Abstract
The past decade has seen intense scientific interest in non-coding RNAs. In particular, the discovery and subsequent exploitation of gene silencing via RNA interference (RNAi) has revolutionized the way in which gene expression is now studied and understood. It is now well established that post-transcriptional gene silencing (PTGS) by the microRNA (miRNA) and other RNAi-associated pathways represents an essential layer of complexity to gene regulation. Gene silencing using RNAi additionally demonstrates huge potential as a therapeutic strategy for eliminating pathogenic gene expression. Yet despite the early promise and excitement of gene-specific silencing, several critical hurdles remain to be overcome before widespread clinical adoption. These include off-target effects, toxicity due to saturation of the endogenous RNAi functions, limited duration of silencing, and effective targeted delivery. In recent years, a range of novel strategies for producing RNA-mediated silencing have been developed that can circumvent many of these hurdles, including small internally segmented interfering RNAs, tandem hairpin RNAs, and pri-miRNA cluster mimics. This review discusses RNA-mediated silencing in light of this recent research, and highlights the benefits and limitations conferred by these novel gene-silencing strategies.
Introduction
RNA research has seen intense growth in recent years since it became clear that the mammalian transcriptome largely comprises nonprotein-coding RNA.1,2,3 In particular, post-transcriptional gene silencing (PTGS) via RNA interference (RNAi) has attracted great interest as a gene regulatory mechanism. RNAi has been implicated in various aspects of animal development and normal physiological function, whereas dysregulation has been linked to several pathologies including viral infections,4 cancers,5 myopathies,6 and neurodegenerative disease.7,8 Excitingly, exploitation of the highly potent and sequence-specific silencing capabilities of RNAi has potential as a promising therapeutic strategy.9,10,11 This review focuses on identifying both the advantages and pitfalls of current RNAi-inducing effectors, before discussing the most recent developments in RNAi effector design that could help to realize the full therapeutic potential of RNA-mediated gene silencing.
RNAi
The endogenous microRNA (miRNA) pathway involves sequential processing of long primary miRNA transcripts into short double-stranded RNA (dsRNA) duplexes of ~19–21 nucleotides (nt) with 2-nt 3′ overhangs, termed short-interfering RNAs (siRNAs)12,13,14 (Figure 1a). The siRNAs associate with an Argonaute (AGO)-containing protein complex to assemble the RNA-induced silencing complex (RISC).15 One “guide” strand of the duplex is preferentially retained based on thermodynamic stability,16 and this activated RISC interacts with mRNA transcripts possessing sequence complementarity. If the antisense guide RNA and sense mRNAs have significant, yet incomplete base pairing, typically seen with endogenous mature miRNAs and their 3′ untranslated region (3′UTR) targets, translation can either be repressed or the mRNA can be destabilized through decapping or deadenylation. For complete base pairing, commonly utilized in synthetic RNAi approaches, the mRNA transcript is degraded.17
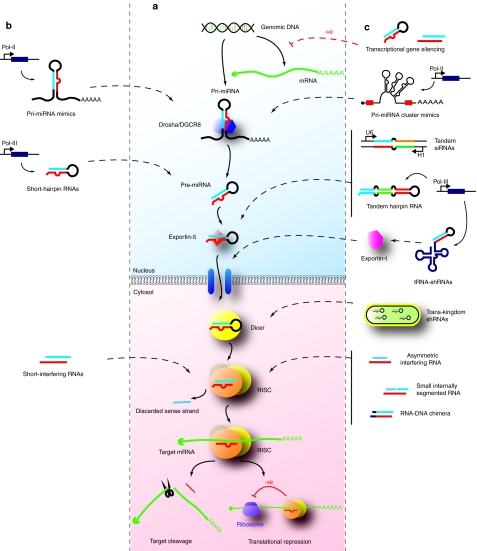
Figure 1.
Endogenous and synthetic methods of RNA induced gene silencing. (a) In the canonical mammalian miRNA pathway, pri-miRNA transcripts are processed by the microprocessor complex into pre-miRNA hairpins that are recognized by exportin-5 for nuclear export. Further processing by Dicer, recruitment into RISC and strand selection result in a mature antisense capable of directing translational repression of imperfectly matched targets, or cleavage of perfectly matched targets. (b) The commonly employed synthetic RNAi effectors (siRNAs, shRNAs, and pri-miRNA mimics) imitate different precursors of the endogenous miRNA pathway. (c) Novel “second-generation” approaches improve on existing strategies. Asymmetric interfering RNAs, small internally segmented interfering RNAs, and RNA-DNA chimeric duplexes reduce off-target effects of siRNAs by limiting sense strand activity and/or off-target effects of antisense seed regions. Trans-kingdom shRNAs and tRNA-shRNAs limit exportin-5-mediated toxicity of shRNAs by using alternate delivery mechanisms of shRNAs to the cytosol. Tandem siRNAs, tandem hairpin RNAs, and miRNA cluster mimics can be used when targeting multiple sites in the same gene or when simultaneously targeting multiple targets is desired. RNAi effectors may be additionally used to induce transcriptional gene silencing that offers the ability to induce long-lasting silencing through epigenetic changes with minimal repeated delivery.
Recent findings have led to an appreciation that natural variations in the biogenesis of RNAi, including the mirtron and endogenous siRNA (endo-siRNA) pathways, add complexity to RNAi regulation within the cells.18 Mirtrons are a novel class of splicing-dependent miRNAs identified in Drosophila and C. elegans,19,20 and are predicted to be present in mammals.21,22 Biogenesis involves removal of short introns with hairpin-forming potential to form pre-miRNA-like hairpins that bypass Drosha/DGCR8 processing. In contrast, 21–22-nt endo-siRNAs have multiple origins. For example, a long antisense transcript (>100-nt), produced from an antisense promoter either in the same gene or a homologue, can hybridize with an mRNA and be processed by unknown mechanisms involving Dicer and Tar repeat-binding protein-2 (TRBP2) into multiple small RNAs capable of RNAi. Long inverted repeat hairpins that are of increased length to canonical Drosha substrates and shRNAs can also be processed to produce active endo-siRNAs.23,24,25,26 Further study of these two still relatively unexplored pathways will no doubt increase our knowledge of RNA-based gene regulation mechanisms and result in future therapeutic opportunities.
Exogenous gene-silencing strategies
The simplicity, specificity, and potency of mimicking precursors of the RNAi pathway have made this an exciting approach to target genes for both research and therapeutic purposes. Exploitation of RNAi has most commonly been achieved using one of the three strategies (Figure 1b).
1. Synthetic siRNAs that enter the RNAi pathway after nuclear export12
2. Post-Drosha pre-miRNA precursor mimics, termed short-hairpin RNAs (shRNAs)27
3. Primary-miRNA (pri-miRNA) mimics28
Each approach has been applied in vitro and in vivo with clinical trials ongoing for, among others, intraocular delivery of siRNAs in age-related macular degeneration, nasal siRNA delivery against respiratory syncytial virus, encapsulated siRNAs for solid tumors, and an shRNA directed against the HIV tat and rev genes as part of a trimeric treatment for AIDS lymphoma.29,30 However, each current strategy has potential disadvantages that may limit its therapeutic usage.
The use of synthetic siRNAs requires delivery of dsRNA duplexes that are rapidly degraded in cells and have limited half-lives in serum, thus necessitating repeated delivery for prolonged in vivo use (Figure 2a). Serum stability and nuclease resistance can be significantly improved by modifications to the dsRNA duplex such as the addition of 2′O-methyl or locked nucleic acids (reviewed in ref. 31). Some modifications have further been shown to reduce activation of the Toll-like receptors and type I interferon response.32 Together, these modifications have made the use of siRNAs a distinct possibility for several diseases. However, some caution must be taken. Even some heavily stabilized siRNAs have been shown through mass spectrometry analysis to be eventually degraded within a matter of hours in serum,33 whereas excessive modification has the potential to reduce siRNA potency34 suggesting an optimal balance must be found.
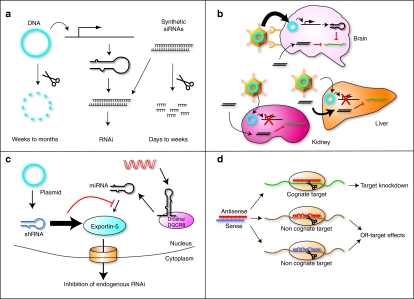
Figure 2.
