Abstract
Red clover (Trifolium pratense L.) is a diploid (2n = 14), self-incompatible legume that is widely cultivated as a forage legume in cold geographical regions. Because it is a short-lived perennial species, improvement of plant persistency is the most important objective for red clover breeding. To develop a marker-assisted selection (MAS) approach for red clover, we identified candidate QTLs related to plant persistency. Two full-sib mapping populations, 272 × WF1680 and HR × R130, were used for QTL identification. Resistance to Sclerotinia trifoliorum and Fusarium species, as well as to winter hardiness, was investigated in the laboratory and in field experiments in Moscow region (Russia), and Sapporo (Japan). With the genotype data derived from microsatellite and other DNA markers, candidate QTLs were identified by simple interval mapping (SIM), Kruskal–Wallis analysis (KW analysis) and genotype matrix mapping (GMM). A total of 10 and 23 candidate QTL regions for plant persistency were identified in the 272 × WF1680 and the HR × R130 mapping populations, respectively. The QTLs identified by multiple mapping approaches were mapped on linkage group (LG) 3 and LG6. The significant QTL interactions identified by GMM explained the higher phenotypic variation than single effect QTLs. Identification of haplotypes having positive effect QTLs in each parent were first demonstrated in this study for pseudo-testcross mapping populations in plant species using experimental data.
Electronic supplementary material
The online version of this article (doi:10.1007/s00122-009-1253-5) contains supplementary material, which is available to authorized users.
Introduction
Red clover (Trifolium pratense L.) is a diploid (2n = 14), self-incompatible legume that is widely cultivated as a forage legume in moist and cool regions of the world, due to its high seedling vigor and adaptability to acidic and humid conditions. Because it is a short-lived perennial species, the use of red clover often results in a reduction in the quality and quantity of forage (Taylor and Quesenberry 1996). Therefore, most red clover breeding programs to date have primarily emphasized persistency, i.e., individual plant longevity (Taylor 2008). However, improvement of plant persistency has been a challenging objective for conventional red clover breeding programs, because it is a complex trait and requires lengthy studies for trait investigation. Marker-assisted selection (MAS) using DNA markers is expected to make an important contribution to red clover breeding programs that aim to enhance persistency.
Although several factors affect plant persistency in red clover, the main factor, which leads to a significant reduction in red clover green matter (from 30 to 80%), is the biotic stress caused by pathogen infections (Novosiolova 2002). One of the most destructive diseases is Sclerotinia crown and root rot, which is caused by the fungal pathogen Sclerotinia trifoliorum Erikson (Dijkstra 1964). The pathogen infects plants in the autumn and develops in the plant under snow cover during the winter. The disease symptoms rapidly appear on the plant once temperatures rise and the snow melts. Another important disease is Fusarium root rot, which is most frequently caused by Fusarium species, such as F. oxysporum, F. solani and F. roseum (Pederson et al. 1980). Because the root rot pathogens tend to accumulate in the soil, it is difficult to control root rot in red clover (Taylor and Quesenberry 1996). Once the pathogen infects the plant, root rot develops throughout plant growth.
Along with biotic stresses, abiotic stresses have an impact on plant persistency in red clover. Because red clover is predominantly produced in cold regions, a low temperature in the winter is generally considered the most severe abiotic stress. Plant vigor and/or survival rate after winter is often used in red clover breeding programs as an index of winter hardiness. Winter hardiness is considered to be a composite trait, consisting of a mixture of freezing tolerance and disease resistance under snow (Larsen 1994).
Several studies have analyzed quantitative trait loci (QTL) related to S. sclerotiorum resistance in sunflower (Helianthus annuus, Bert et al. 2004; Micic et al. 2004; Rönicke et al. 2005; Yue et al. 2008), rapeseed (Brassica napus, Zhao et al. 2003, 2006), soybean (Glycine max, Kim and Diers 2000; Arahana et al. 2001; Vuong et al. 2008) and common bean (Phaseolus vulgaris, Miklas 2007; Park et al. 2001), but no reports have focused on QTLs related to S. trifoliorum resistance. Most candidate QTLs identified for S. sclerotiorum resistance do not explain the large amount of phenotypic variation, which ranges from 10 to 18% in sunflower, 22–23% in rapeseed, and 10–12% in soybean. Miklas (2007) reported the identification and marker-assisted backcrossing of two QTLs associated with partial resistance to S. sclerotiorum in the common bean. Each candidate QTL explained 30–52% of the phenotypic variation for disease symptoms in a greenhouse test and accounted for 10% of the variation in a BC3F4:6 population grown in the field. QTLs for Fusarium root rot resistance explained 5–53% of the total phenotypic variability (Román-Avilés and Kelly 2005; Schneider et al., 2001). These reports imply that Sclerotinia and Fusarium resistance are controlled by multiple genes and pyramiding genes by MAS may therefore be an efficient means to improve QTL identification.
Although QTL identification has been performed in various plant species for the past two decades, investigation of QTLs in self-incompatible species has lagged behind examination of self-compatible species, because of the difficulty in developing inbred lines for segregation analysis. The most popular approach for identifying QTLs in self-incompatible species is to develop pseudo-testcross populations for segregation analysis, and then identify QTLs by interval mapping (Brown et al. 2007; Liebhard et al. 2003; Jansen and Stam 1994; Sim et al. 2007; Studer et al. 2007). In red clover, QTLs have been identified for seed yield, plant persistency, and morphological traits using a pseudo-testcross population (Herrmann et al. 2006, 2008). Herrmann et al. (2008) investigated plant persistency based on seasonal plant vigor and identified several QTLs related to plant persistency by multiple QTL mapping (MQM). The QTL on linkage group (LG)3 which was identified on the basis of one seasonal vigor score (average in the third year) as well as three different indices of persistency was the most potent QTL and explained 7.0–12.2% of the observed phenotypic variability.
