with activated monocytes and DC (data not shown). Monocyte-derived DC was generated as previously described [43].
Rationale of selected bacteria
E. coli was chosen because it is part of the normal gut microflora. The specific strain that we selected (MG1635) was used as representative E. coli in the bacterial genome project. Salmonella typhimurium was chosen to assess the potential relationship between the Th17 pathway and TL1A induction. Salmonella typhimurium causes a localized enteric infection in immunocompetent individuals and the gene expression profile induced by this organism is mainly Th17 response [44]. Listeria monocytogenes was chosen to evaluate the potential interaction between the Th1 response, the innate immune response, and the TL1A signaling pathway. Listeria monocytogenes is a pathogen that has been used as a model to study innate immunity and adaptive immunity, specifically the Th1 response [45]. Staphylococcus epidermidis was selected because in contrast to listeria and salmonella, it is usually nonpathogenic and is not part of normal enteric flora. Clostridium A4 strain was selected because it has a flagellin gene that is highly similar to the FlaX flagellin, to which CD patients have seroreactivity. In addition, the FlaX flagellin is a dominant antigen causing colitis in mice [36]. Campylobacter jejuni was chosen to evaluate the potential interaction between the innate inflammatory response and the Th1-polarized adaptive immune response. It is different from listeria in that campylobacter is a microaerophilic organism. Bacteroides thetaiotaomicron was chosen because it is part of the gut microflora. It is different from E. coli in that Bacteroides is anaerobic organism. Bifidobacterium breve and E. coli Nissle 1917 were used because they are probiotics and reported to suppress colitis.
Bacterial culture
E. coli (strain K19, MG1635, purchased from University of Wisconsin-Madison), E. coli Nissle 1917 [46] (gift from Cohen Lab, University of Rhode Island), Bifidobacterium breve (purchased from ATCC, number 15700), Listeria monocytogenes (strain 10403-S), Staphylococcus epidermis (Miller Lab, UCLA), and Salmonella typhimurium (SL1344) were recovered from frozen stocks and streaked on the appropriate media plates (BHI supplemented with streptomycin 100 μg/mL for L. monocytogenes; trypticase soy agar for S. epidermidis; LB with streptomycin 200 ug/mL for S. typhimurium). Single colonies were picked and grown overnight in corresponding liquid culture. E. coli and E. coli Nissle 1917 are grown overnight at 37 degrees Celsius. Campylobacter jejuni (strain 81–176) was harvested from a fresh overnight lawn grown microaerophilically on Blaser plates at 42 degrees Celsius and resuspended in PBS. Bacteriodes thetaiotaomicron (typed strain VPI-5482) was inoculated directly from frozen stocks into BHI media supplemented with 500 μg/mL cysteine and 5 μg/mL hemin and grown anaerobically for 48 h. Bifidobacterium breve were grown according to ATCC instructions. Bacterial concentrations were estimated by optical density and appropriate dilutions were made for incubation with cells. Culturing conditions for Clostridium A4 (gift from Dr. C. Elson, University of Alabama) were described previously [37]. These bacteria were washed and lyophilized prior to use in experiments. Sonication of bacteria was done in 1 mL overnight sample using Sonic Dismembrator 60 from Fisher Scientific. Samples were sonicated continuously at microtip setting 8 on ice for 10 s, cooled for 30 s, and repeated 5 times. Successful lysis was confirmed by clearing of turbidity and examination under light microscopy. Bacteria were heat lysed for 5 min at 96 degrees Celsius.
Stimulation/inhibition of monocytes and DC
Monocytes and monocyte-derived DC were cultured in RPMI at 1–1.5 million cells/mL in either 12- or 24-well plates. Cells were stimulated from 2 to 24 h with IC, Clostridium A4 (5 μg/mL), microbial organisms (E. coli, S. typhimurium, L. monocytogenes, S. epidermidis, C. jejuni, B. thetaiotaomicron, or Bifidobacterium breve) at MOI of 50 bacteria per monocyte as determined by prior MOI titration of 10, 50, 100 (data not shown), Pam3CSK4 (InvivoGen, 300 ng/mL), HKLM (InvivoGen, 105 cells/mL), Poly(I:C) (InvivoGen, 10 μg/mL), LPS (InvivoGen, 100 ng/ml), flagellin (InvivoGen, 2 μg/mL), FSL1 (InvivoGen, 500 ng/mL), imiquimod (InvivoGen, 200 ng/mL), ssRNA40 (200 ng/mL), CpG ODN2006 and control (InvivoGen, 2 μM).
