Abstract
We describe a high-throughput scanning optical microscope for detecting small-molecule compound microarrays on functionalized glass slides. It is based on measurements of oblique-incidence reflectivity difference and employs a combination of a y-scan galvometer mirror and an x-scan translation stage with an effective field of view of 2 cm×4 cm. Such a field of view can accommodate a printed small-molecule compound microarray with as many as 10,000 to 20,000 targets. The scanning microscope is capable of measuring kinetics as well as endpoints of protein-ligand reactions simultaneously. We present the experimental results on solution-phase protein reactions with small-molecule compound microarrays synthesized from one-bead, one-compound combinatorial chemistry and immobilized on a streptavidin-functionalized glass slide.
Keywords: microarrays, high-throughput, label-free detection, in situ detection, oblique-incidence reflectivity difference, immobilization strategies
Introduction
Microarrays of surface-bound macromolecules and small molecule compounds are powerful tools for highly parallel in vitro analysis of biochemical binding affinity.1, 2 They are most useful in study of genomics, proteomics, glycomics, and even cytomics, as the variety of structures and functions of proteins, glycans, cells, and their enabling∕inhibiting ligands are large. Small-molecule compound microarrays on functionalized solid supports have been used successfully to characterize protein-ligand interaction in a high-throughput manner by exposing the solution-phase protein of interest to the microarray and reading out the endpoints of the reaction simultaneously.3, 4, 5 A notable challenge in the development and application of small-molecule compound microarrays is finding robust methods for attaching small molecules with diverse structures (and in turn, functionalities) to a specifically functionalized solid surface. Detection of a solution-phase molecular probe such as proteins to small-molecule microarrays is another major challenge, particularly when cost, versatility, and effect of detection method on probe-target interaction call for concerns.6, 7, 8 The fluorescence-based detection of microarrays is widely used for superior sensitivity and maturity. However, fluorescence labeling a protein probe inevitably changes the innate properties of the protein in often unknown ways. High cost, variation in labeling efficiency, photobleaching, and extra steps in assay preparation are other undesirable attributes of fluorescence-based detection. As a result, label-free optical detection methods with suitable sensitivity complement the fluorescence-based detection.9, 10
In this paper, we describe (1) an oblique-incidence reflectivity difference (OI-RD) scanning microscope capable of high-throughput, label-free detection of small-molecule microarrays with as many as 10,000 to 20,000 immobilized targets, which has the potential to measure over 100,000 protein-ligand binding reactions in one day; (2) a set of experimental measurements of endpoints and binding curves of protein reactions with biotin-conjugated small-molecule compounds immobilized on a streptavidin-functionalized glass surface; and (3) one-bead, one-compound (OBOC) combinatorial synthesis and OI-RD microscopy detection of a large synthetic small-molecule compound library immobilized through a common “handle” (biotin) on a streptavidin-functionalized glass surface.
High-Throughput Hybrid Scanning Optical Microscope
Based on measuring oblique-incidence reflectivity difference (OI-RD), we developed a hybrid scanning optical microscope capable of detecting over 10,000 molecular targets printed on a functionalized glass slide without fluorescence labeling. The microscope can be used to measure binding curves as well as reaction endpoints of a solution-phase probe to all or a selected set of the printed targets. Compared to imaging surface plasmon resonance (SPR) scanners,10, 11, 12, 13 this microscope does not rely on gold-coated substrates for detection and has a large field of view (presently 2 cm×4 cm) and thus offers a much higher throughput than an SPR sensor. Compared to an imaging ellipsometer based on a polarizer-compensator-sample-analyzer (PCSA) scheme,14, 15, 16 this OI-RD scanning microscope is inherently more sensitive to a surface-bound change (e.g., thickness, density, etc.) by more than one order of magnitude.17, 18, 19
The optical arrangement of a hybrid scanning OI-RD microscope is sketched in Fig. 1. It is a dual-axis mechanically scanned microscope. A linear translation stage holds an assembly of a microarray-bearing glass slide and a fluidic chamber and moves along the x axis for the x-scan. A combination of a galvanometer y-scan mirror and an f-theta lens provides the y-scan. The f-theta lens focuses a monochromatic illumination beam to a small spot on a microarray-covered glass surface at an oblique incidence angle. The binding reaction of a solution-phase probe with the microarray changes the thickness and mass density in the printed target region, and in turn alter the ratio of complex reflection coefficients for p-polarized and s-polarized components of the illumination beam, rp∕rs=tan ψ×exp(iδ).14 In the absence of absorption by the targets, the solution-phase probe, and the glass slide, the binding reaction mainly alters δ. An OI-RD microscope directly measures the change in rp∕rs as follows.17
Figure 1.
