Abstract
Observed phenotype often fails to correspond with genotype. Although it is well established that uncontrolled genetic modifier effects and environmental variability can affect phenotype, stochastic variation in gene expression can also contribute to phenotypic differences. Here we examine recent work that has provided insights into how fundamental physical properties of living cells, and the probabilistic nature of the chemical reactions that underlie gene expression, introduce noise. We focus on instances in which a stochastic decision initiates an event in the development of a multicellular organism and how that decision can be subsequently fixed. We present an example indicating that a similar interplay between an initial stochastic decision and subsequent fixation may underlie the regulation of reproduction in social insects. We argue, therefore, that stochasticity affects biological processes from the single-gene scale through to the complex organization of an ant colony, and represents a largely neglected component of phenotypic variation and evolution.
Understanding the nature of phenotypic variation is central to developmental and evolutionary studies. Yet, we know relatively little about molecular and developmental causes underlying variation in the phenotype. The field of quantitative genetics has provided developmental and evolutionary biologists with a useful framework to classify the different origins of phenotypic variation and how they relate to each other. Phenotypic variation (VP) has been defined (Falconer, 1981) as the sum of the squared deviations from the mean of a population and is partitioned into two basic variance components: genetic (VG) and environmental (VE). Thus, VP=VG+VE. VG is the variance of all “genotypic values” and can be broken down further into three components: additive (VA), dominance (VD), and interaction (VI) variance. VA represents the “breeding value” and is the main determinant of resemblance between relatives. In contrast, VD and VI represent nonadditive sources of genetic variance and are a consequence of the dominance of particular alleles at a locus (VD), or variance that results when two or more loci interact epistatically with each other (VI).
Traditionally, quantitative genetic studies emphasize VA as the most important component for understanding the evolution of traits (Lande, 1975; Falconer, 1981; Roff, 2007). Because VA determines heritability or resemblance between relatives, it is also thought to be sufficient to determine the response of a population to natural selection from one generation to the next. Another component of phenotypic variance, VE, is all variance that is attributed to an environmental nongenetic origin. Nonadditive sources of genetic variance (VD and VI), as well as the different components (nutrition, climate, and maternal effects) of environmental variance (VE), are much harder to estimate and have generally received less attention in evolutionary studies (Roff, 2007).
Evolutionary biologists who focus on mechanisms of development of multicellular organisms, however, have argued that nonadditive and nongenetic sources of variation can be important determinants of phenotypic variation and evolution (Wilkins, 2002; West-Eberhard, 2003; Gilbert and Epel, 2009), and that not all variation can be attributed to genetics and the environment (Leamy and Klingenberg, 2005). An early elaboration of this concept (Falconer, 1981) includes developmental sources of variation under a category, called “intangible” variation, which cannot be experimentally controlled and whose origin was unknown. An important component of intangible variation is caused by the stochastic variation in levels of particular macromolecules, which influences the kinetics of how they interact with one another (Kaern et al., 2005). Inherent stochastic variation in gene expression, sometimes termed molecular noise, was first implicated in biological processes over 50 years ago (Novick and Weiner, 1957). In recent years it has become apparent from work in several systems that regulatory pathways often include bistable loops that take advantage of molecular noise (Blake et al., 2003; Raser and O’Shea, 2005). Stochastic variation does not arise as a consequence of genetic variance (VG) or environmental variance (VE), but is inherent and demonstrable even within a single cell (Spudich and Koshland, 1976; Elowitz et al., 2002). Because stochastic variation is an inherent property of molecular interactions between genes and is not of genetic or environmental origin, we propose that it represents formally a new component of phenotypic variance that we call, VS. Thus, the simplest form of the equation that partitions phenotypic variation would now be VP=VG+VE+VS.
In this article, we will try to define the nature of VS by presenting different examples of stochastic variation and to discuss how VS impacts development and evolution. Our examples will span different levels of the biological hierarchy, from single cells to colonies of social insects, to highlight how stochastic variation is likely to have been exploited in the evolution of stable phenotypic switches.
SOURCES OF STOCHASTIC VARIATION
There are many ways stochastic molecular mechanisms can give rise to biological variation. Stochastic variation itself can arise because of the very small number of macromolecules involved in certain biological processes, such that both the randomness of molecular encounters and the fluctuations in the transitions between the conformational states of a macromolecule, become important (Magnasco, 2007). Transcription, translation, chromatin structure, and associations of regulatory factors with their response elements are all subject to this randomness because their chemical events rely on stochastic collisions between molecules. Noise in biological systems has been mathematically modeled (Kepler and Elston, 2001; Kaern et al., 2005; Krishna et al., 2005), and the underlying theory has been reviewed (Paulsson, 2004; Kaern et al., 2005).