Plate bound, cross-linked human IgG (plate IC) was prepared as described elsewhere [28]. For inhibitory experiments, p38 MAPK inhibitors (SB-203580, Biomol, 1μM; SB-239063, Tocris bioscience, 200 nM) and NF-κB inhibitors (MG-132, Biomol, 10 μM; IKK-NBD or IKK-control, 100 γM; QNZ, Biomol, 20 nM) were introduced 15 min prior to the addition of the stimulatory agonists.
Co-culture of CD4+ T cells with monocytes and DC
After a 12 h stimulation of CD4+ T cells with IL-12 and IL-18, medium was replaced with fresh medium supplemented with IL-12 and IL-18 and the cells were cultured with autologous monocytes or monocyte-derived DC. Before co-cultures, monocytes and DC were incubated with IC, E. coli, Clostridium A4, B. thetaiotaomicron, or Bifidobacterium breve for 6 h (this time point was chosen because it is the average maximal TL1A mRNA stimulation between monocytes (4 h) and DC (8 h) by E. coli. 15 min before co-culturing CD4+ T cells with autologous monocytes, TL1A blocking Ab (IgG2b, Teva Biopharmaceuticals, 10 or 50 mg/mL) [2] or IgG2b isotype control Ab (eBioscience, 10 or 50 mg/mL) were added to the autologous monocytes. Co-cultures were incubated for 48 h and supernatants were harvested and analyzed for IFN-γ production by ELISA. These co-culture conditions were determined in previous titration experiments that demonstrated the highest augmentation of CD4+ T cell activation effects (measured by fold induction of IFN-γ) when co-cultured with activated monocytes and DC ([28].
PCR and real-time PCR analysis
Total RNA was isolated as previously described [28]. Two micrograms of total RNA was used in each RT reaction using the Omniscript kit (Qiagen), with oligo(dT) as primer. PCR condition, TL1A and β-actin transcripts were amplified by quantitative real-time RT-PCR as previously described [28]. Duplicates differing by less than one cycle were averaged and amount of transcript was analyzed and expressed as percentage of β-actin.
Analysis of TL1A and IFN-γ by ELISA
TL1A was quantified in supernatants collected from stimulated monocytes using ELISA as described previously [28]. IFN-γ was quantified as previously reported [2].
Western blot analyses
At various times post-stimulation, culture supernatant was removed and monocytes were directly lysed in cell lysis buffer contained in p38 MAPK assay kit (Cell Signaling Technology) according to manufacturer's protocol. Samples were boiled, and proteins were separated by SDS-15% PAGE (Lonza Rockland, Inc). Proteins were transferred onto PDVF membranes (Invitrogen), and the immunoblots were processed with phospho-specific Ab and respective HRP-conjugated secondary Ab contained in a p38 MAPK assay kit (Cell Signaling Technology) according to manufacturer's protocol.
TACE activity assay
Protein extract were prepared from either IC, E. coli, or Clostridium A4 stimulated monocytes using CytoBuster Protein Extraction Reagent (EMD, Gibbstown, NJ) according to manufacturer's protocol. TACE activities were measured using InnoZyme™ TACE Activity Kit (EMD, Gibbstown, NJ) per manufacturer's protocol.
p38 MAPK assay
The activity of p38 MAPK was analyzed through the use of a p38 MAPK assay kit (Cell Signaling Technology) according to manufacturer's protocol. The kinase reaction was analyzed by Western blotting with a phospho-specific anti-ATF-2 Ab.