Though candidate QTLs have been identified in several self-incompatible species, there have been few attempts to develop strategies to use MAS with the identified QTLs. Complex factors, such as haplotype phase, haplotype combination, high heterozygosity and QTL interactions, may make it difficult to interpret the results of interval mapping and to use the identified QTLs for MAS. Recently, Isobe et al. (2007) developed a new QTL mapping approach, genotype matrix mapping (GMM), which searches for QTL interactions in genetic variation. While this demonstrated that multiple QTL interactions could be identified in unrelated individual populations, we considered that the GMM approach would also be efficient for QTL identification in a family population. The identification of multiple QTL interactions by GMM will greatly advance our understanding of the complex QTL expression in a self-incompatible species.
In the present study, we sought to identify QTLs for three stress tolerance traits related to plant persistency in red clover, namely S. trifoliorum resistance, Fusarium resistance, and winter hardiness. In order to multilaterally discuss the characteristics of the identified QTLs, we performed QTL detection using three calculation approaches, simple interval mapping (SIM), Kruskal–Wallis (KW) analysis, and GMM. SIM is a parametric mapping approach, while KW analysis and GMM are non-parametric mapping approaches (Lehmann 1975; Isobe et al., 2007). SIM and KW analysis identify single locus effects whereas GMM considers multiple locus effects, as described above. In addition, identification of haplotypes having QTLs with positive effects in each heterozygous parental plant was first demonstrated by this study. The study was performed with two pseudo-testcross mapping populations and two phenotypic investigation sites (Moscow region, Russia and Sapporo, Japan), in order to enhance the possibility of capturing various QTLs.
Materials and methods
Plant materials for QTL mapping
Two previously reported red clover mapping populations, 272 × WF1680 and HR × R130, were used in this study. The 272 × WF1680 population is a BC1F1 population consisting of 167 individuals, in which the ‘272’ parent was a single F1 plant arising from a cross between ‘1588’, a wild germplasm collected in the Arhangelsk region of Russia, and ‘WF1680’, which originated from a central Russian variety “Srednerusskiy” (Isobe et al. 2003). HR × R130 is a one-way pseudo-testcross mapping population consisting of 188 individuals, in which the female parent, ‘HR’, originated from the Japanese variety ‘Hokuseki’, and the male parent, ‘R130’, was a progeny of the 272 × WF1680 population (Sato et al. 2005). Each of the mapping progenies was clonally propagated by in vitro culture (for the tests in Russia) or by cuttings (for the tests in Japan), and was transplanted into the field in the All-Russian Williams Fodder Crop Research Institute (WFCRI; Moscow region, Russia) or the National Agricultural Research Center for Hokkaido (NARCH; Sapporo, Japan) to evaluate the traits that relate to plant persistency. The investigated traits and the duration of the experiment, the number of tested progeny, and the number of replications performed are summarized in Table 1.
Table 1.
Experimental design of the investigated traits related to plant persistency
| Population and trait | Trait name | Test field | Location | Season | No. of tested genotypes | No. of replications | |
|---|---|---|---|---|---|---|---|
| Transplanting | Investigation | ||||||
| 272 × WF1680 | |||||||
| S. trifoliorum resistance | |||||||
| Laboratory experiment (damage rate, %) | SRL | Laboratory | WFCRIa | – | 2003 | 36 | 2 |
| Field experiment (extent of plant disease, %) | SRR | Non-artificial | Moscow region | 2001 May | 2002 April | 59 | 6 |
| Fusarium resistance | |||||||
| Field experiment (intensity of disease development, %) | FRR | Non-artificial | Moscow region | 2001 May | 2002 April | 59 | 6 |
| Winter hardiness | |||||||
| Green part ratio of crown (%) | WHJ | Non-artificial | Sapporo | 2001 July | 2002 April | 139 | 1 |
| HR × R130 | |||||||
| S. trifoliorum resistance | |||||||
| Field experiment (extent of plant disease, %) | SRR | Artificial | Moscow region | 2004 July | 2005 April | 96 | 4–13 |
| Field experiment (1: very sensitive; 9: very resistant) | SRJ | Non-artificial | Sapporo | 2003 October | 2005 April | 177 | 5 |
| Fusarium resistance | |||||||
| Field experiment (intensity of disease development, %) | FRR05 | Artificial | Moscow region | 2004 July | 2005 April | 96 | 4-13 |
| Field experiment (intensity of disease development, %) | FRR07 | Artificial | Moscow region | 2006 June | 2007 October | 61 | 4–10 |
| Winter hardiness | |||||||
| Survival rate after winter (%) | WHR | Artificial | Moscow region | 2004 July | 2005 Apr | 96 | 1 |
| Winter hardiness (1: very weak; 9: very hardy) | WHJ | Non-artificial | Sapporo | 2003 October | 2004 Apr | 178 | 5 |
aAll-Russian Williams Fodder Crop Research Institute
Laboratory test on resistance to S. trifoliorum
Resistance to S. trifoliorum was determined in the laboratory at WFCRI to develop a rapid testing method for early diagnosis of infection in red clover, as described by Razgulayeva et al. (1999). S. trifoliorum inoculates were cultured on liquid clover-glucose medium at 16°C to form young, white sclerotiums. After 15–20 days, the mycelia were filtered, dried, minced, and resuspended in sterile water. The leaves of the test plant clones were placed in Petri dishes on moist cotton wool containing 0.004% benzimidazole. Two drops of the pathogen suspension were pipetted onto each leaflet. Two replicates were performed for each genotype and for the control variety ‘VIK-7’, which has an intermediate level of resistance to S. trifoliorum. The inoculated plant material was then transferred into a growth chamber and maintained at 16°C with a 16 h light/8 h dark photoperiod under fluorescent light (5,000 lux). At 7 days post-inoculation, the degree of damage was assessed by visual inspection of symptoms and rated on a scale of 0–3, with 0 representing healthy leaflets and 3 representing severely damaged leaflets. On the base of this rank scale, the damage rate (R%) was calculated using the following formula, according to Ephimova (1981):
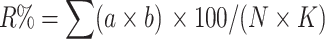 |
where a is the number of injured leaves; b the level of damage; N the total number of leaves in the analysis; and K is the highest level of damage.