Experimental methods for studying stochastic variation have been most powerful in the investigation of transcription and translation. An important step has been the transformative use of quantitative fluorescence microscopy of single cells to monitor expression of a green fluorescent protein (GFP)-tagged protein, as the use of techniques that employ populations of cells to extract RNA or protein levels mask cell-to-cell variations (Skotheim et al., 2008). Similar fluorescence microscopy methods have recently been extended to mRNAs, enabling accurate counts of the number of molecules of a particular mRNA species (Raj et al., 2008). The concentration of a particular protein in a population of genetically identical cells differs from cell to cell due to stochastic processes (reviewed in McAdams and Arkin, 1999; Kaern et al., 2005; Samoilov et al., 2006; Kaufmann and van Oudenaarden, 2007) and usually has a coefficient of variation (standard deviation divided by the mean) in the range 0.1–1.0 (Elowitz et al., 2002; Ozbudak et al., 2002; Blake et al., 2003; Raser and O’Shea, 2004). That is, cell-to-cell variations are on the order of tens of percents of the mean. Such a magnitude of variation can clearly influence phenotypic output.
The cell-to-cell distribution of protein numbers has been shown to be multiplicative (Krishna et al., 2005), indicating processes with several multiplicative stochastic steps that propagate in a cascade of catalytic processes. For example, transcription and translation contribute multiplicatively to the amount of a protein ultimately expressed from a given gene. Variability in gene expression can be affected by several factors. Rapidly degraded proteins have narrower temporal distributions than do stable proteins (Krishna et al., 2005). Noise can be amplified by regulatory cascades, such as kinase pathways, as each step in the cascade receives variability from its upstream regulator (Chang and Karin, 2001). Because each step in the pathway usually amplifies noise in the previous steps (Alon, 2006), the position of the noisiest step in a pathway can affect the overall noise dramatically (McAdams and Arkin, 1997; Raser and O’Shea, 2004). For example, a mechanism by which a very small number of mRNA molecules are made and translated to a large number of proteins, on average, will have much larger fluctuations in protein production than a mechanism by which many mRNAs are made, and each is translated rarely (Thattai and van Oudenaarden, 2001). In addition to extrinsic noise, there are also gene-specific intrinsic stochastic variations in expression. Protein fluctuations depend on a burst of protein expression from low copy-number mRNAs, as was predicted theoretically (McAdams and Arkin, 1997) and has been demonstrated experimentally using single-cell fluorescence at varying rates of transcription and translation (Ozbudak et al., 2002).
Subsequent studies have extended this work to the proteome scale in single yeast cells (Newman et al., 2006) and to cultured human cells (Sigal et al., 2006). The overall conclusions from these and earlier studies (Elowitz et al., 2002; Swain et al., 2002; Raser and O’Shea 2004; Colman-Lerner et al., 2005; Pedraza and van Oudenaarden, 2005; Rosenfeld et al., 2005) are that both extrinsic and intrinsic factors contribute to noise in gene expression and that cell-to-cell variability in expression of a particular protein is proportional to expression level (Bar-Even et al., 2006), typically in the range of 15–30% (Sigal et al., 2006; Rausenberger and Kollmann, 2008), but proteins in some functional pathways are expressed with more or less noise than is the norm (Bar-Even et al., 2006; Newman et al., 2006). For example, stress-related genes exhibit high degrees of noise in their expression, while genes involved in the proteosome pathway or in protein synthesis show much less variability in expression level. These findings imply that mechanisms exist to buffer noise in protein expression and that different optimal levels of tuning have evolved for different genetic pathways.
While it may be advantageous to retain the distribution of outcomes arising from stochastic variation to maximize potential phenotypic variability, variability in gene expression is kept in check and shaped into predictable outcomes by regulatory circuits. Such circuits contain thresholds and nonlinearities; for example, protein levels can be made to fluctuate less by means of negative feedback loops or autorepression. Alternatively, positive autoregulation can increase cell-to-cell variability, providing a means for increasing the phenotypic variability in a population of cells. Strong positive feedback can even lead to bistability, which often leads to a bimodal distribution with two cell populations having high and low expression (Novick and Weiner, 1957; Elowitz et al., 2002), or high and low kinase activity in a regulatory cascade. These nonlinearities confer limitations for biological systems and delimit the effects of noise.