NF-κB activity assay
Nuclear extracts were prepared from either IC or E. coli stimulated monocytes using the nuclear extract kit (Active Motif, Carlsbad, CA) according to manufacturer's protocol. NF-κB activities from 5 μg nuclear protein were measured using the TransAM NF-κB ELISA kit (Active Motif, Carlsbad, CA) per manufacturer's protocol.
Flow cytometry
After monocytes were stimulated for 16 h, they were stained with monoclonal Ab against TL1A (mAb 04H08, IgG2b, Teva Biopharmaceuticals), washed, stained with biotinylated goat anti-mouse IgG2b (CALTAG), washed again, and stained with APC conjugated streptavidin (CALTAG). 16 h were chosen because it yielded the highest surface TL1A in time-course experiments [28]. Cells fixed with 2% paraformaldehyde were analyzed on a Cyan flow cytometer (Dako-Cytomation).
Statistical analysis
Data are presented as the mean ± SEM values. Comparison between two groups was performed by a two tailed, unpaired Student's t-tests. A value of p<0.05 was considered significant.
Acknowledgments
The authors thank Teva Biopharmaceuticals (Rockville, MD) for generously providing us with anti-TL1A Ab. We also acknowledge members of the Targan, Liu, and Underhill labs for helpful discussions. We are grateful to Patricia Lin for help with flow cytometry. This work was supported in part by grants from the NIH Gastroenterology training grant (T32 DK07180), Specialty Training and Advanced Research (STAR) Program at UCLA, and investigator initiated award from Procter and Gamble (DQS) and NIH grants DK056328 and DK046763 (SRT).
Footnotes
Conflict of interest Then authors declare no financial or commercial conflict of interest.
References
- 1.Migone TS, Zhang J, Luo X, Zhuang L, Chen C, Hu B, Hong JS, Perry JW, Chen SF, Zhou JX, Cho YH, Ullrich S, Kanakaraj P, Carrell J, Boyd E, Olsen HS, Hu G, Pukac L, Liu D, Ni J, Kim S, Gentz R, Feng P, Moore PA, Ruben SM, Wei P. TL1A is a TNF-like ligand for DR3 and TR6/DcR3 and functions as a T cell costimulator. Immunity. 2002;16:479–492. doi: 10.1016/s1074-7613(02)00283-2. [DOI] [PubMed] [Google Scholar]
- 2.Prehn JL, Mehdizadeh S, Landers CJ, Luo X, Cha SC, Wei P, Targan SR. Potential role for TL1A, the new TNF-family member and potent costimulator of IFN-gamma, in mucosal inflammation. Clinical Immunology. 2004;112:66–77. doi: 10.1016/j.clim.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 3.Papadakis KA, Prehn JL, Landers C, Han Q, Luo X, Cha SC, Wei P, Targan SR. TL1A synergizes with IL-12 and IL-18 to enhance IFN-gamma production in human T cells and NK cells. Journal of Immunology. 2004;172:7002–7007. doi: 10.4049/jimmunol.172.11.7002. [DOI] [PubMed] [Google Scholar]
- 4.Kunkel EJ, Campbell JJ, Haraldsen G, Pan J, Boisvert J, Roberts AI, Ebert EC, Vierra MA, Goodman SB, Genovese MC, Wardlaw AJ, Greenberg HB, Parker CM, Butcher EC, Andrew DP, Agace WW. Lymphocyte CC chemokine receptor 9 and epithelial thymus-expressed chemokine (TECK) expression distinguish the small intestinal immune compartment: Epithelial expression of tissue-specific chemokines as an organizing principle in regional immunity. Journal of Experimental Medicine. 2000;192:761–768. doi: 10.1084/jem.192.5.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Papadakis KA, Zhu D, Prehn JL, Landers C, Avanesyan A, Lafkas G, Targan SR. Dominant role for TL1A/DR3 pathway in IL-12 plus IL-18-induced IFN-gamma production by peripheral blood and mucosal CCR9+ T lymphocytes. Journal of Immunology. 2005;174:4985–4990. doi: 10.4049/jimmunol.174.8.4985. [DOI] [PubMed] [Google Scholar]
