Abstract
We describe the potential of 5-aminolevulinic acid (ALA)–induced protoporphyrin IX (PpIX) fluorescence as a source of contrast for margin detection in commonly diagnosed breast cancer subtypes. Fluorescence intensity of PpIX in untreated and ALA-treated normal mammary epithelial and breast cancer cell lines of varying estrogen receptor expression were quantitatively imaged with confocal microscopy. Percentage change in fluorescence intensity integrated over 610–700 nm (attributed to PpIX) of posttreated compared to pretreated cells showed statistically significant differences between four breast cancer and two normal mammary epithelial cell lines. However, a direct comparison of post-treatment PpIX fluorescence intensities showed no differences between breast cancer and normal mammary epithelial cell lines due to confounding effects by endogenous fluorescence from flavin adenine dinucleotide (FAD). Clinically, it is impractical to obtain pre- and post-treatment images. Thus, spectral imaging was demonstrated as a means to remove the effects of endogenous FAD fluorescence allowing for discrimination between post-treatment PpIX fluorescence of four breast cancer and two normal mammary epithelial cell lines. Fluorescence spectral imaging of ALA-treated breast cancer cells showed preferential PpIX accumulation regardless of malignant phenotype and suggests a useful contrast mechanism for discrimination of residual cancer at the surface of breast tumor margins.
Keywords: 5-aminolevulinic acid, breast cells, confocal microscopy, fluorescence intensity, protoporphyrin IX, spectroscopy
Introduction
Women with early stage breast cancer are eligible for breast conserving surgery (BCS). The goal of BCS is to remove the entire tumor while minimizing removal of surrounding normal tissue. The commonly accepted pathologic criterion for a negative margin is a 2-mm rim of malignancy-free tissue.1 There are no widely adopted intraoperative tools to assess tumor margins, and all decisions are made postoperatively by a pathologist. If the margin is positive, then a secondary surgical procedure is performed to excise additional tissue and prevent local recurrence of cancer. On average, four out of ten women undergoing BCS return for a reexcision surgery.2, 3, 4, 5, 6, 7, 8, 9, 10 Thus, a critical need exists for new tools that can be used intraoperatively to determine if a tumor margin is positive or negative at the time of the first surgery.
Optical techniques that exploit exogenous contrast agents, such as antibody-conjugated gold nanoparticles11, 12 or fluorescence molecules,13, 14 are currently being investigated to enhance a surgeon’s ability to identify malignancy, with one possible use being intraoperative margin assessment. A contrast agent such as 5-aminolevulinic acid (ALA) is one potential candidate for margin assessment. ALA has been extensively tested in cells, animal models, and humans15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41 and is widely used today for detection and treatment of malignancy. Addition of exogenous ALA causes a preferential accumulation of protoporphyrin IX (PpIX) in cancerous cells when compared to normal cells.15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 33, 34, 35, 36, 39, 40 PpIX is naturally produced by all nucleated cells during the heme cycle and meticulously controlled to prevent its natural accumulation.37 This negative feedback system is thought to be modified in cancerous tissues due to (i) enzymatic defects that lead to an increase in protoporphyrinogen IX oxidase and∕or (ii) reduced activity of ferrochelatase.35
ALA and its derivatives have shown great potential to photodynamically detect and treat cancer in a wide variety of organs and applications. Clinical approval of ALA derivatives has been granted in the European Union for photodynamic diagnosis of cancer of the bladder, brain, and skin, but has only been approved for use on skin by the Food and Drug Administration in the United States.22 Preferential accumulation of PpIX in tumor cells, specifically residual glioma cells within the excision cavity,37, 42 followed by photodynamic therapy of those cells, demonstrates the potential of ALA for use in a “see-and-treat” paradigm in breast cancer.22, 37, 43 ALA-induced PpIX has been shown to have a higher accumulation in cancerous breast cells as compared to prostate, ovarian, and brain cancer cell lines, which implies higher contrast and effective visualization of cancerous cells.18, 30 ALA-induced phototherapy in breast cancer cells (MDA-MB-231) has demonstrated high photoefficiency, further demonstrating the potential for the see-and-treat paradigm within breast tissue.43
Previous studies illustrate that ALA-induced PpIX has the capability to differentially detect breast cancer in multiple biological model systems.18, 20, 29, 30, 36, 40 Fluorescence quantification of PpIX in breast cancer cell lysates treated with ALA was higher as compared to patient-matched normal cell lysates treated with ALA in explanted cell cultures from five breast cancer patients.28 Studies on an in vivo animal model have shown that ALA-induced fluorescence can be used to enhance early detection of neoplastic and metastatic tissue from normal tissue.36 Transgenic mice with induced ductal carcinoma in situ, the earliest form of breast cancer, were shown to have an increase in 635 nm fluorescence (attributed to PpIX) in cancerous tissues over the surrounding normal tissues, after tail-vein injection of ALA 60–75 min prior to imaging.36 In a clinical study, ALA-induced PpIX fluorescence was significantly greater in 13 ex vivo primary breast tumors relative to surrounding normal tissue.24 Metastatic axillary and sentinel lymph node tissues imaged ex vivo also showed an increase in mean PpIX fluorescence as compared to nonmetastatic lymph nodes in seven breast cancer patients.15
The clinical applicability of ALA-induced PpIX has been demonstrated by the early clinical work of Ladner et al.,24 but before diagnosis of breast cancer with ALA is widely accepted, more fundamental cellular, animal, and clinical studies will be required to characterize the effects of breast cancer subtype on uptake and contrast. Breast cancer is highly heterogeneous and has been shown to exhibit large variations in characteristics that may potentially affect metabolism of contrast agents, such as ALA-induced PpIX.