STOCHASTIC PROCESSES IN DEVELOPMENT OF MULTICELLULAR ORGANISMS: FIXATION OF A RANDOM DECISION BY SUBSEQUENT LATERAL INHIBITION
Quantitative studies of stochastic noise in gene expression have thus far been mostly limited to single-cell systems, yet it is clear that stochasticity influences development of multicellular organisms, sometimes driving specific cell fate decisions. Often, a random decision initiates a developmental process, which is then reinforced and made permanent through lateral inhibition mechanisms (Losick and Desplan, 2008). There are several well-documented examples of this. For instance, in the early Drosophila embryo, all cells in proneural clusters initially have the capability of differentiating into neuroblasts. However, once one cell in the cluster differentiates into a neuroblast, all other cells in the cluster must become epidermal cells or else lethality ensues. It is believed that the initial decision to differentiate into a neuroblast is stochastic: one cell in the cluster randomly expresses more Delta protein than the others. Delta is a membrane-bound ligand that activates a cell surface receptor, Notch, in adjacent cells that activates a positive feedback loop whereby the Notch expression is increased in those cells, which in turn increases their sensitivity to Delta (Heitzler and Simpson, 1991). The signaling cell, meanwhile, represses the Notch expression and becomes a neuroblast, while the others adopt the epidermal fate. Importantly, ablation of the presumptive neuroblast abolishes the lateral inhibition and disrupts the bistable loop, in turn, allowing another random cell to activate the Delta expression, to re-initiate the process, and to differentiate into a neuroblast. A similar Notch-dependent mechanism to canalize an initially stochastic decision has been documented for the decision of either of two uncommitted precursor cells to choose between the anchor cell or ventral uterine precursor cell fates in C. elegans (Kimble, 1981; Greenwald et al., 1983; Seydoux and Greenwald, 1989), although this process may not be entirely stochastic but biased by the birth order of the cells, with the first born more likely to become the ventral uterine precursor (Karp and Greenwald, 2003). Stochastic activation coupled with negative feedback regulation also establishes the expression of one and only one olfactory receptor in each sensory neuron in the mouse (Serizawa et al., 2003; Lomvardas et al., 2006).
In some instances, the contribution of stochasticity to developmental decisions is masked among multiple pathways. In Drosophila, the compound eye is composed of 750–800 simple eyes called ommatidia, each consisting of eight photoreceptor cells (numbered R1–R8) and 14 accessory cells (Wolff and Ready, 1993). Precursors to R1, R6, or R7 that express Notch assume the R7 fate; lacking Notch, they become R1 or R6 (Cooper and Bray, 2000; Tomlinson and Struhl, 2001). Notch is believed to promote R7 fate by repressing Seven-up (svp), a nuclear hormone receptor (Cooper and Bray, 2000; Tomlinson and Struhl, 2001). Surprisingly, however, svp mutant R1∕R6 precursors are not simply transformed into R7 precursors but, in fact, make a random choice between R7 and R8 fates with approximately equal likelihood (Miller et al., 2008). In larval development these cells express both R7- and R8-specific markers, but by the pupal stage nearly all svp mutant photoreceptor cells express either R7 or R8 markers but not both. The role of Notch then extends beyond repression of svp; it must also prevent cells from assuming the R8 fate or actively promote the R7 fate. The former possibility appears more likely as Notch represses R8 fate by lateral inhibition during the initial specification of R8 neurons (Ligoxygakis et al., 1998; Jafar-Nejad et al., 2003).
Stochasticity also underlies more complex developmental decisions underlying the patterning of the compound eye of Drosophila. Usually, only one rhodopsin gene is expressed in a given photoreceptor cell (Mazzoni et al., 2008). Photoreceptors R1–R6 all express the same rhodopsin, Rh1, which is sensitive to a broad wavelength spectrum of light (Chou et al., 1996). However, ommatidia differ in which rhodopsins are expressed in R7 and R8; in approximately 30% of ommatidia called pale ommatidia, Rh3 rhodopsin, which is sensitive to ultraviolet light, is expressed in R7, and Rh5 rhodopsin, which is sensitive to blue light, is expressed in R8 (Chou et al., 1996; Papatsenko et al., 1997). In the remaining 70% of ommatidia called yellow ommatidia, Rh4, a different ultraviolet-sensitive rhodopsin, is expressed in R7 while a green-sensitive rhodopsin (Rh6) is expressed in R8 (Huber et al., 1997; Chou et al., 1999). The distribution of pale and yellow ommatidia within the compound eye is random, and results from stochastic expression of a transcription factor, Spineless, in uncommitted photoreceptor cells (Wernet et al., 2006). The spineless expression is both necessary and sufficient to produce the yellow subtype, and those cells that never activate the gene adopt the pale subtype.
FIXATION OF STOCHASTIC DECISIONS THROUGH SUBSEQUENT INHIBITION AND FEEDBACK MECHANISMS IN COLONIES OF SOCIAL INSECTS
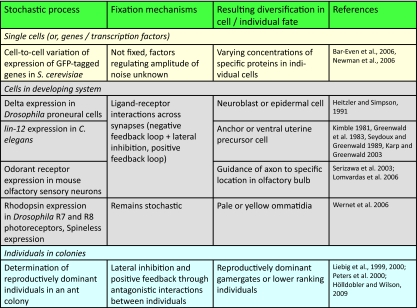
The mechanisms that have evolved to exploit stochastic variation at the level of single cells or whole tissues during development may also operate at much higher levels of biological organization in colonial organisms such as the social insects (ants, bees, and wasps). Table 1 illustrates how stochastic variability might be exploited at different levels of biological organization. Over the last 30 years, there has been a resurgence of the “superorganism concept,” the idea that the social insect colony as a whole is analogous to a single unitary organism (Wheeler, 1911; Wilson and Sober, 1989; Reeve and Hölldobler, 2007; Yang, 2007; Hölldobler and Wilson, 2009). A social insect colony is typically composed of a queen and a small or large number of workers (Hölldobler and Wilson, 1990). The queen is analogous to the germ line of a single organism as it performs most of the reproduction, while the workers are analogous to the soma as their reproduction is either suppressed, or they are sterile, and they perform most of the tasks of the colony such as foraging and brood rearing (Hölldobler and Wilson, 2009).
