Abstract
Cardiac myosin binding protein-C (cMyBP-C) is an accessory protein found in the A-bands of vertebrate sarcomeres and mutations in the cMyBP-C gene are a leading cause of familial hypertrophic cardiomyopathy. The regulatory functions of cMyBP-C have been attributed to the N-terminus of the protein, which is composed of tandem immunoglobulin (Ig)-like domains (C0, C1, and C2), a region rich in proline and alanine residues (the Pro-Ala rich region) that links C0 and C1, and a unique sequence referred to as the MyBP-C motif, or M-domain, that links C1 and C2. Recombinant proteins that contain various combinations of the N-terminal domains of cMyBP-C can activate actomyosin interactions in the absence of Ca2+, but the specific sequences required for these effects differ between species; the Pro-Ala region has been implicated in human cMyBP-C whereas the C1 and M-domains appear important in mouse cMyBP-C. To investigate whether species-specific differences in sequence can account for the observed differences in function, we compared sequences of the Pro-Ala rich region in cMyBP-C isoforms from different species. Here we report that the number of proline and alanine residues in the Pro-Ala rich region varies significantly between different species and that the number correlates directly with mammalian body size and inversely with heart rate. Thus, systematic sequence differences in the Pro-Ala rich region of cMyBP-C may contribute to observed functional differences in human versus mouse cMyBP-C isoforms and suggest that the Pro-Ala region may be important in matching contractile speed to cardiac function across species.
Keywords: Myosin binding protein-C, MyBP-C, Sarcomere, Proline-alanine
Introduction
Myosin binding protein-C (MyBP-C) is a regulatory and structural protein located in the A-bands of vertebrate striated muscle sarcomeres. The N-terminus of cardiac (c) MyBP-C is important for mediating the regulatory functions of cMyBP-C and is composed of 3 immunoglobulin (Ig)–like domains (C0, C1, and C2), a region rich in Pro and Ala residues (the Pro-Ala rich region) that links C0 and C1, and the unique MyBP-C motif (M-domain) that links the C1 and C2 domains. The N-terminal domains act to regulate actomyosin interactions by accelerating cross-bridge cycling rates (Stelzer et al. 2007) through phosphorylation-mediated interactions potentially between the M-domain and myosin S2 (Kunst et al. 2000) and/or actin (Shaffer et al. 2009). In addition, recombinant N-terminal domains of cMyBP-C can activate tension generation and increase the rate of force redevelopment (ktr) in permeabilized myocytes in the absence of Ca2+ (Herron et al. 2006; Razumova et al. 2008).
Whereas the ability of the N-terminus of cMyBP-C to influence actomyosin interactions has been reported by several groups (Kulikovskaya et al. 2003; Herron et al. 2006; Razumova et al. 2006; Stelzer et al. 2007; Razumova et al. 2008), the specific domains and mechanisms required for functional effects are more controversial. For example, Herron et al. (2006) showed that the recombinant human cMyBP-C protein C0C1 (which includes C0, the Pro-Ala rich region, and C1 domains) activated tension and increased ktr in skinned human ventricular myocytes in the absence of Ca2+ and concluded that the Pro-Ala rich region was required for the activating effects. By contrast, Razumova et al. (2008) demonstrated that mouse C1C2 (which includes the C1, M, and C2 domains) activated tension and increased ktr in skinned rat trabeculae, whereas C0C1 (including the Pro-Ala rich region) had no effect. These authors concluded that the C1 and M domains were essential for the activating effects.
