Abstract
Human papillomavirus (HPV) natural history has several characteristics that, at least from a statistical perspective, are not often encountered elsewhere in infectious disease and cancer research. There are, for example, multiple HPV types, and infection by each HPV type may be considered separate events. While concurrent infections are common, the prevalence, incidence, duration/persistence of each individual HPV can be separately measured. However, repeated measures involving the same subject tend to be correlated. The probability of detecting any given HPV type, for example, is greater among individuals who are currently positive for at least one other HPV type. Serial testing for HPV over time represents a second form of repeated measures. Statistical inferences that fail to take these correlations into account would be invalid. However, methods that do not use all the data would be inefficient. Marginal and mixed effects models can address these issues, but are not frequently utilized in HPV research. The current paper provides an overview of these methods, and then uses HPV data from a cohort of HIV-positive women to illustrate how they may be applied, and compare their results. The findings show the greater efficiency of these models compared with standard logistic regression and Cox models. Because mixed effects models estimate subject-specific associations, they sometimes gave much higher effect estimates than marginal models, which estimate population-averaged associations. Overall, the results demonstrate that marginal and mixed effects models are efficient for studying HPV natural history, but also highlight the importance of understanding how these models differ.
Keywords: statistical methods, cervical neoplasia, human papillomavirus, HPV, HIV, frailty models, mixed effects models, WLW models, frailty models
INTRODUCTION
Human papillomavirus (HPV), a sexually transmitted virus, is the central etiologic agent in the development of cervical cancer and precancerous cervical lesions(1). There are more than 40 types of HPV which commonly infect the cervical epithelium as well as other anogenital tissues. At least 13 of these anogenital HPV types are considered oncogenic(2). The incidence of HPV infection is very high, with 3-year cumulative incidence rates reported to be as much as 43% amongst sexually active college aged-women, and the oncogenic HPV types cause the majority of these infections(3). Most cervical HPV infections, however, including those with oncogenic HPV types, resolve spontaneously within two years(4). Only a minority of oncogenic HPV infections persists and leads to clinically significant cervical disease.
While much has been learned regarding the natural history of HPV, there is currently a growing focus on type-specific HPV associations rather than data grouped by broad categories (e.g., any oncogenic HPV type), which was common in the past. Randomized clinical trials, for example, are using the incident development of a persistent infection with a vaccine-related HPV type as an intermediate endpoint(5). Observational studies are also increasingly studying the natural history of HPV on a type-specific basis, whether to compare vaccinated versus unvaccinated women (in non-clinical trial settings), or to investigate other exposures while accounting for possible type-specific differences in effect estimates. The standard logistic regression and Cox model approaches commonly employed in HPV research up until now may not always be optimal for such analyses. More efficient statistical methods may need to be adopted.
The choice of statistical method(s) must address several aspects of HPV natural history that are not often encountered elsewhere in infectious disease and cancer research. For example, while co-infection by more than one HPV type is common(3,6,7,8,9), prior studies suggest that they infect separate cells, causing separate foci of infection(8). Therefore, the prevalence, incidence, clearance/persistence, progression, of each HPV type-specific infection can be examined as distinct events/outcomes. On the other hand, because of shared risk factors, these separate HPV infections may be correlated. The probability of detecting any given HPV type, for example, is greater among individuals who are currently positive for at least one other HPV type(3,6,7,8,9). Another type of correlation in HPV natural history data relates to repeated (serial) testing of the same women over time. If these two kinds of correlations are ignored, it can affect p-values and confidence intervals, leading to an incorrect statistical inference. On the other hand, statistical models that do not use all the available data (e.g., across all the visits and HPV types examined) may be inefficient.
The current paper provides an overview of selected statistical methods that can be used to address these issues. We then illustrate how these methods may be applied to HPV natural history data, and compare their results. The example dataset is from the Women’s Interagency HIV Study (WIHS), a large prospective cohort of HIV-infected and HIV-uninfected women. It is worth noting that these analyses involve more than twice the total person-visits of data available at the time of an earlier report regarding HPV type-specific infection in HIV-positive women based in the WIHS(8) and represent an update of those earlier findings.
MATERIAL AND METHODS
Subjects
Data were obtained from the Women’s Interagency HIV Study (WIHS) cohort. Details of this cohort have been previously reported(6,7,8). Briefly, between October, 1994 and November, 1995, 2,058 HIV-seropositive and 568 HIV-seronegative women older than 13 years were enrolled in the WIHS from similar clinical and outreach sources in Brooklyn/Manhattan/Bronx, NY; Chicago, IL; Los Angeles and San Francisco, CA; and Washington, DC. In 2002, an additional 738 HIV-seropositive and 406 HIV-seronegative women were similarly enrolled. On a semiannual basis in this ongoing cohort subjects undergo a structured interview and a physical examination that includes a gynecologic examination. Following a Pap smear, a cervicovaginal lavage (CVL) is collected for HPV DNA testing, as are blood samples. At the time of this analysis, there was a median follow-up of 14 visits among all subjects, with 9,475 persons-years of observation among HIV-positive women, and 2,708 person-years among HIV-negative women.
Laboratory Testing/Data
As described in detail elsewhere(6,7,8,9), HPV testing was conducted at every visit using an MY09/MY11/HMB01 polymerase chain reaction (PCR) assay. Amplification of a 268 bp cellular β-globin DNA fragment was included in each assay as an internal control. Following amplification, the presence of HPV DNA was assessed using filters hybridized with biotinylated oligonucleotide probes for specific HPV types, as well as a general probe mixture able to detect most anogenital HPV. For this analysis, oncogenic HPV types were defined as types 16/18/31/33/35/39/45/51/52/56/58/59/66.
Blood specimens were tested for CD4+ T-cell count and HIV RNA levels in laboratories participating in the AIDS Clinical Trials Quality Assurance Program. CD4+ count was categorized into three strata (i.e., >500, 200–500, and <200 CD4+ cells/mm3), and plasma HIV RNA level into four strata (i.e., <4000; 4000–20,000; 20,001–100,000; and >100,000 copies/mL); a total of 13 combined CD4+ / HIV RNA strata that were found to be relevant in prior studies(4,6), with HIV-negative women as the reference group.
OVERVIEW OF STATISTICAL METHODS
1. Analysis of HPV Prevalence
Standard Logistic Regression
It is common in HPV studies to use multivariable logistic regression to cross-sectionally analyze the factors associated with “any HPV”, “any oncogenic HPV type”, or an individual HPV type, at a given study visit. In such analyses, each woman contributes a single outcome. Notationally, let Yi=1 represent the detection of HPV16 infection for the ith person at the selected study visit, and 0 otherwise. We model Pi = P(Yi = 1) by
| (1.1) |
where Zi is our exposure variable of interest (e.g., a vector of indicators of 12 separate CD4+ / HIV RNA strata) and Wi is the vector of other adjusted variables, including age, race, lifetime of number of male sex, etc. for the ith person. The primary parameter of interest is β , defined as the log odds ratio (OR) of HPV16 infection between women in a certain CD4+ / HIV RNA stratum and HIV-negative women.