(a) Top view of a hybrid scanning OI-RD microscope with a combination of a galvometer y-scan mirror (RM) and an x-scan by a linear translation stage. The microarray-bearing glass slide is assembled with a fluid chamber so that the microarray is in contact with an aqueous solution. (b) Side view of the scanning microscope that illustrates the y-scan. PEM: photoelastic modulator at modulation frequency Ω=50 kHz. PS: phase shifter. FM: fixed mirror. OBJ: objective lens. A: analyzer. PD: photodiode detector.
As illustrated in Fig. 1a, a linearly polarized continuous-wave Nd:YAG laser beam at λ=532 nm passes through a photoelastic modulator (PEM) so that the polarization of the outgoing beam changes from p- to s-polarization at a frequency Ω=50 kHz. The beam passes through a phase shifter (PS) that adds a variable but static phase ΦPS between the p- and s-polarized components. Using a combination of a galvometer y-scan mirror (RM) and an f-theta lens (FTL), we focus the beam into a 10-μm spot on the microarray-bearing glass surface at incident angle θ=34.66 deg inside the glass slide (with optical dielectric constant ε0.) The microarray-bearing surface is in contact with an aqueous solution (with optical dielectric constant εs.) The reflected beam from the illuminated spot passes through an analyzer (A) with its transmission axis set at θA (45 deg in the present study) from s-polarization and is imaged with an objective lens onto a single-element photodiode (PD). The first-harmonic (in modulation frequency Ω=50 kHz) of the detected light intensity is given by I(Ω)=Iinc|rprs|sin(2θA)sin(ηsys+δ+ΦPS).ηsys is the phase difference between the p- and s-polarized components introduced by optical elements in the beam path other than the microarray-bearing surface; δ is the phase difference due to reflection from the surface. Let δ0 be the phase difference for a bare glass slide surface. We initially adjust ΦPS so that ηsys+δ0+ΦPS=0 and thus I(Ω)=0 on the bare glass surface (nulling ellipsometry). When a thin layer of molecules such as solution-phase probes is subsequently added to the bare surface or when the focused beam is scanned from the bare surface to the region of the surface covered with immobilized target (with optical dielectric constant εd), I(Ω) becomes Iinc|rprs|sin(2θA)sin(δ−δ0). By independently measuring Iinc|rprs|sin(2θA), we directly extract Δδ=δ−δ0. When the layer thickness of the targets or targets plus probes d is much less than the optical wavelength λ, Δδ varies linearly with d:17, 19, 20
| (1) |
Θ is the coverage of the targets. An image of a microarray is obtained by scanning the illumination beam along the y direction with the encoded y-scan mirror and moving the sample assembly along the x direction with the encoded translation stage.
To demonstrate the performance of the new microscope, we printed a bovine serum albumin (BSA) microarray with 10,804 spots covering an area of 2 cm×4 cm on an epoxy-functionalized glass slide (ArrayIt, Sunnyvale, California), using an OmniGrid100 contact-printing arrayer (Digilab, Holliston, Massachusetts). The printed BSA spots have an average diameter of 100 μm and are separated center-to-center by 250 μm. The BSA molecules covalently bind to the glass slide through exothermic reaction between amine residues on the exposed surface of BSA and free epoxy groups on the glass slide. The printed glass slide was assembled with a fluid chamber and washed with 1× phosphate buffer saline (PBS; pH=7.4) to remove the excess (unreacted) BSA molecules. The microarray in contact with 1×PBS was imaged with the OI-RD scanning microscope at scan steps of 20 μm in the x direction and 18.7 μm in the y direction (i.e., each pixel measures 20 μm×18.7 μm). Figure 2 shows the optical image in Δδ. The acquisition took 90 min.
