Abstract
Chromatin structure is a powerful tool to regulate eukaryotic transcription. Moreover, nucleosomes are constantly remodeled, disassembled, and reassembled in the body of transcribed genes. Here we propose a general model that explains, in quantitative terms, how transcription elongation affects nucleosome structure at a distance as a result of the positive torque the polymerases create as they translocate along DNA templates.
It is nowadays fully recognized that chromatin structure serves two major purposes: to compact the large amount of DNA into the confines of the eukaryotic nucleus and to participate in control of transcription and other processes that use DNA as a template. Apart from this “control” function, chromatin must undergo dramatic changes during these processes by the simple need to have DNA exposed to the machineries that act to replicate, transcribe, repair, and recombine genetic information. We now know that chromatin is in constant flux with nucleosomes being remodeled or totally removed from the path of nuclear enzymes and being quickly reassembled in the wake of these enzymes. However, our knowledge of how chromatin structure changes during these processes and the molecular mechanisms behind these changes are poorly understood. Here, we make use of the abundant information on transcription elongation of Drosophila heat-shock genes to attempt to uncover the mechanisms responsible for chromatin changes. We specifically focus on the recent publication from John Lis laboratory (Petesch and Lis, 2008) since it reveals some unexpected features of nucleosome loss on transcription activation. We propose a model that not only explains the Petesch and Lis observations but applies to transcription elongation through chromatin in general.
THE HSP70 GENE IN DROSOPHILA: A MODEL SYSTEM TO STUDY TRANSCRIPTION THROUGH CHROMATIN
The Hsp70 gene under non-heat shock conditions
Heat-shock genes in Drosophila provide a useful system to study regulation of transcription of inducible eukaryotic genes. These genes constitute a family of genes whose expression is rapidly (within seconds) and robustly (∼500 fold increase in mRNA levels) induced under a variety of stress conditions including heat-shock. The individual members of the gene family are spread over the genome but all undergo highly synchronous changes in chromatin structure on heat-shock. These changes are recognized cytologically as appearance of decondensed “puffs” in the polytene chromosomes that exist in the cells of the salivary glands (polytene chromosomes arise from successive rounds of DNA replication that are not followed by cell divisions; the chromatids in these chromosomes remain aligned along their lengths, giving rise to giant chromosome easily identifiable by light microscopy even in interphase cells).
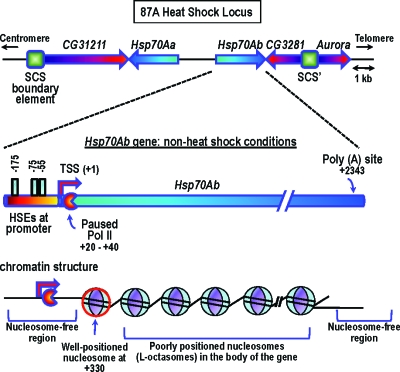
The genomic structure of one heat-shock locus, 87A, is depicted in Fig. 1. The Hsp70Ab gene possesses three heat-shock elements in its promoter region; these elements provide the binding sites for the heat-shock transcription factor (HSF), the critical activator ofheat-shock genes. The HSF is not bound under these conditions; however, a number of other proteins, including GAGA transcription factor, TATA-binding protein TBP, Spt5, and negative elongation factor (NELF) are present on the gene. Poly (ADP-ribose) polymerase 1 (PARP1) is also present near the 5′ end of the gene (Petesch and Lis, personal communication).
Figure 1. The HS87A heat-shock locus under non-heat shock conditions (further details given in text).
Even under non-heat shock conditions, the gene harbors a paused molecule of RNA polymerase II (Pol II) at position +20 to +40. Pol II stalling at this position occurs even after the gene is induced; however, the residence time of the stalled Pol II dramatically decreases on gene activation, as reviewed in Saunders et al. (2006), Lis (2007), and Nechaev and Adelman (2008). The existence of this promoter-proximal pausing has been considered an idiosyncratic feature of heat-shock genes. Recent genomewide studies have, however, revealed that promoter-proximal pausing may be rather widespread affecting approximately 10–15% of all Drosophila genes (Lis, 2007; Nechaev and Adelman, 2008). Genomewide Pol II localization studies in human cells indicate a similar widespread occurrence of promoter-proximal stalling; for further references, see Lis (2007).