Table 1.
Summary of framework and terminology for proposed corresponding mechanisms by which stochastic variation operates at different levels of biological complexity.
 |
Ants are a remarkably successful group of social insects in terms of their ecological dominance, global biomass, and evolution (Hölldobler and Wilson, 1990). A key to their success lies in the diversity of colony-level mechanisms that ants have evolved to regulate the reproductive division of labor between queens and workers (Hölldobler and Wilson, 2009). In the eusocially ancestral ant, Harpagnathos saltator, queens and workers are morphologically similar, the colony size is small (mean=40–50 individuals), and the level of conflict over who reproduces is high (Peeters et al., 2000). Interestingly, it appears that Harpagnathos has evolved social mechanisms that exploit stochastic variation to regulate colony reproduction in a manner that is analogous to the mechanism by which cell fate is determined in Drosophila proneural clusters—an initially stochastic decision is reinforced and made permanent through lateral inhibition and feedback mechanisms.
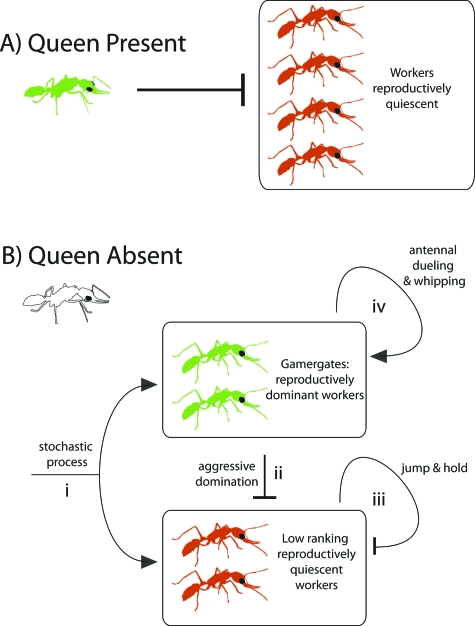
The work by Liebig and his colleagues has shown that reproductive dominance hierarchies in Harpagnathos are established through antagonistic interactions among nestmates (Hölldobler and Wilson, 2009). Although both queens and workers can reproduce, workers do not have the same reproductive capacity as the queen. Thus, while the queen is alive and fecund, most of the workers in the colony are reproductively quiescent [Peeters et al., 2000; Fig. 1A]. When the queen’s fecundity starts to decrease or when she dies, however, a small group of workers called “gamergates” become established as the top-ranking, reproductively dominant, individuals in the colony (Peeters et al., 2000). Because gamergates do not have the same reproductive capacity as the queen, several gamergate individuals are required to replace her. The exact number of individuals that replace the queen, however, is highly variable. Several experimental attempts have been made to try and uncover the deterministic rules that govern which individuals in the colony will become established as gamergates, but all attempts have thus far failed to reveal any rules (J Liebig, personal communication 2009). Although requiring empirical confirmation, the process that governs exactly which workers in the colony will emerge to become gamergates appears to be stochastic (J Liebig, personal communication 2009). Once new gamergates are stochastically determined, they establish and maintain their reproductive dominance by inhibiting any reproduction of lower-ranking individuals through three types of antagonistic interactions (Hölldobler and Wilson, 2009), as illustrated [Fig. 1B].
Figure 1. Network diagram of a potentially stochastic decision in social insects subsequently reinforced through inhibition and feedback mechanisms.
The underlying experiments have been described (Hölldobler and Wilson, 2009). Reproductively active queens and workers are green, while reproductively quiescent workers are red. (A) When a reproductively active queen is present in the colony, she inhibits the workers from reproducing. (B) When the queen is absent, new gamergates (reproductively dominant workers) emerge in what appears to be a stochastic process (i). Once gamergates are established, they are fixed by lateral inhibition through aggressive domination (ii), negative feedback through jump and hold policing behavior (iii), and positive feedback through antennal whipping and dueling (iv).