Field experiments in the Moscow region
The field tests on resistance to S. trifoliorum in the Moscow region were carried out in the test fields at WFCRI. The 272 × WF1680 population traits were examined in normal field conditions (non-artificial infection), while the HR × R130 population was cultivated in an infected field, in which the dose of inoculum cultivated with malt was 25 g/m2 of soil. The propagated plants and the control variety ‘VIK-7’ were transplanted into the field in replicates. The damage caused by S. trifoliorum infection was evaluated 2 weeks after spring plant regrowth started. The extent of plant disease (P%) was estimated using the following formula according to Chumackov et al. (1974):
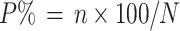 |
where n is the number of plants damaged by S. trifoliorum and N is the total number of plants evaluated.
In order to investigate Fusarium resistance in the infected field, the propagated seedlings of the mapping population and a control variety ‘VIK-7’ were transplanted into a field infected with a mixture of six Fusarium fungus species, which occur widely in the Moscow region, namely F. oxysporum Schl. em. Snyd. et Hans, F. culmorum (Sm.) Sacc., F. solani (Mart.) App. Et Wr., F. avenaceum (Fr.) Sacc., F. gibbosum App. et Wr. em. Bilai, F. sambucinum Fuck. The dose of inocula was 60 g/m2 of soil. For analysis of resistance, the roots of each plant were removed, and the degree of damage was assessed using a rank scale of 0 (no symptoms) to 5 (more than 75% of the root tissue damaged) at the end of the second year after planting. Based on this scale, the intensity of disease development (R%) was estimated using the following formula (Putsa et al. 1999):
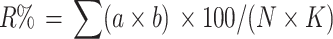 |
where a is the total number of injured plants; b the level of damage; N the total number of plants examined; and K is the highest scale of the investigation. The survival rate after winter was estimated in the HR × R130 population using data of estimation in the field in 2004–2005.
Field experiments in Sapporo
The progeny of the 272 × WF1680 mapping population were transplanted to a non-artificial field in July 2001 with no replication. No plants showed symptoms of S. trifoliorum infection in the early spring of 2002. Each plant was photographed 2 weeks after the disappearance of snow, and the numbers of green color pixels to non-green color pixels of digitized photographs of the crown were evaluated using Microsoft Photo Editor (Microsoft Corporation, Redmond, WA, USA).
To estimate the winter hardiness of the HR × R130 mapping population, plant cuttings were transplanted to the non-artificial field at NARCH in October 2003 with five replications. None of the plants showed symptoms of S. trifoliorum infection in the spring of the second year (2004). Winter hardiness was then scored on a scale of 1 (very weak) to 9 (very hardy). Symptoms of S. trifoliorum infection were observed on the plants 2 weeks after the disappearance of snow in the spring of the third year (2005), and Sclerotinia resistance was scored on a scale of 1 (very sensitive) to 9 (very resistant).
Marker analysis and linkage map construction
Combined genetic linkage maps of 272 × WF1680 and HR × R130 were reconstructed based on previously reported restriction fragment length polymorphism (RFLP), microsatellite, morphological, random amplified polymorphic DNA (RAPD), and sequence tagged site (STS) markers (Herrmann et al. 2006; Isobe et al. 2003, 2009; Sato et al. 2005). Markers designated a single ‘C’ or ‘OPB’ label represent RFLP and RAPD markers, respectively, while ‘RCS’ and ‘TPSSR’ followed by a number indicate microsatellite markers. For map construction, parent-specific maps were first developed using a combination of the color map method (Kiss et al. 1998) and JoinMap® 4 (Van Ooijen 2006), as previously reported (Isobe et al. 2003). These data were then combined with a genetic linkage map using the ‘Join’ module of JoinMap® 4.
Statistical analysis and QTL detection
The normality of the phenotypic data distributions was confirmed using the D’Agostino–Pearson K 2 test with the Data Transformation program (http://www.tuat.ac.jp/~ethology/Columbo/Stat/Data_Trans). If the phenotypic data sets did not have a normal distribution, they were transformed to fit a normal distribution. After data transformation, correlations between phenotypic data were assessed using the Pearson correlation coefficients. The broad sense heritability of each trait was calculated as follows:
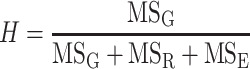 |
where MSG is the mean square for genotype MSR the mean square for replication and MSE is the mean square for error.
For identification of candidate QTL regions, interval mapping and nonparametric mapping were performed, and then the corresponding QTLs were confirmed. SIM was performed using MapQTL® 5 (Van Ooijen 2004). The significant LOD (logarithm of odds) threshold was determined at 5.0% via a permutation test module (10,000 replications). Although MQM is implemented in MapQTL® 5, this mapping method was not employed in this study, because the run could not be completed by the software due to the large number of data points.
Nonparametric mapping was performed following the two approaches: KW analysis (Lehman, 1975) using MapQTL® 5, and GMM (Isobe et al. 2007) using the GMM ver. 2.1 program (http://www.kazusa.or.jp/GMM). KW analysis identifies markers related to single dose QTLs, while GMM is a method that searches for QTL interactions that have an effect on genetic variation. For KW analysis, significant statistics adjusted for ties, which are indicated by K*, were employed, and their error probability was set to 0.005. For GMM mapping, single effect QTLs, as well as double and triple locus interactions, were examined within a default searching range, which was set by the program, based on the input data, and the minimum number of corresponding individuals was set to 1.
Two different approaches were employed to identify the haplotypes contributing to the positive effects of QTLs: one for SIM, and the other for KW analysis and GMM. By performing SIM on MapQTL®, phenotypic mean values of ‘ac’, ‘ad’, ‘bc’ and ‘bd’ genotypes on each locus were estimated in a ‘cp’ population, i.e. a pseudo-testcross population, in which maternal and paternal genotype were coded as ‘ab’ and ‘cd’, respectively. Therefore, the mean values between ‘ac’ and ‘ad’, ‘bc’ and ‘bd’, ‘ac’ and ‘bc’, and ‘ad’ and ‘bd’ were calculated for the phenotypic mean value of each haplotype of ‘a’ and ‘b’ (two maternal haplotypes), and ‘c’ and ‘d’ (two paternal haplotypes), respectively. Positive effect haplotypes were then identified by comparison of the mean values between maternal haplotypes, ‘a’ and ‘b’, and paternal haplotypes, ‘c’ and ‘d’. KW analysis and GMM calculate the phenotypic mean value of each genotype code for which the haplotype phases are unknown. Thus, the haplotype phase of each genotype code on the candidate QTLs was confirmed based on the results of color mapping, which identifies the haplotype phase by graphical genotypes. The positive effect haplotype in each parent was then identified by comparing the mean values of haplotypes.