Repeated Observations Over Time
However, if HPV16 infection were assessed repeatedly over several visits it would not be appropriate to use model (1.1) to incorporate all the available data: model (1.1) would naively treat repeated observations of each individual over time as if they are independent (i.e., as if they were from different subjects), in essence amplifying the sample size. This can result in underestimation of the variation in the estimate of β and its P-value, as well as a confidence interval that is too narrow(10). To avoid these problems, a generalized estimating equation (GEE)(11) can be incorporated in the logistic regression model.
Generalization Estimating Equation (GEE) Models
Let Yij represent the HPV16 infection status (0/1) for the ith person at jth semiannual visit, we model Pij = P(Yij = 1) by
| (1.2) |
where Zijand Wijare defined similarly as in (1.1). The difference with standard logistic regression is in how the parameters are estimated. Specifically, GEE uses an estimating equation to estimate β in which a particular correlation structure (referred to as the working correlation) is assumed between repeated measures of Y. Several choices of working correlations are available in most statistical packages, including “independent” (suitable when a small correlation between data at different visits is assumed), and “exchangeable correlation” (suitable when the correlation between all pairs of observations can be assumed to be similar; e.g., visits 1 and 2 have the same correlation as visits 3 and 14), as well as other working correlations. Often we do not know how the repeated HPV outcomes are correlated with each other, but a useful feature of the GEE approach is that even if the working correlation is mis-specified, the estimate of β is still unbiased. Further, because a robust variance is used the confidence interval for β is correct for large sample size. The main benefit of properly specifying the correlation between repeated observations is that if the working correlation chosen is close to the actual one the model has greater efficiency(11,12). In practice, it is recommended that several working correlations be assessed. The one that produces the smallest standard error for the estimate of β is generally accepted to be the one that is most consistent to the true correlation. In many cases, though, the results will be similar across different correlation assumptions. Time-dependent covariates can be incorporated.
In model (1.2) β has the same interpretation as that in the logistic regression model (model(1.1)). Since β describes the average risk difference between two groups/populations, it is often referred to as the “population-averaged effect”.
Multiple HPV Types
GEE models, furthermore, can be used to incorporate data across multiple types of HPV. In a single GEE logistic regression model we can treat each of the several different oncogenic HPV types as separate endpoints, and then estimate the exposure effect on oncogenic HPV as a whole. We note that this common effect can also be examined with “any oncogenic HPV” as a single binary outcome. However, greater efficiency is obtained for grouped HPV data since each HPV type separately contributes data to the analysis, and the final result is a weighted average across HPV types. This source of efficiency is in addition to that discussed above, of being able to assess repeated visits over time.
We can also assess HPV type-specific relationships with the exposure variables using GEE models. Compared with conducting separate standard logistic regression models, GEE models still obtain greater efficiency since we reduce the number of covariate coefficients by assuming a common effect for “nuisance variables”. For example, only one “life-time number of sexual partners” coefficient is required if all individual oncogenic HPV are studied in the same GEE model, whereas the sexual partners variable would require a separate coefficient in each separate standard logistic regression for each HPV type. Further, GEE models allow us to statistically evaluate if the exposure-disease association is common across types.
To illustrate how GEE models can be used to model multiple types of HPV measured across multiple visits, let Yijk represent the infection of HPV type k for the ith person at jth semiannual visit. To assess an overall effect estimate across oncogenic HPV types, a type-specific baseline prevalence is assumed for each type of infection but a common odds ratio is assumed across the types, the infection rate of Yijk,Pijk , is modeled by
| (1.3) |
When sufficient events are available to study each HPV type individually, we can further allow the association with HIV & CD4+ to be different across HPV types. In this case, equation (1.3) becomes
| (1.4) |
In this model, the association with CD4+ / HIV RNA stratum is modeled by βk so that it can vary by HPV type k. Model (1.4) also allows us to test if βk’s are the same across HPV types.
Note that two types of correlations exist in the above GEE models: correlation between different types of HPV infection, and correlation between the same types of HPV infections at different visits. Standard GEE methods do not easily enable construction of different correlation structures for these two types of correlation(13,14): only one working correlation structure can usually be used in a single model. Based on prior experience(8), we recommend using either an independent or exchangeable working correlation as an approximation for both correlations.
ORs are a good approximation of relative risk when prevalence rates are low (e.g., less than 10%)(15), but when prevalence rates are high ORs overestimate relative risk. Therefore, an additional advantage of the GEE logistic regression approach proposed under model (1.3) is that because it estimates associations “across oncogenic HPV types” (the weighted average of the effects for individual types) the OR is likely to be a good estimate of relative risk; i.e., since the prevalence of oncogenic HPV on a type-specific basis is usually less than 10% even in high risk populations. In contrast, “any oncogenic HPV” as defined under model (1.1) is binary (detection of one or more oncogenic HPV types) which can involve high prevalence in high risk populations. When a situation arises in which the prevalence of HPV is substantially greater than 10% direct estimation of prevalence ratios may be preferred to the use of an OR. This can be accomplished in GEE (or mixed effects models; see below) using log link instead of the logit link used in logistic regression (i.e, ORs), but log link is more likely than logit link to have problems with non-convergence(16,17).
Note that there are a number of links available for studying binary HPV outcomes such as probit, complementary log-log link or identity links. Alternative links provide different measures of exposure–disease associations: for example, an identity link provides an estimate of the difference in prevalence (i.e., attributable risk). These other links can sometimes give different results than logit or log links. The decision regarding which link functions are correct depends on the underlying biologic relationship and usually can not be determined solely on a statistical basis. However, the logit link (standard logistic regression) is the most common link function used in HPV research.
All the above GEE models can be implemented using PROC GENMOD in SAS. Details to implementing model (1.4) are given in the appendix.
Mixed Effects Models
A mixed effects model(18,19) is an alternative approach for incorporating repeated HPV prevalence data across HPV types and across visits involving the same woman. A mixed effects model for repeated assessments of HPV16 infection can be expressed as follows:
| (1.5) |
which allows each subject to have her own baseline rate of infection modeled by µ + αi where µ is the population average baseline rate of infection and αi represents the ith subject’s departure from the population average and is modeled as a random effect. The random effect is unobserved and characterized by a distribution function (typically a normal distribution centered at 0 with variation σ2), while Z and W are observed covariates and referred to as fixed effects. Model (1.5) is therefore called a mixed effects model. The parameter δ has a different interpretation from β in GEE: it is the log OR of HPV 16 infection in a woman in a certain CD4+ / HIV RNA stratum compared with her risk if she were HIV-negative. Since δ describes the change in risk within a person were her risk factor level to change, it is often referred to as the “subject-specific effect”(20,21). In addition to δ and η, the variation in the random effect σ can also be estimated from the model. The variation in the random effect is a measure of the amount of heterogeneity (i.e., the level of intra-subject correlation) in the population, if σ =0, the model reduces to a fixed effects model. Under a normal assumption, the variation in the random effect can be interpreted as follow: 95% of the subjects in the population will not depart from the population mean μ by more than 1.96σ.