The goal of the study reported here was to evaluate ALA-induced PpIX fluorescence of a variety of breast cancer cell lines of varying phenotypes that are most commonly seen clinically44 and compare their ALA-induced PpIX fluorescence to those of normal mammary epithelial cells.23 Specifically, confocal microscopy at 405 nm excitation was utilized to image ALA-induced PpIX fluorescence in a panel of six different breast cell lines—two normal, two estrogen receptor negative (ER−), and two ER positive (ER+)—before and after treatment with ALA. This is the first study to establish the sources of contrast in different breast cancer and normal epithelial cells after application of PpIX. Confocal microscopy is an ideal modality for cell studies due to its high-resolution capabilities. Imaging of ALA-induced PpIX fluorescence for intraoperative margin assessment will likely need full-field imaging techniques, with a larger field of view and higher speed than confocal fluorescence imaging.
Methods and Materials
Cell Culture
Six breast cell lines were used in this study; two were normal mammary epithelial cells [MCF10A and human mammary epithelial cells (HMEC)] and four were breast cancer (MDA-MB-231, MDA-MB-361, MDA-MB-435, and MCF7). All cell lines, except for HMEC, were obtained from the American Type Culture collection (Manassas, Virginia). HMEC primary cells were obtained from Lonza (Basel, Switzerland) and infected with a retrovirus encoding human telomerase reverse transcriptase for immortalization. All cells remained free of contaminants and were propagated by adherent culture according to established protocols.44 All breast cancer cell lines were cultured in α-MEM (Minimum Essential Media, Gibco, Carlsbad, California) supplemented with 6% fetal bovine serum, 1% hepes, 1% nonessential amino acids, 1% of 100 mM sodium pyruvate, 10 μg∕mL insulin, 10 μg∕mL hydrocortisone, and 5 μg∕mL epidermal grown factor (EGF). All normal mammary epithelial cell lines were cultured in mammary epithelial basal media (MEBM) (Lonza, Basel, Switzerland) with 0.4% bovine pituitary extract, 0.01% hydrocortisone, insulin, and human EGF. Plated cells were incubated at 5% CO2 and cultured every 3–4 days. Cells were double washed in 3 mL of phosphate buffer saline and detached from flasks with 0.25% trypsin before centrifugation.
After 3 min of centrifugation at 120 relative centrifugal force (rcf), a standard hemocytometer was used to count the number of cells per milliliter. Approximately 200,000 cells were plated on a 35-mm coverslip dish (Mat-Tek, Ashland, Massachusetts) to be tested.
Confocal Microscope and Imaging Parameters
All confocal images were collected at the Duke University Light Microscopy Core Facility on a Leica SP5 laser scanning confocal microscope (Wetzlar, Germany). All cells were imaged in a temperature-controlled (37 °C) live cell chamber. A 405-nm diode laser source with a mean power of 2.4 mW at the sample plane and scanning rate of 400 Hz was coupled to an inverted Leica DM16000CS microscope with a 40X oil-immersion objective (Leica Plan NeoFluor, NA=1.25). The imaging field of view was 242×242 μm with 512×512 pixels per image. An acousto-optic beamsplitter (AOBS) served as a dichroic mirror to allow the 405-nm excitation light to reach the sample and to allow selection of emission wavelengths. Cooled photomultiplier tubes (PMTs) were used to measure fluorescence through a 121-μm pinhole in a confocal arrangement with a theoretical axial resolution of 2 μm.
The emission bandpass for integrated fluorescence intensity measurements was set to 610–700 nm, which was chosen from preliminary spectroscopy data on cells that showed a fluorescence peak at 630 nm, corresponding to PpIX, as published in the literature.42 Images were taken at two separate fields of view and line averaged (16X per line) for improved image quality. Each fluorescence intensity image was acquired in ∼20 s per image (1∕400 per second 16 averages per line×512 lines per image). Spectrally resolved fluorescence images at 405 nm excitation were collected at each of 34 emission wavelengths between 420 and 750 nm with a line average of 1. The AOBS had a collection bandwidth of 20 nm. A prism with a sliding mirror placed in the optical path prior to the PMT was scanned every 10 nm to ensure Nyquist sampling was fulfilled across all wavelengths. Total acquisition time was 43.5 s for all wavelengths in each spectral image (1∕400 per second 1 average per line×512 lines×34 wavelengths per spectral image). Leica LAS AF 1.8.2 software (Wetzlar, Germany) was used for data acquisition. All images (intensity and spectral) were acquired with a gain of 1200 V, offset 0.7% and zoom of 1.6.
Prior to imaging, a calibration intensity image of standard fluorescent beads was obtained (FocalCheck Fluorescence Microscope Test Slide No. 2, Invitrogen, Carlsbad, California) to correct for daily variations in microscope throughput. The mean fluorescence intensity over all experiments was 208±5 units. Integrated fluorescence intensity images were normalized by dividing the measured intensity by the intensity of the calibration standard measured on the same day.
Cell Imaging
Twenty-four hours after plating, the original cell media was removed from the culture dish, and cell lines were treated either with 500 μg∕mL of ALA in standard cell media or standard media alone and incubated for 2 h prior to imaging. Because ALA is not fluorescent, it was not removed before imaging. The concentration of ALA (data not shown) was chosen experimentally. At 500 μg∕mL of ALA, no phototoxic effects were seen in any cell line tested for this paper. Cell viability was verified by a trypan blue exclusion assay as determined by a previously tested protocol.45 Viable cell concentration was then determined by counting the ratio of cells that were still viable (unstained) to all cells, and 89%±6% of ALA-treated and 85%±8% of untreated cells were found to be viable. Finally, 500 μg∕mL of ALA was found to be within the range of concentrations used in previous cell culture studies.18, 40
To determine the optimal time to measure the fluorescence of PpIX (data not shown), a subset of cells [normal mammary epithelial MCF10A, MDA-MB-435 (ER−), and MCF7 (ER+)] was tested. The fluorescence intensity increased linearly from 0 to 6 h post-ALA treatment, as seen in previous studies,40 and therefore, the peak intensity window was assumed to be >6 h post-ALA treatment for all cell lines. The 2-h time point was chosen because it is clinically practical and showed distinct differences between normal mammary epithelial and breast cancer cell lines (see Sec. 3).
The sample size for each cell line included 12 treated and seven untreated plates. Fluorescence intensity images were obtained from six of the treated and six of the untreated plates. Confocal spectral imaging was completed on six additional treated plates and a single untreated plate for each cell line. Sample size calculations for imaging of integrated fluorescence and confocal spectral imaging are summarized in Table 1.