- 6.Fang L, Adkins B, Deyev V, Podack ER. Essential role of TNF receptor superfamily 25 (TNFRSF25) in the development of allergic lung inflammation. J Exp Med. 2008 doi: 10.1084/jem.20072528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pappu BP, Borodovsky A, Zheng TS, Yang X, Wu P, Dong X, Weng S, Browning B, Scott ML, Ma L, Su L, Tian Q, Schneider P, Flavell RA, Dong C, Burkly LC. TL1A-DR3 interaction regulates Th17 cell function and Th17-mediated autoimmune disease. J Exp Med. 2008 doi: 10.1084/jem.20071364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meylan F, Davidson TS, Kahle E, Kinder M, Acharya K, Jankovic D, Bundoc V, Hodges M, Shevach EM, Keane-Myers A, Wang EC, Siegel RM. The TNF-family receptor DR3 is essential for diverse T cell-mediated inflammatory diseases. Immunity. 2008;29:79–89. doi: 10.1016/j.immuni.2008.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bamias G, Martin C, 3rd, Marini M, Hoang S, Mishina M, Ross WG, Sachedina MA, Friel CM, Mize J, Bickston SJ, Pizarro TT, Wei P, Cominelli F. Expression, localization, and functional activity of TL1A, a novel Th1-polarizing cytokine in inflammatory bowel disease. Journal of Immunology. 2003;171:4868–4874. doi: 10.4049/jimmunol.171.9.4868. [DOI] [PubMed] [Google Scholar]
- 10.Bamias G, Mishina M, Nyce M, Ross WG, Kollias G, Rivera-Nieves J, Pizarro TT, Cominelli F. Role of TL1A and its receptor DR3 in two models of chronic murine ileitis. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:8441–8446. doi: 10.1073/pnas.0510903103. see comment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei B, Velazquez P, Turovskaya O, Spricher K, Aranda R, Kronenberg M, Birnbaumer L, Braun J. Mesenteric B cells centrally inhibit CD4+ T cell colitis through interaction with regulatory T cell subsets. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:2010–2015. doi: 10.1073/pnas.0409449102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brewer S, McPherson M, Fujiwara D, Turovskaya O, Ziring D, Chen L, Takedatsu H, Targan SR, Wei B, Braun J. Molecular imaging of murine intestinal inflammation with 2-deoxy-2-[18F]fluoro-D-glucose and positron emission tomography. Gastroenterology. 2008;135:744–755. doi: 10.1053/j.gastro.2008.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takedatsu H, Michelsen KS, Wei B, Landers CJ, Thomas LS, Dhall D, Braun J, Targan SR. TL1A (TNFSF15) regulates the development of chronic colitis by modulating both T-helper 1 and T-helper 17 activation. Gastroenterology. 2008;135:552–567. doi: 10.1053/j.gastro.2008.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmada MM, Bitton A, Dassopoulos T, Datta LW, Green T, Griffiths AM, Kistner EO, Murtha MT, Regueiro MD, Rotter JI, Schumm LP, Steinhart AH, Targan SR, Xavier RJ, Libioulle C, Sandor C, Lathrop M, Belaiche J, Dewit O, Gut I, Heath S, Laukens D, Mni M, Rutgeerts P, Van Gossum A, Zelenika D, Franchimont D, Hugot JP, de Vos M, Vermeire S, Louis E, Cardon LR, Anderson CA, Drummond H, Nimmo E, Ahmad T, Prescott NJ, Onnie CM, Fisher SA, Marchini J, Ghori J, Bumpstead S, Gwilliam R, Tremelling M, Deloukas P, Mansfield J, Jewell D, Satsangi J, Mathew CG, Parkes M, Georges M, Daly MJ. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamazaki K, McGovern D, Ragoussis J, Paolucci M, Butler H, Jewell D, Cardon L, Takazoe M, Tanaka T, Ichimori T, Saito S, Sekine A, Iida A, Takahashi A, Tsunoda T, Lathrop M, Nakamura Y. Single nucleotide polymorphisms in TNFSF15 confer susceptibility to Crohn's disease. Human Molecular Genetics. 2005;14:3499–3506. doi: 10.1093/hmg/ddi379. [DOI] [PubMed] [Google Scholar]