Problems associated with synthetic RNAi approaches. (a) DNA-based approaches offer long-term induction of RNAi, whereas siRNAs are short-lived effectors requiring repeat delivery. (b) Promoter and/or viral specificity can allow spatially restricted control of DNA-encoded approaches to areas not readily accessible by siRNAs due to restricted access. (c) DNA-based approaches can compete with endogenous miRNAs for different components of the miRNA pathway, including exportin-5-mediated nuclear export, leading to associated toxicities. (d) In addition to desired silencing of targets, off-target effects can result from antisense or sense strands binding nonspecifically to other mRNAs.
The absence of promoter regulation and delivery specificity that is conferred by delivery vectors to DNA-based approaches additionally limits delivery approaches for siRNAs (Figure 2b) (reviewed in refs. 29,35). An exception to this is the targeted delivery of therapeutics to the liver, the site siRNAs accumulate after systemic intravenous delivery. Indeed, considerable progress has been made using siRNAs to target hepatitis B and C viruses in vivo, suggesting intravenous delivery to liver is a viable approach.36,37,38 Success will continue to be reported for cases in which unique biology can be exploited. This includes delivery to immunoprivileged sites such as the eye, or when using efficient targeting peptides or delivery vehicles.36,39,40,41,42 However, the limited ability of existing targeting peptides and vehicles to target many tissues deep within the body and the substantial costs that would be required for gram quantities of clinical-grade siRNA for human systemic delivery38 dictates that the likely application of siRNAs may be for repeated local delivery to easily accessible tissues such the eye, various epithelia, or skin.35 Consequently, other organs including the brain and heart will unlikely be suitable for siRNA delivery due to the invasive nature of access for repeat delivery and lack of cell-specific uptake.
RNAi effectors based on shRNA and pri-miRNA mimics are delivered as DNA vectors, enabling viral delivery approaches. When used in conjunction with site-specific promoters and viral vectors with specific tropisms, targeting of organs via systemic administration is possible.43,44 However, high levels of expression can outcompete the endogenous RNAi pathway leading to fatal toxicity in vivo due to exportin-5 saturation and off-target effects due to high levels of antisense RNAs45,46 (Figure 2c). The ubiquitous U6 and H1 promoters used for shRNA expression were originally selected for their relative ease of producing short RNA transcripts defined by well-characterized start and termination sequences.27,47 However, both drive high levels of expression of small RNAs that can elicit toxicity. Pri-miRNA mimics, discussed in detail later, mitigate this problem as they can utilize low-expression RNA polymerase-II (pol-II) promoters. However, it is clear that the endogenous RNAi pathway can be saturated and the risk of this should be minimized.
A major hurdle common to all aforementioned RNAi-based approaches is off-target effects (Figure 2d). Recent microarray data have revealed that complementary base pairing with as little as 8-nt homology is sufficient to cause translational repression of nontarget genes when multiple target matches are present within their 3′UTRs.48,49 Indiscriminate strand selection by RISC can subsequently lead to combined activity of both antisense and “passenger” sense strands resulting in considerable dysregulation of many mRNA transcripts.50 Much work therefore needs to be done to optimize selective loading of the antisense strand into RISC to minimize this possibility. In many cases, this is achievable with both careful consideration of the thermodynamic stability across the dsRNA duplexes and stringent testing.51 This can be further reduced with careful, specific delivery and expression in regions requiring therapeutic intervention.43,52
New generation of RNA-based silencing approaches
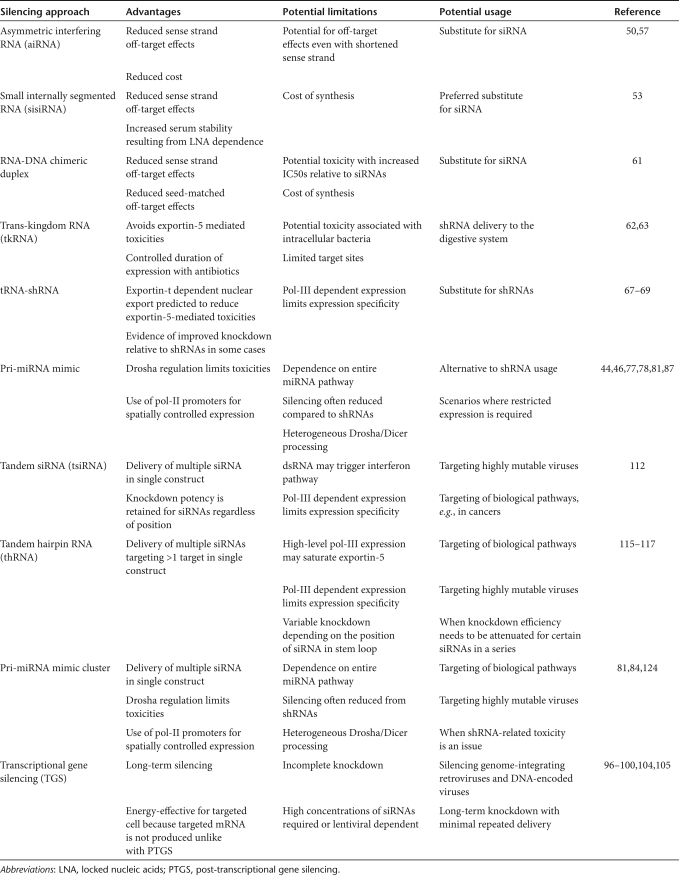
The limitations of current RNAi strategies have been acknowledged in recent years,30,50,53 and novel approaches that circumvent these have been developed (Table 1, Figure 1c). These “second-generation” RNAi effectors discussed here each have individual merits for specific uses that will hopefully drive RNAi closer to the clinic.
Table 1.
Advantages and limitations of novel RNA-dependent gene silencing approaches
Asymmetric interfering RNA. Although using thermodynamic stability to bias strand selection can reduce off-target silencing, infrequent sense strand incorporation can still lead to off-target effects.54 Atypical overhangs have been demonstrated to further improve strand bias in siRNAs, with unilateral 2-nt 3′overhangs on the antisense strand improving antisense selection.55,56 An alternative strategy is to use siRNAs composed of regular antisense strands combined with shortened sense strands that are no longer RISC substrates. Indeed, Sun et al. have shown that 19-nt antisense strands carrying 2-nt 5′ overhangs paired with 15-nt sense strands achieved efficient RISC loading and gene silencing comparable to that of siRNAs with full-length sense strands.50 Excitingly, one such asymmetric interfering RNA (aiRNA) demonstrated a nearly twofold reduction in the number of genes downregulated when looking at microarray data of 38,500 human mRNA transcripts in HeLa cells, relative to an antisense matched siRNA.
This work is supported by Chu et al. who demonstrated that aiRNAs consisting of either a 16-nt core duplex or 15-nt core duplex were potent RNAi molecules.57 Although off-target silencing was not determined, deletions to either 5′ or 3′ regions of the sense strand were readily accommodated by RISC, implying flexibility in aiRNA design. However, they reported that deletions could additionally be made to antisense strands. Interestingly, 16-nt antisense strands were capable of mediating RNAi. Furthermore, when combined with 16-nt sense strands, the silencing was improved beyond that of “parent” 19-nt siRNAs, from which truncated duplexes were derived due to an increase in the rate of RISC-mediated cleavage. If this finding is confirmed, it will significantly influence designs of aiRNAs. The results imply that 16-nt sense strands have the potential to act as RISC substrates and still mediate off-target effects. They also report a 50% reduction in silencing by a 15-nt duplex. This finding is supported by reverse asymmetry data provided by Sun et al. that showed that 15-nt antisense strands paired with 21-nt sense strands failed to silence.50 Taken together, the evidence suggests that 15-nt sense strands may have the desired potential for reducing off-target silencing in aiRNAs. Although these precise design details need to be finalized, the reports suggest aiRNAs should be investigated as improved alternatives to siRNAs due to their retention of potent activity and reduced off-target effects. Asymmetry may even be possible in DNA-encoded approaches because sense and antisense strands can be generated from separate pol-III expression cassettes to generate functional siRNAs.58,59
Small internally segmented RNAs. An equally impressive challenge to current understanding of RNAi substrates was made by Bramsen et al. who hypothesized that the undesired activity of sense strands could be eliminated by segmentation of their 22-nt sense sequence into two smaller sequences.53 These small internally segmented interfering RNAs (sisiRNAs), held tightly together by locked nucleic acid modifications, were highly potent RNAi effectors capable of reducing both synthetic and endogenous targets to levels equal to unsegmented 22-nt siRNAs. Furthermore, off-target silencing of perfectly complementary sense strand synthetic targets was abolished by segmentation of sense strands, implying neither sense segment is active. A screen of multiple sisiRNA variants suggests that for optimal silencing, contiguous sense strands are required, whereas the position of the break can be flexible and not necessarily mimic the natural cleavage site of the target. A preliminary preference for 10+12-nt mini-sense strands was suggested by the authors, but it is further suggested that in preliminary designs, a variety of sequences should be tested.