Table 1.
Experimental sample size for confocal microscopy.
| Sample size definition | No. Images∕ plate | No. plates | Total No. of samples | ||
|---|---|---|---|---|---|
| Integrated fluorescence intensity | Treated | 1 Sample=Average intensity of all cells per image | 6 | 2 | 12 per cell line |
| Untreated | 1 Sample=Average intensity of all cells per image | 6 | 2 | 12 per cell line | |
| Spectrally resolved fluorescence | Treated | 1 Sample=3 spectra averaged from a cluster of cells (3–5 cells) per image | 6 | 2 | 12 per cell line |
| Untreated | 1 Sample=3 spectra averaged from a cluster of cells (3–5 cells) per image | 1 | 1 | 1 per cell line | |
Analysis of Integrated Fluorescence Intensity Images
Integrated fluorescence intensity images were analyzed using NIH ImageJ software. Integrated fluorescence intensity for each breast cell line was determined by manually segmenting each cell in the image and then computing the average fluorescence intensity. Background intensity was subtracted, and the resulting data normalized by the fluorescence of the calibration was standard. The background was defined as an area without cells that was approximately equivalent to the size of an entire cell (2000–5000 pixels, depending on the cell line). Fluorescence intensity values for all cells within each image were then averaged to represent one sample (each image had approximately 10–50 cells within a field of view, depending on the cell line). A total of n=12 fields of view (two images per sample) were imaged, from which the average integrated fluorescence intensity per cell line was derived for both treated and untreated cells.
Analysis of Spectrally Resolved Fluorescence Images
For spectrally resolved fluorescence images, a spectrum was calculated by the Leica software from an area of 3–5 adjacent cells (6000–15,000 pixels, depending on the cell line). Three groups of adjacent cells were manually segmented, and all spectra were averaged to obtain a single spectrum (three per image). A background spectrum was obtained from a cell-free region in each image (2000–90,000 pixels, depending on the cell-line growth pattern) and subtracted from each of the three spectra. These three spectra were imported into MATLAB (Mathworks, Inc., Natick, Massachusetts) to calculate Fractional PpIX contribution (FPC). (See Section 3 for further discussion of this calculation). The three FPC values from each image were averaged to obtain a single FPC per image. A total of n=12 FPCs (two images per sample) was calculated for all six treated cell plates, from which the average FPC for the cell line was calculated.
Statistics
All statistics were computed with JMP (SAS, Cary, North Carolina) software. ANOVAs and t-tests were completed with a Tukey–Kramer correction for multiple comparisons to determine statistical significance. Exact two-sided p-values were computed, and all p<0.05 were considered significant. All calculated correlations are reported with a Pearson coefficient.
Results
Integrated Fluorescence Intensity Images
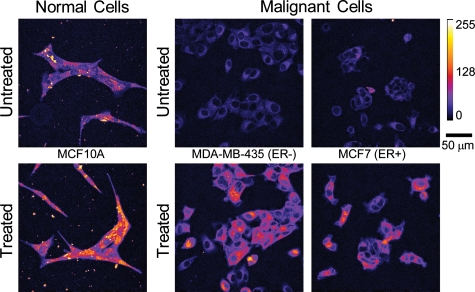
Figure 1 shows representative integrated fluorescence intensity images of untreated and treated cell lines with different ER phenotypes. The untreated normal mammary epithelial cell line image has much higher pretreatment endogenous fluorescence than either breast cancer cell line, which is likely due to the endogenous fluorophore, FAD. Bright specks outside of the cells in the normal column are cellular debris caused by plating on untreated glass and were not included in subsequent analyses. Untreated ER− and ER+ breast cancer cell lines qualitatively show similar distributions of endogenous fluorescence, which is diffusely distributed throughout the breast cancer cells prior to treatment and a few areas of high intensity around the nucleus. All cells showed an increase in PpIX fluorescence after 2 h of ALA treatment, as seen in the “Treated” row in Fig. 1. The normal mammary epithelial cell line shows bright points of PpIX fluorescence within an even distribution of cellular fluorescence. Breast cancer cell lines show brighter fluorescence in the perinuclear cytoplasm, and although the edges of the cells are clearly visible in the post-treatment images, the edges are not as intense. PpIX fluorescence is only in the cellular cytoplasm (as opposed to the nucleus) in all cell lines.
Figure 1.
Representative confocal integrated fluorescence intensity images (uncalibrated) of normal mammary epithelial (Normal) and breast cancer (Malignant) cells treated with ALA for 2 h. Excitation was at 405 nm, and emission was collected between 610 and 700 nm.
Cellular Fluorescence Spectra
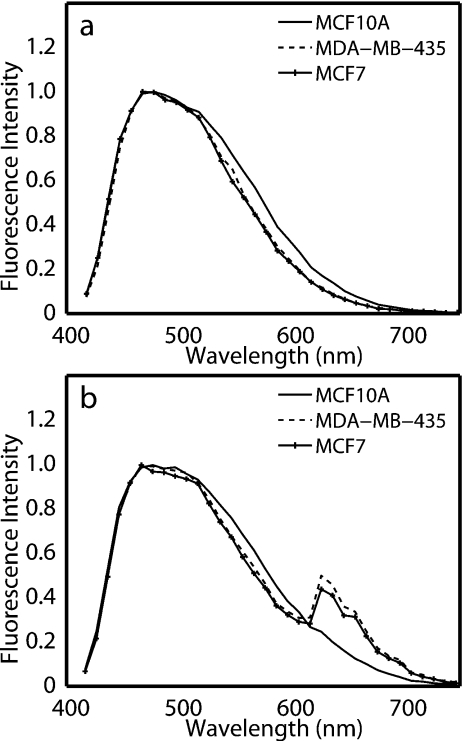
Figure 2 shows normalized representative fluorescence spectra from one untreated [Fig. 2a] and one ALA-treated plate [Fig. 2b] of normal mammary epithelial, ER−, and ER+ cell lines at 405 nm excitation. Excitation at 405 nm is expected to elicit fluorescence from FAD and PpIX.46 Normalized representative spectra in Fig. 2 show a single broad emission, which is due to FAD, and spectra from treated cells show an additional distinct ALA-induced PpIX emission peak at 630 nm with a shoulder at 700 nm [Fig. 2b], similar to that observed by others.46 It should be noted that the tail of the FAD fluorescence spectrum overlaps with the PpIX fluorescence between 610 and 700 nm [Fig. 2b], and therefore, it is assumed to contribute to integrated fluorescence intensity images collected over a bandpass of 610–700 nm.