Results
Phenotypic data analysis
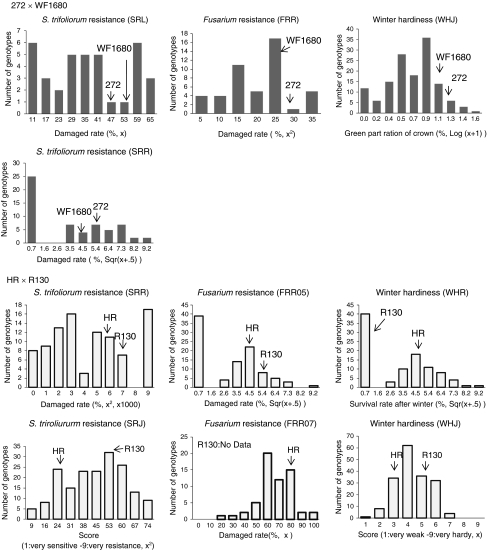
Continuous distributions were observed in all ten of the traits investigated, all of which were related to S. trifoliorum resistance, Fusarium resistance, and winter hardiness (Fig. 1). According to the D’Aogostino–Pearson K 2 test, three of the ten traits (SRL in the 272 × WF1680 population, and FRR07 and WHJ in the HR × R130 population) showed normal distributions, while the seven remaining traits did not have normal distributions (Table 2). These latter seven traits were transformed using square, square root, or Log functions to fit the normal distribution. Broad sense heritability ranged from 0.05 to 0.40, with the lowest and highest heritability values being observed for FRR05 and SRR, respectively, from the HR × R130 population. The coefficient of variance ranged from 5.8% (SRR in the 272 × WF1680 population) to 60.0% (FRR in the 272 × WF1680 population).
Fig. 1.
Frequency distributions of the traits relating to plant persistency in red clover populations 272 × WF1680 and HR × R130. Parental values of phenotypes are indicated by arrows. The transform functions used to convert the data to normal distributions are described in parentheses on the X-axis title
Table 2.
Mean, CV(%), LSD and heritability of the investigated traits related to plant persistency for the mapping progenies and the parents
| Traitsa | 272 × WF1680 | |||||
|---|---|---|---|---|---|---|
| SRL | SRR | FRR | WHJ | |||
| Transform function | x | Sqr (x + 0.5) | x 2 | Log (x + 1) | ||
| Mean | 38.15 | 2.13 | 18.53 | 0.78 | ||
| 272 | 42.49 | 5.78 | 28.44 | 1.33 | ||
| WF1680 | 50.00 | 4.78 | 25.00 | 1.11 | ||
| CV (%) | 38.3 | 5.8 | 60.0 | – | ||
| LSD (0.05) | 29.60 | 0.47 | 21.87 | – | ||
| Heritability | 0.30 | 0.12 | 0.13 | – | ||
| Traitsa | HR × R130 | |||||
|---|---|---|---|---|---|---|
| SRR | SRJ | FRR05 | FRR07 | WHR | WHJ | |
| Transform function | x 2 | x 2 | Sqr (x + 0.5) | x | Sqr (x + 0.5) | x |
| Mean | 4,136 | 48.20 | 4.06 | 64.10 | 4.20 | 4.02 |
| HR | 5,098 | 18.77 | 3.80 | 72.00 | 3.84 | 3.00 |
| R130 | 6,400 | 49.00 | 4.53 | nd | 0.70 | 5.00 |
| CV (%) | 14.7 | 55.2 | 33.2 | 41.5 | – | 56.2 |
| LSD (0.05) | 2,061 | 47.5 | ns | ns | – | 7.03 |
| Heritability | 0.40 | 0.37 | 0.05 | 0.32 | – | 0.37 |
CV coefficient of variance, LSD least significant difference, nd no data, ns no significant
aTransform function: function for transformation of phenotypic value to fit normal distribution
The range of phenotypic values of the mapping progenies exceeded the differences between the parents in all traits (Fig. 1). In the 272 × WF1680 population, the 272 parent, showed lower resistance to S. trifoliorum in the Russian field tests (SRR) than the WF1680 parent, while no phenotypic differences between the parents were observed in other traits (Table 2). In the HR × R130 population, no significant differences were observed between the parents for all the traits investigated. In the 272 × WF1680 population, a significant correlation was observed only between SRR and SRL (Table 3). In the HR × R130 population, significant correlations were observed between SRR and FRR05, and between SRR and WHR.
Table 3.
Correlation coefficients between resistance to S. trifoliorum, Fusarium and winter hardiness in 272 × WF1680 and HR × R130
| Trait | |||||
|---|---|---|---|---|---|
| SRR | FRR | WHJ | |||
| 272 × WF1680 | |||||
| SRL | 0.365* | −0.113 | 0.015 | ||
| SRR | −0.160 | −0.168 | |||
| FRR | 0.110 | ||||
| Trait | |||||
|---|---|---|---|---|---|
| SRJ | FRR05 | FRR07 | WHR | WHJ | |
| HR × R130 | |||||
| SRR | −0.015 | −0.624** | 0.082 | −0.698** | −0.165 |
| SRJ | −0.077 | −0.004 | 0.061 | 0.010 | |
| FRR05 | −0.203 | −0.073 | 0.164 | ||
| FRR07 | 0.059 | 0.013 | |||
| WHR | 0.077 | ||||
* and ** indicate significant correlation at p < 0.05 and p < 0.001, respectively
Linkage map construction
A total of 437 loci, including 249 RFLPs, 182 microsatellites, two STSs, four RAPDs and one morphological locus, were mapped onto seven linkage groups of the 272 × WF1680 integrated map. The total length of the linkage map was 817 cM and the average distance between two markers was 1.9 cM. The HR × R130 integrated map consisted of a total of 1502 loci, including 1352 RFLP and 150 RFLP loci, spanning seven linkage groups. The total length of the map was 801 cM and the average distance between two markers was 0.53 cM. Between the two combined linkage maps, 88 RFLP and 167 microsatellite markers were commonly mapped. The locus orders on the two maps correspond to those of the accession-specific maps reported by Isobe et al. (2009).