The presence of locked nucleic acid modifications to increase stability appears essential to sisiRNA design. However, these modifications have an additional benefit. No evidence was found for induction of the interferon response, in agreement with existing data on locked nucleic acid–based siRNAs,60 whereas serum stability was strongly increased from 90 minutes to over 13 hours. With abrogated sense strand activity and increased stability, sisiRNAs are on a par with aiRNAs at mitigating off-target effects and should again be considered as improved alternatives in siRNA-based therapies.
RNA–DNA chimeras. A final approach for reducing off-target effects of siRNAs is to substitute parts of the dsRNA duplex with cognate deoxyribonucleotides, in addition to the commonly utilized deoxythymidine 3′ overhangs.12,61 Potent-silencing activity is retained in chimeras in which the 5′ end of RNA antisense strands are replaced with DNA, or in which all of the nucleotides of the 8-nt dsRNA region at 5′ ends of antisense strands, together with 3′ overhangs of sense strands, are replaced with DNA.61 Despite overlapping with the essential seed region of antisense strands, the latter substitution has been confirmed with 17 functional siRNAs demonstrating little loss of efficacy implying that this is a reproducible modification. Furthermore, the method of gene silencing appears indistinguishable from that of unmodified siRNAs with an AGO-2- and RISC-dependent mechanism resulting in cleavage of target mRNA across from positions 10 and 11 of antisense strands.
The most intriguing aspect of this design is that off-target effects of the modified siRNAs are dramatically reduced from the parental siRNAs. Unlike unmodified siRNAs, the expression of perfectly complementary sense strand targets is unaffected with all chimeras reported. Even more encouraging is that both in synthetic assays and microarray profiling, there is a significant reduction in repression of seed-match targets in which just positions 2–8 of the antisense strand are paired to the target. This is important because recent microarray profiling reveals unintended silencing is predominantly associated with such seed-matches.49 It seems likely that the reduction seen in repression with these RNA–DNA chimeras is due to reduced thermodynamic stability of the resulting DNA seed region paired to RNA target compared to the RNA-duplex pairing such that RISC no longer initiates repression. Together, the results demonstrate a promising alternative to siRNAs and may circumvent the ambiguity of whether shortened sense strands can still remain active with the aiRNAs.50,57 One limiting factor that does need to be investigated before widespread adoption is the increased IC50 values of these chimeras.61 Therapeutic concentrations of chimeric duplexes will need to be determined with each construct, but whether an increased working concentration leads to further detrimental effects will need to be stringently investigated in each specific case.
Trans-kingdom RNAi. The commonly employed DNA-encoded RNAi effectors rely heavily on the endogenous miRNA pathway for processing of synthetic precursors that can result in toxic effects due to competition with endogenous miRNAs.45,46 One means of bypassing parts of the endogenous miRNA biogenesis pathway is to produce active RNAi effectors outside of mammalian cells. In proof-of-principle experiments, Xiang et al. have demonstrated that shRNA-like molecules expressed from bacteriophage T7 promoters in intracellular bacteria can effect RNAi on host cells.62,63 They termed this effect trans-kingdom RNAi (tkRNAi). Incorporation of the hemolysin gene allows the shRNA to be exported out of the endolysosomal compartment in which the bacteria resided and into the cytoplasm following transcription. However, unlike U6 promoter–driven shRNA, the 3′ end of the shRNA incorporates a T7 terminator sequence. The product thus appears to be uncleaved in the bacteria, and may require further processing to form pre-miRNA stem loops. In addition, no interferon response was detected in vitro and not reported in vivo. This is perhaps surprising because T7 transcribed sequences include 5′ terminal triphosphate groups that are a potent trigger for retinoic acid inducible gene (RIG)-1, a key protein involved in the initiation of innate antiviral responses.64,65 Further experiments will thus be needed in order to establish the precise interaction of the shRNA with the host cell.
Regardless of processing steps prior to the formation of the shRNA loop, tkRNAi has been shown to result in the cleavage of target mRNA via 5′ RACE and is likely to enter the RNAi pathway at the level of Dicer processing. Oral administration of the bacteria resulted in knockdown of the CTTNB1 gene in intestinal epithelium cells by over 60%, which demonstrated impressively that tkRNAi can achieve therapeutic levels of knockdown in vivo in spite of its distinct biogenesis pathway. As intracellular bacteria can be mercifully sacrificed with antibiotics, tkRNAi confers the ability to turn off the production of shRNA when required, especially in cases when the prolonged expression of shRNA can result in toxicity. However, the limited number of cell types permissible to intracellular bacteria may limit potential of this technology. In this example, nonpathogenic Escherichia coli was orally or intravenously delivered, and knockdown reported in the intestines and colon cancer xenografts. This suggests that such an approach could be used as an alternative delivery method to these regions to viral delivery of DNA-encoded shRNAs.
tRNA-shRNAs. A different solution for avoiding cellular toxicity with shRNAs is to use alternative pol-III promoters. One such family under investigation is the well-characterized tRNA promoters that drive expression of tRNAs with characteristic cloverleaf secondary structures and cargoes covalently linked at the 3′ acceptor stem. Following previous reports of extension of tRNAs into active ribozyme transcripts,66 tRNA promoters have been successfully modified to drive shRNA expression.67,68,69 Importantly, extension and development of the acceptor stem into shRNA-like hairpins have been demonstrated to allow for highly efficient processing into RNAi effectors with comparable or even improved efficacy to matched siRNAs and U6/H1-driven shRNAs.67,68,69 It remains to be determined whether Dicer is responsible for this separation of tRNA and shRNA in a similar manner to the separation of analogous aptamer-linked siRNAs formulated to enhance delivery specificity of siRNAs.70 An alternative is that the endoribonuclease responsible for removing endogenous 3′ trailers from pre-tRNAs, tRNase ZL, is required.69,71
It appears that potent tRNA-shRNAs may still be in association with the tRNAVal promoter in the cytosol. In this example, northern blotting has revealed a cytoplasmic-specific transcript that is considerably larger than the identical hairpin sequence transcribed off a U6 promoter, suggestive of a tRNA–shRNA chimera being exported to the cytosol.67 Furthermore, unlike U6-driven shRNAs, tRNA-shRNAs are not retained in the nucleus which could explain their improved potency over U6 promoters in multiple cases.67,68 When considered with the fact that products of tRNA promoters are recognized by the nuclear karyopherin, exportin-t, for nuclear export,67,72,73,74,75 it suggests an efficient alternate nuclear export route may be in use that could potentially circumvent toxicities associated with exportin-5. However, the authors did not investigate either issue, and it will be important to see in future whether the toxic competition for exportin-5 could be bypassed by an exportin-t-dependent tRNA diversion.
Evidence from Xenopus laevis oocytes implies that nuclear export of tRNAs may not be completely exportin-5 free.76 In this model, the introduction of high concentrations of unlabeled tRNAs is sufficient to compete and reduce the nuclear export of radiolabeled tRNAs, a block that can be relieved by co-injection of either exportin-5 or exportin-t. Yet redundancy of the two exportins would still be expected to be beneficial in alleviating exportin-5-mediated toxicities due to splitting of tRNA-shRNA export between the two nuclear export pathways. If proven, as expected, then the use of tRNA-shRNAs would be a considerable improvement over existing shRNA strategies. Although absence of cell type/tissue specificity of these promoters would remain limiting, potent silencing together with reduced toxicity of tRNA-shRNAs could be beneficial in therapies where complete gene knockdown is desired.