Figure 2.
Representative normalized fluorescence spectra at 405-nm excitation of (a) untreated cells and (b) ALA-treated cells. Spectra were normalized to the peak intensity.
Quantitative Integrated Fluorescence Intensity Measurements
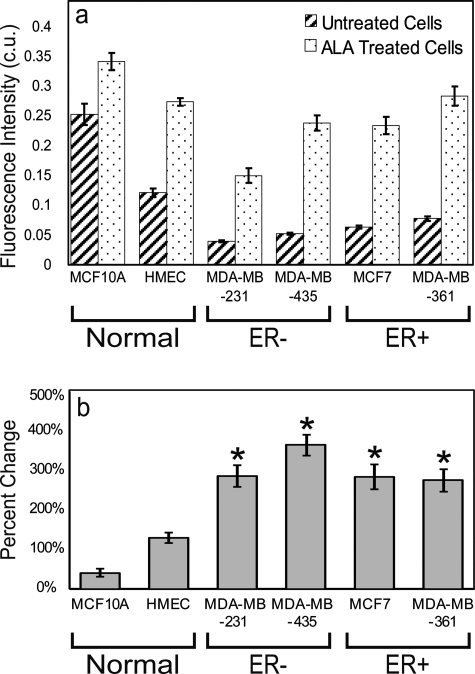
In Fig. 3, the six cell lines were grouped in pairs by ER expression (normal mammary epithelial, malignant ER−, and malignant ER+). All cell lines exhibited significantly greater fluorescence intensity following treatment (p<0.05). The normal mammary epithelial cell lines (MCF10A and HMEC) untreated fluorescence intensity was significantly greater than all untreated breast cancer (MDA-MB-231, MDA-MB-435, MCF7, and MDA-MB-361) cell lines [Fig. 3a, p<0.01]. Calibrated fluorescence intensity of the ALA-treated HMEC cell line was significantly greater than the treated MDA-MB-231 breast cancer cell line, but not significantly different compared to the other ALA-treated breast cancer cell lines. Figure 3b shows that the percentage change in PpIX fluorescence intensity following treatment was significantly greater in all breast cancer cell lines as compared to both normal mammary epithelial cell lines by at least 150% [Fig. 3b, p<0.05]. No significant differences in the percent change of fluorescence intensity were observed between or within the ER− and ER+ breast cancer cell lines.
Figure 3.
Average and standard error of the (a) calibrated integrated fluorescence intensity (c.u.) per cell line (n=12) and (b) percentage change in integrated fluorescence intensity of treated compared to the untreated controls (n=12). Asterisks denote that each ALA-treated breast cancer cell line has a statistically higher percentage change in integrated fluorescence compared to ALA-treated normal mammary epithelial cell lines (Normal) (p<0.05).
Quantitative analysis of the fluorescence spectral data
Although the breast cancer cells demonstrated a significant percentage increase in fluorescence following ALA treatment, it may be clinically impractical to obtain a pretreatment measurement. Post-treatment intensity measurements alone were not useful in discriminating normal mammary epithelial cell lines from breast cancer due to cell-to-cell variability in endogenous FAD fluorescence. A method for differentiating between normal mammary epithelial and breast cancer cells using only post-treatment measurements would be desirable. Fluorescence spectral images that capture both FAD and PpIX fluorescence have the potential to directly address this problem as presented below.
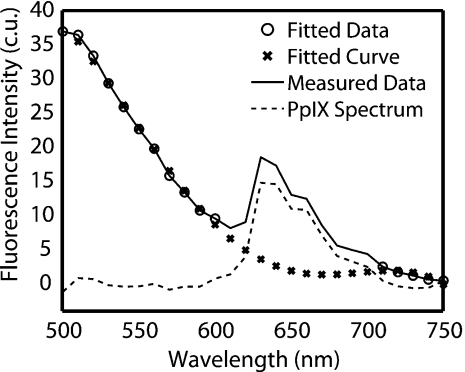
The contribution of FAD fluorescence to the PpIX fluorescence was quantified and removed as shown in Fig. 4. First, the spectrum between 520–600 nm and 700–750 nm (○) was fit with a fourth-order polynomial to approximate an untreated spectrum (×). All polynomials were shown to have a goodness-of-fit coefficient (r) of 0.93. The fitted curve (×) was then subtracted from the measured data (—), which resulted in an FAD-subtracted PpIX spectrum (--). The area under the curve of the resulting PpIX spectrum was calculated over 610–700 nm and represents integrated PpIX fluorescence intensity. The area under the curve of the fitted FAD spectrum was calculated to represent the integrated baseline FAD fluorescence intensity. Then, FPC was calculated as the ratio of integrated PpIX and FAD fluorescence intensities.
Figure 4.
Representative spectral data (—), spectral data used in fit (○), polynomial fit (×), and the resulting PpIX spectrum (--).
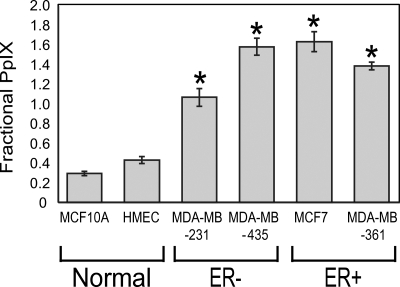
Figure 5 shows the FPC for each cell line. All breast cancer cell lines have a significantly higher FPC than normal mammary epithelial cell lines. It is not surprising to note that the FPC and percentage change in fluorescence intensity, (Figs. 35), are significantly and positively correlated (Pearson coefficient=0.92, p<0.05). No differences in FPC were observed between ER+ and ER− cell lines. Therefore, it can be said that PpIX fluorescence is preferentially accumulated in all breast cancer cells studied here, regardless of ER expression.