Because repeated observations of HPV16 infection share a common αi they are correlated. The simple mixed effects model (1.5) above specifically assumes an exchangeable correlation structure, analogous to that described for GEE (above); that is, it assumes that any pair of repeated observations of HPV within the same woman (e.g., at visits 1 and 5 versus at visits 3 and 14) has the same correlation. However, other correlation structures can be assumed in mixed effects models, and in general mixed effects models attempt to more accurately model the correlations rather than rely on the use of robust variance as in the GEE models. If the correlation is correctly specified, a mixed effects model is more efficient than a GEE model. However, if it is not correctly specified, the results can be biased(18).
To incorporate repeated observations across multiple HPV types in addition to repeated observations across multiple visits, a mixed effects model of Pijk (which assumes a common OR across the types) can be defined as
| (1.6) |
where the type-specific baseline prevalence rate represented by γ k, like αi, is assumed be a random effect. This model assumes that correlations among repeated infections of the same type are higher than correlations between different types of HPV, consistent with empiric observations. We can further expand model (1.6) to allow HPV type-specific odds ratios (details are not provided here). Mixed effects models for binary outcome are in general more computationally intensive than GEE and can sometimes have difficulty in convergence (i.e., the models can not be fit and no result is obtained) especially with small datasets.
The mixed effects model can be implemented using PROC NLMIXED in SAS
Comparison between GEE and Mixed Effects Models
Numerically, compared to β in the GEE model, δ in the mixed effects model is in general larger in magnitude(20), but because the standard error for the estimate of δ is also larger their significance levels (p-value) are similar. The interpretations of these estimators also differ, and mixed effects models may be of greater clinical relevance. For example, if a doctor wants to describe to an individual patient how much her risk of HPV16 infection will change if her CD4+ count changes, the subject-specific estimator δ more directly addresses this. The population-averaged estimator given by the GEE is of greater interest from a public health perspective because it describes on average how two groups of women with different CD4+ levels will differ in prevalence of HPV infection. For a more detailed discussion on the comparison of β and δ, see Hu(22).
Furthermore, on a practical basis, certain types of analyses that can be conducted using mixed effects models can not be conducted using GEE. For example, a mixed effects model but not GEE can be used to contrast rates of HPV infection before compared with after the initiation of highly active anti-retroviral therapy (HAART), using women as their own comparison group; an instance when it is the within-individual comparison that is of interest.
Power and Sample Size
Because GEE and mixed effects models make use of all available data they generally produce a more efficient estimate and have greater statistical power than standard logistic regression, which involves a single study visit and a single HPV endpoint. It is, however, often difficult to determine an exact power estimate for GEE and mixed effects models. Statistical simulations are sometimes necessary to estimate power for such studies. Here we discuss a simplified approach, which may be used as a first order approximation(23,24). To obtain a “ball park” estimate of power for correlated binary outcomes, we first assume an average correlation (ρ) among repeated observations within a subject. If on average each subject contributes J observations and there are a total of n subjects, then the “effective size” (i.e., the equivalent size if each subject only contributes one observation) is nJ/(1+(J-1)ρ). For example, if ρ=0.3 and there are J=5 multiple observations per subject and 100 subjects, the power based on these 100 subjects is equivalent to the power based on 100*5/(1+(5−1)*0.3)=227 subjects with only one observation per subject. Standard software packages for power/sample size calculation, for example, PASS(25), can then be used to calculation the power for the latter case (see Results).
2. Analysis of Incident Detection and/or Persistence of HPV
Standard Cox Models
A standard approach for analyzing time to first incident detection of HPV of a particular type, such as HPV16, is to use a Cox proportional hazards model. The Cox model is defined as follows:
| (1.7) |
where λ0(t) is the baseline hazard function and exp(β) is the hazard ratio of HPV16 infection between women in a certain CD4+ / HIV RNA stratum and HIV-negative women, while holding all other factors constant. Both the primary covariate Z and adjusting covariates W can be time-dependent, i.e., the values of Z and W are allowed to change during the follow-up.
However, model (1.7) can not simultaneously analyze time to first incident detection of several different types of HPV, because it does not address possible correlations between incident HPV infections. Instead, it is often more efficient to use methods that can simultaneously assess multiple types of HPV as separate endpoints in a single model. There are two main statistical methods appropriate for this purpose, as follows.
Wei, Lin Weissfeld (WLW) Method
Wei, Lin and Weissfeld(26) developed an approach that can be used to simultaneously analyze time to first incident detection of several types of HPV either in the same or different clinical visits, taking into account possible correlations between the types. While each HPV type is allowed to have its own baseline hazard function, a common (overall) exposure effect can be assumed in the following WLW model:
| (1.8) |
where λ0k(t) is the baseline hazard function for the kth type of event and exp(β) is the common hazard ratio across oncogenic HPV types. Similar to the parameter estimate in GEE, β has a population-averaged interpretation. Model (1.8) is in essence a Cox model stratified by HPV type and it is estimated under an independent working correlation, with a variance estimator that is robust to possible correlations between events.
Alternatively, if there are an adequate number of endpoints to study each HPV type separately, we can calculate type-specific hazard ratios using the following WLW model:
| (1.9) |
where exp(βk) is the hazard ratio for incident detection of HPV type k for subjects in a certain CD4+ / HIV RNA stratum relative to HIV-negatives.
Model (1.8) has greater power than a standard Cox model since in WLW women contribute multiple time to event endpoints (one for each HPV type) that are incorporated into the overall effect estimate. Greater efficiency is also obtained in the type-specific analyses using WLW in model (1.9), relative to using separate Cox regression models for each type, since we reduce the number of covariate coefficients by assuming a common effect (i.e., γ) for nuisance variables. Further, model (1.9) allows testing of equality in exposure effects across HPV types.
The WLW model can be implemented using the SAS PHREG procedure, selecting the STRATA option to allow different baseline hazards function for each HPV type. A robust variance is requested by choosing the option COVS(AGGREGATE).
Frailty Models
An alternative to the WLW method is to use frailty models to analyze multiple incident HPV infections(27). A frailty model introduces a frailty term, νi, to the Cox model as follows
| (1.10) |
Where νi, treated as a random effect, represents the ith woman’s unobserved individual susceptibility to develop an oncogenic HPV infection relative to the average level in the population. The coefficient δ is interpreted as the log of the hazard ratio for any incident oncogenic HPV detection in a woman at a certain CD4+ / HIV RNA stratum compared with her risk if she were HIV-negative while holding all other factors constant. Frailty models are a form of mixed effects model, and the parameter estimate δ has a subject-specific interpretation. Model (1.10) can also be extended to assess HPV type-specific associations (not shown).