The first and most common type of antagonistic interaction is called aggressive domination. This is an attack behavior, where top-ranked gamergates attack lower-ranking infertile workers and inhibit them from reproducing by standing over them, grasping them, and then vigorously shaking them up and down. Aggressive domination is a linear or lateral inhibition mechanism that maintains the reproductive dominance of reproductive gamergates by inhibiting low-ranking workers who attempt to reproduce. The second type of antagonistic interaction is called jump and hold, where low-ranking infertile workers leap forward about 1–2 cm, and then hold with their jaws other workers that attempt to reproduce (Liebig et al., 1999). Jump and hold is a negative feedback mechanism where low-ranking workers inhibit each other from reproducing. Finally, the third type of antagonistic interaction is called antennal whipping and dueling, which begins with one gamergate lashing and whipping another gamergate with its antennae, followed by reciprocal lashes and whipping by the recipient. This interaction often ends with no further consequences and the individuals walk away. If, however, this interaction occurs between a gamergate and a lower-ranking worker, then the gamergate launches an aggressive domination attack on the lower-ranking worker. Thus, antennal whipping and dueling comprise a positive feedback mechanism to assess social equality and positively reinforce reproductive domination among gamergates. A summary of these three types of interactions and their role in regulating the reproductive dominance hierarchies in Harpagnathos reveals that ant colonies may regulate their societies through an initially stochastic decision that is reinforced through lateral inhibition, as well as positive and negative feedback mechanisms. This is analogous to how stochastic variation has been exploited at lower levels of biological organization.
Furthermore, the potential stochasticity underlying the establishment of new gamergates may itself be driven by underlying stochasticity in gene expression. In Harpagnathos saltator, the fertility or reproductive status of queens and workers is reflected by their cuticular hydrocarbon (CHC) profiles (Liebig et al., 2000). CHC profiles act as fertility signals that allow all individuals in the colony to clearly discriminate those individuals that are reproductive (queens and gamergates) from those that are not (low-ranking workers; Dietemann et al., 2003). CHC profiles can also change dynamically depending on the current reproductive status of the individual, such that when a nonreproductive worker attempts to become a gamergate, there is a small time lag between the change in fertility and the change in CHC profile of this individual (Liebig et al., 2000). These dynamically changing CHC profiles are the consequence of the differential activity of particular enzymes (Schal et al., 1998). Therefore, it may be that the stochastic expression of these enzymes coupled with the short time lag between changes in fertility and changes in CHC profiles underlies the stochastic establishment of gamergates after the queen dies. Stochastic establishment of gamergates may be favored by natural selection because it is a flexible mechanism that can ensure, especially in small colonies, the emergence of reproductive individuals upon the demise of the queen. When the queen or other gamergates die, the colony must maximize its potential for replacing these reproductively dominant individuals, which may otherwise be limited by deterministic rules.
CONCLUDING THOUGHTS
The existence of diffusible morphogens that specify pattern in a one-cell embryo based on concentration gradients, and the concept that genes are expressed in bursts because the nature of the underlying molecular interactions is stochastic, were proposed on theoretical grounds before supporting empirical data were available (Spemann, 1912; Child, 1941; Crick, 1970; McAdams and Arkin, 1997). Mechanisms by which stochastic variation impacts on phenotype are conserved from populations of single-celled organisms to developing multicellular organisms. We propose that similar mechanisms will operate at yet more complex levels of biological organization (Table 1), including not only colonies of social insects but perhaps even whole ecosystems. Interdisciplinary efforts that coordinate theorists and biologists working in many different specialties will be essential to provide experimental tests of this idea.
REFERENCES
- Alon, U (2006). An Introduction to Systems Biology: Design Principles of Biological Circuits, CRC, Boca Raton, FL. [Google Scholar]