- 16.Kakuta Y, Kinouchi Y, Negoro K, Takahashi S, Shimosegawa T. Association study of TNFSF15 polymorphisms in Japanese patients with inflammatory bowel disease. Gut. 2006;55:1527–1528. doi: 10.1136/gut.2006.100297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tremelling M, Berzuini C, Massey D, Bredin F, Price C, Dawson C, Bingham SA, Parkes M. Contribution of TNFSF15 gene variants to Crohn's disease susceptibility confirmed in UK population. Inflamm Bowel Dis. 2008;14:733–737. doi: 10.1002/ibd.20399. [DOI] [PubMed] [Google Scholar]
- 18.Picornell Y, Mei L, Taylor K, Yang H, Targan SR, Rotter JI. TNFSF15 is an ethnic-specific IBD gene. Inflammatory Bowel Diseases. 2007;13:1333–1338. doi: 10.1002/ibd.20223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kugathasan S, Baldassano RN, Bradfield JP, Sleiman PM, Imielinski M, Guthery SL, Cucchiara S, Kim CE, Frackelton EC, Annaiah K, Glessner JT, Santa E, Willson T, Eckert AW, Bonkowski E, Shaner JL, Smith RM, Otieno FG, Peterson N, Abrams DJ, Chiavacci RM, Grundmeier R, Mamula P, Tomer G, Piccoli DA, Monos DS, Annese V, Denson LA, Grant SF, Hakonarson H. Loci on 20q13 and 21q22 are associated with pediatric-onset inflammatory bowel disease. Nat Genet. 2008;40:1211–1215. doi: 10.1038/ng.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shih DQ, Targan SR. Immunopathogenesis of inflammatory bowel disease. World J Gastroenterol. 2008;14:390–400. doi: 10.3748/wjg.14.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim SC, Tonkonogy SL, Albright CA, Tsang J, Balish EJ, Braun J, Huycke MM, Sartor RB. Variable phenotypes of enterocolitis in interleukin 10-deficient mice monoassociated with two different commensal bacteria. Gastroenterology. 2005;128:891–906. doi: 10.1053/j.gastro.2005.02.009. see comment. [DOI] [PubMed] [Google Scholar]
- 22.Kim SC, Tonkonogy SL, Karrasch T, Jobin C, Sartor RB. Dual-association of gnotobiotic IL-10−/− mice with 2 nonpathogenic commensal bacteria induces aggressive pancolitis. Inflammatory Bowel Diseases. 2007;13:1457–1466. doi: 10.1002/ibd.20246. [DOI] [PubMed] [Google Scholar]
- 23.Bohn E, Bechtold O, Zahir N, Frick JS, Reimann J, Jilge B, Autenrieth IB. Host gene expression in the colon of gnotobiotic interleukin-2-deficient mice colonized with commensal colitogenic or noncolitogenic bacterial strains: common patterns and bacteria strain specific signatures. Inflammatory Bowel Diseases. 2006;12:853–862. doi: 10.1097/01.mib.0000231574.73559.75. [DOI] [PubMed] [Google Scholar]
- 24.Dieleman LA, Goerres MS, Arends A, Sprengers D, Torrice C, Hoentjen F, Grenther WB, Sartor RB. Lactobacillus GG prevents recurrence of colitis in HLA-B27 transgenic rats after antibiotic treatment. Gut. 2003;52:370–376. doi: 10.1136/gut.52.3.370. see comment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schultz M, Tonkonogy SL, Sellon RK, Veltkamp C, Godfrey VL, Kwon J, Grenther WB, Balish E, Horak I, Sartor RB. IL-2-deficient mice raised under germfree conditions develop delayed mild focal intestinal inflammation. American Journal of Physiology. 1999;276:G1461–1472. doi: 10.1152/ajpgi.1999.276.6.G1461. [DOI] [PubMed] [Google Scholar]