By incorporating eθk in the model, frailty models assume that the baseline hazard functions are proportional to one another (i.e., the hazard functions have similar shape). If instead the baseline hazard function for one HPV type increases with age while the baseline hazard function for another decreases with age this assumption would be violated. WLW, in contrast, is more flexible, in that the baseline hazard function for each infection can be entirely different from each other. In addition, as with other mixed effects models, frailty models: (i) explicitly model the correlation between multiple events and, therefore, can be more efficient than WLW (a marginal model) if the correlation is correctly specified; but (ii) tend to be more computationally intensive than marginal models, and obtaining model convergence can be problematic, especially with small datasets.
Frailty models can be implemented using a SAS macro %gamfrail.1
Clearance / Persistence of HPV
HPV persistence can be measured as the time to clearance of an HPV type following its initial detection, with clearance defined as the first subsequent negative result (or preferably two sequential negative results) for that HPV type(6,8). The WLW and frailty models described above can be used to assess time to clearance of HPV on a type-specific basis(6,8).
Power and Sample Size
can be estimated for WLW and frailty models using the formula described for GEE and mixed effects models (i.e., nJ/(1+(J−1)ρ) where J is the number of HPV types)(28).
SAS programs for implementing some of the statistical models (above) as well as cautions needed to use these programs are provided on the web2.
RESULTS
WIHS women have been followed on a semiannual basis. Mid-interval (i.e., the midpoint calendar data between two consecutive visits) was used to estimate the time of each incident event. Women were censored for missed visits, and we assumed missing at random. Since our statistical models adjusted for CD4+ count and HIV viral load as well as other risk factors the analyses addressed the main factors that might cause informative censoring in the WIHS cohort. Incident detection of HPV incident detection of type-specific HPV was defined as a positive HPV DNA test result for a specific type in a participant who was negative for that HPV type in all earlier visits (prevalent infections are excluded).
Below we discussed the results for the prevalence data and incidence data.
1. HPV Prevalence
We assessed the prevalent detection of HPV16 and other oncogenic HPV types (across all visits) and their relationships with host immune status (i.e., CD4 / HIV RNA stratum, defined as a time-dependent covariate) using GEE. The prevalence of HPV16 infection varied by host immune status. For example, HPV16 prevalence (model(1.2)) was significantly greater among HIV-positive women with CD4+ count <200 cells/mm3 and HIV viral load >100,000 copies/mL compared with HIV-negative women (OR = 4.66 ,95% confidence interval [CI],2.60–8.35). However, the same effect estimate measured across oncogenic types (model (1.3)) was much stronger (OR = 10.82,95% CI,8.32–14.08) than that for HPV16 alone, suggesting that there may be type-specific differences in these associations.
GEE model (1.4) was, therefore, used to assess HPV type-specific differences in the effects of host immune status. HPV16 was the referent HPV type, and the ratio of the type-specific ORs was estimated. As shown in Table 1, each of the other oncogenic HPV types had stronger associations with host immune status than did HPV16. For example, the OR for HPV31 was 2.62 fold greater (95% CI,1.02–6.71)) and the OR for HPV45 was 3.49 fold greater (95% CI,1.39–8.80) than the OR for HPV16.
Table 1.
The weak association between the prevalence of HPV16 and host immune status relative to that for other oncogenic HPV types.
| HPV Types | Ratio of the OR (95% CI)* |
|---|---|
| HPV16(reference) | 1.00 |
| HPV 18 | 2.05 (0.97–4.34) |
| HPV 31 | 2.62 (1.02–6.71) |
| HPV 33 | 2.97 (1.09–8.13) |
| HPV 35 | 2.66 (1.08–6.55) |
| HPV 39 | 5.24 (1.39–19.7) |
| HPV 45 | 3.49 (1.39–8.80) |
| HPV 51 | 2.56 (1.06–6.19) |
| HPV 52 | 3.03 (1.35–6.80) |
| HPV 56 | 2.55 (1.14–5.72) |
| HPV 58 | 2.70 (1.09–6.68) |
| HPV 59 | 4.42 (1.72–11.4) |
| HPV 66 | 5.85 (2.55–13.5) |
The ratio of odds ratios was estimated using a GEE logistic regression model (model (1.4)) examining the associations of host immune status with the prevalent detection of HPV by type, with HPV16 as the reference type. For convenience, the table shows only shows the contrast between HIV-positive women with CD4+ count <200 cells/mm3 and HIV viral load >100,000 copies/mL versus HIV-negative women. The adjustment variables included age at visit (<30, 30–34, 35–39, 40–44, >=45), race (White, Black, Hispanic, and other), sexual partners in the past 6 months (none, 1(married), 1(single), 2 and >2) and smoking status (none, former smoker, current smoker<10 packs per year and current smoker ≥10pk per year).
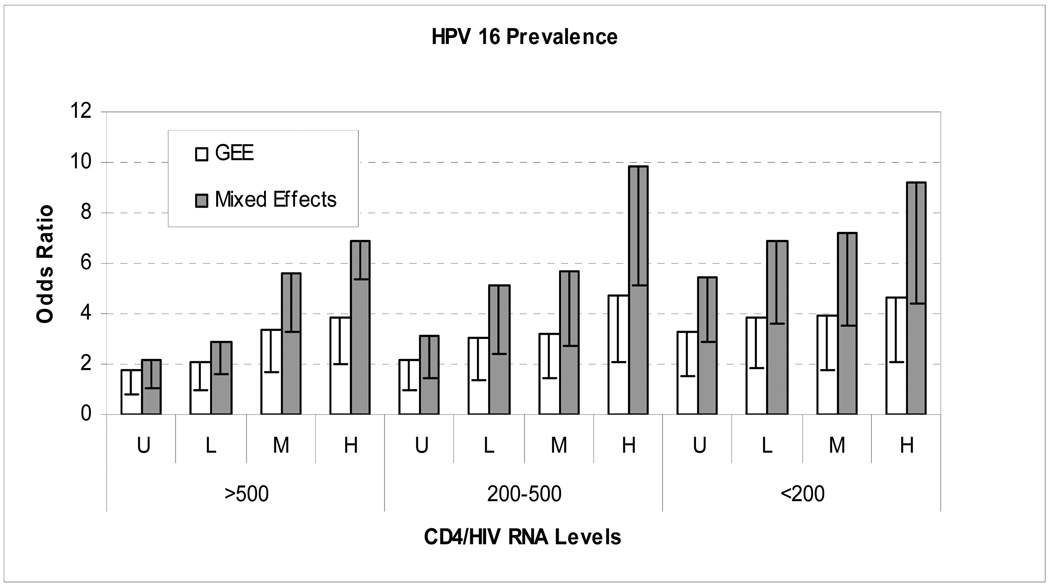
We then compared these GEE results with those obtained using a mixed effects model (model(1.5)). As expected, the mixed effects model gave much higher estimates of association, with wider confidence intervals than did GEE (Figure 1), but similar P-values (P<0.001). For example, the OR (i.e., eδ̄ in model (1.5)) of HPV16 detection associated with having a CD4+ count <200 cells/mm3 and HIV viral load >100,000 copies/mL (the most immunosupressed stratum) relative to being HIV-negative was 9.22 (95% CI,4.84–17.6). This estimate indicates that if an individual HIV-negative woman were to become HIV-positive and her immune status dropped to the most immunosupressed stratum, her risk of having HPV 16 infection would increase approximately 9-fold. In contrast, the OR (i.e., eβ̄ in model(1.3)) from the above GEE model would be interpreted as that on average an HIV-positive woman in the most immunosuppressed stratum has a 4-fold greater risk of HPV16 than an HIV-negative woman. A common limitation of mixed effects models was also revealed. Specifically, mixed effects models to assess HPV type-specific associations with host immune status failed to converge, consistent with the greater data requirement to run these models compared with GEE.