Pri-miRNA mimics. As mentioned previously, high shRNA expression can lead to toxic effects both in vitro and in vivo. Although possible, it is unlikely that the pre-miRNA structure of shRNAs is the inherent problem. Supporting this are side-by-side northern blot comparisons of shRNAs and pri-miRNA mimics carrying the same antisense strands both transcribed from U6 promoters that demonstrate reduced pre-miRNA precursor and mature sequences with pri-miRNA mimics despite comparable levels of silencing.77 The results support a protective role for Drosha processing by restricting the progression of pri-miRNA transcripts into the RNAi pathway. Encouragingly, this shRNA to miRNA-mimic switch was additionally associated with less toxicity including in vivo.46,77,78 However, in several cases, it has been reported that the levels of silencing with pri-miRNA mimics are reduced relative to matched shRNAs and this will influence the choice between them.77,79,80
Importantly, when designing pri-miRNA mimics directed against genes of interest, a consensus has emerged that maintenance of the secondary structure of the parental miRNA is highly desirable where possible for correct processing. This includes the flanking sequences preceding the Drosha processing site, of which maintaining one helical turn appears beneficial,81,82,83 and the incorporation of both stem mismatches and a terminal loop sequence that closely, if not fully, resemble the parental miRNA.81,83,84 In addition, maintenance of the full intron sequence and flanking exonic sequences when mimicking intron-derived miRNAs has been shown to be beneficial, suggesting that the intricate relationship between splicing and miRNA processing can influence the success of synthetic approaches and should be considered.84,85,86
Finally, unlike other DNA-based approaches, pri-miRNAs have the considerable advantage that they can utilize pol-II promoters that can drive tissue-specific expression. The substrate recognition for Drosha is an extended stem loop importantly flanked by single-stranded RNA,82 and it is this flanking region that allows much greater flexibility in substrate length and termination signals permitting pol-II promoters to be used. Examples have already been published demonstrating expression of miR-30-based constructs targeting green fluorescent protein in neurons of the striatum using a neuron-specific enolase promoter,44 and with a miR-30a-based construct targeting the voltage-gated L-type calcium channel specifically in vascular smooth muscle cells in culture with a truncated SM22α promoter.87 Together, the data lend support to selection of pri-miRNA mimics over shRNA variants to deliver therapeutic RNAi where possible.
Transcriptional gene silencing (TGS). Although the aforementioned approaches all depend on PTGS, a recent report suggests that endogenous miRNAs are also capable of TGS. Following demonstration of unexplained nuclear localization of several mature miRNAs,88,89 Kim et al. demonstrated both transcriptional inhibition and epigenetic changes at the promoter region of the cell cycle gene POLR3D following ectopic expression of miR-320, a miRNA encoded within the promoter region of this gene in the antisense orientation.90 In the absence of seed-matched target sites in the 3′UTR of POLR3D for this miRNA, the observed changes, together with the inverse relationship between POLR3D and miR-320 in several cell lines, appear dependent on the perfectly matched miR-320 target site in the promoter region of this gene. This work highlights a previously unknown function of this miRNA, and potentially several others, in which gene silencing can occur at the level of transcription. In addition, an alternative biogenesis pathway for endogenous TGS involves processing of complementary non-coding RNA transcripts by Dicer into short RNAs that associate with DNA methylation of suspected targets to help direct X-chromosome inactivation, again implicating aspects of PTGS machinery within a TGS pathway.91
Interestingly, TGS is a case where reports of biology have come after the reports of synthetic usage. The earliest examples demonstrated induction of TGS via dsRNA-dependent formation of heterochromatin and/or DNA methylation around promoter sequences of targeted genes in several organisms such as plants,92,93 fission yeast,94 and Drosophila.95 It was subsequently found that siRNAs and shRNAs targeting promoter regions of human genes could also direct epigenetic changes required for heterochromatin formation and/or DNA methylation in order to initiate TGS.96,97 As well as involving DNA methyltransferases the mechanism has been demonstrated to be both AGO-1 and TRBP2-dependent,90,98,99 and it may be in association with one or both of these latter proteins that siRNAs are localized to the nucleus to direct TGS. In addition, protein-protein interactions with AGO-1 implicate pol-II in the induction of TGS.97,98 Several reports have now been made of transcription across many promoter regions,3,100,101,102,103 and it is possible that pol-II's involvement in TGS is linked to generation of this promoter-associated RNA. Indeed the integrity of low-level promoter RNA has been found to be essential in some reports of TGS.100,104 Yet how these transcripts are involved, and whether antisense strands interact directly with DNA or RNA remains to be definitively determined.
Despite a less complete understanding than for PTGS approaches, TGS is interesting as a potential therapeutic strategy because both endogonous91 and synthetically-induced104,105 examples of TGS indicate that epigenetic changes responsible for silencing are long lasting. A seven day tet-induced expression of a shRNA targeting the ubiquitin C promoter was sufficient to induce TGS for over a month.104 As such TGS could be ideally suited where long-term gene silencing is needed but where re-adminstration of effectors is problematic. In addition, TGS could be highly promising for genome-integrating retroviruses like HIV and SIV where high replication rates allow escape mutants to evolve against targeted therapies, and for DNA-encoded viruses. Long-term suppression of viral transcription by TGS could mimic viral latency, which is also associated with epigenetic changes, and reduce generation and selection of escape mutants to make infections more manageable. Indeed this principle has been demonstrated for both HIV-1 (ref. 105) and the simian immunodeficiency virus.106 However, selective TGS of genes of interest has yet to be reported in vivo, and it will be exciting to see if this can also be demonstrated to encourage use of TGS as a novel RNA-induced silencing mechanism.
Tandem siRNAs. In several therapeutic scenarios delivery of multiple RNAi effectors may be desired in so-called multiplexing approaches. These include when it may be beneficial to target biological pathways rather than single targets in cancers107,108 or complex genetic diseases, or where high rates of viral mutation dictate that silencing multiple targets is required to avoid viral tolerance of silencing.109,110 Recently, incorporation of gene-specific siRNA sequences in between initiation and termination signals for opposing U6 and H1 promoters has been demonstrated as a DNA-encoded method of generating siRNAs.111 The resulting duplexes resemble Dicer digestion products and can be efficiently incorporated into RISC to silence both synthetic and endogenous targets.111 Encouragingly, this bi-directional siRNA strategy also allows delivery of multiple siRNAs within single constructs.112 Extension of siRNAs from 19-21-nt sequences to 40-42-nt sequences, complete with 2-nt overhangs separating the siRNA sequences, results in two functional siRNAs being produced. Importantly, despite a long dsRNA duplex being formed when using these tandem siRNAs (tsiRNAs), the size appears to avoid interferon response activation in Huh7 cells. However, induction of the interferon response shows cell-type variability, with some lines exhibiting weaker activation.113 It remains to be seen whether avoidance is replicated in cell-lines with more aggressive IFN responses, although a single bi-directional siRNA of 19-21nt would be expected to be tolerated.114
Tandem hairpin RNAs. An alternative method to delivery of multiple effectors is to utilize a hairpin-based system analogous to tsiRNAs. Instead of generating two separate hairpins, tandem hairpin RNAs (thRNAs), which encompass both extended-shRNAs (e-shRNAs)115,116 and specific long-hairpin RNAs (lhRNAs)117 from previous literature, produce single hairpin sequences with multiple siRNAs in tandem. These thRNAs are distinct from the originally described lhRNAs that were designed to incorporate a long contiguous stretch of target mRNA and its complementary sequence in order to achieve single target knockdown, a method adapted from plants to mammalian systems.118,119,120,121,122,123 Instead, within a thRNA up to 4 individual potent siRNA sequences targeting different target sites from the same or multiple genes can be constructed within a single hairpin and retain silencing activity.116
Unlike tandem siRNAs, however, there is considerable difference in efficacy of gene knock down depending on the position relative to the stem loop. A repeated observation is that the most proximal siRNA to the loop exhibits the greatest reduction in both mature species production and knock down when compared to single shRNA controls, particularly when 3 or more siRNA units are included.116,117 This can be both an advantage in terms of regulating the levels of knock down by different siRNAs but a disadvantage if potent silencing by all siRNAs is a necessity. Further careful considerations must also be made when 4 siRNA sequences are included. At these lengths the silencing ability of all siRNA sequences across the thRNA can also be drastically reduced for unknown reasons.116 This is despite the fact that these sequences will otherwise be highly potent in identical positions of double or triple-stacked thRNAs. For maximal levels of silencing, it therefore appears that no more than 2–3 siRNA units should be used.