Figure 2.
OI-RD image of a 10,804-spot BSA microarray in 1×PBS acquired with the scanning microscope shown in Fig. 1. The image covers an area (field of view) of 2 cm×4 cm. The pixel dimension is 20 μm×18.7 μm.
Detection of Protein Mixture Reaction with a Biotin-Conjugated Compound Microarray on Streptavidin-Functionalized Glass Surface
From one-bead, one-compound (OBOC) chemical synthesis (as described in the next section) and a bead-based phage display assay,21 Lam and coworkers identified over 80 small molecules that were strong ligand candidates to intracellular proteins of Jurkat cells. The structures of 20 such molecules are shown in Fig. 3. We measured binding reaction endpoints and real-time binding curves of a Jurkat cell lysate with the 20 molecules using our hybrid scanning OI-RD microscope. To immobilize these synthetic compounds on a streptavidin-functionalized glass slide (ArrayIt, Sunnyvale, California), we conjugated a biotin molecule to the compounds at a common location through a flexible linker, as illustrated by Fig. 4. Using the OmniGrid100 arrayer, we printed these biotinylated compounds into two 100-spot microarrays on a streptavidin-coated glass slide: each contains 10 compounds; duplicates of each compound were printed at five concentrations (1.0 mM, 0.5 mM, 0.25 mM, 0.125 mM, and 0.0625 mM). The printed surface was washed with 1×PBS to remove the excess (unbound) compounds. The microarray was imaged with the hybrid scanning OI-RD microscope before and after the reaction with a Jurkat cell lysate solution. The change in OI-RD image (i.e., δ) due to the reaction is displayed in Fig. 5.
Figure 3.
Chemical structures of 20 small-molecule compounds that are strong ligand candidates to Jurkat cell intracellular proteins. The free amine residues on the lower right of these molecules are used for conjugation to biotin through a flexible linker. The biotin motifs are used for immobilizing the compound on the streptavidin-functionalized glass slide surface.
Figure 4.
Chemical structure of LD2-2 conjugated to a biotin molecule through a linker at the primary amine residue on the original LD2-2. The other 19 compounds in Fig. 3 are conjugated to biotin in the same way.
Figure 5.
Change in OI-RD image of 20 small-molecule compounds printed in microarray format on a streptavidin-functionalized glass slide after they reacted with Jurkat cell lysate. Each column corresponds to one compound but is printed in duplicate at decreasing concentrations from 1 mM to 62.5 μM. The images are obtained by taking the difference between the image after the reaction with Jurkat cell lysate and the image taken before.
All compounds except for LB20-1 (the second compound from the top of the last column in Fig. 3) reacted with the intracellular proteins in the Jurkat cell lysate. The amplitude of the differential optical signal for a compound printed at concentration at or above 62.5 μM is more or less the same, indicating that the printing concentration at or even below 62 μM was sufficient to cover a streptavidin-coated glass surface with a full monolayer of biotin-conjugated compounds in single contact printing (i.e., by tapping the solution-loaded printing pin on the functionalized surface once instead of multiple times, and thus transferring roughly 1 nL of the solution to the surface).
We subsequently measured the binding curves of the Jurkat cell lysate with the same 20 synthetic compounds printed (at concentration of 250 μM) on another streptavidin-coated glass slide. For binding curve measurement, we read out the OI-RD signal from one pixel at or near the center of a printed target spot. By sequentially interrogating all 20 printed targets this way, we obtained the binding (association–dissociation) curves for the cell lysate to all 20 compounds, as shown in Fig. 6. The time interval between two successive scans was chosen to be 60 s in the present experiment. We note that in the binding curve measurement, LB20-1 reacted with the Jurkat cell lysate, in disagreement with the endpoint OI-RD image measurement, indicating that LB20-1 targets were not printed properly on the glass slide on which the endpoint image measurement was performed (see Fig. 5).
