Abstract
We conducted a two-stage genome-wide association study (GWAS) of pancreatic cancer, a cancer with one of the poorest survival rates worldwide. Initially, we genotyped 558,542 single nucleotide polymorphisms in 1,896 incident cases and 1,939 controls drawn from twelve prospective cohorts plus one hospital-based case-control study. In a combined analysis adjusted for study, sex, ancestry and five principal components that included an additional 2,457 cases and 2,654 controls from eight case-control studies, we identified an association between a locus on 9q34 and pancreatic cancer marked by the single nucleotide polymorphism, rs505922 (combined P=5.37 × 10-8; multiplicative per-allele odds ratio (OR) 1.20; 95% CI 1.12-1.28). This SNP maps to the first intron of the ABO blood group gene. Our results are consistent with earlier epidemiologic evidence suggesting that people with blood group O may have a lower risk of pancreatic cancer than those with groups A or B.
Pancreatic cancer shows amongst the highest mortality rates of any cancer, with a five year relative survival rate of less than 5%1,2. There is currently no effective screening test for the malignancy, and by the time of initial diagnosis, metastatic disease is commonly present. Established risk factors include a family history of pancreatic cancer, a medical history of diabetes type II and cigarette smoking3. Studies have also suggested an increased risk of pancreatic cancer within families with hereditary pancreatitis4,5. It has also been estimated that a small proportion of pancreatic cancers are due to highly penetrant germ-line mutations6. These studies suggested genetic contribution to pancreatic cancer, although there has been limited success in resolving common variants associated to this disease. We report here a genome-wide association study (GWAS) to resolve common variants associated to pancreatic cancer.
We conducted a GWAS in 1,896 cases and 1,939 controls of incident pancreatic cancer cases drawn from twelve prospective cohorts plus one hospital-based case-control study (American Cancer Society Cancer Prevention Study-II (CPS II)7 Alpha-Tocopherol, Beta-Carotene Cancer Prevention Study (ATBC)8 European Prospective Investigation into Cancer and Nutrition Study (EPIC)9 CLUE II10 Health Professionals Follow-up Study (HPFS)11 New York University Women’s Health Study (NYUWHS)12 Nurses’ Health Study (NHS)11 Physicians’ Health Study I (PHS)11 Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial (PLCO)13 Shanghai Men’s and Women’s Health Study (SMWHS)14,15 Women’s Health Initiative (WHI)16 the Women’s Health Study (WHS)17 and the Mayo Clinic Molecular Epidemiology of Pancreatic Cancer Study18; Supplemental Table 1). Eight case-control studies participated in the independent ‘Fast-Track’ replication phase of 2,457 cases and 2,654 controls (the University of Toronto19 University of California San Francisco20 Johns Hopkins University, MD Anderson Cancer Center21 PACIFIC Study of Group Health and Northern California Kaiser Permanente, Memorial Sloan-Kettering Cancer Center22 Yale University23 and the Mayo Clinic Molecular Epidemiology of Pancreatic Cancer Study18; Supplemental Table 2).
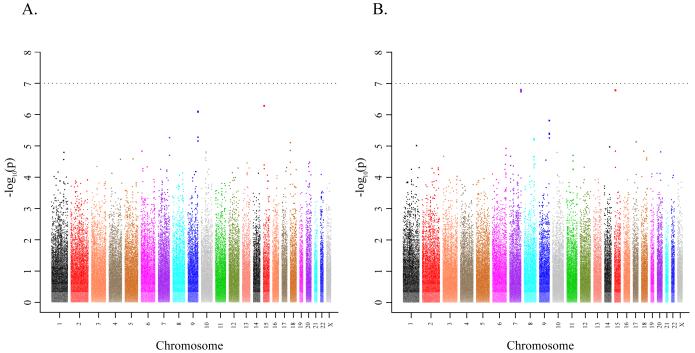
After quality control assessment of genotypes assayed using the HumanHap500 chip (Illumina, San Diego, CA), 558,542 SNPs were available for analysis. A logistic regression model was fit for genotype trend effects (1 d.f.) adjusting for study, age, sex, ancestry and the top five principal components of population stratification (Online Methods). The quantile-quantile plot (QQ plot) does not demonstrate a systematic deviation from the expected distribution, minimizing the likelihood of systematic genotype error or bias due to underlying population substructure (Supplemental Figure 1). The results of the GWAS are shown in Figure 1a. Because of the potential for survivor bias in case-control studies due to rapid mortality, we also analyzed the GWAS for cohort studies only as shown in Figure 1b (i.e., excluding Mayo subjects).