- 26.Sartor RB. Therapeutic manipulation of the enteric microflora in inflammatory bowel diseases: antibiotics, probiotics, and prebiotics. Gastroenterology. 2004;126:1620–1633. doi: 10.1053/j.gastro.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 27.Cong Y, Weaver CT, Lazenby A, Elson CO. Colitis induced by enteric bacterial antigen-specific CD4+ T cells requires CD40-CD40 ligand interactions for a sustained increase in mucosal IL-12. J Immunol. 2000;165:2173–2182. doi: 10.4049/jimmunol.165.4.2173. [DOI] [PubMed] [Google Scholar]
- 28.Prehn JL, Thomas LS, Landers CJ, Yu QT, Michelsen KS, Targan SR. The T cell costimulator TL1A is induced by FcgammaR signaling in human monocytes and dendritic cells. Journal of Immunology. 2007;178:4033–4038. doi: 10.4049/jimmunol.178.7.4033. [DOI] [PubMed] [Google Scholar]
- 29.Inohara N, Ogura Y, Fontalba A, Gutierrez O, Pons F, Crespo J, Fukase K, Inamura S, Kusumoto S, Hashimoto M, Foster SJ, Moran AP, Fernandez-Luna JL, Nunez G. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn's disease. Journal of Biological Chemistry. 2003;278:5509–5512. doi: 10.1074/jbc.C200673200. [DOI] [PubMed] [Google Scholar]
- 30.Cuenda A, Rouse J, Doza YN, Meier R, Cohen P, Gallagher TF, Young PR, Lee JC. SB 203580 is a specific inhibitor of a MAP kinase homologue which is stimulated by cellular stresses and interleukin-1. FEBS Letters. 1995;364:229–233. doi: 10.1016/0014-5793(95)00357-f. [DOI] [PubMed] [Google Scholar]
- 31.Barone FC, Irving EA, Ray AM, Lee JC, Kassis S, Kumar S, Badger AM, White RF, McVey MJ, Legos JJ, Erhardt JA, Nelson AH, Ohlstein EH, Hunter AJ, Ward K, Smith BR, Adams JL, Parsons AA. SB 239063, a second-generation p38 mitogen-activated protein kinase inhibitor, reduces brain injury and neurological deficits in cerebral focal ischemia. J Pharmacol Exp Ther. 2001;296:312–321. [PubMed] [Google Scholar]
- 32.Grisham MB, Palombella VJ, Elliott PJ, Conner EM, Brand S, Wong HL, Pien C, Mazzola LM, Destree A, Parent L, Adams J. Inhibition of NF-kappa B activation in vitro and in vivo: role of 26S proteasome. Methods in Enzymology. 1999;300:345–363. doi: 10.1016/s0076-6879(99)00140-8. [DOI] [PubMed] [Google Scholar]
- 33.Snyder JG, Prewitt R, Campsen J, Britt LD. PDTC and Mg132, inhibitors of NF-kappaB, block endotoxin induced vasodilation of isolated rat skeletal muscle arterioles. Shock. 2002;17:304–307. doi: 10.1097/00024382-200204000-00011. [DOI] [PubMed] [Google Scholar]
- 34.Tobe M, Isobe Y, Tomizawa H, Nagasaki T, Takahashi H, Hayashi H. A novel structural class of potent inhibitors of NF-kappa B activation: structure-activity relationships and biological effects of 6-aminoquinazoline derivatives. Bioorg Med Chem. 2003;11:3869–3878. doi: 10.1016/s0968-0896(03)00438-3. [DOI] [PubMed] [Google Scholar]
- 35.May MJ, D'Acquisto F, Madge LA, Glockner J, Pober JS, Ghosh S. Selective inhibition of NF-kappaB activation by a peptide that blocks the interaction of NEMO with the IkappaB kinase complex. Science. 2000;289:1550–1554. doi: 10.1126/science.289.5484.1550. [DOI] [PubMed] [Google Scholar]