Figure 6.
Real-time binding curves of a Jurkat cell lysate solution to a microarray of the 20 compounds shown in Fig. 3 measured by the hybrid scanning OI-RD microscope. The structures of the 20 compounds are displayed in the same layout in Fig. 3. All 20 compounds reacted with the Jurkat cell lysate with equilibrium dissociation constants in the nM range.
One-Bead, One-Compound Combinatorial Synthesis and OI-RD Detection of Large Small-Molecule Libraries on a Strepatavidin-Functionalized Glass Slide
One-bead, one-compound (OBOC) combinatorial library synthesis, pioneered by Lam and coworkers, enables quick generation of a diverse library of small molecules from a small set of structural motifs. The OBOC combinatorial chemistry has the dual advantages of (1) rapidly producing a large collection of encoded compounds constructed with distinct molecular structures, and (2) enabling addition of a common molecular motif or “special handle” (often through a linker) to the synthesized compounds for surface immobilization or other purposes without unduly changing the activity of the compounds.22, 23, 24, 25
For OBOC synthesis of small-molecule compounds used in the present study, we used TentaGel resin beads (90 μm in diameter with load capacity ∼0.3 mmole∕gm) as the solid support for library synthesis. TentaGel beads were first topologically segregated into two layers where the outer layer with 20% of the full substitution of the beads was protected with Fmoc and the interior core with 80% of the full substitution was protected with Alloc. Upon Fmoc removal, a disulfide-containing linker, N-Fmoc-2-[(2-aminoethyl) disulfanyl] ethylamine monosuccinamide, A(S-S), was tethered to the outer layer, followed by sequential assembly of Fmoc-Lys(Biot)-OH, Fmoc-link-OH [N-Fmoc-2,2′-ethylenedioxy-bis(ethylamine) monosuccinamide] using Fmoc chemistry. Dde was then added to temporarily cap the N-terminus of the outer layer, allowing for a CNBr-cleavable linker to be generated using Palladium chemistry in the interior core after Alloc deprotection. The cleavage linker included four components: methionine (“Met”), arginine (“Arg”), 3-(4-bromophenyl)-β-alanine [“A(Br)”], and 2,2′-ethylenedioxy-bis(ethylamine) monosuccinamide (“Link”). After Fmoc deprotection, a mixture of Fmoc-Osu and Alloc-Osu (1:1) was coupled to the bead interior core. Upon Dde and Fmoc deprotection, a tetrafunctional scaffold, N-Dde-3-(4-fluoro-3-nitrophenyl)-3-aminopropionic acid was conjugated to the beads. The Alloc group was then removed by Palladium chemistry, followed by coupling with a mixture of 4-fluor-3-nitrobenzoic acid and bromoacetic acid using HOBt and DIC. The resulting beads were split into 20 aliquots, and each aliquot reacted with a primary amine in the presence of DIEA (diisopropylethylamine), respectively. The beads were mixed and reduced with tin chloride solution for 4 h, followed by cyclization with 17 aromatic aldehyde molecules after splitting. Upon treating with acetic anhydride, the beads were Dde-deprotected, and 21 acids or isocyanates were acylated to the free amino group using the split-mix approach. The beads were finally treated with a side-chain cleavage cocktail (TFA:water:TIS=10:0.5:0.5) for 2.5 h to furnish the desired library, which contained about 7,000 distinct compounds. After synthesis, individual beads were dispensed into individual wells of polypropylene microtiter plates so that each well with a volume of 50 μL contained only one bead. Figure 7 illustrates the synthetic route for preparing an orthogonal releasable OBOC benzoimidazole library. The attached compounds were released from the beads by cleaving the disulfide bond with a TCEP solution. Roughly 20 picomoles of OBOC compounds were released from the outer layer of a single 90-μm-diam bead. By adding 15 μL of TCEP in H2O, we obtained a target printing solution at 1.3 μM. The biotin group on the released compounds, introduced during the fourth step of the synthesis as Fmoc-Lys(Biot)-OH, was used as the special or universal handle to anchor the OBOC compounds on a streptavidin-functionalized glass surface.