The different conclusions reached in the two studies could potentially be explained by species-specific differences between the N-terminal domains of mouse and human cMyBP-C. Consistent with this, chimeric proteins containing mouse or human N-terminal domains have differential effects on actomyosin interactions (Shaffer et al. 2010). While the C0, C1, M, and C2 domains all share greater than 80% identity between human and mouse isoforms (with the C1, M, and C2 domains each >90% identical), the Pro-Ala rich regions of human and mouse cMyBP-C are less well conserved—only 46% of the amino acids are identical. The major difference between the Pro-Ala rich regions of human and mouse cMyBP-C is the number of proline and alanine residues—the mouse Pro-Ala rich region is composed of 28.2% proline and alanine residues, while the human isoform contains 51.0% (Fig. 1a; Table 1). In addition, the Pro-Ala rich region of human cMyBP-C has been proposed to bind to actin (Squire et al. 2003), while the mouse Pro-Ala rich region may not bind actin or may only bind weakly (Shaffer et al. 2009). Thus, differences in the number of proline and alanine residues in the human and mouse Pro-Ala rich regions and potential differences in actin binding interactions suggest that sequence differences in the Pro-Ala rich regions of cMyBP-C can account for the observed functional differences.
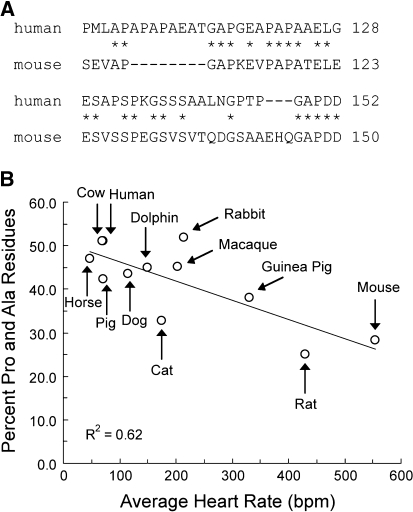
Fig. 1.
a Seqeunce alignment of human and mouse Pro-Ala rich regions. Asterisks denote identical residues. b Relationship between average resting heart rate and the percentage of Pro and Ala residues in the Pro-Ala rich region of mammalian cMyBP-C isoforms. Data are shown for mouse, rat, guinea pig, rabbit, cat, dog, macaque monkey, pig, dolphin, horse, cow, and human species. Heart rate data are from (Spector 1956)
Table 1.
Composition of the cMyBP-C Pro-Ala rich region in different species
| Species | Pro-Ala length | % Pro & Ala | Sequence identifier | Database |
|---|---|---|---|---|
| Human | 51 | 51.0 | NP_000247.2 | Genbank |
| Cow | 49 | 51.0 | NP_001070004.1 | Genbank |
| Horse | 49 | 46.9 | XP_001491151.2 | Genbank* |
| Dolphin | 49 | 44.9 | ENSTTRP00000013421 | Ensembl* |
| Pig | 49 | 42.4 | ENSSSCP00000014064 | Ensembl* |
| Macaque | 51 | 45.1 | ENSMMUP00000020472 | Ensembl* |
| Dog | 55 | 43.6 | NP_001041571.1 | Genbank |
| Cat | 49 | 32.7 | ENSFCAP00000000303 | Ensembl |
| Rabbit | 52 | 51.9 | ENSOCUP00000001825 | Ensembl* |
| Guinea Pig | 48 | 38.0 | ENSCPOP00000012034 | Ensembl* |
| Rat | 52 | 25.0 | NP_001099960.1 | Genbank |
| Mouse | 46 | 28.2 | AF097333 | Genbank |
| Chicken | 41 | 48.8 | NP_990447.1 | Genbank |
| Tropical Frog | 58 | 53.4 | NP_001106379.1 | Genbank |
| Zebrafish | 62 | 34.3 | NP_001037814 | Genbank |
The boundaries for the Pro-Ala rich region were determined by sequence homology to human cMyBP-C using domain boundaries described previously (Gautel et al. 1995). Asterisks denote sequences that were predicted by comparing that organism’s genome to the human genome and have not yet been verified by direct gene cloning
The purpose of the current study was to perform a systematic comparison of the Pro-Ala rich sequences in different species. By comparing cMyBP-C sequence data, we demonstrate that the length of the Pro and Ala region is not conserved and that the Pro-Ala content varies in different species. Furthermore, the number of proline and alanine residues correlates directly with body weight and inversely with heart rate in mammals. These findings raise the intriguing possibility that the Pro-Ala rich region of cMyBP-C scales in concert with mechanisms that fine-tune the speed of contraction to match cardiac function in different species.