QTL detection in the 272 × WF1680 population
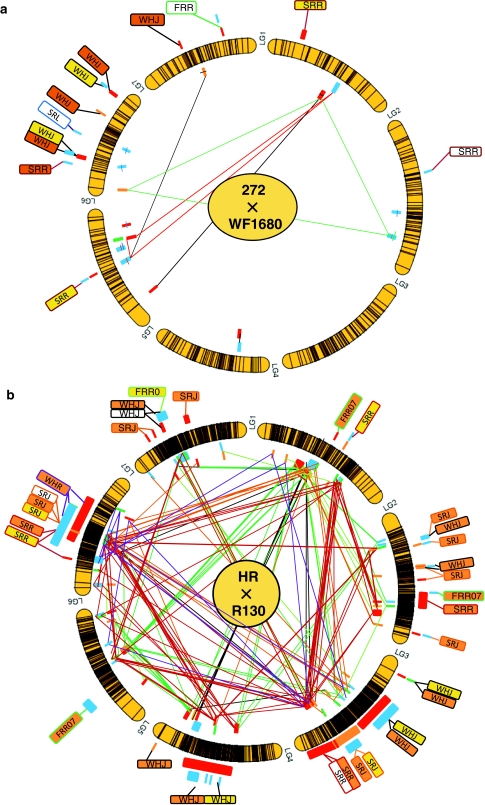
A total of 4, 5 and 3 single effect QTLs were detected in the 272 × WF1680 population by SIM, KW analysis, and GMM, respectively (Fig. 2a and Supplementary Table 1). Single QTL were detected for SRL and FRR, while four QTLs were identified for SRR and WHJ. Of the seven linkage groups present in red clover, most QTLs mapped to the LG6. The positive effect QTLs were identified only in one haplotype in WF1680, but in both haplotypes in 272. The phenotypic variance explained by QTLs detected by SIM ranged from 76.1 to 91.7% for SRR and from 13.5 to 16.2% for WHJ, while variance explained by QTLs, as detected by GMM, was 7.4% for SRL, 10.8% for SRR, and 13.8% for FRR.
Fig. 2.
Mapping position of identified QTLs in the 272 × WF1680 (a) and the HR × R130 (b) populations. The segments of the outer circle indicate the linkage maps consisting of seven linkage groups and the black bars in each segment represent the positions of loci used for QTL detection. Short bars outside the segments of the circle indicate the positions of single effect QTLs detected by SIM, KW analysis, and GMM, while bars inside the circle represent interacting QTLs detected by GMM (two- or three-locus interactions). Red and yellow bars indicate haplotypes with higher resistance or winter hardiness than other haplotypes in 272 or HR, while blue and green bars represent haplotypes with a higher tolerance effect in WF1680 or R130. The haplotype phase can only be confirmed within linkage groups, and not across linkage groups. The letters on boxes indicate trait names explained in Table 1, and yellow, orange and white boxes represent QTLs detected by SIM, KW analysis and GMM, respectively. The lines connecting short bars inside the circles represent combinations of interacting QTLs. Red, green, blue, and black lines in the 272 × WF1680 linkage circle represent interacting QTLs for SRL, SRR, FRR, and WHJ, respectively. Red, orange, light green, green, purple, and black lines in the HR × R130 linkage circle indicate QTLs for SRR, SRJ, FRR05, FRR07, WHR, and WHJ, respectively
The locus combinations representing interacting QTLs were identified by GMM for two- and three-locus combinations (Supplementary Tables 1–3). Significant QTL interactions were observed for each investigated trait: a single two-locus combination for SRL, SRR, and FRR; four–two-locus combinations for WHJ; six–three-locus combinations for SRL, two–three-locus combinations for SRR, one–three-locus combination for FRR and three–three-locus combinations for WHJ. The identified loci were mapped across linkage groups and their combinations were observed both within and between parents (Fig. 2a). In each trait, the highest phenotypic variance explained was observed in the three-locus combinations (Supplementary Tables 1–3).
QTL detection in the HR × R130 population
A total of 10, 191, and 8 single effect QTLs were detected in the HR × R130 population by SIM, KW analysis, and GMM, respectively (Supplementary Table 1). Many QTLs identified by KW analysis and some of the QTLs identified by GMM were sequentially mapped onto LG3, LG4 and LG6 (Fig. 2b). The positive effect QTLs were identified only in one haplotype in R130 with the exception that QTLs for WHJ mapped onto LG3, but in both haplotypes in HR. Most of the QTLs identified in LG3, LG4 and LG6 were detected by multiple approaches, for example, in LG3, WHJ at 5.4 cM (by SIM and KW analysis), WHJ at 27 cM (by SIM and KW analysis), SRJ at 87 cM (by SIM and KW analysis), and SRR at 97 cM (KW analysis and GMM); in LG4, WHJ at 59 cM and 71 cM (SIM and KW analysis); in LG6, SRR at 48 cM (SIM and KW analysis), SRJ at 51 and 61 cM (by SIM and KW analysis and GMM). Co-localized QTLs for different traits were observed in LG2 at 24 cM (by SRJ and WHJ), LG2 at 50 cM (SRJ and WHJ), and LG6 at 45–78 cM (by SRR, SRJ and WHR). QTLs for stress tolerance were mapped to a total of 23 regions, including 5 for SRR, 9 for SRJ, 1 for FRR05, 3 for FRR07, 3 for WHR, and 6 for WHJ. The phenotypic variance explained by QTLs detected by SIM ranged from 70.0 to 94.0% for SRR, 13.8–17.6% for SRJ, 81.6% for FRR05, and 10.7–41.7% for WHJ, while those detected by GMM were 9.1% for SRR, 10.4–11.5% for SRJ, and 7.4–7.7% for WHJ.