Figure 1.
A comparison of the odds ratios (ORs) calculated using mixed effects models (model (1.5))versus logistic regression models that incorporated a generalized estimating equation (model (1.2)). The specific example given is the relationship between HPV16 prevalence and 12 separate CD4+ / HIV RNA strata, with HIV-negative women used as the reference group. The vertical bars represent the ORs, and the line within each bar represents the lower 95% confidence limit of those ORs. The CD4+ / HIV RNA strata are defined as the follows. Reference Group = HIV negative women. CD4+ count: >500, 200–500, <200 cells/mm3. HIV RNA level: Undetectable (U)<4000, Low (L)=4000–20000, Moderate (M)=20001–100000 and High (H)>100000 copies/mL. Both models adjusted for age (<30, 30–34, 35–39, 40–44, >=45), race (White, Black, Hispanic, and other), sexual partners in the past 6 months (none, 1(married), 1(single), 2 and >2) and smoking status (none, former smoker, current smoker<10 packs per year and current smoker ≥10pk per year) and incorporated each individual HPV type as a separate indicator variable. The results show that mixed effects models provide much higher OR estimates than GEE (a form of marginal models) using the same dataset.
Statistical Power
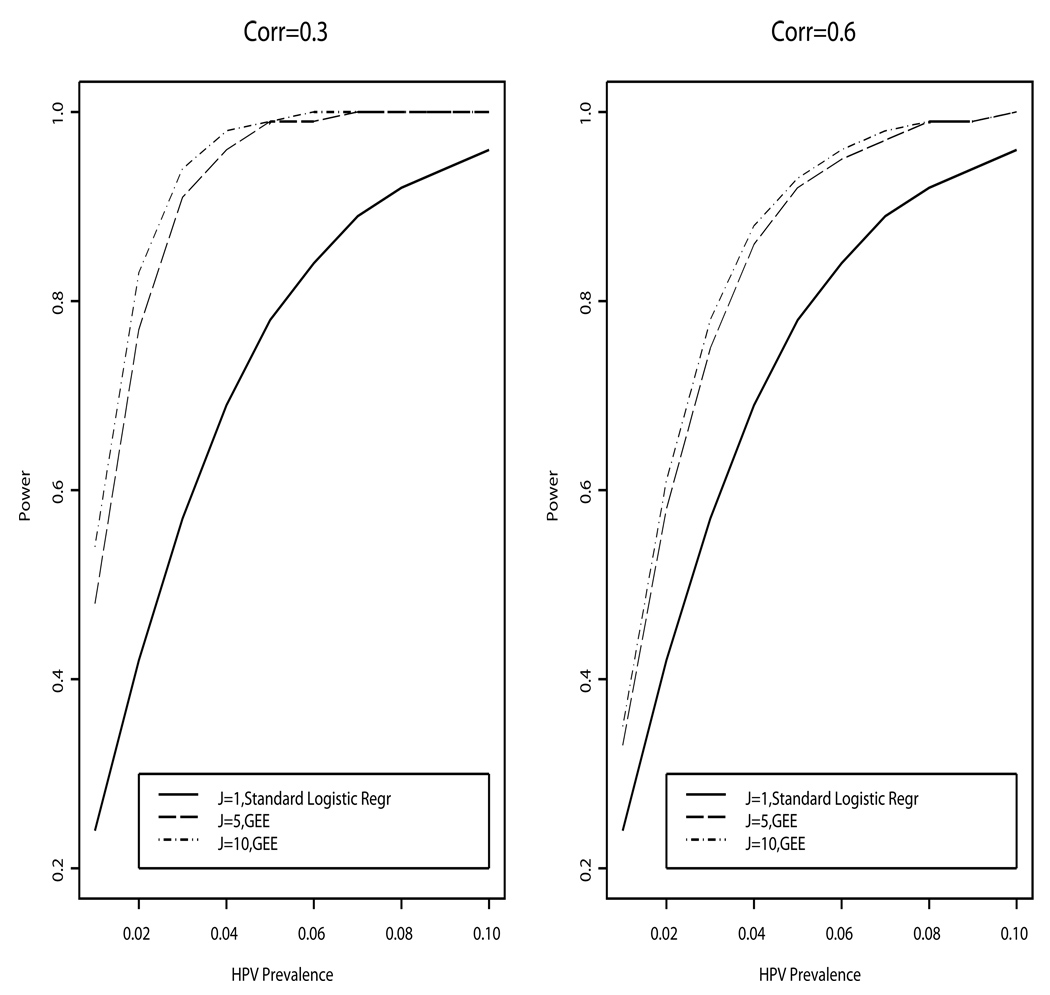
We use data from Strickler et al(8) to illustrate the gain in power with GEE compared with standard logistic regression. Based on 535 women with a CD4+ count <200 cells/mm3 and 458 women with ≥500 cells/mm3, Figure 2 shows the power to detect OR=2.0 for the association of HPV prevalence and CD4+ count with (i) only one observation (e.g., one visit with one HPV type), J=1 (i.e., standard logistic regression), versus (ii) J=5 or =10 (based on GEE) per subject under various assumptions regarding the prevalence rate of HPV infection. In our example, the correlation between repeated observations across HPV types and visits were assumed to be either 0.3 (moderate) or 0.6 (high). As demonstrated in Figure 2, the power when J=5 is substantially larger than the power without repeated observations (J=1). The power when J=10 is only slightly higher than the power with J=5, indicating that while there is a benefit to statistical power with a certain number of repeated observations, there is a diminishing impact on power as the number of repeats increases. Further, the increase in power is greater when the correlation is lower; e.g., ρ=0.3 versus ρ=0.6. This is as expected since the level of correlation is inversely related to effective sample size. Intuitively, when the correlation between repeated observations is low having additional repeats should provide substantial new information, whereas having a high correlation implies that the repeated observations are not greatly independent of one another and therefore the repeats provide limited additional information. If the correlation is unknown a conservative estimate should be used to calculate power/sample size.
Figure 2.