What about the chromatin organization over the Hsp70Ab gene? Under non-heat shock conditions, the gene is characterized by two nucleosome-free regions: one large region at the promoter and the 5′ portion of the coding region (encompassing the stalled Pol II) and another one at the 3′ end, including the Poly(A) site (Fig. 1) (Petesch and Lis, 2008). The first nucleosome is well positioned, centered at +330 (well downstream of the stalled Pol II); the nucleosomes in the body of the gene gradually lose their positioning.
The transcription of the Hsp70 gene under heat-shock conditions
Applying heat-shock to Drosophila cells in culture leads to a prompt and robust transcriptional activation of the gene. HSF binds to the HSEs in the promoter within 5 s of heat-shock, thus constituting the first response to the activation signal (Yao et al., 2006). Interestingly, some HSF bind even under non-induced conditions but the bound molecules undergo rapid exchange with the free pool of HSF in the nucleoplasm; heat-shock induction results in stabilization of the binding with the protein showing very slow exchange once transcriptional activation occurs (Yao et al., 2006). The paused Pol II at the promoter-proximal site is released into productive elongation through the action of the positive transcription elongation factor P-TEFb. P-TEFb is a kinase that phosphorylates serine 2 on the C-terminal domain of the largest subunit of Pol II, NELF, and DSIF; phosphorylated NELF leaves the complex—other subunits join and travel with the polymerase along the gene, as reviewed in Saunders et al. (2006), Lis (2007), and Nechaev and Adelman (2008). Finally, early in induction, new unphosphorylated Pol II molecules enter the promoter. These molecules also pause at the promoter-proximal site but the duration of the stall is dramatically decreased in comparison with the non-heat shock conditions. It is not yet clear what factors modulate the residence time of Pol II at this site.
THE NEW OBSERVATIONS IN PETESCH AND LIS (2008)
Petesch and Lis (2008) reported several important observations concerning the changes in chromatin structure that occur on the transcription activation of the Hsp70Ab gene. The chromatin organization was assessed by classical micrococcal nuclease (MNase) treatment of fibers isolated from formaldehyde-fixed control and heat-shocked cells early during the induction process. The MNase protection pattern was determined by PRC amplification using primer sets that would amplify overlapping 100 bp regions along the gene (the presence of amplified fragments depends on the amount of contiguous DNA between the primers that still remains following MNase digestion). The authors observed a rapid and broad disruption of nucleosome structure; importantly, this disruption occurred faster than the rate on Pol II translocation along the gene. It affected neighboring genes and effectively stopped at the two insulator elements (scs and scs’) that flank the region (Fig. 1). Importantly, chromatin immunoprecipitation (ChIP) experiments indicated a decreased density in histone H3 in the two regions tested (the +330 region that contains the well positioned nucleosome in non-inducing conditions and the +1702 region toward the 3′ end of the gene). Of note, even at 2 min of heat-shock, when the first round of transcription has already occurred, ∼30% of the histones are still present at both these positions.
The authors argue that the initial changes in chromatin architecture are not dependent on ongoing transcription. We will describe these results in some detail since they are crucial to the interpretation. The authors used two chemicals known to reduce the level of transcription: the nucleotide analog DRB (Giardina and Lis, 1993) and sodium salicylate (Winegarden et al., 1996). Indeed, treatment of cells with DRB before applying the heat-shock resulted in normal recruitment of HSF to the promoter and of Pol II to the pause site but no polymerase was detected at the +1702 position, indicating inhibition of elongation. The MNase protection profile of DRB-treated cells at 2 min of heat-shock did show disruption of nucleosomal structure, however the disruption was only partial and much less than the 2 min shocked control (untreated) cells. This “partial” response of the nucleosome profile to heat-shock of DRB-treated cells may reflect incomplete inhibition of elongation by the chemical. Indeed, DRB has been shown to reduce the number of polymerases on the 3′ end of genes (Fraser et al., 1978; Petesch and Lis, 2008) while having little effect on polymerase density in the promoter-proximal region (Fraser et al., 1978).