The Pro-Ala rich region of cMyBP-C varies with heart rate in mammals
Protein sequence data for 15 species isoforms of cMyBP-C were downloaded from GenBank (Benson et al. 2009) or Ensembl (Hubbard et al. 2009) databases and sequences were aligned and compared using CLUSTALX 2.0.10 (Larkin et al. 2007). To compare Pro-Ala regions, the Pro-Ala rich region of human cMyBP-C was used as reference with domain boundaries for the 51 amino acid sequence determined as described by Gautel et al. (1995) and the Universal Protein Resources Databank (UniProt) (Jain et al. 2009). The Pro-Ala rich regions for the remaining sequences were determined using sequence homology to the human isoform. The percentage of Pro and Ala residues in each Pro-Ala rich region was calculated by dividing the number of Pro and Ala residues in the region of homology by the total number of amino acids in the entire Pro-Ala sequence.
Table 1 shows results of sequence comparisons of the cMyBP-C Pro-Ala regions of 15 species, including representative sequences from mammals, aves, amphibians, and fish. The total length of the Pro-Ala region and the percentage of Pro and Ala residues in the sequence were not conserved but varied across all the species examined. The longest Pro-Ala sequence occurred in Zebrafish (62 amino acids), whereas the shortest Pro-Ala region occurred in chicken (41 amino acids). The percentage of Pro-Ala residues varied from ~ 25% (rat) to ~ 53% (Xenopus). Trends across different phyla were not apparent, but the limited number of sequences available from non-mammalian species preclude a more definitive statement. By contrast, a trend in mammals was noted where the percent of proline and alanine residues correlated with body weight such that species with greater body weights had the highest percentage of Pro-Ala residues. Because body weight is inversely correlated with heart rate (Schmidt-Nielsen 1997), we also plotted the percentage of Pro-Ala residues as a function of heart rate. As shown in Fig. 1, the percentage of proline and alanine residues in the Pro-Ala rich region was inversely correlated with heart rate (R 2 = 0.62).
Implications for the role of cMyBP-C in sarcomeres
Pro-Ala rich sequences have been identified in other thick filament regulatory proteins including cardiac and skeletal isoforms of myosin essential light chain (ELC). In these proteins the Pro-Ala sequences modulate cross-bridge cycling rates and shortening velocity (Vmax) through mechanisms that involve binding of the ELCs to actin (Andreev et al. 1999). It is possible that the Pro-Ala rich region of cMyBP-C performs a similar role and contributes to the ability of cMyBP-C to limit myocyte shortening velocity, cross-bridge cycling, and power output (Korte et al. 2003). If so, then graded changes in the content of the Pro-Ala region described here could serve to optimize power output (the product of force and velocity) such that contractile efficiency is maximized in hearts that contract under different hemodynamic loads and at different speeds. Thus, changes in the Pro-Ala rich region of cMyBP-C may act to accelerate cross-bridge cycling kinetics in small mammals or slow cycling rates in larger mammals and thereby fine tune changes in cross-bridge cycling kinetics that occur from isoform shifts in other contractile proteins such as in myosin heavy chain that shifts from fast α-MHC (high ATPase activity and crossbridge cycling) to slow β-MHC (Pope et al. 1980; Schwartz et al. 1981; Morano et al. 1988; Hamilton and Ianuzzo 1991).