- Bar-Even, A, Paulsson, J, Maheshri, N, Carmi, M, O’Shea, E, Pilpel, Y, and Barkai, N (2006). “Noise in protein expression scales with natural protein abundance.” Nat. Genet. 38, 636–643. 10.1038/ng1807 [DOI] [PubMed] [Google Scholar]
- Blake, W J, Kærn, M, Cantor, C R, and Collins, J J (2003). “Noise in eukaryotic gene expression.” Nature (London) 422, 633–637. 10.1038/nature01546 [DOI] [PubMed] [Google Scholar]
- Chang, L, and Karin, M (2001). “Mammalian MAP kinase signaling cascades.” Nature (London) 410, 37–40. 10.1038/35065000 [DOI] [PubMed] [Google Scholar]
- Child, C M (1941). Patterns and Problems of Developmental Biology, Chicago University Press, Chicago, IL. [Google Scholar]
- Chou, W H, Hall, K J, Wilson, D B, Wideman, C L, Townson, S M, Chadwell, L V, and Britt, S G (1996). “Identification of a novel Drosophila opsin reveals specific patterning of the R7 and R8 photoreceptor cells.” Neuron 17, 1101–1115. 10.1016/S0896-6273(00)80243-3 [DOI] [PubMed] [Google Scholar]
- Chou, W H, Huber, A, Bentrop, J, Schulz, S, Schwab, K, Chadwell, L V, Paulsen, R, and Britt, S G (1999). “Patterning of the R7 and R8 photoreceptor cells of Drosophila: evidence for induced and default state specification.” Development 126, 607–616. [DOI] [PubMed] [Google Scholar]
- Colman-Lerner, A, Gordon, A, Serra, E, Chin, T, Resnekov, O, Endy, D, Pesce, C G, and Brent, R (2005). “Regulated cell-to-cell variation in a cell-fate decision system.” Nature (London) 437, 699–706. 10.1038/nature03998 [DOI] [PubMed] [Google Scholar]
- Cooper, M T, and Bray, S J (2000). “R7 photoreceptor specification requires Notch activity.” Curr. Biol. 10, 1507–1510. 10.1016/S0960-9822(00)00826-5 [DOI] [PubMed] [Google Scholar]
- Crick, F (1970). “Diffusion in embryogenesis.” Nature (London) 225, 420–422. 10.1038/225420a0 [DOI] [PubMed] [Google Scholar]
- Dietemann, V, Peeters, C, Liebig, J, Thivet, V, and Hölldobler, B (2003). “Cuticular hydrocarbons mediate discrimination of reproductives and nonreproductives in the ant Myrmecia gulosa.” Proc. Natl. Acad. Sci. U.S.A. 100, 10341–10346. 10.1073/pnas.1834281100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz, M B, Levine, A J, Siggia, E D, and Swain, P S (2002). “Stochastic gene expression in a single cell.” Science 297, 1183–1186. 10.1126/science.1070919 [DOI] [PubMed] [Google Scholar]
- Falconer, D S, (1981). Introduction to Quantitative Genetics, Longman, London, UK. [Google Scholar]
- Gilbert, S F, and Epel, D (2009). Ecological Developmental Biology, Sinauer, Sunderland, MA. [Google Scholar]
- Greenwald, I S, Sternberg, P W, and Horvitz, H R (1983). “The lin-12 locus specifies cell fates in Caenorhaebditis elegans.” Cell 34, 435–444. 10.1016/0092-8674(83)90377-X [DOI] [PubMed] [Google Scholar]
- Heitzler, P, and Simpson, P (1991). “The choice of cell fate in the epidermis of Drosophila.” Cell 64, 1083–1092. 10.1016/0092-8674(91)90263-X [DOI] [PubMed] [Google Scholar]
- Hölldobler, B, and Wilson, E (1990). The Ants, Harvard University Press, Cambridge, MA. [Google Scholar]
- Hölldobler, B, and Wilson, E (2009). The Superorganism, Norton, New York. [Google Scholar]
- Huber, A, Schulz, S, Bentrop, C, Groell, U, Wolfrum, U, and Paulsen, R (1997). “Molecular cloning of Drosophila Rh6 rhodopsin: the visual pigment of a subset of R8 photoreceptor cells.” FEBS Lett. 406, 6–10. 10.1016/S0014-5793(97)00210-X [DOI] [PubMed] [Google Scholar]
- Jafar-Nejad, H, Acar, M, Nolo, R, Lacin, H, Pan, H, Parkhurst, S M, and Bellen, H J (2003). “Senseless acts as a binary switch during sensory organ precursor selection.” Genes Dev. 17, 2966–2978. 10.1101/gad.1122403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaern, M, Elston, T C, Blake, W J, and Collins, J J (2005). “Stochasticity in gene expression: from theories to phenotypes.” Nat. Rev. Genet. 6, 451–464. 10.1038/nrg1615 [DOI] [PubMed] [Google Scholar]
- Karp, X, and Greenwald, I (2003). “Post-transcriptional regulation of the E/Daughterless ortholog HLH-2, negative feedback, and birth order bias during the AC/VU decision in C. elegans.” Genes Dev. 17, 3100–3111. 10.1101/gad.1160803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann, B B, and van Oudenaarden, A (2007). “Stochastic gene expression: from single molecules to the proteome.” Curr. Opin. Genet. Dev. 17, 107–112. 10.1016/j.gde.2007.02.007 [DOI] [PubMed] [Google Scholar]
- Kepler, T B, and Elston, T C (2001). “Stochasticity in transcriptional regulation: origins, consequences and mathematical representations.” Biophys. J. 81, 3116–3136. 10.1016/S0006-3495(01)75949-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble, J (1981). “Alterations in cell lineage following laser ablation of cells in the somatic gonad of Caenorhaebditis elegans.” Dev. Biol. 87, 286–300. 10.1016/0012-1606(81)90152-4 [DOI] [PubMed] [Google Scholar]