The second agent used as an inhibitor of elongation, salicylate, does induce HSF binding under non-heat shock conditions (Winegarden et al., 1996; Petesch and Lis, 2008) and heat-shock puffs (Winegarden et al., 1996) but fails to induce transcription as assayed by primer extension (Winegarden et al., 1996). However, primer extension assays only reveal the presence of full length transcripts, and as discussed by Winegarden et al. (1996), the salicylate-induced puffs represent a localized decondensation brought about by HSF binding and early transcription events. Of note, the amount of Pol II at the pause site in salicylate-treated (non-shocked and shocked) cells is significant (comparable to the non-shocked untreated cells) and may represent the steady state level between polymerases released from the pause and those that enter the pause site from the newly recruited enzymes. Finally, in both cases of drug inhibition of elongation, the lack of Pol II occupancy of the +1702 site does not necessarily indicate lack of transcription at the beginning of the gene; actually in both cases, some transcription does seem to occur. The significance of this statement will become obvious later during this discussion.
Finally, Petesch and Lis (2008) reported another set of very interesting, and potentially important, observations. Using selective RNAi screens, the authors identified absolute requirements for HSF, GAGA factor, and PARP1 for the changes in the chromatin landscape occurring on heat-shock. PARP’s enzymatic activity seemed to be required for the loss of nucleosomes. At least in vitro, the automodification reaction (the attachment of long, branched poly (ADP-ribose) polymers to numerous sites of the enzyme) is needed to release PARP from the nucleosome, potentially leading to transcriptional activation (Kim et al., 2004). Thus, PARP1 release may be a prerequisite for the further chromatin changes (nucleosome loss) observed.
It may be worthwhile taking a broader look at the involvement of PARP1 in transcription. Recent work has implicated PARP and∕or its enzymatic activity in numerous processes that regulate transcription, as reviewed in Kraus (2008). PARP interacts with nucleosomes at the site where linker DNA enters and exits the nucleosome, very similar to the way that linker histones interact with the particle (Kim et al., 2004). PARP1 and linker histones compete for binding to nucleosomes in vitro and reside in distinct nucleosome fractions in vivo (Kim et al., 2004). Genomewide and gene-specific localization studies at gene promoters have also revealed reciprocal binding of PARP1 and linker histones (Krishnakumar et al., 2008). Importantly, despite the similar nucleosome-binding properties of PARP1 and linker histones, they define distinct transcriptional outcomes in vivo, stimulating or repressing transcription, respectively, (Krishnakumar et al., 2008). The possible functional differences between the two proteins have been recently attributed to the distinct dynamic properties of nucleosomes conferred by the slightly different binding of these two proteins to the nucleosome particle; for further discussion, see Zlatanova et al. (2008). In this view, binding of linker histones creates particles in which the spontaneous thermally driven motions of the nucleosomal DNA ends are prohibited giving rise to particles almost impenetrable to the transcribing polymerase. On the other hand, the binding of PARP1 may allow some “breathing” motions of nucleosomal DNA, allowing for a more dynamic particle, easier to be transcribed. This scenario does not take into account the automodification activity of PARP, which may lead to even greater access to nucleosomal DNA by simply dissociating PARP; nucleosomes that do not have any proteins bound to the DNA entry-exit point will be free to frequently breathe (or “open,”Zlatanova et al., 2008), thus making nucleosomal DNA easily transcribable.
THE POSITIVE SUPERHELICAL STRESS ACCUMULATING IN FRONT OF THE ELONGATING POL II CREATES A WAVE OF R-OCTASOMES
What are the molecular mechanisms that may account for the apparent loss of nucleosomes in front of the transcribing polymerase through an action at a distance? Before suggesting a possible mechanism, we would first recall the state-of-the-art biophysical knowledge of transcription elongation.
The twin-supercoiled domain model
Transcription elongation requires relative rotation of Pol II around the DNA. In the so-called twin-supercoiled domain model (Liu and Wang, 1987) Pol II is assumed to be fixed, anchored to some nuclear structure, e.g., a transcription factory (Cook, 1999), so that the DNA has to screw inside it. This suppresses RNA transcript entangling around the DNA but creates positive supercoiling in the downstream portion of the template and negative supercoiling in the upstream portion. Transcription-coupled negative supercoiling has been observed in vivo, even in the presence of topoisomerase activity, e.g., Matsumoto and Hirose (2004). Moreover, recent experiments (Kouzine et al., 2008) gave the firstin vivo evidence for torque generation by elongating Pol II in eukaryotes.
We therefore incorporate the twin-supercoiled domain model in our further analysis. Relevant to our discussion, supercoiling is confined to individual chromatin domains (loci, chromatin loops) by two insulator elements that flank any given domain; the insulators clamp the DNA at both ends of the domain, thus topologically constraining the chromatin fiber.