The power to detect an odds ratio of 2.0 for the association between prevalent HPV detection and host immune status, comparing HIV-positive women with CD4+ count <200 (n=535) versus >500 cells/mm3 (n=458) under various conditions: (i) using data from only a single observation (J=1; equivalent to standard logistic regression), (ii) using data from 5 or 10 repeated study visits per observations (J=5, J=10), and (iii) assuming the correlation between repeated observations is moderate (corr=0.3) or very high (corr=0.6). The figure shows greater statistical power for models that incorporate a generalized estimating equation (GEE), and use data from multiple observations, compared with standard logistic regression models, which can only use data from a single observation.)
2. Incident HPV Detection
A WLW model of incident oncogenic HPV detection by type (Model(1.8)) showed that women with a CD4+ count <200 cells/mm3 and HIV RNA > 100,000 copies/mL had significantly increased risk of incident oncogenic HPV detection (HR = eβ̄ = 4.9; 95% CI,3.8–6.3) relative to HIV-negative women (Table 2). Further analysis involving measurement of type-specific associations (Model(1.9)), using HPV16 as the referent HPV type, showed that most oncogenic HPV types, with the exception of HPV18 and HPV56, had stronger associations with host immune status than HPV16 (Table 3). While on an individual basis these two-fold or greater differences did not reach significance, the power to study incident HPV detection is less than to study HPV prevalence; in our WLW model of incident HPV detection each person contributed one time-to-event result per HPV type, whereas in our GEE logistic regression model on HPV prevalence the dataset involved repeated observations of each HPV type at each visit. That said, when assessed as a group, the incident detection of all other oncogenic HPV types excluding HPV18 and HPV56 (model (1.9)) had a three-fold (HR = eβ̄otheronc / eβ̂16 = 3.1) greater (95% CI,1.4–7.2) association with CD4+ count than HPV16.
Table 2.
Hazard ratios for the incident detection of oncogenic HPV infection as estimated using a Wei Lin Weissfeld (WLW) Model*
| CD4 T-cell count, cells per mm3 | HIV RNA, copies per mL | Hazard Ratio (95% CI) |
|---|---|---|
| >500 | <4000 | 1.61(1.23,2.10) |
| 4001–20000 | 2.03(1.39,2.97) | |
| 20001–100000 | 3.06(1.96,4.76) | |
| >100000 | 3.40(1.82,6.32) | |
| 200–500 | <4000 | 2.58(2.02,3.30) |
| 4001–20000 | 3.15(2.37,4.18) | |
| 20001–100000 | 4.02(3.02,5.35) | |
| >100000 | 4.77(3.47,6.56) | |
| <200 | <4000 | 4.55(3.33,6.21) |
| 4001–20000 | 5.10(3.70,7.03) | |
| 20001–100000 | 4.55(3.38,6.12) | |
| >100000 | 4.87(3.75,6.34) |
Table 3.
The weak association between the incident detection of HPV16 and host immune status relative to that for other oncogenic HPV types.
| HPV Types | Ratio of HRs (95% CI) |
|---|---|
| HPV16 (reference) | 1.00 |
| HPV 18 | 1.12 (0.37–3.39) |
| HPV 31 | 2.02 (0.68–6.03) |
| HPV 33 | 5.28 (1.53–18.2) |
| HPV 35 | 1.71 (0.43–6.72) |
| HPV 39 | 3.65 (0.84–15.9) |
| HPV 45 | 2.05 (0.63–6.70) |
| HPV 51 | 2.15 (0.72–6.41) |
| HPV 52 | 5.54 (1.81–17.0) |
| HPV 56 | 0.97 (0.32–2.93) |
| HPV 58 | 2.42 (0.82–7.11) |
| HPV 59 | 3.69 (1.12–12.1) |
| HPV 66 | 6.29 (2.07–19.1) |
The ratio of hazard ratios (HR) was estimated using a WLW model (model (1.9)) examining the associations of host immune status with the incident detection of HPV by type. The adjustment variables included all those shown in Figure 2. For convenience, the table shows only shows the contrast between HIV-positive women with CD4+ count <200 cells/mm3 and HIV viral load >100,000 copies/mL versus HIV-negative women. Although most of the oncogenic types did not on an individual basis have a significantly different HR than that for HPV16, all other oncogenic types when assessed as a group (excluding HPV 18 and 56) did have a significantly higher HR than HPV16 (P-value=0.008). The findings were also significant if we included HPV 18 and 56 in the analysis.
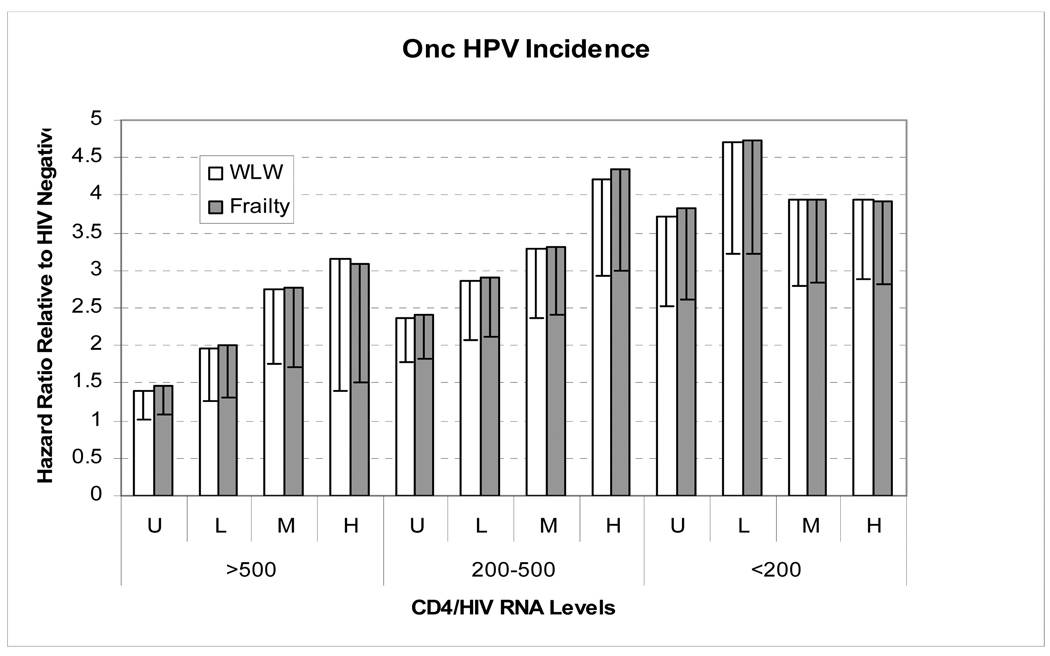
Lastly, we compared WLW and frailty model (model (1.10)) results. Figure 3 shows the findings of an analysis involving the incident detection of 7 common oncogenic HPV, namely, HPV16/18/31/33/35/45/58 selected since the frailty model had problem converging when less common HPV types were analyzed (most likely because of insufficient occurrences to define their baseline hazards). Interestingly, unlike the comparison of mixed effects and GEE models, effect estimates obtained using the frailty model were only slightly larger than those obtained using WLW. Frailty models to assess type-specific associations failed to converge.
Figure 3.