Figure 5.
Average and standard error of the FPC. Asterisks denote that the FPC of each breast cancer cell line is significantly higher than those of both normal mammary epithelial cell lines (p<0.05).
Discussion
This study demonstrates the ability of ALA-induced PpIX to discriminate breast cancer cell lines from normal mammary epithelial cells. Breast cancer cell lines used in this study comprised a variety of phenotypes, including ER− and ER+ (ER is expressed in ∼60% of all breast cancers).47 The research showed that fluorescence of ALA-treated cells (two normal mammary epithelial and four breast cancer cell lines) was significantly greater than the equivalent untreated cell lines. Also, breast cancer cell lines could be easily discriminated from normal mammary epithelial cell lines by determining the percentage change in fluorescence intensity after PpIX treatment. However, the high endogenous FAD fluorescence present in untreated normal mammary epithelial cells resulted in overall greater post-treatment fluorescence in normal mammary epithelial cells compared to breast cancer cells and demonstrated the need for a method to account for the cell-to-cell variability in FAD fluorescence in the absence of pretreatment images. Spectroscopy provided a means of separating endogenous fluorescence contribution from ALA-induced PpIX fluorescence and enabled discrimination of normal mammary epithelial and breast cancer cells based on a single post-treatment measurement.
Ratiometric methods have been used previously for ALA studies in oral, bladder, and head and neck tissues31, 41, 48 to account for the contribution of endogenous fluorescence. Fluorometric ratios of red fluorescence to green or blue fluorescence were able to detect malignancies in these various organ sites.31, 41, 48 However, implementing these methods to account for endogenous fluorescence in the present study did not produce statistically significant differences in the breast cancer cell lines compared to the normal mammary epithelial lines. The probable reason for the lack of significant differences in this study was that the high endogenous fluorescence in the green or blue wavelength range overwhelms the red PpIX contribution. Instead of calculating the ratio in the blue or green, decoupling PpIX fluorescence from the overall remitted fluorescence to compute fractional PpIX fluorescence contribution was required for discrimination of normal mammary epithelium from all breast cancer cell lines and could be completed with a single post-treatment spectroscopic measurement.
Spectroscopic approaches similar to that presented here for separating PpIX fluorescence from endogenous tissue fluorescence has been previously reported by Klinteberg et al.,21 who used a similar technique for separation of PpIX fluorescence from tissue autofluorescence in photodynamic therapy of basal cell carcinoma. The endogenous fluorescence was removed by exponentially fitting the FAD fluorescence and then subtracting the FAD signal out to determine the photoproducts after photodynamic therapy. Also, spectroscopic deconvolution of the endogenous signal from PpIX spectra has been previously shown by Gibbs-Strauss et al.49 Comparison of the detected signals to a liquid tissue phantom containing PpIX allowed for delineation of the PpIX signal from nonspecific fluorescence transmitted through normal and cancerous tissue in a mouse brain.49 The results from the study by Gibbs-Strauss et al.49 provided evidence of high endogenous fluorescence background when measuring PpIX fluorescence from ALA-treated cancer.
The breast cancer cell lines in this study exhibited a significant increase in fluorescence intensity compared to normal mammary epithelial cells after administration of ALA, as seen previously in vitro.16, 23, 29 Only one other in vitro study has compared PpIX production in a breast cancer and corresponding normal cell line. Specifically, Rodriguez et al.29 showed that HB4-A R-ras breast cancer cell lines had higher PpIX fluorescence compared to immortalized cells not transfected with the Ras oncogene.
The cellular origin of the MDA-MB-435 cell line has recently been questioned. It has previously been shown that the MDA-MB-435 has a gene-expression profile consistent with M14 melanoma cells.50 However, more recently it has been argued that the MDA-MB-435 cell line is indeed of breast cancer origin.51 The MDA-MB-435 originated from a female, and the original M14 melanoma line was reported to be derived from a male patient.51 The current M14 melanoma line stock does not contain a Y chromosome, which indicates that the M14 was most likely compromised.51 In spite of the controversy surrounding the origin of the MDA-MB-435 cell line, the results from this study demonstrate concordance between MDA-MB-435 and the other breast cancer cell lines studied in terms of its uptake of ALA.
This paper presents an examination of the diagnostic potential of PpIX with a panel of normal mammary epithelial and breast cancer cell lines. It was found that the relative change in PpIX fluorescence intensity could discriminate breast cancer from normal mammary epithelial cell lines and was not affected by varying ER expression in cell lines. The ability to discriminate cells post-ALA treatment shows the potential for the use of spectroscopic methods in margin assessment of breast cancer with PpIX fluorescence, regardless of ER status. The MDA-MB-361 cell line tested here also expressed the HER2+∕NEU phenotype model, but did not have a significantly different PpIX fluorescence intensity as compared to all other cancer cells tested. Therefore, HER2+∕NEU cell lines were not further explored by our group, and we believe PpIX would have similar results in other phenotypes, as shown by the lack of difference in the HER2+∕NEU phenotype MDA-MB-361 cells.
This study is an important initial step in characterizing PpIX fluorescence expression within different biological subtypes of breast cancer cell lines and comparing them to normal mammary epithelial cell lines. Because of the limitations of an in vitro study, it was not possible to evaluate the possible effects of vascular transport of ALA to the target tissues and the corresponding effect on uptake of ALA and conversion to PpIX. Thus, further work is required to address this issue, and the work presented here is a first step toward realizing the utility of ALA for breast cancer delineation. In future studies, ALA-induced PpIX fluorescence will be examined in xenograft breast cancer models, including ER and HER2+∕NEU phenotypes.
Acknowledgments
The authors acknowledge the support of the NIH Imaging Training Grant and DOD Grant No. W81XWH-05-1-0363. The authors also acknowledge the intellectual input and financial support of GE Global Research Center and state that no conflict of interest exists.