Nucleosome dynamics in response to torsional stress
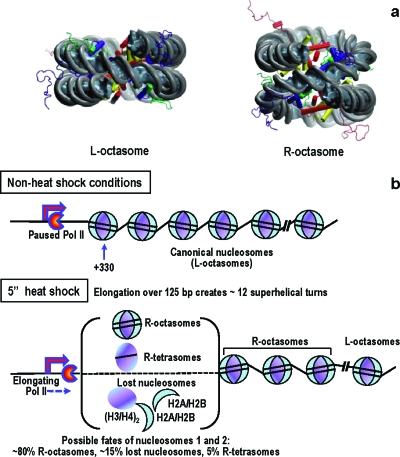
Recent magnetic tweezers studies evaluated the response of a single chromatin fiber to applied torsion (Bancaud et al., 2006 and 2007). The authors showed that nucleosomal arrays can absorb a significant amount of positive stress, without major changes to fiber length. This behavior can be explained by conformational transitions in nucleosomes: due to thermal fluctuations the entry∕exit linkers alternate their relative orientation, passing from the canonical “negatively” crossed configuration (as observed in crystals) to a “positively” crossed state back and forth, possibly going through an open state (in which the linkers do not cross at all). Applying positive torsional stress shifts the equilibrium toward the positively crossed state. When the torsional stress reaches a critical value (see Supplementary Information), the nucleosomes undergo a structural transition, practically turning inside out through a chiral transition of the DNA superhelix (going from negatively to positively wrapped) while preserving the whole histone octamer; for further in-depth discussion, see Zlatanova et al. (2009) [Fig. 2a]. In such a structural transition, the linking number per nucleosome changes from −1 to +1 so that every nucleosome absorbs up to two turns when subjected to high levels of positive torsional stress; in other words, the L-octasome to R-octasome transition acts as a topological buffer.
Figure 2. L-octasomes and R-octasomes and changes occurring in nucleosomes 5 s after heat-shock.
(a) All-atoms structures of the canonical nucleosome (L-octasome) and of the particle produced by the chiral transition (R-octasome). The docking domain of H2A that binds the dimer H2A/H2B to the tetramer (H3∕H4)2 in L-octasomes does not interact with the tetramer in the R-octasome, thus leading to a more open structure. The R-octasome is a metastable particle that is stabilized by the positive torsional stress applied on the fiber by elongating Pol II. (b) Model depicting the changes in nucleosome structure occurring on induction of Hsp70Ab gene (5 s of heat-shock treatment). Elongation of over 125 bp creates ∼12 superhelical turns, which is enough to convert six L-octasomes into R-octasomes at a rate by far exceeding the rate of Pol II translocation. MNase protection assays and histone H3 ChIP indicate changes in the first two nucleosomes as indicated. Similar changes occur with downstream nucleosomes later during Hsp70Ab gene expression.
Proposed physical model for nucleosome disruption
We are now ready to propose a physical model for the extensive and fast nucleosome disruption observed by Petesch and Lis (2008). We hypothesize that HSF binding to HSEs triggers a rapid productive elongation phase during which Pol II translocates over some genomic distance. The movement of Pol II in a topologically constrained environment creates positive torque in the downstream portion of the template, high enough to convert, at a distance, a fraction of L-octasomes (canonical nucleosomes) into R-octasomes (see Supplementary Information); this first productive elongation phase is too fast for topoisomerases to come into play. Before HS, the fiber throughout the locus is compact (heterochromatin) so that nucleosomes can be turned into R-octasomes only gradually through a domino-like effect. This generates a wave downstream that acts long before nucleosomes are physically disrupted by the invading polymerase. R-octasomes are expected to be much less stable and to easily lose H2A∕H2B dimers to form hexasomes or tetrasomes. The all-atom model clearly predicts no strong interactions between the C-terminal portion of H2A (the so-called “docking domain” in canonical L-octasomes) and H3, for example. This is reflected by the much more open structure shown in Fig. 2. Some R-octasomes (or their derivative R-tetrasomes or R-hexasomes) may be lost altogether because H3∕H4 tetramers prefer to bind negatively supercoiled DNA (Zlatanova et al., 2009). Thus, the positive torque in front of the advancing RNAP will produce a complex (random) mixture of R-octasomes, R-hexasomes and R-tetrasomes, or will disrupt the nucleosome particles altogether [Fig. 2b].