Importantly, thRNAs produce multiple siRNAs from a single promoter in a single hairpin suggesting that exportin-5-mediated export is only utilized once per set of siRNAs. This contrasts with similar multi-effector technologies such as miRNA clusters and expression of multiple shRNAs. This property may result in reduced competition with the endogenous miRNA pathway and ultimately, reduce the potential toxicity for multiple siRNA delivery. Encouragingly thRNAs also fail to trigger an interferon response which tsiRNAs may be susceptible to and which delivered dsRNA duplexes are known to activate.115,116. Taken together, these points encourage further development of this approach which has primarily been utilized in the development of gene therapies with impressive results against viruses in preventing the generation of escape mutants.
Pri-miRNA clusters. Like natural miRNA clusters, multiple pri-miRNA mimics can also be generated within a single transcript. The approach can be used to create multiple copies of the same pri-miRNA mimic or pri-miRNA mimics targeting different genes.81,84,124 For instance, Chung et al. have demonstrated that two or more identical miR-155 mimic sequences targeting luciferase, processed from the same intron, show a positive correlation between the number of mimics in a transcript and the level of knockdown.124 Encouragingly, it was also shown that expressing up to two miR-155 mimics targeting different synthetic or endogenous genes in series can effectively reduce both targets to levels comparable to single pri-miRNA mimics. This work suggests that multiplexing of miRNAs is possible and is an attractive new approach to target multiple genes from a single transcript.
As discussed previously, adherence to natural context for pri-miRNA mimics is beneficial for activity,81,82,83 and it is predicted that this will additionally be applicable to miRNA clusters. Support for this comes from Aagaard et al. who explored a tri-cistronic miRNA cluster that expresses three miRNAs (miR-106b, miR-93, and miR-25) from an intron of <800 nt.84 Thus, the intron, being relatively short, can easily be incorporated into viral vectors. Expression of the natural miRNA sequences present in this cluster showed a clear dependence on the flanking structures as expected. Deletion of the region from the base of the miRNA stem up to the final nucleotide adjacent to the mature antisense sequence abolishes all activity of the modified miRNA, whereas removal or shortening of the sequences flanking each miRNA stem was also sufficient to abrogate activity. In addition, the most significant knockdown achieved with miRNA mimics was seen when the full natural miRNA secondary structure of each hairpin was conserved. This agrees with previous data for pri-miRNA mimics and stresses the importance of maintaining recognition signals for efficient Drosha and Dicer processing. When considered together with the advantages of using pri-miRNA mimics in general, the use of pri-miRNA cluster mimics is a highly promising method for future studies or therapeutics in which multiplexing is desirable.
Future Perspectives
The discussed approaches exemplify significant improvements on the original synthetic RNAi technologies to circumvent associated problems. Recent advances in the understanding of RNAi biology mean that there are yet further strategies to be explored. The recent characterization of mirtrons and endo-siRNAs paves the way for exploitation of the inherent advantages of these endogenous systems. For example, the dependence on splicing rather than Drosha processing for pre-miRNA hairpin formation suggests that mirtrons would be less dependent on the canonical miRNA biogenesis pathway. Artificial mirtrons could thus represent a new class of RNAi effectors that would be ideally suited for the co-delivery of protein-coding genes and multiple RNAi molecules in a multiplexing approach.
Exploitation of the endo-siRNA pathway in mammalian cells with the expression of complementary long antisense mRNA transcripts may be a highly potent alternative to shRNA and miRNA pathways. Interestingly, the pathway utilized for generation of endo-siRNAs, although overlapping on some level with those of the miRNA and siRNA pathways, uses a set of homologous, yet distinct regulatory genes to those used in the miRNA pathway in Drosophila.125 If replicated in mammalian cells, then several problems associated with saturation of the miRNA pathway may be avoidable through use of this unique pathway. Furthermore, as long antisense transcripts utilized in this biogenesis pathway are likely to be produced and exported as an mRNA and could therefore be used therapeutically with pol-II promoters, it is unlikely to result in saturation of the endogenous miRNA pathways while still retaining the capability for tissue-specific expression.
Conclusion
At present, it is apparent that none of the aforementioned technologies resolves all issues raised for the therapeutic application of RNAi. However, careful consideration of the problem that is to be addressed with a gene silencing approach alongside an understanding of the advantages and disadvantages associated with each technology will improve decision making on which optimized effector should be used and make therapeutic success more likely. For example, several skin conditions remain candidates for siRNA-mediated approaches due to the ease of access and suitability for repeat delivery, and clinical trials are due for the rare autosomal-dominant skin disease Pachyonychia congenital.126,127 Adoption of one of aiRNA, sisiRNA, or chimeric RNA–DNA duplex methods as opposed to traditional siRNAs may considerably reduce the expected associated off-target effects and make such therapies safer in the long term.
Although the effectors discussed are significant improvements on the first-generation effectors and we encourage widespread adoption of these approaches, progress in this field is set to continue, and several unique novel approaches will certainly be reported in future. However, considerable gains have been made with the effectors discussed, and combining two or more of the above approaches could have benefits that may be additive in “third-generation” gene silencing effectors. For example, thRNAs have the advantage of delivering multiple siRNAs from one construct, yet the disadvantage of requiring pol-III-dependent expression therefore making them susceptible to potential toxicities associated with saturating the endogenous pathway as seen with shRNAs. Two viable solutions to limiting this disadvantage would be to pair thRNAs with a tRNA promoter or deliver as a trans-kingdom thRNA in order to limit the exportin-5 saturation. Similarly, TGS has the advantage of long-term silencing at the transcriptional level, yet current induction commonly uses very high doses of siRNAs or shRNAs that can lead to considerable toxicities and off-target effects both through TGS and PTGS mechanisms. High doses of an aiRNA, sisiRNA, or chimeric RNA–DNA duplexes may still be a necessity, but the reduced off-target effects that are associated with these approaches may improve this technology with regard to therapeutic safety.
In summary, RNA-induced gene silencing has yet to be fully explored, and there are still many areas to this field in which understanding is lacking. Several novel RNAi effectors discussed here demonstrate that our understanding of the substrate recognition of RNAi is incomplete. By developing new understanding to circumvent current limitations, several highly promising RNAi effectors have been designed and tested that offer enhanced therapeutic potential. Although further improvements are expected, the stage is now set for some of these to become accepted by the mainstream in order to push RNA-induced silencing methods closer to therapeutic usefulness.
Acknowledgments
We thank Janine Scholefield, Thomas Roberts, and Marc Weinberg for comments and critical reading of the manuscript. C.R.S. acknowledges funding support from the UK MRC and the UK Parkinson's Disease Society, Y.S. from the Agency for Science, Technology and Research (Singapore), and M.J.A.W. from the UK MRC, and Biotechnology and Biological Sciences Research Council (BBSRC), The Wellcome Trust, the UK Parkinson's Disease Society, the Muscular Dystrophy Campaign, and Action Duchenne.