Number of significant locus combinations were identified by GMM: for two-locus combinations, 2 were detected for SRR, 10 for SRJ, 7 for FRR05, 10 for FRR07 and 5 for WHJ; for three-locus combinations, 39 were detected for SRR, 10 for SRJ, 7 for FRR05, 11 for FRR07, 6 for WHR, and 2 for WHJ (Supplementary Tables 1–3). The interacting loci were mapped across linkage groups and their combinations were observed both within and between parents (Fig. 2b). The interacting QTLs for multiple traits were identified in LG1 at 31–58 cM (for SRJ, FRR05, FRR07 and WHJ), LG3 at 79–119 cM (for SRR, SRJ, FRR05, WHR and WHJ) and LG6 at 46–60 cM (for SRR, SRJ, FRR07, WHR and WHJ). The phenotypic variance explained of two-QTLs combinations ranged 23.8–24.7% for SRR, 13.4–25.8% for SRJ, 12.9–23.8% for FRR05, 13.7–21.3% for FRR07, and 16.0–23.4% for WHJ, while those of three QTLs combinations were 18.5–47.5% for SRR, 22.6–36.8% for SRJ, 31.3–41.2% for FRR05, 46.0–56.0% for FRR07, 25.4–50.6% for WHR and 25.6–26.57% for WHJ (Supplementary Tables 1–3).
Discussion
Controlling Sclerotinia and Fusarium field resistance, as well as winter hardiness, in red clover is difficult, because the sources of stress develop in the soil and/or beneath snow-covered ground (Dijkstra 1964; Taylor and Quesenberry 1996). The coefficient of variance of the traits investigated in this study was generally high and the broad sense heritabilities were low and variant. Significant QTL interactions were observed between SRL and SRR in the 272 × WF1680 population, and between SRR and FRR05, and SRR and WHR, in the HR × R130 population. However, no other significant interactions were observed. Because vegetative propagation of red clover is difficult, we performed a limited number of phenotypic investigations with a limited number of replications in this study. The difficulty in controlling stress tolerances and the low number of trials may not be sufficient to accurately determine the genotype effect of the populations in this study. However, the low and variant heritability, and the few significant interactions between the traits investigated suggest an absence of major genes controlling the stress tolerances investigated.
A total of 10 and 23 regions were identified where single effect QTLs related to plant persistency mapped in the 272 × WF1680 and the HR × R130 populations, respectively. Because the QTL identification was based on statistical approaches, the possibility of identifying false positive and false negative QTLs for the thresholds and mapping approaches used still exists (McElroy et al. 2006; Mackay and Powell 2007). However, if QTLs were identified by multiple approaches, the reliability of the QTLs is likely to be enhanced. The QTLs observed on LG6 in the 272 × WF1680 population for WHJ, and on LG3, LG4 and LG6 in the HR × WF1680 population for SRR, SRJ and WHJ, were identified by multiple approaches and can be regarded as reliable QTLs. The reliability of QTLs identified for winter hardiness and Sclerotinia resistance on LG6 is also suggested by the overlap of QTLs identified in the two mapping populations. Thus, we conclude that LG6 is one of the most significant linkage groups with regard to stress tolerance. Herrmann et al. (2008) recently identified a QTL which was associated with the largest effect on plant persistency in red clover on LG3. Because LG3 contains three candidate QTL regions detected by multiple approaches in this study, we propose that LG3 is also a significant linkage group for red clover plant persistency.
A greater number of QTLs was identified in the HR × R130 population than in the 272 × WF1680 population. Because more DNA markers were used in the HR × R130 population, the power of QTL detection was higher in the HR × R130 population than in the 272 × WF1680 population. Of the three approaches used in this study, KW analysis identified the most single effect QTLs. Along with the differences in algorithms used to detect QTLs in these approaches, the different levels of severity of the thresholds used to identify significant QTLs had a major effect on the number of identified QTLs. The thresholds in SIM (LOD value) and the searching range in GMM were defined in every trait, in consideration of the specific properties of the data, while the thresholds in the KW analysis were uniformly defined for all traits and populations with an error probability of 0.005. Thus, the KW threshold used in this study may be less stringent than the SIM thresholds or the GMM searching range.
The five QTLs identified by SIM (two SRR QTLs in the 272 × WF1680 population, and two SRR QTLs and one FRR05 QTL in the HR × R130 population) were assessed as large effect QTLs that explained more than 70% of the phenotypic variance of each single QTL. However, because the broad sense heritability of the SRR and FRR QTLs was low (0.12–0.40 in SRR and 0.05–0.32 in FRR), and the QTLs were not identified by the other two approaches, the reliability of the estimation of the magnitude of phenotypic variance explained by these QTLs may be questionable. In SIM, significant QTLs are explored at genotyped loci and each ‘mapping step’ point which configured by operators. For the configured ‘mapping step’ points, genotypes are predicted according to the nearest genotyped loci, and then the existence of the QTLs is assessed based on the predicted genotypes. However, when a pseudo-testcross population is used for SIM, the nearest genotyped loci often provide genotype information only for one parent, because the other parent does not have polymorphisms in the loci. Therefore, the reliability of SIM in a pseudo-testcross is generally regarded as low, relative to that of a self-crossing population. Thus, we concluded that the magnitude of QTLs identified by SIM in this study was doubtful and sometimes overestimated.
In this study, combinations of interacting loci that affect the traits investigated were identified by GMM (two and three locus interactions). More interacting QTLs were identified in the HR × R130 population, which may reflect the greater number of genotyped loci. The highest number of interactions was observed for SRR in the HR × R130 population. Although the interacting QTLs were identified across genomes, several ‘hub’ regions for the interactions were observed in the HR × R130 population: for example, LG1 at 31-58 cM (for SRJ, FRR05, FRR07 and WHJ), LG1 at 83–91 cM (for SRR), LG3 at 79–119 cM (for SRR, SRJ, FRR05, WHR and WHJ), and LG6 at 46–60 cM (for SRR, SRJ, FRR07, WHR and WHJ). Interestingly, the numbers of significant QTL interactions identified by GMM were more than that of single effect QTLs. In addition, the percentages of the phenotypic variance explained by the QTL combinations were higher than those of the single effect QTLs. These results suggest that there were many QTLs for plant persistency that had a small effect as single QTLs, but indicate a large effect with a specific locus combination.