- 36.Lodes MJ, Cong Y, Elson CO, Mohamath R, Landers CJ, Targan SR, Fort M, Hershberg RM. Bacterial flagellin is a dominant antigen in Crohn disease. Journal of Clinical Investigation. 2004;113:1296–1306. doi: 10.1172/JCI20295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duck LW, Walter MR, Novak J, Kelly D, Tomasi M, Cong Y, Elson CO. Isolation of flagellated bacteria implicated in Crohn's disease. Inflamm Bowel Dis. 2007;13:1191–1201. doi: 10.1002/ibd.20237. [DOI] [PubMed] [Google Scholar]
- 38.Papadakis KA, Landers C, Prehn J, Kouroumalis EA, Moreno ST, Gutierrez-Ramos JC, Hodge MR, Targan SR. CC chemokine receptor 9 expression defines a subset of peripheral blood lymphocytes with mucosal T cell phenotype and Th1 or T-regulatory 1 cytokine profile. J Immunol. 2003;171:159–165. doi: 10.4049/jimmunol.171.1.159. [DOI] [PubMed] [Google Scholar]
- 39.Papadakis KA, Prehn J, Nelson V, Cheng L, Binder SW, Ponath PD, Andrew DP, Targan SR. The role of thymus-expressed chemokine and its receptor CCR9 on lymphocytes in the regional specialization of the mucosal immune system. J Immunol. 2000;165:5069–5076. doi: 10.4049/jimmunol.165.9.5069. [DOI] [PubMed] [Google Scholar]
- 40.Papadakis KA, Prehn J, Moreno ST, Cheng L, Kouroumalis EA, Deem R, Breaverman T, Ponath PD, Andrew DP, Green PH, Hodge MR, Binder SW, Targan SR. CCR9-positive lymphocytes and thymus-expressed chemokine distinguish small bowel from colonic Crohn's disease. Gastroenterology. 2001;121:246–254. doi: 10.1053/gast.2001.27154. [DOI] [PubMed] [Google Scholar]
- 41.Smythies LE, Sellers M, Clements RH, Mosteller-Barnum M, Meng G, Benjamin WH, Orenstein JM, Smith PD. Human intestinal macrophages display profound inflammatory anergy despite avid phagocytic and bacteriocidal activity. J Clin Invest. 2005;115:66–75. doi: 10.1172/JCI19229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flores-Langarica A, Meza-Perez S, Calderon-Amador J, Estrada-Garcia T, Macpherson G, Lebecque S, Saeland S, Steinman RM, Flores-Romo L. Network of dendritic cells within the muscular layer of the mouse intestine. Proc Natl Acad Sci U S A. 2005;102:19039–19044. doi: 10.1073/pnas.0504253102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chapuis F, Rosenzwajg M, Yagello M, Ekman M, Biberfeld P, Gluckman JC. Differentiation of human dendritic cells from monocytes in vitro. Eur J Immunol. 1997;27:431–441. doi: 10.1002/eji.1830270213. [DOI] [PubMed] [Google Scholar]
- 44.Raffatellu M, Santos RL, Verhoeven DE, George MD, Wilson RP, Winter SE, Godinez I, Sankaran S, Paixao TA, Gordon MA, Kolls JK, Dandekar S, Baumler AJ. Simian immunodeficiency virus-induced mucosal interleukin-17 deficiency promotes Salmonella dissemination from the gut. Nat Med. 2008;14:421–428. doi: 10.1038/nm1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zenewicz LA, Shen H. Innate and adaptive immune responses to Listeria monocytogenes: a short overview. Microbes Infect. 2007;9:1208–1215. doi: 10.10110/2/076/j.micinf.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Autieri SM, Lins JJ, Leatham MP, Laux DC, Conway T, Cohen PS. L-fucose stimulates utilization of D-ribose by Escherichia coli MG1655 DeltafucAO and E. coli Nissle 1917 DeltafucAO mutants in the mouse intestine and in M9 minimal medium. Infect Immun. 2007;75:5465–5475. doi: 10.1128/IAI.00822-07. [DOI] [PMC free article] [PubMed] [Google Scholar]