References
- Cabioglu N., Hunt K., Sahin A., and Kuerer H., “Role for intraoperative margin assessment in patients undergoing breast-conserving surgery,” Ann. Surg. Oncol. 14, 1458–1471 (2007). 10.1245/s10434-006-9236-0 [DOI] [PubMed] [Google Scholar]
- Bani M. R., Lux M. P., Heusinger K., Wenkel E., Magener A., Schulz-Wendtland R., Beckmann M. W., and Fasching P. A., “Factors correlating with reexcision after breast-conserving therapy,” Eur. J. Surg. Oncol. 35(1), 32–37 (2009). [DOI] [PubMed] [Google Scholar]
- Dillon M. F., Mc Dermott E. W., O’Doherty A., Quinn C. M., Hill A. D., and O’Higgins N., “Factors affecting successful breast conservation for ductal carcinoma in situ,” Ann. Surg. Oncol. 14(5), 1618–28 (2007). 10.1245/s10434-006-9246-y [DOI] [PubMed] [Google Scholar]
- Florentine B. D., Kaushal N. K., Puig M. R., Sarda N., Senofsky G., Cooper B. T., Black A., Zimmerman R., and Barstis J., “Breast-conservation treatment outcomes: a community hospital’s experience,” Breast J. 15(1), 76–84 (2009). 10.1111/j.1524-4741.2008.00674.x [DOI] [PubMed] [Google Scholar]
- Huston T. L., Pigalarga R., Osborne M. P., and Tousimis E., “The influence of additional surgical margins on the total specimen volume excised and the reoperative rate after breast-conserving surgery,” Am. J. Surg. 192(4), 509–512 (2006). 10.1016/j.amjsurg.2006.06.021 [DOI] [PubMed] [Google Scholar]
- Kurniawan E. D., Wong M. H., Windle I., Rose A., Mou A., Buchanan M., Collins J. P., Miller J. A., Gruen R. L., and Mann G. B., “Predictors of surgical margin status in breast-conserving surgery within a breast screening program,” Ann. Surg. Oncol. 15(9), 2542–2549 (2008). 10.1245/s10434-008-0054-4 [DOI] [PubMed] [Google Scholar]
- McLaughlin S. A., Ochoa-Frongia L. M., Patil S. M., H. S.Cody, 3rd, and Sclafani L. M., “Influence of frozen-section analysis of sentinel lymph node and lumpectomy margin status on reoperation rates in patients undergoing breast-conservation therapy,” J. Am. Coll. Surg. 206(1), 76–82 (2008). 10.1016/j.jamcollsurg.2007.07.021 [DOI] [PubMed] [Google Scholar]
- Mendez J. E., Lamorte W. W., de Las Morenas A., Cerda S., Pistey R., King T., Kavanah M., Hirsch E., and Stone M. D., “Influence of breast cancer margin assessment method on the rates of positive margins and residual carcinoma,” Am. J. Surg. 192(4), 538–540 (2006). 10.1016/j.amjsurg.2006.06.009 [DOI] [PubMed] [Google Scholar]
- Menes T. S., Tartter P. I., Bleiweiss I., Godbold J. H., Estabrook A., and Smith S. R., “The consequence of multiple re-excisions to obtain clear lumpectomy margins in breast cancer patients,” Ann. Surg. Oncol. 12(11), 881–885 (2005). 10.1245/ASO.2005.03.021 [DOI] [PubMed] [Google Scholar]
- Waljee J. F., Hu E. S., Newman L. A., and Alderman A. K., “Predictors of re-excision among women undergoing breast-conserving surgery for cancer,” Ann. Surg. Oncol. 15(5), 1297–1303 (2008). 10.1245/s10434-007-9777-x [DOI] [PubMed] [Google Scholar]
- Lin A. W., Lewinski N. A., West J. L., Halas N. J., and Drezek R. A., “Optically tunable nanoparticle contrast agents for early cancer detection: model-based analysis of gold nanoshells,” J. Biomed. Opt. 10(6), 064035 (2005). 10.1117/1.2141825 [DOI] [PubMed] [Google Scholar]
- Sokolov K., Nida D., Descour M., Lacy A., Levy M., Hall B., Dharmawardhane S., Ellington A., Korgel B., and Richards-Kortum R., “Molecular optical imaging of therapeutic targets of cancer,” Adv. Cancer Res. 96, 299–344 (2007). 10.1016/S0065-230X(06)96011-4 [DOI] [PubMed] [Google Scholar]
- Veiseh M., Gabikian P., Bahrami S. B., Veiseh O., Zhang M., Hackman R. C., Ravanpay A. C., Stroud M. R., Kusuma Y., Hansen S. J., Kwok D., Munoz N. M., Sze R. W., Grady W. M., Greenberg N. M., Ellenbogen R. G., and Olson J. M., “Tumor paint: a chlorotoxin:Cy5.5 bioconjugate for intraoperative visualization of cancer foci,” Cancer Res. 67(14), 6882–6888 (2007). 10.1158/0008-5472.CAN-06-3948 [DOI] [PubMed] [Google Scholar]
- Uzgiris E., Sood A., Bove K., Grimmond B., Lee D., and Lomnes S., “A multimodal contrast agent for preoperative MR imaging and intraoperative tumor margin delineation,” Technol. Cancer Res. Treat. 5(4), 301–309 (2006). [DOI] [PubMed] [Google Scholar]
- Frei K. A., Bonel H. M., Frick H., Walt H., and Steiner R. A., “Photodynamic detection of diseased axillary sentinel lymph node after oral application of aminolevulinic acid in patients with breast cancer,” Br. J. Cancer 90(4), 805–809 (2004). 10.1038/sj.bjc.6601615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallegos E. R., DeLeon Rodriguez I., Martinez Guzman L. A., and Perez Zapata A. J., “In vitro study of biosynthesis of protoporphyrin IX induced by delta-aminolevulinic acid in normal and cancerous cells of the human cervix,” Arch. Med. Res. 30(3), 163–170 (1999). 10.1016/S0188-0128(99)00013-5 [DOI] [PubMed] [Google Scholar]