Figure 7.
Synthetic route to OBOC benzoimidazole combinatorial library. A(S-S): N-Fmoc-2-[(2-aminoethyl) disulfanyl]ethylamine monosuccinamide; Link: N-Fmoc-2,2′-ethylenedioxy-bis(ethylamine) monosuccinamide; A(Br): 3-(4-bromophenyl)-β-alanine; CL1: Link-K(Biotin)-A(S-S)-; CL2: Link-A(Br)-R-M-.
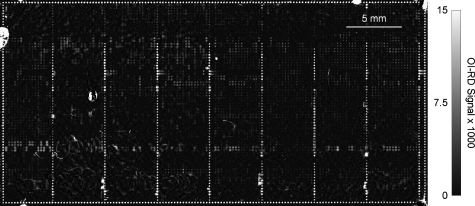
To demonstrate that we can detect, using the hybrid OI-RD scanning microscope, large OBOC compound microarrays with over 10,000 printed targets on a single glass slide, we printed 4608 biotinylated OBOC compounds in pairs (in the vertical direction in Fig. 8) on a streptavidin-coated glass slide. Using a printing concentration of 1.33 μM (limited by the amount of synthetic compounds released from a single 90-μm bead), we needed to print twice at each spot to ensure immobilization of a full monolayer of the OBOC compounds on the streptavidin-functionalized glass slide. In addition to 4608 OBOC compounds printed in pairs, we also printed replicates of 36 biotinylated control compounds such as biotin-conjugated BSA, biotin-linker-biotin, and biotinylated peptides. The entire microarray had 10,804 targets with 74 rows and 146 columns. The layout of the microarray is as follows: (1) the border of the microarray consists of 436 biotin-conjugated bovine serum albumin (BSA), and on each BSA molecule there are more than five conjugated biotin molecules (one biotin is used to immobilize the conjugate to streptavidin-coated surface, the remaining biotin molecules are available as antigen to antibodies raised against biotin); (2) 4608 OBOC compounds printed in pairs form eight 72×16 subarrays; and (3) on the right side of each OBOC subarray are a column of 36 “control” compounds printed in pairs (72×1) and an empty column (72×1). Table 1 lists the 36 control compounds in the order of printing from top to bottom (see Fig. 8). The printed surface was washed with 1×PBS to remove the excess printed targets. Figure 8 shows the OI-RD image in Δδ of the compound microarray in contact with 1×PBS. The outermost border consists of biotin-conjugated BSA. The lone biotin molecular residue on each of 4608 biotinylated OBOC compounds is used to anchor the molecule on the streptavidin-coated glass surface; thus, it is not available subsequently as an antigen to antibodies raised against biotin.
Figure 8.
The OI-RD image of a 10,804 biotinylated OBOC compound microarray printed on a streptavidin-functionalized glass slide after the excess (unbound) target materials were washed off. The image was acquired with the microarray in contact with 1×PBS. The average size of the printed spots is 100 μm, with center-to-center separation of 250 μm. The outermost border consists of biotin-conjugated bovine serum albumin (BSA). (See the main text for details of the microarray layout.)
Table 1.
36 control compounds that are printed along with 4608 OBOC compounds in pairs to form the 10,804-spot target microarray, as shown in Fig. 8.