- Krishna, S, Banerjee, B, Ramakrishnan, T V, and Shivashankar, G V (2005). “Stochastic simulations of the origins and implications of long-tailed distributions in gene expression.” Proc. Natl. Acad. Sci. U.S.A. 102, 4771–4776. 10.1073/pnas.0406415102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande, R (1975). “Maintenance of genetic-variability by mutation in a polygenic character with linked loci.” Genet. Res. 26, 221–235. 10.1017/S0016672300016037 [DOI] [PubMed] [Google Scholar]
- Leamy, L J, and Klingenberg, C P (2005). “The genetics and evolution of fluctuating asymmetry.” Annu. Rev. Ecol. Evol. Syst. 36, 1–21. 10.1146/annurev.ecolsys.36.102003.152640 [DOI] [Google Scholar]
- Liebig, J, Peeters, C, and Hölldobler, B (1999). “Worker policing limits the number of reproductives in a ponerine ant.” Proc. R. Soc. London, Ser. B 266, 1865–1870. 10.1098/rspb.1999.0858 [DOI] [Google Scholar]
- Liebig, J, Peeters, C, Oldman, N J, Markstädter, C, and Hölldobler, B (2000). “Are variations in cuticular hydrocarbons of queens and workers a reliable signal of fertility in the ant Harpegnathos saltator?” Proc. Natl. Acad. Sci. U.S.A. 97, 4124–4131. 10.1073/pnas.97.8.4124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligoxygakis, P, Yu, S Y, Delidakis, C, and Baker, N E (1998). “A subset of notch functions during Drosophila eye development require Su(H) and the E(Spl) gene complex.” Development 125, 2893–2900. [DOI] [PubMed] [Google Scholar]
- Lomvardas, S, Barnea, G, Pisapia, D J, Mendelsohn, M, Kirkland, J, and Axel, R (2006). “Interchromosomal interactions and olfactory receptor choice.” Cell 126, 403–413. 10.1016/j.cell.2006.06.035 [DOI] [PubMed] [Google Scholar]
- Losick, R, and Desplan, C (2008). “Stochasticity and cell fate.” Science 320, 65–68. 10.1126/science.1147888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnasco, M (2007). “Measuring variability.” HFSP J. 1, 147–151. 10.2976/1.2784546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzoni, E O, Celik, A, Wernet, M F, Vasiliauskas, D, Johnston, R J, Cook, T A, Pichaud, F, and Desplan, C (2008). “Iroquois complex genes induce co-expression of rhodopsins in Drosophila.” PLoS Biol. 6, 0825–0835. 10.1371/journal.pbio.0060097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAdams, H H, and Arkin, A (1997). “Stochastic mechanisms in gene expression.” Proc. Natl. Acad. Sci. U.S.A. 94, 814–819. 10.1073/pnas.94.3.814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAdams, H H, and Arkin, A (1999). “It’s a noisy business! Genetic regulation at the nanomolar scale.” Trends Genet. 15, 65–69. 10.1016/S0168-9525(98)01659-X [DOI] [PubMed] [Google Scholar]
- Miller, A C, Seymour, H, King, C, and Herman, T G (2008). “Loss of seven-up from Drosophila R1/R6 photoreceptors reveals a stochastic fate choice that is normally biased by Notch.” Development 135, 707–715. 10.1242/dev.016386 [DOI] [PubMed] [Google Scholar]
- Newman, J R, Ghaemmaghami, S, Ihmels, J, Breslow, D K, Noble, M, DeRisi, J L, and Weissman, J S (2006). “Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise.” Nature (London) 441, 840–846. 10.1038/nature04785 [DOI] [PubMed] [Google Scholar]
- Novick, A, and Weiner, M (1957). “Enzyme induction as an all-or-none phenomenon.” Proc. Natl. Acad. Sci. U.S.A. 43, 553–566. 10.1073/pnas.43.7.553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozbudak, E M, Thattai, M, Kurtser, I, Grossman, A D, and van Oudenaarden, A (2002). “Regulation of noise in the expression of a single gene.” Nat. Genet. 31, 69–73. 10.1038/ng869 [DOI] [PubMed] [Google Scholar]
- Papatsenko, D, Sheng, G, and Desplan, C (1997). “A new rhodopsin in R8 photoreceptors of Drosophila: evidence for coordinate expression with Rh3 in R7 cells.” Development 124, 1665–1673. [DOI] [PubMed] [Google Scholar]
- Paulsson, J (2004). “Summing up noise in gene networks.” Nature (London) 427, 415–418. 10.1038/nature02257 [DOI] [PubMed] [Google Scholar]
- Pedraza, J M, and van Oudenaarden, A (2005). “Noise propagation in gene networks.” Science 307, 1965–1969. 10.1126/science.1109090 [DOI] [PubMed] [Google Scholar]
- Peeters, C, Liebig, J, and Hölldobler, B (2000). “Sexual reproduction by both queens and workers in the ponerine ant Harpegnathos saltator.” Insectes Soc. 47, 325–332. 10.1007/PL00001724 [DOI] [Google Scholar]