A comparison of the hazard ratios (HRs) calculated using frailty models (model(1.10)) versus Wei Lin Weisfeld (WLW) Cox models (model (1.8)). The specific example given is the relation between the incident detection of seven common oncogenic HPV (i.e., 16/18/31/33/35/45/58) and 12 separate CD4+ / HIV RNA strata, with HIV-negative women used as the reference group. The vertical bars represent the HRs, and the line within each bar represents the lower 95% confidence limit of those HRs. The CD4+ / HIV RNA strata are defined as the follows. Reference Group = HIV negative women. CD4+ count: >500, 200–500, <200 cells/mm3. HIV RNA level: Undetectable (U)<4000, Low (L)=4000–20000, Moderate (M)=20001–100000 and High (H)>100000 copies/mL. The adjustment variables are the same as reported in Figure 1. Both the frailty and WLW models incorporated HPV type-specific baseline hazard functions while assuming a common association with CD4+ count / HIV RNA stratum. Overall, the results show that frailty models (a form of mixed effects models) provide similar HR estimates as WLW (a form of marginal models) using the same HPV dataset. As discussed in the text, the close agreement between WLW and frailty models in our data may reflect small correlation between the incident detections of different types of HPV, whereas the large differences observed between the GEE and mixed effects model estimates (Fig 1) likely reflects strong correlations between the cross-sectional detection of the same HPV types over serial semi-annual visits (i.e., the high frequency of persistent and recurrent infections).
DISCUSSION
This paper shows that marginal and mixed effects models are appropriate, efficient methods for the analysis of HPV natural history data. While it has been common to use standard logistic regression for the analysis of the prevalent (cross-sectional) detection of HPV, and standard Cox models for time to incident detection (or clearance) of HPV, these common approaches do not make use of all the available data. That is, standard logistic and Cox regression models can only consider a single HPV type, and logistic regression can also only consider a single visit or a single time period. Marginal and mixed effects models, in contrast, can simultaneously address multiple oncogenic HPV types as well as multiple visits over time. As shown in paper, they consequently have greater statistical power in the analysis of HPV natural history data. In addition, these models allow comparison of exposure-disease associations between HPV types. Thus, marginal and mixed effects models could have many useful applications to HPV research, both in randomized clinical trials and epidemiologic studies.
There are, however, important differences in the estimates obtained from marginal and mixed effects models. Our paper examined the major current forms of these models relevant to the analysis of HPV data. The marginal models were GEE for the analysis of HPV prevalence, and WLW for the incident detection or clearance of HPV. Marginal models estimate the average relative difference in risk between two groups of subjects with different levels of risk factor exposure. This is often referred to as the “population-averaged effect”. The mixed effects models examined were frailty models for analyzing the incident detection (or clearance) of HPV, and mixed effects models for analyzing HPV prevalence. Mixed effects models estimate the change in risk that would occur within a person were risk factor exposure to change. This is often referred to as the “subject-specific effect”.
The effect estimates obtained using mixed effects models tend to be considerably larger than those obtained using marginal models but, since the standard errors in mixed effect models are also larger, the significance levels obtained are often similar in both types of models. Interestingly, in our dataset we found that the results of GEE and mixed effects models had these expected differences, but the WLW and frailty models provided very similar effect estimates. Close agreement between WLW and frailty models in our data may reflect small correlation between the incident detections of different types of HPV, whereas the large differences observed between the GEE and mixed effects model estimates likely reflects strong correlations between the cross-sectional detection of the same HPV types over serial semi-annual visits (i.e., the high frequency of persistent and recurrent infections).
As an important practical matter, though, mixed effects models are much more computationally intensive than the marginal models and are more likely to have problems with convergence; i.e., mixed effects models require more extensive data. For this reason, marginal models may be seen as having an advantage over mixed effects models in many clinical trials and epidemiologic studies, or whenever data are limited. As mentioned, though, certain types of analysis require mixed effects models. In general, we recommend a biostatistician be involved when implementing and interpreting results from mixed effects models.
While not our main focus, this paper also served to further demonstrate that the prevalent and incident detection of HPV16 (the HPV type that accounts for half of all cervical cancers) is more weakly associated with host immune status than other oncogenic HPV types, as we and others have previously reported(8). The relative independence of HPV16 infection from host immune status has been interpreted as evidence that HPV16 may be better able to avoid the effects of immune surveillance than other HPV types, and that this ability might help partly explain the predominance of HPV16 in cervical disease. The current study represents an update of the prior data, involving more than double the person-years of observation available at the time of our earlier report, and approximately a third of the women were new to the analysis.
In summary, while the statistical methods discussed in this paper are well established and highly efficient, their application to HPV research has been limited(29). To our knowledge, this is the first formal discussion of these methods in the context of HPV natural history data. However, even if the greater use of these methods were to represent an advance, the analysis of the natural history of HPV will continue to be a challenge. It is hoped, therefore, that this paper will not only spur interest in current marginal and mixed effects models, but will additionally interest biostatisticians in the development of new and better methods for the analysis of HPV natural history data.
Acknowledgments
Financial support: HPV DNA testing is funded through R01-CA-085178. All specimens and other data in this study were collected by the Women’s Interagency HIV Study (WIHS) Collaborative Study Group with centers (principal investigators) at New York City/Bronx Consortium (Kathryn Anastos, MD); Brooklyn, NY (Howard Minkoff, MD); Washington, DC Metropolitan Consortium (Mary Young, MD); The Connie Wofsy Study Consortium of Northern California (Ruth Greenblatt, MD); Los Angeles County/Southern California Consortium (Alexandra Levine, MD); Chicago Consortium (Mardge Cohen, MD); Data Coordinating Center (Stephen Gange, PhD). The WIHS is funded by the National Institute of Allergy and Infectious Diseases with supplemental funding from the National Cancer Institute and the National Institute on Drug Abuse (U01-AI-35004, U01-AI-31834, U01-AI-34994, U01-AI-34989, U01-AI-34993, and U01-AI-42590). Funding is also provided by the National Institute of Child Health and Human Development (U01-HD-32632) and the National Center for Research Resources (M01-RR-00071, M01-RR-00079, M01-RR-00083).
Appendix
Model (1.4)
is implemented by including indicator variables (Type ijk for k=1, …, K) for each of the other oncogenic HPV types (with HPV16 as a reference) and their interactions with CD4+ / HIV RNA stratum in the model. The model becomes
so that β represents the relationship of HPV16 detection with host immune status and φk represents the difference between HPV type k and HPV16 in relation to host immune status, i.e., the log ratio of the ORs. Type-specific analyses for other models (mixed effects, WLW, frailty) (such as models (1.9)) are implemented using the same approach.
Footnotes
Potential conflicts of interest: The authors do not have a commercial or other association that might pose a conflict of interest.