The proline-alanine rich regions of different ELC isoforms are thought to slow cross-bridge kinetics either by binding directly to actin (Trayer, Trayer et al. 1987) or by functioning as a rigid spacer arm that extends an actin binding site located near the N-terminus of the ELC out toward the thin filament (Timson and Trayer 1997). In both cases, interactions with actin serve to slow shortening velocity and limit cross-bridge cycling (Sweeney 1995), possibly by creating a drag that limits filament sliding. Consistent with this idea, atrial myocytes that express an atrial ELC isoform with reduced affinity for actin have greater Vmax and maximal power output than ventricular myocytes expressing an ELC that binds to actin with increased affinity (Schaub et al. 1998). It remains to be determined whether changes in the proline-alanine content of cMyBP-C described here also affect actin binding affinity, but available data from different studies suggests that proteins containing the human Pro-Ala sequence bind actin with greater affinity (Kulikovskaya et al. 2003) than those containing the mouse sequence (Shaffer et al. 2009). If so, reduced binding of the Pro-Ala sequence to actin could contribute to accelerated cycling kinetics in mouse hearts, whereas increased binding may slow cross-bridge kinetics in human hearts or hearts from larger mammals. Similarly, it will be of interest to determine whether the actin binding affinity of fast skeletal light chain (MLC1F) varies by species since Bicer and Reiser (Bicer and Reiser 2007) found that the molecular weight of MLC1F increased with increasing body mass in species from the shrew to the elephant due to changes in the Pro-Ala rich sequence.
Conclusions
The goal of this study was to perform a sequence comparison of the Pro-Ala rich region of cMyBP-C in different species. Results demonstrate that the sequence is not conserved but varies in length and in the number of proline and alanine residues. In mammals, the percentage of proline and alanines correlated inversely with species resting heart rate. By analogy with the proline-alanine rich sequences in myosin essential light chains, we propose that the Pro-Ala region of cMyBP-C slows cardiac cross-bridge cycling kinetics. Species-specific differences in the Pro-Ala region could thus account for functional differences observed in proteins containing mouse or human Pro-Ala sequences (Herron et al. 2006; Razumova et al. 2008; Shaffer et al. 2010) and thereby fine-tune cross-bridge speed to match cardiac function in hearts of different species.
Acknowledgments
The authors thank Todd Gillis for helpful discussions and careful reading of an earlier version of the manuscript.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Andreev OA, et al. Interaction of the N-terminus of chicken skeletal essential light chain 1 with F-actin. Biochemistry. 1999;38(8):2480–2485. doi: 10.1021/bi981706x. [DOI] [PubMed] [Google Scholar]
- Benson DA, et al. GenBank. Nucleic Acids Res. 2009;37(Database issue):D26–D31. doi: 10.1093/nar/gkn723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicer S, Reiser PJ. Variations in apparent mass of mammalian fast-type myosin light chains correlate with species body size, from shrew to elephant. Am J Physiol Regul Integr Comp Physiol. 2007;292(1):R527–R534. doi: 10.1152/ajpregu.00098.2006. [DOI] [PubMed] [Google Scholar]
- Gautel M, et al. Phosphorylation switches specific for the cardiac isoform of myosin binding protein-C a modulator of cardiac contraction. EMBO J. 1995;14(9):1952–1960. doi: 10.1002/j.1460-2075.1995.tb07187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton N, Ianuzzo CD. Contractile and calcium regulating capacities of myocardia of different sized mammals scale with resting heart rate. Mol Cell Biochem. 1991;106(2):133–141. doi: 10.1007/BF00230179. [DOI] [PubMed] [Google Scholar]
- Herron TJ, et al. Activation of myocardial contraction by the N-terminal domains of myosin binding protein-C. Circ Res. 2006;98(10):1290–1298. doi: 10.1161/01.RES.0000222059.54917.ef. [DOI] [PubMed] [Google Scholar]