| Compound | Full name | Compound | Full name | Compound | Full name |
|---|---|---|---|---|---|
| TG1 | HHHHHH-linker- biotin | B6 | HKRTLGLGG- linker-biotin | LC5-1 | (See Fig. 3) |
| TG2 | EQKLISEEDL- linker-biotin | B7 | ELDKWAGGG- linker-biotin | LC7-1 | (See Fig. 3) |
| TG3 | YPYDVPDYA- linker-biotin | B-B | Biotin-linker-biotin | LC11-1 | (See Fig. 3) |
| TG4 | YTDIEMNRLGK- linker-biotin | LC22-1 | (See Fig. 3) | LC12-1 | (See Fig. 3) |
| TG5 | QPELAPEDPED- linker-biotin | L20-1 | (See Fig. 3) | LC13-1 | (See Fig. 3) |
| TG6 | DYKDDDK-linker- biotin | LA12-2 | (See Fig. 3) | LC14-2 | (See Fig. 3) |
| Biotin-DNP | DNP-linker- biotin | LA22 | (See Fig. 3) | LC15-1 | (See Fig. 3) |
| B1 | NSNNPDWDF- linker-biotin | LA24 | (See Fig. 3) | LC21-1 | (See Fig. 3) |
| B2 | IRSWFGLPI-linker- biotin | LB12-1 | (See Fig. 3) | LC22-2 | (See Fig. 3) |
| B3 | PNPLGFFPG- linker-biotin | LB13-2 | (See Fig. 3) | LD1-2 | (See Fig. 3) |
| B4 | KYDEVARKG- linker-biotin | LB20-1 | (See Fig. 3) | LD2-2 | (See Fig. 3) |
| B5 | LITALFGLG- linker-biotin | LC2-2 | (See Fig. 3) | Linker-B | Linker- biotin |
We performed a positive control experiment by exposing the 10,804-spot compound microarray for 1 h with a solution of unlabeled monovalent Fab fragments of mouse IgG against biotin at concentration of 0.013 mg∕ml or 260 nM in 1×PBS. Afterward, the solution of the mouse IgG was replaced with 1×PBS again. The Fab fragments were purchased from Jackson ImmunoResearch Laboratories (West Grove, Pennsylvania). Figure 9 shows the change in OI-RD image of the microarray in 1×PBS obtained by taking the difference between the image taken after the reaction and the image shown in Fig. 8. Fab fragments of the mouse IgG reacted with biotin-BSA (spots along the perimeter) as expected and with the control compounds (8 single-file columns) except for DNP-linker-biotin. Some of the 4608 OBOC compounds also reacted with the mouse IgG. The mean and the standard deviation of the optical signal in response to the reaction of the mouse antibiotin IgG with 436 biotin-conjugated BSA spots are Δδ=0.022±0.0075. Table 2 lists the mean and standard deviation of the optical signals due to reaction with the mouse antibiotin IgG with 8 replicates of each of the 36 “control” compounds (see Table 1). The variation in optical signal for a given immobilized compound target comes mainly from the variation in target density (common in microarray-based studies) and partially from the noise in the optical detection process.
Figure 9.
Change in OI-RD image after the 10,804 biotinylated OBOC compound microarray (shown in Fig. 8) reacted with unlabeled monovalent Fab fragment of mouse IgG against biotin (in 1×PBS at concentration of 260 nM) for 1 h. Fab fragments of the mouse IgG reacted with biotin-BSA (spots along the perimeter) and with the control compounds (8 single-file columns) except for DNP-linker-biotin.
Table 2.
Optical signals from replicates (8) of 36 immobilized “control” compounds after reaction with Fab fragments of the mouse anti-biotin IgG.
| Compound | Optical signal | Compound | Optical signal | Compound | Optical signal |
|---|---|---|---|---|---|
| TG1 | 0.0057±0.0057 | B6 | 0.017±0.0033 | LC5-1 | 0.0082±0.0016 |
| TG2 | 0.0022±0.0021 | B7 | 0.015±0.0020 | LC7-1 | 0.0097±0.0017 |
| TG3 | 0.0025±0.0020 | B-B | 0.0032±0.0021 | LC11-1 | 0.0090±0.0027 |
| TG4 | 0.0042±0.0020 | LC22-1 | 0.011±0.0021 | LC12-1 | 0.011±0.0035 |
| TG5 | 0.0019±0.0008 | L20-1 | 0.0055±0.0009 | LC13-1 | 0.0057±0.0032 |
| TG6 | 0.022±0.0013 | LA12-2 | 0.0081±0.0013 | LC14-2 | 0.011±0.0027 |
| Biotin-DNP | 0 | LA22 | 0.0075±0.0023 | LC15-1 | 0.011±0.0023 |
| B1 | 0.013±0.0023 | LA24 | 0.010±0.0022 | LC21-1 | 0.0091±0.0055 |
| B2 | 0.011±0.0026 | LB12-1 | 0.011±0.0014 | LC22-2 | 0.011±0.0043 |
| B3 | 0.015±0.0034 | LB13-2 | 0.011±0.0020 | LD1-2 | 0.013±0.0057 |
| B4 | 0.015±0.0027 | LB20-1 | 0.0079±0.0026 | LD2-2 | 0.020±0.0043 |
| B5 | 0.014±0.0030 | LC2-2 | 0.0076±0.0012 | Linker-B | 0.0096±0.0031 |
We developed a hybrid scanning oblique-incidence reflectivity difference (OI-RD) microscope for high-speed, label-free, in situ detection of a large small-molecule compound microarray on a functionalized glass slide. We demonstrated the capability of such a new scanning microscope for detection of endpoints and real-time binding curves of a protein probe to an OBOC synthesized compound library in the form of microarrays with over 10,000 immobilized targets on a single streptavidin-coated glass slide.