Available at http://www.biostat.mcw.edu/software/SoftMenu.html
Reference
- 1.IARC. IARC Monographs on the Evaluation of Carcinogenic Risks to Human Papillomaviruses. vol. 90. Lyon: IARC; 2007. [PMC free article] [PubMed] [Google Scholar]
- 2.IARC. IARC Monographs on Carcinogenicity of Human Papillomaviruses. vol. 6. Lyon: IARC; 2005. [Google Scholar]
- 3.Ho GY, Bierman R, Beardsley L, Chang CJ, Burk RD. Natural history of cervicovaginal papillomavirus infection in young women. N Engl J Med. 1998;338(7):423–428. doi: 10.1056/NEJM199802123380703. [DOI] [PubMed] [Google Scholar]
- 4.Rodríguez AC, Schiffman M, Herrero R, et al. Rapid clearance of human papillomavirus and implications for clinical focus on persistent infections. J Natl Cancer Inst. 2008;100(7):513–517. doi: 10.1093/jnci/djn044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jenkins D. A review of cross-protection against oncogenic HPV by an HPV-16/18 AS04-adjuvanted cervical cancer vaccine: importance of virological and clinical endpoints and implications for mass vaccination in cervical cancer prevention. Gynecol Oncol. 2008;110 supp:S18–S25. doi: 10.1016/j.ygyno.2008.06.027. [DOI] [PubMed] [Google Scholar]
- 6.Strickler HD, Burk RD, Fazzari M, et al. HPV Natural History and Possible HPV Reactivation in HIV-Positive Women. J Natl Cancer Inst. 2005;97:577–586. doi: 10.1093/jnci/dji073. [DOI] [PubMed] [Google Scholar]
- 7.Barkan SE, Melnick SL, Preston-Martin S, et al. WIHS Collaborative Study Group. The Women’s Interagency HIV Study. Epidemiology. 1998;9:117–125. [PubMed] [Google Scholar]
- 8.Strickler HD, Palefsky JM, Shah KV, et al. Human papillomavirus 16 and immune status in human immunodeficiency virus-seropositive women. J Natl Cancer Inst. 2003;95:1062–1071. doi: 10.1093/jnci/95.14.1062. [DOI] [PubMed] [Google Scholar]
- 9.Palefsky JM, Minkoff H, Kalish LA, et al. Cervicovaginal human papillomavirus infection in human immunodeficiency virus-1 (HIV)-positive and high-risk HIV-negative women. J Natl Cancer Inst. 1999;91(3):226–236. doi: 10.1093/jnci/91.3.226. [DOI] [PubMed] [Google Scholar]
- 10.Diggle PJ, Heagerty P, Liang KY, Zeger SL. Analysis of Longitudinal Data. second edition. Oxford: Oxford University Press; 2002. [Google Scholar]
- 11.Liang KY, Zeger SL. Longitudinal Data Analysis Using Generalized Linear Models. Biometrika. 1986;73:13–22. [Google Scholar]
- 12.Wang YG, Carey VJ. Working correlation misspecification, estimation and covariate design: implications for generalized estimating equation performance. Biometrika. 2003;90:29–41. [Google Scholar]
- 13.Galecki AT. General class of covariance structures for two or more repeated factors in longitudinal data analysis. Communications in Statistics-Theory and Methods. 1994;23:3105–3120. [Google Scholar]
- 14.Shults J, Whitt MC, Kumanyika S. Analysis of data with multiple sources of correlation in the framework of generalize estimating equations. Statistics in Medicine. 2004;23(20):3209–3226. doi: 10.1002/sim.1887. [DOI] [PubMed] [Google Scholar]
- 15.Zhang J, Yu KF. What’s the relative risk? A method of correcting the odds ratio in cohort studies of common outcomes. JAMA. 1998;280(19):1690–1691. doi: 10.1001/jama.280.19.1690. [DOI] [PubMed] [Google Scholar]
- 16.Wacholder S. Binomial regression in GLIM: estimating risk ratios and risk differences. Am J Epidemiol. 1986;123:174–184. doi: 10.1093/oxfordjournals.aje.a114212. [DOI] [PubMed] [Google Scholar]
- 17.McNutt LA, Wu C, Xue X, Hafner JP. Estimating the relative risk in cohort studies and clinical trials of common outcomes. Am J Epidemiol. 2003;157:940–943. doi: 10.1093/aje/kwg074. [DOI] [PubMed] [Google Scholar]
- 18.Goldstein H. Multilevel Statistical Models. London: Halstead Press; 1995. [Google Scholar]
- 19.Fitzmaurice GM, Laird NM, Ware JH. Applied Longitudinal Analysis. 1st ed. John Wiley and Sons; 2004. [Google Scholar]
- 20.Neuhaus JM, Kalbfleisch JD, Hauck WW. A Comparison of Cluster-Specific and Population-Averaged Approaches for Analyzing Correlated Binary Data. International Statistical Review / Revue Internationale de Statistique. 1991;vol. 59(1):25–35. [Google Scholar]
- 21.Pendergast JF, Gange SJ, Newton MA, Lindstrom MJ, Palta M, Fisher MR. A Survey of Methods for Analyzing Clustered Binary Response Data. International Statistical Review / Revue Internationale de Statistique. 1996;vol. 64(1):89–118. [Google Scholar]
- 22.Hu FB, Goldberg J, Hedeker D, Flay BR, Pentz MA. Comparison of populaton-averaged and subject-specific approaches for analyzing repeated binary outcomes. Am J Epidemiol. 1998;147:694–703. doi: 10.1093/oxfordjournals.aje.a009511. [DOI] [PubMed] [Google Scholar]
- 23.Pan W. Sample size and power calculations with correlated binary data. Controlled Clinical Trials. 2001;22:211–227. doi: 10.1016/s0197-2456(01)00131-3. [DOI] [PubMed] [Google Scholar]
- 24.Xue X, Hoover D. Statistical Methods in Cancer Epidemiologic Studies. In: Verma M, editor. Cancer Epidemiology. vol. 1. New York: Humana Press; 2009. [Google Scholar]
- 25.Hintze JL. PASS - Power and Sample Size Software East Kaysville Utah. 2001 [Google Scholar]
- 26.Wei LJ, Lin DY, Weissfeld L. Regression Analysis of Multivariate Incomplete Failure Time Data by Modeling Marginal Distribution. Journal of the American Statistical Association. 1989;84:1065–1073. [Google Scholar]
- 27.Hougaard P. Analysis of Multivariate Survival Data. New York: Springer; 2000. [Google Scholar]
- 28.Xie T, Waksman J. Design and sample size estimation in clinical trials with clustered survival times as the primary endpoint. Statistics in Medicine. 2003;133:2835–2846. doi: 10.1002/sim.1536. [DOI] [PubMed] [Google Scholar]
- 29.Duffy SW, Rohan TE, McLaughlin JR. Design and analysis considerations in a cohort study involving repeated measurements of both exposure and outcome: the association between genital papillomavirus and risk of cervical intraepithelial neoplasia. Statistics in Medicine. 1994;13:379–390. doi: 10.1002/sim.4780130405. [DOI] [PubMed] [Google Scholar]