- Hubbard TJ, et al. Ensembl 2009. Nucleic Acids Res. 2009;37(Database issue):D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain E, et al. Infrastructure for the life sciences: design and implementation of the UniProt website. BMC Bioinformatics. 2009;10:136. doi: 10.1186/1471-2105-10-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korte FS, et al. Loaded shortening, power output, and rate of force redevelopment are increased with knockout of cardiac myosin binding protein-C. Circ Res. 2003;93(8):752–758. doi: 10.1161/01.RES.0000096363.85588.9A. [DOI] [PubMed] [Google Scholar]
- Kulikovskaya I, et al. Effect of MyBP-C binding to actin on contractility in heart muscle. J Gen Physiol. 2003;122(6):761–774. doi: 10.1085/jgp.200308941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst G, et al. Myosin binding protein C, a phosphorylation-dependent force regulator in muscle that controls the attachment of myosin heads by its interaction with myosin S2. Circ Res. 2000;86(1):51–58. doi: 10.1161/01.res.86.1.51. [DOI] [PubMed] [Google Scholar]
- Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Morano I, et al. Skinned fibers of human atrium and ventricle: myosin isoenzymes and contractility. Circ Res. 1988;62(3):632–639. doi: 10.1161/01.res.62.3.632. [DOI] [PubMed] [Google Scholar]
- Pope B, et al. The ATPase activities of rat cardiac myosin isoenzymes. FEBS Lett. 1980;118(2):205–208. doi: 10.1016/0014-5793(80)80219-5. [DOI] [PubMed] [Google Scholar]
- Razumova MV, et al. Effects of the N-terminal domains of myosin binding protein-C in an in vitro motility assay: evidence for long-lived cross-bridges. J Biol Chem. 2006;281(47):35846–35854. doi: 10.1074/jbc.M606949200. [DOI] [PubMed] [Google Scholar]
- Razumova MV, et al. Contribution of the myosin binding protein C motif to functional effects in permeabilized rat trabeculae. J Gen Physiol. 2008;132(5):575–585. doi: 10.1085/jgp.200810013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub MC, et al. Modulation of contractility in human cardiac hypertrophy by myosin essential light chain isoforms. Cardiovasc Res. 1998;37(2):381–404. doi: 10.1016/S0008-6363(97)00258-7. [DOI] [PubMed] [Google Scholar]
- Schmidt-Nielsen K. Animal physiology: adaptation and environment. Cambridge: Cambridge University Press; 1997. [Google Scholar]
- Schwartz K, et al. Myosin isoenzymic distribution correlates with speed of myocardial contraction. J Mol Cell Cardiol. 1981;13(12):1071–1075. doi: 10.1016/0022-2828(81)90297-2. [DOI] [PubMed] [Google Scholar]
- Shaffer JF, et al. The myosin-binding protein C motif binds to F-actin in a phosphorylation-sensitive manner. J Biol Chem. 2009;284(18):12318–12327. doi: 10.1074/jbc.M808850200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer JF et al (2010) Functional differences between the N-terminal domains of mouse and human myosin binding protein-C. J Biomed Biotech 2010: Article ID 789798 [DOI] [PMC free article] [PubMed]
- Spector WS, editor. Handbook of biological data. Philadelphia: W.B. Saunders Company; 1956. [Google Scholar]
- Squire JM, et al. Structural evidence for the interaction of C-protein (MyBP-C) with actin and sequence identification of a possible actin-binding domain. J Mol Biol. 2003;331(3):713–724. doi: 10.1016/S0022-2836(03)00781-2. [DOI] [PubMed] [Google Scholar]
- Stelzer JE, et al. Differential roles of cardiac myosin-binding protein C and cardiac troponin I in the myofibrillar force responses to protein kinase A phosphorylation. Circ Res. 2007;101(5):503–511. doi: 10.1161/CIRCRESAHA.107.153650. [DOI] [PubMed] [Google Scholar]
- Sweeney HL. Function of the N terminus of the myosin essential light chain of vertebrate striated muscle. Biophys J. 1995;68(4 Suppl):112S–118S. [PMC free article] [PubMed] [Google Scholar]
- Timson DJ, Trayer IP. The role of the proline-rich region in A1-type myosin essential light chains: implications for information transmission in the actomyosin complex. FEBS Lett. 1997;400(1):31–36. doi: 10.1016/S0014-5793(96)01314-2. [DOI] [PubMed] [Google Scholar]
- Trayer IP, et al. Evidence that the N-terminal region of A1-light chain of myosin interacts directly with the C-terminal region of actin. A proton magnetic resonance study. Eur J Biochem. 1987;164(1):259–266. doi: 10.1111/j.1432-1033.1987.tb11019.x. [DOI] [PubMed] [Google Scholar]