How does the proposed model accommodate the Petesch and Lis (2008) data? Our model explains straightforwardly the following major features of nucleosome disruption, as observed by Petesch and Lis (2008). Nucleosome disruption (1) is a cis-acting process, (2) is a rapid, much faster than the rate of Pol II elongation; the wave velocity is ∼200 bp∕s (see Supplementary Information), (3) occurs over the entire locus, and (4) is restricted to the locus: scs and scs’ boundary elements act as barriers to the spread of the disruption wave.
In addition, the model bears on some other observations of Petesch and Lis (2008).
(1) High-resolution MNase protection assays: primer sets are designed to amplify 100 bp regions; thus, protected fragments smaller than 100 bp are not amplified. Therefore, tetrasomes and hexasomes will not be detected by the MNase assay because less than 100 bp (∼50 bp in tetrasomes and ∼95 in hexasomes (Zlatanova et al., 2009) are protected in these particles. Thus, the MNase assay depicts the behavior of intact nucleosomes only but not of tetrasomes or hexasomes (Zlatanova et al., 2009).
The MNase protection profiles at 30 s and 60 s of heat-shock (Petesch and Lis, 2008) represent the nucleosome structure of the locus after the superhelical stress wave (but before Pol II passage). In our model, all nucleosomes would be converted to R-octasomes after only 20 s, i.e., before the time these curves are obtained (30 s and 60 s of heat-shock). Since complete R-octasomes will be detected by the MNase assay exactly as are intact canonical nucleosomes (the histone octamers provide protection over 147 bp in both L-octasomes and R-octasomes), it must be concluded that the apparent disruption of nucleosomes as inferred from this assay does not necessarily reflect complete nucleosome loss; rather loss of one or both H2A∕H2B dimers from some R-octasomes could have occurred.
(2) Histone H3 ChIP assay: the H3 ChIP results reveal the behavior of the first (+330) nucleosome and a nucleosome further downstream (+1702, which corresponds, according to simple calculations, to the seventh nucleosome). In 5 s, the superhelical wave should affect the first but not the seventh nucleosome (see Supplementary Information), which is exactly what is observed. A particle containing more than 100 bp—hence an intact nucleosome—is still present at +330 bp in ∼80% of the population (these measurements are averaged over the entire cell population) (MNase protection assay). Three-fourths of the remaining 20% cells have lost their H3 (ChIP), i.e., the nucleosome has disintegrated; the final 1∕4 is not protected against MNase but has retained its H3—these are most probably R-hexasomes or R-tetrasomes [Fig. 2b].
(3) Nucleosome disruption under drug inhibition of transcription elongation: the most puzzling observation is the nucleosome pattern disruption seen when elongation is inhibited by chemicals. However, as discussed above, neither of the two treatments used inhibits elongation completely. Because the superhelical wave is so fast and so potent (see Supplementary Information), only very limited transcription is needed to cause the observed behavior.
CONCLUSION
Chromatin is a highly dynamic structure, and processes that need access to the underlying DNA template by necessity cause profound changes in chromatin. Two factors can affect nucleosome structure during transcription elongation: the physical “invasion” of the nucleosome by the translocating RNA polymerase and the creation of positive superhelical stress in front of the enzyme. This second factor can act at a distance, as long as the chromatin fiber is topologically constrained, as is the case in vivo. The wave of positive stress is extremely fast and can create conditions in which nucleosomes are highly destabilized to help the polymerase overcome the nucleosomal barrier.
ACKNOWLEDGMENTS
J.Z. is supported in part by NSF Grant No. 0504239; J.M.V. by ANR Grant No. 05-NANO-062-03. The authors are indebted to Hua Wong for the all-atom models of Fig. 2a.
References
- Bancaud, A, et al. (2007). “Nucleosome chiral transition under positive torsional stress in single chromatin fibers.” Mol. Cell 27, 135–147. 10.1016/j.molcel.2007.05.037 [DOI] [PubMed] [Google Scholar]
- Bancaud, A, Conde e Silva, N, Barbi, M, Wagner, G, Allemand, J F, Mozziconacci, J, Lavelle, C, Croquette, V, Victor, J M, Prunell, A, and Viovy, J L (2006). “Structural plasticity of single chromatin fibers revealed by torsional manipulation.” Nat. Struct. Mol. Biol. 13, 444–450. 10.1038/nsmb1087 [DOI] [PubMed] [Google Scholar]
- Cook, P R (1999). “The organization of replication and transcription.” Science 284, 1790–1795. 10.1126/science.284.5421.1790 [DOI] [PubMed] [Google Scholar]
- See EPAPS Document No. E-HJFOA5-3-004907 for supplemental material. This document can be reached through a direct link in the online’s HTML reference section or via the EPAPS homepage (http://www.aip.org/pubservs/epaps.html).