- Gamarra F., Wagner S., Al-Batran S., Maier I., Castro M., Hautmann H., Bergner A., Baumgartner R., and Huber R. M., “Kinetics of 5-aminolevulinic acid-induced fluorescence in organ cultures of bronchial epithelium and tumor,” Respiration 69(5), 445–450 (2002). 10.1159/000064008 [DOI] [PubMed] [Google Scholar]
- Gibbs S. L., Chen B., O’Hara J. A., Hoopes P. J., Hasan T., and Pogue B. W., “Protoporphyrin IX level correlates with number of mitochondria, but increase in production correlates with tumor cell size,” Photochem. Photobiol. 82(5), 1334–1341 (2006). 10.1562/2006-03-11-RA-843 [DOI] [PubMed] [Google Scholar]
- Hirschberg H., Sorensen D., Angell-Petersen E., Peng Q., Tromberg B., Sun C., Spetalen S., and S M., “Repetitive photodynamic therapy of malignant brain tumors,” J. Environ. Pathol. Toxicol. Oncol. 25(1–2), 261–279. (2006). [DOI] [PubMed] [Google Scholar]
- Ji Z., Yang G., Vasovic V., Cunderlikova B., Suo Z., Nesland J. M., and Peng Q., “Subcellular localization pattern of protoporphyrin IX is an important determinant for its photodynamic efficiency of human carcinoma and normal cell lines,” J. Photochem. Photobiol., B 84(3), 213–220 (2006). 10.1016/j.jphotobiol.2006.03.006 [DOI] [PubMed] [Google Scholar]
- Klinteberg C., Enejder A., Wang I., Andersson-Engels S., Svanberg S., and Svanberg K., “Kinetic fluorescence studies of 5-aminolaevulinic acid-induced protoporphyrin IX accumulation in basal cell carcinomas,” J. Photochem. Photobiol. B: Biol. 49, 120–128 (1999). 10.1016/S1011-1344(99)00045-7 [DOI] [PubMed] [Google Scholar]
- Krammer B. and Plaetzer K., “ALA and its clinical impact from benchside to bedside,” Photochem. Photobiol. Sci. 7(3), 283–289 (2008). 10.1039/b712847a [DOI] [PubMed] [Google Scholar]
- Krieg R. C., Messmann H., Rauch J., Seeger S., and Knuechel R., “Metabolic characterization of tumor cell-specific protoporphyrin IX accumulation after exposure to 5-aminolevulinic acid in human colonic cells,” Photochem. Photobiol. 76(5), 518–525 (2002). [DOI] [PubMed] [Google Scholar]
- Ladner D. P., Steiner R. A., Allemann J., Haller U., and Walt H., “Photodynamic diagnosis of breast tumours after oral application of aminolevulinic acid,” Br. J. Cancer 84(1), 33–37 (2001). 10.1054/bjoc.2000.1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leunig A., Rick K., Stepp H., Gutmann R., Alwin G., Baumgartner R., and Feyh J., “Fluorescence imaging and spectroscopy of 5-aminolevulinic acid induced protoporphyrin IX for the detection of neoplastic lesions in the oral cavity,” Am. J. Surg. 172(6), 674–677 (1996). 10.1016/S0002-9610(96)00312-1 [DOI] [PubMed] [Google Scholar]
- Messmann H., Knuchel R., Baumier W., Holstege A., and Scholmerich J., “Endoscopic fluorescence detection of dysplasia in patients with Barrett’s esophagus, ulcerative colitis, or adenomatous polyps after 5-aminolevulinic acid-induced protoporphyrin IX sensitization,” Gastrointest. Endosc. 49(1), 1–5 (1999). 10.1016/S0016-5107(99)70453-0 [DOI] [PubMed] [Google Scholar]
- Na R., Stender I., and Wulf H., “Can autofluorescence demarcate basal cell carcinoma from normal skin? A comparison with protoporphyrin IX fluorescence,” Acta Derm Venereol 81, 246–249 (2001). 10.1080/00015550152572859 [DOI] [PubMed] [Google Scholar]
- Navone N., Polo C., Frisardi A., Andrade N., and Battle A., “Heme biosynthesis in human breast cancer—mimetic in vitro studies and some heme enzymic activity levels,” Int. J. Biochem. 22(12), 1407–1411 (1990). 10.1016/0020-711X(90)90230-Z [DOI] [PubMed] [Google Scholar]
- Rodriguez L., Divenosa G., Batlle A., Macrobert A., and Casas A., “Response to ALA-based PDT in an immortalised normal breast cell line and its counterpart transformed with the Ras oncogene,” Photochem. Photobiol. Sci. 6(12), 1306–1310 (2007). 10.1039/b704235c [DOI] [PubMed] [Google Scholar]
- Sailer R., Strauss W. S., Wagner M., Emmert H., and Schneckenburger H., “Relation between intracellular location and photodynamic efficacy of 5-aminolevulinic acid-induced protoporphyrin IX in vitro. Comparison between human glioblastoma cells and other cancer cell lines,” Photochem. Photobiol. Sci. 6(2), 145–151 (2007). 10.1039/b611715e [DOI] [PubMed] [Google Scholar]
- Sharwani A., Jerjes W., Salih V., MacRobert A. J., El-Maaytah M., Khalil H. S., and Hopper C., “Fluorescence spectroscopy combined with 5-aminolevulinic acid-induced protoporphyrin IX fluorescence in detecting oral premalignancy,” J. Photochem. Photobiol., B 83(1), 27–33 (2006). 10.1016/j.jphotobiol.2005.11.007 [DOI] [PubMed] [Google Scholar]
- Moesta K. T., Ebert B., Handke T., Nolte D., Nowak C., Haensch W. E., Pandey R. K., Dougherty T. J., Rinneberg H., and Schlag P. M., “Protoporphyrin IX occurs naturally in colorectal cancers and their metastases,” Cancer Res. 61(3), 991–999 (2001). [PubMed] [Google Scholar]