REFERENCES
- Mercer TR, Dinger ME., and , Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, ENCODE Project Consortium; NISC Comparative Sequencing Program; Baylor College of Medicine Human Genome Sequencing Center; Washington University Genome Sequencing Center; Broad Institute; Children's Hospital Oakland Research Institute et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fejes-Toth K, Sotirova V, Sachidanandam R, Assaf G, Hannon GJ, Kapranov P, et al. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature. 2009;457:1028–1032. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen BR. Viral and cellular messenger RNA targets of viral microRNAs. Nature. 2009;457:421–425. doi: 10.1038/nature07757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg I, Eran A, Nishino I, Moggio M, Lamperti C, Amato AA, et al. Distinctive patterns of microRNA expression in primary muscular disorders. Proc Natl Acad Sci USA. 2007;104:17016–17021. doi: 10.1073/pnas.0708115104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, et al. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WX, Rajeev BW, Stromberg AJ, Ren N, Tang G, Huang Q, et al. The expression of microRNA miR-107 decreases early in Alzheimer's disease and may accelerate disease progression through regulation of beta-site amyloid precursor protein-cleaving enzyme 1. J Neurosci. 2008;28:1213–1223. doi: 10.1523/JNEUROSCI.5065-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garofalo M, Condorelli G., and , Croce CM. MicroRNAs in diseases and drug response. Curr Opin Pharmacol. 2008;8:661–667. doi: 10.1016/j.coph.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Martínez MA. Progress in the therapeutic applications of siRNAs against HIV-1. Methods Mol Biol. 2009;487:343–368. doi: 10.1007/978-1-60327-547-7_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barik S. Treating respiratory viral diseases with chemically modified, second generation intranasal siRNAs. Methods Mol Biol. 2009;487:331–341. doi: 10.1007/978-1-60327-547-7_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K., and , Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA., and , Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Lendeckel W., and , Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A., and , Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Yi R., and , Cullen BR. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc Natl Acad Sci USA. 2003;100:9779–9784. doi: 10.1073/pnas.1630797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg MS., and , Wood MJ. Short non-coding RNA biology and neurodegenerative disorders: novel disease targets and therapeutics. Hum Mol Genet. 2009;18 R1:R27–R39. doi: 10.1093/hmg/ddp070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan CH., and , Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Hagen JW, Duan H, Tyler DM., and , Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E, Chung WJ, Willis J, Cuppen E., and , Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiarz JE, Ruby JG, Wang Y, Bartel DP., and , Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, et al. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008;320:1077–1081. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus MT, Petersen CP, Haines BB, Chen J., and , Sharp PA. Gene silencing using micro-RNA designed hairpins. RNA. 2002;8:842–850. doi: 10.1017/s1355838202024032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin KJ, Wall EA, Zavzavadjian JR, Santat LA, Liu J, Hwang JI, et al. A single lentiviral vector platform for microRNA-based conditional RNA interference and coordinated transgene expression. Proc Natl Acad Sci USA. 2006;103:13759–13764. doi: 10.1073/pnas.0606179103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Fougerolles A, Vornlocher HP, Maraganore J., and , Lieberman J. Interfering with disease: a progress report on siRNA-based therapeutics. Nat Rev Drug Discov. 2007;6:443–453. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanotto D., and , Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457:426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behlke MA. Chemical modification of siRNAs for in vivo use. Oligonucleotides. 2008;18:305–319. doi: 10.1089/oli.2008.0164. [DOI] [PubMed] [Google Scholar]
- Robbins M, Judge A, Liang L, McClintock K, Yaworski E., and , MacLachlan I. 2'-O-methyl-modified RNAs act as TLR7 antagonists. Mol Ther. 2007;15:1663–1669. doi: 10.1038/sj.mt.6300240. [DOI] [PubMed] [Google Scholar]
- Zou Y, Tiller P, Chen IW, Beverly M., and , Hochman J. Metabolite identification of small interfering RNA duplex by high-resolution accurate mass spectrometry. Rapid Commun Mass Spectrom. 2008;22:1871–1881. doi: 10.1002/rcm.3561. [DOI] [PubMed] [Google Scholar]
- Czauderna F, Fechtner M, Dames S, Aygün H, Klippel A, Pronk GJ, et al. Structural variations and stabilising modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res. 2003;31:2705–2716. doi: 10.1093/nar/gkg393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead KA, Langer R., and , Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey DV, Lockridge JA, Shaw L, Blanchard K, Jensen K, Breen W, et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat Biotechnol. 2005;23:1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- McCaffrey AP, Meuse L, Pham TT, Conklin DS, Hannon GJ., and , Kay MA. RNA interference in adult mice. Nature. 2002;418:38–39. doi: 10.1038/418038a. [DOI] [PubMed] [Google Scholar]
- Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- Aouadi M, Tesz GJ, Nicoloro SM, Wang M, Chouinard M, Soto E, et al. Orally delivered siRNA targeting macrophage Map4k4 suppresses systemic inflammation. Nature. 2009;458:1180–1184. doi: 10.1038/nature07774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crombez L, Morris MC, Dufort S, Aldrian-Herrada G, Nguyen Q, Mc Master G, et al. Targeting cyclin B1 through peptide-based delivery of siRNA prevents tumour growth. Nucleic Acids Res. 2009;37:4559–4569. doi: 10.1093/nar/gkp451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrams MT, Koser ML, Seitzer J, Williams SC, Dipietro MA, Wang W, et al. 2009Evaluation of efficacy, biodistribution, and inflammation for a potent siRNA nanoparticle: effect of dexamethasone co-treatment Mol Therepub ahead of print). [DOI] [PMC free article] [PubMed]
- Bartlett DW, Su H, Hildebrandt IJ, Weber WA., and , Davis ME. Impact of tumor-specific targeting on the biodistribution and efficacy of siRNA nanoparticles measured by multimodality in vivo imaging. Proc Natl Acad Sci USA. 2007;104:15549–15554. doi: 10.1073/pnas.0707461104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hommel JD, Sears RM, Georgescu D, Simmons DL., and , DiLeone RJ. Local gene knockdown in the brain using viral-mediated RNA interference. Nat Med. 2003;9:1539–1544. doi: 10.1038/nm964. [DOI] [PubMed] [Google Scholar]
- Nielsen TT, Marion I, Hasholt L., and , Lundberg C. Neuron-specific RNA interference using lentiviral vectors. J Gene Med. 2009;11:559–569. doi: 10.1002/jgm.1333. [DOI] [PubMed] [Google Scholar]
- Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- Boudreau RL, Martins I., and , Davidson BL. Artificial microRNAs as siRNA shuttles: improved safety as compared to shRNAs in vitro and in vivo. Mol Ther. 2009;17:169–175. doi: 10.1038/mt.2008.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JY, DeRuiter SL., and , Turner DL. RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc Natl Acad Sci USA. 2002;99:6047–6052. doi: 10.1073/pnas.092143499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, et al. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L, et al. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA. 2006;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Rogoff HA., and , Li CJ. Asymmetric RNA duplexes mediate RNA interference in mammalian cells. Nat Biotechnol. 2008;26:1379–1382. doi: 10.1038/nbt.1512. [DOI] [PubMed] [Google Scholar]
- Pei Y., and , Tuschl T. On the art of identifying effective and specific siRNAs. Nat Methods. 2006;3:670–676. doi: 10.1038/nmeth911. [DOI] [PubMed] [Google Scholar]
- Decherf S, Hassani Z., and , Demeneix BA. In vivo siRNA delivery to the mouse hypothalamus shows a role of the co-chaperone XAP2 in regulating TRH transcription. Methods Mol Biol. 2008;433:355–366. doi: 10.1007/978-1-59745-237-3_21. [DOI] [PubMed] [Google Scholar]