Acknowledgments
This work was supported by NIH under NIH-R01-HG003827-04.
References
- MacBeath G., “Protein microarrays and proteomics,” Nat. Genet. 32, 526–532 (2002). 10.1038/ng1037 [DOI] [PubMed] [Google Scholar]
- Schena M., Microarray Analysis, Wiley, Hoboken, NJ: (2003). [Google Scholar]
- MacBeath G., Koehler A. N., and Schreiber S. L., “Printing small molecules as microarrays and detecting protein-ligand interactions en masse,” J. Am. Chem. Soc. 121, 7967–7968 (1999). 10.1021/ja991083q [DOI] [Google Scholar]
- Uttamchandani M., Walsh D. P., Yao S. Q., and Chang Y. T., “Small molecule microarrays: recent advances and applications,” Curr. Opin. Chem. Biol. 9, 4–13 (2005). 10.1016/j.cbpa.2004.12.005 [DOI] [PubMed] [Google Scholar]
- Falsey J. R., Renil M., Park S., Li S., and Lam K. S., “Peptide and small molecule microarray for high-throughput cell adhesion and functional assays,” Bioconjugate Chem. 12, 346–353 (2001). 10.1021/bc000141q [DOI] [PubMed] [Google Scholar]
- Zhu H., Bilgin M., Bangham R., Hall D., Casamayor A., Bertone P., Lan N., Jansen R., Bidingmaier S., Houfek T., Mitchell T., Miller P., Dean R. A., Gerstein M., and Synder M., “Global analysis of protein activities using proteome chips,” Science 293, 2101–2105 (2001). 10.1126/science.1062191 [DOI] [PubMed] [Google Scholar]
- Mitchell P., “A perspective on protein microarrays,” Nat. Biotechnol. 20, 225–229 (2002). 10.1038/nbt0302-225 [DOI] [PubMed] [Google Scholar]
- Shliom O., Huang M., Sachais B., Kuo A., Weisel J. W., Nagaswami C., Nassar T., Bdeir K., Hiss E., Gawlak S., Harris S., Mazar A., and Higazi A. A., “Novel interactions between urokinase and its receptor,” J. Biol. Chem. 275, 24304–24312 (2000). 10.1074/jbc.M002024200 [DOI] [PubMed] [Google Scholar]
- Karlsson R., “SPR for molecular interaction analysis: a review of emerging application areas,” J. Mol. Recognit. 17, 151–161 (2004). 10.1002/jmr.660 [DOI] [PubMed] [Google Scholar]
- Singh B. K. and Hillier A. C., “Surface plasmon resonance imaging of biomolecular interactions on a grating-based sensor array,” Anal. Chem. 78, 2009–2018 (2006). 10.1021/ac0519209 [DOI] [PubMed] [Google Scholar]
- Boozer C., Kim G., Cong S., Guan H., and Londergan T., “Looking towards label-free biomolecular interaction analysis in a high-throughput format: a review of new surface plasmon resonance technologies,” Curr. Opin. Chem. Biotechnol.17, 400–405 (2006). [DOI] [PubMed]
- Shumaker-Parry J. S. and Campbell C. T., “Quantitative methods for spatially resolved adsorption∕desorption measurements in real time by surface plasmon resonance microscopy,” Anal. Chem. 76, 907–917 (2004). 10.1021/ac034962a [DOI] [PubMed] [Google Scholar]
- Usui-Aoki K., Shimada K., Nagano M., Kawai M., and Koga H., “A novel approach to protein expression profiling using antibody microarrays combined with surface plasmon resonance technology,” Proteomics 5, 2396–2401 (2005). 10.1002/pmic.200401171 [DOI] [PubMed] [Google Scholar]