- Andersson-Engels S., Canti G., Cubeddu R., Eker C., Klinteberg C., Pifferi A., Svanberg K., Svanberg S., Taroni P., Valentini G., and Wang I., “Preliminary evaluation of two fluorescence imaging methods for the detection and the delineation of basal cell carcinomas of the skin,” Lasers Surg. Med. 26(1), 76–82 (2000). [DOI] [PubMed] [Google Scholar]
- Betz C. S., Stepp H., Janda P., Arbogast S., Grevers G., Baumgartner R., and Leunig A., “A comparative study of normal inspection, autofluorescence and 5-ALA-induced PPIX fluorescence for oral cancer diagnosis,” Int. J. Cancer 97(2), 245–252 (2002). 10.1002/ijc.1596 [DOI] [PubMed] [Google Scholar]
- Chen J. Y., Mak N. Q., Cheung N. H., Leung R. N., and Peng Q., “Endogenous production of protoporphyrin IX induced by 5-aminolevulinic acid in leukemia cells,” Acta Pharmacol. Sin. 22(2), 163–168 (2001). [PubMed] [Google Scholar]
- Dorward A. M., Fancher K. S., Duffy T. M., Beamer W. G., and Walt H., “Early neoplastic and metastatic mammary tumours of transgenic mice detected by 5-aminolevulinic acid-stimulated protoporphyrin IX accumulation,” Br. J. Cancer 93(10), 1137–1143 (2005). 10.1038/sj.bjc.6602840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotinos N., Campo M. A., Popowycz F., Gurny R., and Lange N., “5-Aminolevulinic acid derivatives in photomedicine: Characteristics, application and perspectives,” Photochem. Photobiol. 82(4), 994–1015 (2006). 10.1562/2006-02-03-IR-794 [DOI] [PubMed] [Google Scholar]
- Berger Y., Chapuis Bernasconi C., Schmitt F., Neier R., and Juillerat-Jeanneret L., “Determination of intracellular prolyl∕glycyl proteases in intact living human cells and protoporphyrin IX production as a reporter system,” Chem. Biol. 12(8), 867–872 (2005). 10.1016/j.chembiol.2005.05.016 [DOI] [PubMed] [Google Scholar]
- Alvarez M. G., Lacelli M. S., Rivarola V., Batlle A., and Fukuda H., “5-aminolevulinic acid-mediated photodynamic therapy on Hep-2 and MCF-7c3 cells,” J. Environ. Pathol. Toxicol. Oncol. 26(2), 75–82 (2007). [DOI] [PubMed] [Google Scholar]
- Wu S., Ren Q., Zhou M., Wei Y., and Chen J., “Photodynamic effects of 5-aminolevulinic acid and its hexylester on several cell lines,” Sheng Wu Hua Xue Yu Shen Wu Wu Li Xue Bao 35(7), 655–660 (2003). [PubMed] [Google Scholar]
- Zaak D., Frimberger D., Stepp H., Wagner S., Baumgartner R., Schneede P., Siebels M., Knuchel R., Kriegmair M., and Hofstetter A., “Quantification of 5-aminolevulinic acid induced fluorescenc improves the specificity of bladder cancer detection,” J. Urol. (Baltimore) 166(5), 1665–1668 (2001). [PubMed] [Google Scholar]
- European Public Assessment Report: Gliolan Available from ⟨http://www.emea.europa.eu/humandocs/PDFs/EPAR/gliolan/H-744-en1.pdf⟩ (2007).
- Tsai T., Ji H. T., Chiang P. C., Chou R. H., Chang W. S., and Chen C. T., “ALA-PDT results in phenotypic changes and decreased cellular invasion in surviving cancer cells,” Lasers Surg. Med. 41(4), 305–315 (2009). 10.1002/lsm.20761 [DOI] [PubMed] [Google Scholar]
- Anandappa S., Sibson R., Platt-Higgins A., Winstanley J., Rudland P., and Barraclough R., “Variant estrogen receptor alpha mRNAs in human breast cancer specimens,” Int. J. Cancer 88, 209–216 (2000). [DOI] [PubMed] [Google Scholar]
- ⟨www.protocol-online.org/biology-forums/posts/5763.htm⟩ (2008).
- DaCosta R. S., Andersson H., and Wilson B. C., “Molecular fluorescence excitation-emission matrices relevant to tissue spectroscopy,” Photochem. Photobiol. 78(4), 384–392 (2003). [DOI] [PubMed] [Google Scholar]
- Dietze E. C., Troch M. M., Bean G. R., Heffner J. B., Bowie M. L., Rosenberg P., Ratliff B., and Seewaldt V. L., “Tamoxifen and tamoxifen ethyl bromide induce apoptosis in acutely damaged mammary epithelial cells through modulation of AKT activity,” Oncogene 23(21), 3851–3862 (2004). 10.1038/sj.onc.1207480 [DOI] [PubMed] [Google Scholar]
- Wang I., Clemente L., Pratas R., Cardoso E., Clemente M., Montan S., Svanberg S., and Svanberg K., “Fluorescence diagnostics and kinetics studies in the head and neck region utilizing low-dose d-aminolevulinic acid sensitization,” Cancer Lett. 135(11–9), 1–8 (1999). [DOI] [PubMed] [Google Scholar]
- Gibbs-Strauss S. L., O’Hara J. A., Hoopes P. J., Hasan T., and Pogue B. W., “Noninvasive measurement of aminolevulinic acid-induced protoporphyrin IX fluorescence allowing detection of murine glioma in vivo,” J. Biomed. Opt. 14(1), 014007 (2009). 10.1117/1.3065543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae J. M., Creighton C. J., Meck J. M., Haddad B. R., and Johnson M. D., “MDA-MB-435 are derived from M-14 melanoma cells-a loss for breast cancer, but a boon for melanoma research,” Breast Cancer Res. Treat. 104(1), 13–19 (2007). 10.1007/s10549-006-9392-8 [DOI] [PubMed] [Google Scholar]
- Chambers A. F., “MDA-MB-435 and M14 cell lines: Identical but not M14 melanoma?” Cancer Res. 69(13), 5292–5293 (2009). 10.1158/0008-5472.CAN-09-1528 [DOI] [PubMed] [Google Scholar]