- Bramsen JB, Laursen MB, Damgaard CK, Lena SW, Babu BR, Wengel J, et al. Improved silencing properties using small internally segmented interfering RNAs. Nucleic Acids Res. 2007;35:5886–5897. doi: 10.1093/nar/gkm548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei JX, Yang J, Sun JF, Jia LT, Zhang Y, Zhang HZ, et al. Both strands of siRNA have potential to guide posttranscriptional gene silencing in mammalian cells. PLoS ONE. 2009;4:e5382. doi: 10.1371/journal.pone.0005382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano M, Sierant M, Miyagishi M, Nakanishi M, Takagi Y., and , Sutou S. Effect of asymmetric terminal structures of short RNA duplexes on the RNA interference activity and strand selection. Nucleic Acids Res. 2008;36:5812–5821. doi: 10.1093/nar/gkn584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen A, Behlen L, Reynolds A, Wolfson A, Marshall WS, Karpilow J, et al. The contributions of dsRNA structure to Dicer specificity and efficiency. RNA. 2005;11:674–682. doi: 10.1261/rna.7272305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CY., and , Rana TM. Potent RNAi by short RNA triggers. RNA. 2008;14:1714–1719. doi: 10.1261/rna.1161908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishi M., and , Taira K. U6 promoter-driven siRNAs with four uridine 3′ overhangs efficiently suppress targeted gene expression in mammalian cells. Nat Biotechnol. 2002;20:497–500. doi: 10.1038/nbt0502-497. [DOI] [PubMed] [Google Scholar]
- Lee NS, Dohjima T, Bauer G, Li H, Li MJ, Ehsani A, et al. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat Biotechnol. 2002;20:500–505. doi: 10.1038/nbt0502-500. [DOI] [PubMed] [Google Scholar]
- Dahlgren C, Wahlestedt C., and , Thonberg H. No induction of anti-viral responses in human cell lines HeLa and MCF-7 when transfecting with siRNA or siLNA. Biochem Biophys Res Commun. 2006;341:1211–1217. doi: 10.1016/j.bbrc.2006.01.085. [DOI] [PubMed] [Google Scholar]
- Ui-Tei K, Naito Y, Zenno S, Nishi K, Yamato K, Takahashi F, et al. Functional dissection of siRNA sequence by systematic DNA substitution: modified siRNA with a DNA seed arm is a powerful tool for mammalian gene silencing with significantly reduced off-target effect. Nucleic Acids Res. 2008;36:2136–2151. doi: 10.1093/nar/gkn042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang S, Keates AC, Fruehauf J, Yang Y, Guo H, Nguyen T, et al. In vitro and in vivo gene silencing by TransKingdom RNAi (tkRNAi) Methods Mol Biol. 2009;487:147–160. doi: 10.1007/978-1-60327-547-7_7. [DOI] [PubMed] [Google Scholar]
- Xiang S, Fruehauf J., and , Li CJ. Short hairpin RNA-expressing bacteria elicit RNA interference in mammals. Nat Biotechnol. 2006;24:697–702. doi: 10.1038/nbt1211. [DOI] [PubMed] [Google Scholar]
- Kim DH, Longo M, Han Y, Lundberg P, Cantin E., and , Rossi JJ. Interferon induction by siRNAs and ssRNAs synthesized by phage polymerase. Nat Biotechnol. 2004;22:321–325. doi: 10.1038/nbt940. [DOI] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzózka K, Jung A, Kato H, et al. 5'-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- Kuwabara T, Warashina M, Orita M, Koseki S, Ohkawa J., and , Taira K. Formation of a catalytically active dimer by tRNA(Val)-driven short ribozymes. Nat Biotechnol. 1998;16:961–965. doi: 10.1038/nbt1098-961. [DOI] [PubMed] [Google Scholar]
- Kawasaki H., and , Taira K. Short hairpin type of dsRNAs that are controlled by tRNA(Val) promoter significantly induce RNAi-mediated gene silencing in the cytoplasm of human cells. Nucleic Acids Res. 2003;31:700–707. doi: 10.1093/nar/gkg158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boden D, Pusch O, Lee F, Tucker L, Shank PR., and , Ramratnam B. Promoter choice affects the potency of HIV-1 specific RNA interference. Nucleic Acids Res. 2003;31:5033–5038. doi: 10.1093/nar/gkg704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer LJ, Frank R., and , Rossi JJ. Optimization and characterization of tRNA-shRNA expression constructs. Nucleic Acids Res. 2007;35:2620–2628. doi: 10.1093/nar/gkm103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara JO, 2nd, Andrechek ER, Wang Y, Viles KD, Rempel RE, Gilboa E, et al. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat Biotechnol. 2006;24:1005–1015. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- Shibata HS, Takaku H, Takagi M., and , Nashimoto M. The T loop structure is dispensable for substrate recognition by tRNase ZL. J Biol Chem. 2005;280:22326–22334. doi: 10.1074/jbc.M502048200. [DOI] [PubMed] [Google Scholar]
- Kutay U, Lipowsky G, Izaurralde E, Bischoff FR, Schwarzmaier P, Hartmann E, et al. Identification of a tRNA-specific nuclear export receptor. Mol Cell. 1998;1:359–369. doi: 10.1016/s1097-2765(00)80036-2. [DOI] [PubMed] [Google Scholar]
- Arts GJ, Kuersten S, Romby P, Ehresmann B., and , Mattaj IW. The role of exportin-t in selective nuclear export of mature tRNAs. EMBO J. 1998;17:7430–7441. doi: 10.1093/emboj/17.24.7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara T, Warashina M, Sano M, Tang H, Wong-Staal F, Munekata E, et al. Recognition of engineered tRNAs with an extended 3′ end by Exportin-t (Xpo-t) and transport of tRNA-attached ribozymes to the cytoplasm in somatic cells. Biomacromolecules. 2001;2:1229–1242. doi: 10.1021/bm0101062. [DOI] [PubMed] [Google Scholar]
- Koseki S, Tanabe T, Tani K, Asano S, Shioda T, Nagai Y, et al. Factors governing the activity in vivo of ribozymes transcribed by RNA polymerase III. J Virol. 1999;73:1868–1877. doi: 10.1128/jvi.73.3.1868-1877.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calado A, Treichel N, Müller EC, Otto A., and , Kutay U. Exportin-5-mediated nuclear export of eukaryotic elongation factor 1A and tRNA. EMBO J. 2002;21:6216–6224. doi: 10.1093/emboj/cdf620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride JL, Boudreau RL, Harper SQ, Staber PD, Monteys AM, Martins I, et al. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc Natl Acad Sci USA. 2008;105:5868–5873. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholefield J, Greenberg LJ, Weinberg MS, Arbuthnot PB, Abdelgany A., and , Wood MJ. Design of RNAi hairpins for mutation-specific silencing of ataxin-7 and correction of a SCA7 phenotype. PLoS ONE. 2009;4:e7232. doi: 10.1371/journal.pone.0007232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreau RL, Monteys AM., and , Davidson BL. Minimizing variables among hairpin-based RNAi vectors reveals the potency of shRNAs. RNA. 2008;14:1834–1844. doi: 10.1261/rna.1062908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Lin X, Khvorova A, Fesik SW., and , Shen Y. Defining the optimal parameters for hairpin-based knockdown constructs. RNA. 2007;13:1765–1774. doi: 10.1261/rna.599107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Xia XG., and , Xu Z. An RNA polymerase II construct synthesizes short-hairpin RNA with a quantitative indicator and mediates highly efficient RNAi. Nucleic Acids Res. 2005;33:e62. doi: 10.1093/nar/gni061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, et al. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Yi R., and , Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 2005;24:138–148. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aagaard LA, Zhang J, von Eije KJ, Li H, Saetrom P, Amarzguioui M, et al. Engineering and optimization of the miR-106b cluster for ectopic expression of multiplexed anti-HIV RNAs. Gene Ther. 2008;15:1536–1549. doi: 10.1038/gt.2008.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK., and , Kim VN. Processing of intronic microRNAs. EMBO J. 2007;26:775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlando M, Ballarino M, Gromak N, Pagano F, Bozzoni I., and , Proudfoot NJ. Primary microRNA transcripts are processed co-transcriptionally. Nat Struct Mol Biol. 2008;15:902–909. doi: 10.1038/nsmb.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee SW, Stimers JR, Wang W., and , Pang L. Vascular smooth muscle-specific knockdown of the noncardiac form of the L-type calcium channel by microRNA-based short hairpin RNA as a potential antihypertensive therapy. J Pharmacol Exp Ther. 2009;329:775–782. doi: 10.1124/jpet.108.148866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang HW, Wentzel EA., and , Mendell JT. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315:97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- Politz JC, Zhang F., and , Pederson T. MicroRNA-206 colocalizes with ribosome-rich regions in both the nucleolus and cytoplasm of rat myogenic cells. Proc Natl Acad Sci USA. 2006;103:18957–18962. doi: 10.1073/pnas.0609466103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Saetrom P, Snøve O., Jr, and , Rossi JJ. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci USA. 2008;105:16230–16235. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa Y, Sun BK., and , Lee JT. Intersection of the RNA interference and X-inactivation pathways. Science. 2008;320:1336–1341. doi: 10.1126/science.1157676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mette MF, Aufsatz W, van der Winden J, Matzke MA., and , Matzke AJ. Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 2000;19:5194–5201. doi: 10.1093/emboj/19.19.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T, Vijn I, Rebocho A, van Blokland R, Roelofs D, Mol JN, et al. Transcriptional and posttranscriptional gene silencing are mechanistically related. Curr Biol. 2001;11:436–440. doi: 10.1016/s0960-9822(01)00116-6. [DOI] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI., and , Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra M, Bhadra U., and , Birchler JA. RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Mol Cell. 2002;9:315–327. doi: 10.1016/s1097-2765(02)00440-9. [DOI] [PubMed] [Google Scholar]