- Fraser, N W, Sehgal, P B, and Darnell, J E (1978). “DRB-induced premature termination of late adenovirus transcription.” Nature (London) 272, 590–593. 10.1038/272590a0 [DOI] [PubMed] [Google Scholar]
- Giardina, C, and Lis, J T (1993). “Polymerase processivity and termination on Drosophila heat shock genes.” J. Biol. Chem. 268, 23806–23811. [PubMed] [Google Scholar]
- Kim, M Y, Mauro, S, Gevry, N, Lis, J T, and Kraus, W L (2004). “NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1.” Cell 119, 803–814. 10.1016/j.cell.2004.11.002 [DOI] [PubMed] [Google Scholar]
- Kouzine, F, Sanford, S, Elisha-Feil, Z, and Levens, D (2008). “The functional response of upstream DNA to dynamic supercoiling in vivo.” Nat. Struct. Mol. Biol. 15, 146–154. 10.1038/nsmb.1372 [DOI] [PubMed] [Google Scholar]
- Kraus, W L (2008). “Transcriptional control by PARP-1: chromatin modulation, enhancer-binding, coregulation, and insulation.” Curr. Opin. Cell Biol. 20, 294–302. 10.1016/j.ceb.2008.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar, R, Gamble, M J, Frizzell, K M, Berrocal, J G, Kininis, M, and Kraus, W L (2008). “Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes.” Science 319, 819–821. 10.1126/science.1149250 [DOI] [PubMed] [Google Scholar]
- Lis, J T (2007). “Imaging Drosophila gene activation and polymerase pausing in vivo.” Nature (London) 450, 198–202. 10.1038/nature06324 [DOI] [PubMed] [Google Scholar]
- Liu, L F, and Wang, J C (1987). “Supercoiling of the DNA template during transcription.” Proc. Natl. Acad. Sci. U.S.A. 84, 7024–7027. 10.1073/pnas.84.20.7024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto, K, and Hirose, S (2004). “Visualization of unconstrained negative supercoils of DNA on polytene chromosomes of Drosophila.” J. Cell Sci. 117, 3797–3805. 10.1242/jcs.01225 [DOI] [PubMed] [Google Scholar]
- Nechaev, S, and Adelman, K (2008). “Promoter-proximal Pol II: when stalling speeds things up.” Cell Cycle 7, 1539–1544. [DOI] [PubMed] [Google Scholar]
- Petesch, S J, and Lis, J T (2008). “Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci.” Cell 134, 74–84. 10.1016/j.cell.2008.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders, A, Core, L J, and Lis, J T (2006). “Breaking barriers to transcription elongation.” Nat. Rev. Mol. Cell Biol. 7, 557–567. 10.1038/nrm1981 [DOI] [PubMed] [Google Scholar]
- Winegarden, N A, Wong, K S, Sopta, M, and Westwood, J T (1996). “Sodium salicylate decreases intracellular ATP, induces both heat shock factor binding and chromosomal puffing, but does not induce hsp 70 gene transcription in Drosophila.” J. Biol. Chem. 271, 26971–26980. 10.1074/jbc.271.43.26971 [DOI] [PubMed] [Google Scholar]
- Yao, J, Munson, K M, Webb, W W, and Lis, J T (2006). “Dynamics of heat shock factor association with native gene loci in living cells.” Nature (London) 442, 1050–1053. 10.1038/nature05025 [DOI] [PubMed] [Google Scholar]
- Zlatanova, J, Bishop, T C, Victor, J M, Jackson, V, and van Holde, K (2009). “The nucleosome family: dynamic and growing.” Structure (London) 17, 160–171. 10.1016/j.str.2008.12.016 [DOI] [PubMed] [Google Scholar]
- Zlatanova, J, Seebart, C, and Tomschik, M (2008). “The linker-protein network: control of nucleosomal DNA accessibility.” Trends Biochem. Sci. 33, 247–253. 10.1016/j.tibs.2008.04.001 [DOI] [PubMed] [Google Scholar]