In a pseudo-testcross mapping population, polymorphic QTLs are identified by segregation analysis between two haplotypes in each parent. However, little attention has hitherto been paid to the identification of the haplotype phase associated with positive (or negative) effects of the QTLs. Therefore, the direction of identified QTLs has not been described in past reports that used pseudo-testcross mapping populations for QTL detection. In this study, we first performed an identification of the haplotypes that were associated with positive effect QTLs related to plant persistency. The single dose QTLs that were associated with a positive effect had a tendency to identify with either haplotype of each parent, especially in the HR × R130 population, while the QTL interactions were observed across the four haplotypes. This result implied that it may not be effective to improve plant persistency by pyramiding only single effect QTLs, because the positive QTL interactions would be lost. Therefore, we concluded that the accumulation of both single effect QTLs and interacting QTLs is necessary for the improvement of the traits. However, because the genetic diversity of the breeding populations might be much higher than that of the mapping parents used in this study, there would be other QTL combinations as well as single effect QTLs in each breeding populations. Therefore, the existence of the targeted QTLs in the breeding populations must be confirmed before performing the MAS approach. The QTL regions identified in this study, especially LG1 at 31–58 cM, LG3 at 79–119 cM, and LG6 at 46–60 cM, where interacting QTLs for multiple traits related to plant persistency were located and single effect QTLs also mapped, will be potential candidate regions for the re-investigation of QTLs in red clover breeding populations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work was supported by the Kazusa DNA Research Institute Foundation, the All-Russian Williams Fodder Crop Research Institute Foundation, the National Agricultural Research Center for the Hokkaido Region, and the Ministry of Agriculture, Forestry and Fisheries (Rice Genome Project DM-2102 and Low Temperature Stress Project). The authors are grateful to members of the Biotechnology Department (WFCRI), Dr Agafodorova M., Dr. Solodkaya L. and Dr. Lapotishkina L., for helpful advice and technical assistance in propagation and maintenance of the mapping populations in in vitro culture.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Arahana VS, Graef GL, Specht JE, Steadman JR, Eskridge KN. Identification of QTLs for Resistance to Sclerotinia sclerotiorum in Soybean. Crop Sci. 2001;41:180–188. [Google Scholar]
- Bert PF, Dechamp-Guillaume G, Serre F, Jouan I, de Labrouhe DT, Nicolas P, Vear F. Comparative genetic analysis of quantitative traits in sunflower (Helianthus annuus L.) 3. Characterisation of QTL involved in resistance to Sclerotinia sclerotiorum and Phoma macdonaldi. Theor Appl Genet. 2004;109(4):865–874. doi: 10.1007/s00122-004-1701-1. [DOI] [PubMed] [Google Scholar]
- Brown JS, Phillips-Mora W, Power EJ, Krol C. Cuauhtemoc Cervantes-Martinez, Juan Carlos Motamayor, and Raymond J. Schnell Mapping QTLs for resistance to frosty pod and black pod diseases and horticultural traits in Theobroma cacao L. Crop Sci. 2007;47:1851–1858. doi: 10.2135/cropsci2006.11.0753. [DOI] [Google Scholar]
- Chumackov AE, Minkevich II, Vlasov UI, Gavrilov EA (1974) The basic methods of phytopathological investigations. Kolos, Moscow, p 189
- Dijkstra J. Inoculation with ascospores of Sclerotinia trifoliorum for detection of clover rot resistant red clover. Euphytica. 1964;13:314–329. [Google Scholar]
- Ephimova G. Methods for estimation of red clover resistance to Sclerotinia. Breed Seed Prod. 1981;10:18–19. [Google Scholar]
- Herrmann D, Boller B, Studer B, Widmer F, Kölliker R. QTL analysis of seed yield components in red clover (Trifolium pratense L) Theor Appl Genet. 2006;112(3):536–545. doi: 10.1007/s00122-005-0158-1. [DOI] [PubMed] [Google Scholar]
- Herrmann D, Boller B, Studer B, Widmer F, Kölliker R. Improving persistence in red clover: insights from QTL analysis and comparative phenotypic evaluation. Crop Sci. 2008;48:269–277. doi: 10.2135/cropsci2007.03.0143. [DOI] [Google Scholar]
- Isobe S, Klimenko I, Ivahuta S, Gau M, Kozlov NN. First RFLP linkage map of red clover (Trifolium pratense L.) based on cDNA probes and its transferability to other red clover germplasms. Theor Appl Genet. 2003;108:105–112. doi: 10.1007/s00122-003-1412-z. [DOI] [PubMed] [Google Scholar]
- Isobe S, Nakaya A, Tabata S. Genotype matrix mapping: searching for quantitative trait loci interactions in genetic variation in complex traits. DNA Res. 2007;14(5):217–225. doi: 10.1093/dnares/dsm020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isobe S, Kölliker R, Hisano H, Sasamoto S, Wada T, Klimenko I, Okumura K, Tabata S. Construction of a consensus linkage map for red clover (Trifolium pratense L) BMC Plant Biol. 2009;9:57. doi: 10.1186/1471-2229-9-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RC, Stam P. High resolution of quantitative traits into multiple loci via interval mapping. Genetics. 1994;136:1447–1455. doi: 10.1093/genetics/136.4.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Diers BW. Inheritance of partial resistance to sclerotinia stem rot in soybean. Crop Sci. 2000;40:55–61. [Google Scholar]