- Azzam R. M. A. and Bashara N. M., Ellipsometry and Polarized Light, Elsevier Science, New York: (1987). [Google Scholar]
- Jin G., Jansson R., and Arwin H., “Imaging ellipsometry revisited: development for visualization of thin transparent layers on silicon substrates,” Rev. Sci. Instrum. 67, 2930–2936 (1996). 10.1063/1.1147074 [DOI] [Google Scholar]
- Wang Z. H. and Jin G., “A label-free multi-sensing immuno-sensor based on imaging ellipsometry,” Anal. Chem. 75, 6119–6123 (2003). 10.1021/ac0347258 [DOI] [PubMed] [Google Scholar]
- Thomas P., Nabighian E., Bartelt M. C., Fong C. Y., and Zhu X. D., “An oblique-incidence optical reflectivity difference and LEED study of rare-gas growth on a lattice-mismatched metal substrate,” Appl. Phys. A 79, 131–137 (2004). 10.1007/s00339-003-2261-y [DOI] [Google Scholar]
- Landry J. P., Zhu X. D., and Gregg J. P., “Label-free detection of microarrays of biomolecules by oblique-incidence reflectivity difference microscopy,” Opt. Lett. 29, 581–583 (2004). 10.1364/OL.29.000581 [DOI] [PubMed] [Google Scholar]
- Zhu X. D., Landry J. P., Shun Y. S., Gregg J. P., Lam K. S., and Guo X. W., “Oblique-incidence reflectivity difference microscope for label-free high-throughput detection of biochemical reactions in a microarray format,” Appl. Opt. 46, 1890–1895 (2007). 10.1364/AO.46.001890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry J. P., Sun Y. S., Guo X. W., and Zhu X. D., “Protein reaction with surface-bound molecular targets detected by oblique-incidence reflectivity difference microscopes,” Appl. Opt. 47, 3275–3288 (2008). 10.1364/AO.47.003275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q., Miyamoto S., and Lam K. S., “A novel approach to chemical microarray using ketone-modified macromolecular scaffolds: application in micro cell-adhesion assay,” Mol. Divers. 8, 301–310 (2004). 10.1023/B:MODI.0000036238.42200.3d [DOI] [PubMed] [Google Scholar]
- Lam K. S., Saknibm S. E., Hersh E. M., Hruby V. J., Kazmierski W. M., and Knapp R. J., “A new type of synthetic peptide library for identifying ligand-binding activity,” Nature 354, 82–84 (1991). 10.1038/354082a0 [DOI] [PubMed] [Google Scholar]
- Lam K. S., Lebl M., and Krchnak V., “The one-bead-one-compound combinatorial library method,” Chem. Rev. (Washington, D.C.) 97, 411–448 (1997). [DOI] [PubMed] [Google Scholar]
- Liu R., Marik J., and Lam K. S., “A novel peptide-based encoding system for one-bead one-compound peptidomimetic and small molecule combinatorial libraries,” J. Am. Chem. Soc. 124, 7678–7680 (2002). 10.1021/ja026421t [DOI] [PubMed] [Google Scholar]
- Song A., Zhang J., Lebrilla C. B., and Lam K. S., “A novel and rapid encoding method based on mass spectrometry for one-bead-one-compound small molecule combinatorial libraries,” J. Am. Chem. Soc. 125, 6180–6188 (2003). 10.1021/ja034539j [DOI] [PubMed] [Google Scholar]