- Morris KV, Chan SW, Jacobsen SE., and , Looney DJ. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–1292. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- Weinberg MS, Villeneuve LM, Ehsani A, Amarzguioui M, Aagaard L, Chen ZX, et al. The antisense strand of small interfering RNAs directs histone methylation and transcriptional gene silencing in human cells. RNA. 2006;12:256–262. doi: 10.1261/rna.2235106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Villeneuve LM, Morris KV., and , Rossi JJ. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat Struct Mol Biol. 2006;13:793–797. doi: 10.1038/nsmb1142. [DOI] [PubMed] [Google Scholar]
- Janowski BA, Huffman KE, Schwartz JC, Ram R, Nordsell R, Shames DS, et al. Involvement of AGO1 and AGO2 in mammalian transcriptional silencing. Nat Struct Mol Biol. 2006;13:787–792. doi: 10.1038/nsmb1140. [DOI] [PubMed] [Google Scholar]
- Han J, Kim D., and , Morris KV. Promoter-associated RNA is required for RNA-directed transcriptional gene silencing in human cells. Proc Natl Acad Sci USA. 2007;104:12422–12427. doi: 10.1073/pnas.0701635104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ., and , Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, et al. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, et al. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- Hawkins PG, Santoso S, Adams C, Anest V., and , Morris KV. Promoter targeted small RNAs induce long-term transcriptional gene silencing in human cells. Nucleic Acids Res. 2009;37:2984–2995. doi: 10.1093/nar/gkp127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Shijuuku T, Fukamachi T, Zaunders J, Guillemin G, Cooper D, et al. Prolonged transcriptional silencing and CpG methylation induced by siRNAs targeted to the HIV-1 promoter region. J RNAi Gene Silencing. 2005;1:66–78. [PMC free article] [PubMed] [Google Scholar]
- Lim HG, Suzuki K, Cooper DA., and , Kelleher AD. Promoter-targeted siRNAs induce gene silencing of simian immunodeficiency virus (SIV) infection in vitro. Mol Ther. 2008;16:565–570. doi: 10.1038/sj.mt.6300380. [DOI] [PubMed] [Google Scholar]
- O'Grady M, Raha D, Hanson BJ, Bunting M., and , Hanson GT. Combining RNA interference and kinase inhibitors against cell signalling components involved in cancer. BMC Cancer. 2005;5:125. doi: 10.1186/1471-2407-5-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tummalapalli P, Spomar D, Gondi CS, Olivero WC, Gujrati M, Dinh DH, et al. RNAi-mediated abrogation of cathepsin B and MMP-9 gene expression in a malignant meningioma cell line leads to decreased tumor growth, invasion and angiogenesis. Int J Oncol. 2007;31:1039–1050. [PMC free article] [PubMed] [Google Scholar]
- ter Brake O, Konstantinova P, Ceylan M., and , Berkhout B. Silencing of HIV-1 with RNA interference: a multiple shRNA approach. Mol Ther. 2006;14:883–892. doi: 10.1016/j.ymthe.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Grimm D., and , Kay MA. Combinatorial RNAi: a winning strategy for the race against evolving targets. Mol Ther. 2007;15:878–888. doi: 10.1038/sj.mt.6300116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Liu J, Batalov S, Zhou D, Orth A, Ding S, et al. An approach to genomewide screens of expressed small interfering RNAs in mammalian cells. Proc Natl Acad Sci USA. 2004;101:135–140. doi: 10.1073/pnas.2136685100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D, Lee H, Kim SI, Yoon Y., and , Kim M. Optimization of linear double-stranded RNA for the production of multiple siRNAs targeting hepatitis C virus. RNA. 2009;15:898–910. doi: 10.1261/rna.1268209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A, Anderson EM, Vermeulen A, Fedorov Y, Robinson K, Leake D, et al. Induction of the interferon response by siRNA is cell type- and duplex length-dependent. RNA. 2006;12:988–993. doi: 10.1261/rna.2340906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manche L, Green SR, Schmedt C., and , Mathews MB. Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol Cell Biol. 1992;12:5238–5248. doi: 10.1128/mcb.12.11.5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YP, Haasnoot J., and , Berkhout B. Design of extended short hairpin RNAs for HIV-1 inhibition. Nucleic Acids Res. 2007;35:5683–5693. doi: 10.1093/nar/gkm596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YP, von Eije KJ, Schopman NC, Westerink JT, Brake O, Haasnoot J, et al. Combinatorial RNAi against HIV-1 using extended short hairpin RNAs. Mol Ther. 2009;17:1712–1723. doi: 10.1038/mt.2009.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saayman S, Barichievy S, Capovilla A, Morris KV, Arbuthnot P., and , Weinberg MS. The efficacy of generating three independent anti-HIV-1 siRNAs from a single U6 RNA Pol III-expressed long hairpin RNA. PLoS ONE. 2008;3:e2602. doi: 10.1371/journal.pone.0002602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi H, Miyagishi M, Yokota T, Watanabe T, Hino T, Nishina K, et al. Escape from the interferon response associated with RNA interference using vectors that encode long modified hairpin-RNA. Mol Biosyst. 2005;1:382–390. doi: 10.1039/b510159j. [DOI] [PubMed] [Google Scholar]
- Konstantinova P, de Vries W, Haasnoot J, ter Brake O, de Haan P., and , Berkhout B. Inhibition of human immunodeficiency virus type 1 by RNA interference using long-hairpin RNA. Gene Ther. 2006;13:1403–1413. doi: 10.1038/sj.gt.3302786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitsuji H, Kohara M, Kannagi M., and , Masuda T. Effective suppression of human immunodeficiency virus type 1 through a combination of short- or long-hairpin RNAs targeting essential sequences for retroviral integration. J Virol. 2006;80:7658–7666. doi: 10.1128/JVI.00078-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano M, Li H, Nakanishi M., and , Rossi JJ. Expression of long anti-HIV-1 hairpin RNAs for the generation of multiple siRNAs: advantages and limitations. Mol Ther. 2008;16:170–177. doi: 10.1038/sj.mt.6300298. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Sudoh M, Miyagishi M, Akashi H, Arai M, Inoue K, et al. Intracellular-diced dsRNA has enhanced efficacy for silencing HCV RNA and overcomes variation in the viral genotype. Gene Ther. 2006;13:883–892. doi: 10.1038/sj.gt.3302734. [DOI] [PubMed] [Google Scholar]
- Weinberg MS, Ely A, Barichievy S, Crowther C, Mufamadi S, Carmona S, et al. Specific inhibition of HBV replication in vitro and in vivo with expressed long hairpin RNA. Mol Ther. 2007;15:534–541. doi: 10.1038/sj.mt.6300077. [DOI] [PubMed] [Google Scholar]
- Chung KH, Hart CC, Al-Bassam S, Avery A, Taylor J, Patel PD, et al. Polycistronic RNA polymerase II expression vectors for RNA interference based on BIC/miR-155. Nucleic Acids Res. 2006;34:e53. doi: 10.1093/nar/gkl143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Hotta I, Denli AM, Hong P, Perrimon N., and , Hannon GJ. Comparative analysis of argonaute-dependent small RNA pathways in Drosophila. Mol Cell. 2008;32:592–599. doi: 10.1016/j.molcel.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickerson RP, Smith FJ, Reeves RE, Contag CH, Leake D, Leachman SA, et al. Single-nucleotide-specific siRNA targeting in a dominant-negative skin model. J Invest Dermatol. 2008;128:594–605. doi: 10.1038/sj.jid.5701060. [DOI] [PubMed] [Google Scholar]
- Leachman SA, Hickerson RP, Hull PR, Smith FJ, Milstone LM, Lane EB, et al. Therapeutic siRNAs for dominant genetic skin disorders including pachyonychia congenita. J Dermatol Sci. 2008;51:151–157. doi: 10.1016/j.jdermsci.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]