- Kiss GB, Kereszt A, Kiss P, Endre G. Colormapping: a non-mathematical procedure for genetic mapping. Acta Biol Hung. 1998;19:125–142. [Google Scholar]
- Larsen A. Breeding winter hardy grasses. Euphytica. 1994;77:231–237. doi: 10.1007/BF02262635. [DOI] [Google Scholar]
- Lehmann EL. Nonparametrics. New York: McGraw-Hill; 1975. [Google Scholar]
- Liebhard R, Kellerhals M, Pfammatter W, Jertmini M, Gessler C. Mapping quantitative physiological traits in apple (Malus x domestica Borkh) Plant Mol Biol. 2003;52(3):511–526. doi: 10.1023/A:1024886500979. [DOI] [PubMed] [Google Scholar]
- Mackay I, Powell W. Methods for linkage disequilibrium mapping in crops. Trends Plant Sci. 2007;12(2):57–63. doi: 10.1016/j.tplants.2006.12.001. [DOI] [PubMed] [Google Scholar]
- McElroy JP, Zhang W, Koehler KJ, Lamont SJ, Dekkers JC. Comparison of methods for analysis of selective genotyping survival data. Genet Sel Evol. 2006;38(6):637–655. doi: 10.1186/1297-9686-38-6-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micic Z, Hahn V, Bauer E, Schön CC, Knapp SJ, Tang S, Melchinger AE. QTL mapping of Sclerotinia midstalk-rot resistance in sunflower. Theor Appl Genet. 2004;109(7):1474–1484. doi: 10.1007/s00122-004-1764-z. [DOI] [PubMed] [Google Scholar]
- Miklas PN. Marker-assisted backcrossing QTL for partial resistance to sclerotinia white mold in dry bean. Crop Sci. 2007;47:935–942. doi: 10.2135/cropsci2006.06.0410. [DOI] [Google Scholar]
- Novosiolova A. Increasing of red clover resistance to disease and pests. Voronej: Clover in Russia; 2002. p. 126. [Google Scholar]
- Park SO, Coyne DP, Steadman JR, Skroch PW. Mapping of QTL for resistance to white mold disease in common bean. Crop Sci. 2001;41:1253–1262. [Google Scholar]
- Pederson GA, Hill RR, Jr, Leath KT. Host-pathogen variability for Fusarium-caused root rot in red clover. Crop Sci. 1980;20:787–789. [Google Scholar]
- Putsa N, Razgulayeva N, Kostenko N (1999) Methodological recommendations on investigations of forage crops resistance to fungous pathogens at the artificial specialized infected field. Moscow, Russian Academy of Agriculture, All-Russian Williams Fodder Research Institute, p 38
- Razgulayeva N, Kostenko N, Putsa N (1999) Methods of earlier diagnostic of red clover and brome grass on resistance to diseases. Theor and Appl Basis of Resources Saving in Agric, Russia, Tyumen, pp 115–116
- Román-Avilés B, Kelly JD. Identification of quantitative trait loci conditioning resistance to fusarium root rot in common bean. Crop Sci. 2005;45:1881–1890. doi: 10.2135/cropsci2005.0028. [DOI] [Google Scholar]
- Rönicke S, Hahn V, Vogler A, Friedt W. Quantitative trait loci analysis of resistance to Sclerotinia sclerotiorum in sunflower. Phytopathology. 2005;95(7):834–839. doi: 10.1094/PHYTO-95-0834. [DOI] [PubMed] [Google Scholar]
- Sato S, Isobe S, Asamizu E, Ohmido N, Kataoka R, Nakamura Y, Kaneko T, Sakurai N, Okumura K, Klimenko I, Sasamoto S, Wada T, Watanabe A, Kohara M, Fujishiro T, Tabata S. Comprehensive structural analysis of the genome of red clover (Trifolium pratense L.) DNA Res. 2005;12:301–364. doi: 10.1093/dnares/dsi018. [DOI] [PubMed] [Google Scholar]
- Schneider KA, Grafton KF, Kelly JD. QTL analysis of resistance to fusarium root rot in bean. Crop Sci. 2001;41:535–542. [Google Scholar]
- Sim S, Diesburg K, Casler M, Jung G. Mapping and comparative analysis of QTL for crown rust resistance in an Italian x Perennial ryegrass population. Phytopathology. 2007;97(6):767–776. doi: 10.1094/PHYTO-97-6-0767. [DOI] [PubMed] [Google Scholar]
- Studer B, Boller B, Bauer E, Posselt UK, Widmer F, Kölliker R. Consistent detection of QTLs for crown rust resistance in Italian ryegrass (Lolium multiflorum Lam.) across environments and phenotyping methods. Theor Appl Genet. 2007;115(1):9–17. doi: 10.1007/s00122-007-0535-z. [DOI] [PubMed] [Google Scholar]
- Taylor NL. A century of clover breeding development in the United States. Crop Sci. 2008;48:1–13. doi: 10.2135/cropsci2007.08.0446. [DOI] [Google Scholar]
- Taylor NL, Quesenberry KH. Red clover science. Dordrecht: Kluwer Academic Publishers; 1996. [Google Scholar]
- Van Ooijen JW. MapQTL® 5, Software for the mapping of quantitative trait loci in experimental populations. Wageningen: Kyazma BV; 2004. [Google Scholar]
- Van Ooijen JW. JoinMAP® 4, Software for the calculation of genetic linkage maps in experimental populations. Wageningen: Kyazma BV; 2006. [Google Scholar]
- Vuong TD, Diers BW, Hartman GL. Identification of QTL for resistance to sclerotinia stem rot in soybean. Crop Sci. 2008;48:2209–2214. doi: 10.2135/cropsci2008.01.0019. [DOI] [Google Scholar]
- Yue B, Radi SA, Vick BA, Cai X, Tang S, Knapp SJ, Gulya TJ, Miller JF, Hu J. Identifying quantitative trait loci for resistance to Sclerotinia head rot in two USDA sunflower germplasms. Phytopathology. 2008;98(8):926–931. doi: 10.1094/PHYTO-98-8-0926. [DOI] [PubMed] [Google Scholar]
- Zhao J, Meng J. Genetic analysis of loci as sociated with partial resistance to Sclerotinia sclerotiorum in rapeseed (Brassica napus L.) Theor Appl Genet. 2003;106:759–764. doi: 10.1007/s00122-002-1171-2. [DOI] [PubMed] [Google Scholar]
- Zhao J, Udall JA, Quijada PA, Grau CR, Meng J, Osborn TC. Quantitative trait loci for resistance to Sclerotinia sclerotiorum and its association with a homeologous non-reciprocal transposition in Brassica napus L. Theor Appl Genet. 2006;112(3):509–516. doi: 10.1007/s00122-005-0154-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.