- Raj, A, van den Bogaard, P, Rifkin, S, van Oudenaarden, A, and Tyagi, S (2008). “Imaging individual mRNA molecules using multiple singly labeled probes.” Nat. Methods 5, 877–879. 10.1038/nmeth.1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raser, J M, and O’Shea, E K (2004). “Control of stochasticity in eukaryotic gene expression.” Science 304, 1811–1814. 10.1126/science.1098641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raser, J M, and O’Shea, E K (2005). “Noise in gene expression: origins, consequences, and control.” Science 309, 2010–2013. 10.1126/science.1105891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausenberger, J, and Kollmann, M (2008). “Quantifying origins of cell-to-cell variations in gene expression.” Biophys. J. 95, 4523–4528. 10.1529/biophysj.107.127035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve, H K, and Hölldobler, B (2007). “The emergence of a superorganism through intergroup competition.” Proc. Natl. Acad. Sci. U.S.A. 104, 9736–9740. 10.1073/pnas.0703466104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roff, D A (2007). “A centennial celebration for quantitative genetics.” Evolution (Lawrence, Kans.) 61, 1017–1032. 10.1111/j.1558-5646.2007.00100.x [DOI] [PubMed] [Google Scholar]
- Rosenfeld, N, Young, J W, Alon, U, Swain, P S, and Elowitz, M N (2005). “Gene regulation at the single-cell level.” Science 307, 1962–1965. 10.1126/science.1106914 [DOI] [PubMed] [Google Scholar]
- Samoilov, M S, Price, G, and Arkin, A P (2006). “From fluctuations to phenotypes: the physiology of noise.” Sci. STKE 366, re17. 10.1126/stke.3662006re17 [DOI] [PubMed] [Google Scholar]
- Schal, C, Sevela, V L, Young, H P, and Bachmann, J AS (1998). “Sites of synthesis and transport pathways of insect hydrocarbons: cuticle and ovary as target tissues.” Am. Zool. 38, 382–393. [Google Scholar]
- Serizawa, S, Miyamichi, K, Nakatani, H, Suzuki, M, Saito, M, Yoshihara, Y, and Sakano, H (2003). “Negative feedback regulation ensures the one receptor-one olfactory neuron rule in mouse.” Science 302, 2088–2094. 10.1126/science.1089122 [DOI] [PubMed] [Google Scholar]
- Seydoux, G, and Greenwald, I (1989). “Cell autonomy of lin-12 function in a cell fate decision in C. elegans.” Cell 57, 1237–1245. 10.1016/0092-8674(89)90060-3 [DOI] [PubMed] [Google Scholar]
- Sigal, A, Milo, R, Cohen, A, Geva-Zatorsky, N, Klein, Y, Liron, Y, Rosenfeld, N, Danon, T, Persov, N, and Alon, U (2006). “Variability and memory of protein levels in human cells.” Nature (London) 444, 643–646. 10.1038/nature05316 [DOI] [PubMed] [Google Scholar]
- Skotheim, J M, DiTalia, S, Siggia, E D, and Cross, F R (2008). “Positive feedback of G1 cyclins ensure coherent cell cycle entry.” Nature (London) 454, 291–296. 10.1038/nature07118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spemann, H (1912). “Zur Entwicklung des Wilbeltierauges.” Zool Jahrb Abt Allg Zool Physiol. Tiere. 32, 1–98. [Google Scholar]
- Spudich, J L, and Koshland, D E, Jr. (1976). “Non-genetic individuality: chance in the single cell.” Nature (London) 262, 467–471. 10.1038/262467a0 [DOI] [PubMed] [Google Scholar]
- Swain, P S, Elowitz, M B, and Siggia, E D (2002). “Intrinsic and extrinsic contributions to stochasticity in gene expression.” Proc. Natl. Acad. Sci. U.S.A. 99, 12795–12800. 10.1073/pnas.162041399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thattai, M, and van Oudenaarden, A (2001). “Intrinsic noise in gene regulatory networks.” Proc. Natl. Acad. Sci. U.S.A. 98, 8614–8619. 10.1073/pnas.151588598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson, A, and Struhl, G (2001). “Delta/Notch and Boss/Sevenless signals act combinatorially to specify the Drosophila R7 photoreceptor.” Mol. Cell 7, 487–495. 10.1016/S1097-2765(01)00196-4 [DOI] [PubMed] [Google Scholar]
- Wernet, M F, Mazzoni, E O, Çelik, A, Duncan, D M, Duncan, I, and Desplan, C (2006). “Stochastic spineless expression creates the retinal mosaic for colour vision.” Nature (London) 440, 174–180. 10.1038/nature04615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West-Eberhard, M J (2003). Developmental Plasticity and Evolution, Oxford University Press, Oxford, UK. [Google Scholar]
- Wheeler, W M (1911). “The ant-colony as an organism.” J. Morphol. 22, 307–325. 10.1002/jmor.1050220206 [DOI] [Google Scholar]
- Wilkins, A S (2002). The Evolution of Developmental Pathways, Sinauer, Sunderland, MA. [Google Scholar]
- Wilson, D S, and Sober, E (1989). “Reviving the superorganism.” J. Theor. Biol. 136, 337–356. 10.1016/S0022-5193(89)80169-9 [DOI] [PubMed] [Google Scholar]
- Wolff, T, and Ready, D F (1993). “Pattern formation in the Drosophila retina.” The Development of Drosophila melanogaster, Bate, M, and Ashburner, M, eds., pp. 1277–1325, Cold Spring Harbor Laboratory, Plainview, NY. [Google Scholar]
- Yang, A S (2007). “Thinking outside the embryo: the superorganism as a model for evo-devo studies.” Biol. Theory 2, 398–408. 10.1162/biot.2007.2.4.398 [DOI] [Google Scholar]