Figure 1. Manhattan plot of the P values in the pancreatic cancer GWAS.
The association with pancreatic cancer is shown for the entire GWAS (12 cohort studies, and the Mayo case-control study, see Online Methods) (A), and the results of the GWAS including only the 12 cohorts studies (B). Association was assessed using unconditional logistic regression adjusted for study, arm, age, sex, ancestry and the top five principal components of the population stratification analysis. The x axis represents chromosomal locations and the y axis shows P values on a logarithmic scale.
We conducted a rapid follow-up scan, or “Fast Track”, of SNPs from three regions in eight case-control studies (four hospital based and four population based). At least two SNPs per region were ranked among the lowest 25 p-values in the initial GWAS; 1.) chromosome 9q34, which includes the ABO gene (rs505922, rs495828, rs657152 and rs630014; ranked 2, 3, 8 and 17); 2.) chromosome 7q36, which includes Sonic Hedgehog (SHH), (rs167020, rs172310, and rs288746; ranked 6, 10 and 89); and 3.) a gene desert on chromosome 15q14 (rs8028529, rs4130461 and rs4459505, ranked 1, 18 and 26) (Table 1).
Table 1. Association of SNPs on chromosomes 9q34, 7q36 and 15q14 to risk of pancreatic cancer.
The results from the unconditional logistic regression of the genotypes generated in the initial GWAS and the follow-up studies in a total of 3,891 pancreatic cancer cases and 4,001 controls. The analysis was adjusted for age in ten-year categories, sex, study, arm, ancestry and five principal components of population stratification.
| Markera, Allelesb, Chrc, Locationc and Gened |
Subsete | MAFf | Subjectsg | χ2h | P valueh | ORHet (95% CI) | ORHom (95% CI)i |
|---|---|---|---|---|---|---|---|
| rs505922 (T, C) | Stage 1 Cohorts | 0.357|0.417 | 1462|1406 | 21.11 | 4.33E-06 | 1.29 (1.16-1.43) | 1.66 (1.33-2.05) |
| 9q34 | Stage 1 All | 0.357|0.411 | 1805|1771 | 22.18 | 2.48E-06 | 1.26 (1.14-1.39) | 1.59 (1.31-1.92) |
| 135139050 | Stage 2 | 0.343|0.375 | 2127|2120 | 9.50 | 2.06E-03 | 1.15 (1.05-1.26) | 1.32 (1.11-1.58) |
| ABO | Stage 1 + 2 | 0.349|0.392 | 3932|3891 | 29.58 | 5.37E-08 | 1.20 (1.12-1.28) | 1.44 (1.26-1.63) |
| rs495828 (G, T) | Stage 1 Cohorts | 0.192|0.236 | 1423|1362 | 18.11 | 2.08E-05 | 1.35 (1.18-1.55) | 1.82 (1.38-2.41) |
| 9q34 | Stage 1 All | 0.194|0.236 | 1755|1717 | 21.37 | 3.78E-06 | 1.34 (1.18-1.51) | 1.79 (1.40-2.29) |
| 135144688 | Stage 2 | 0.223|0.238 | 1786|1718 | 2.10 | 1.47E-01 | 1.09 (0.97-1.21) | 1.18 (0.94-1.47) |
| ABO | Stage 1 + 2 | 0.209|0.237 | 3541|3435 | 17.93 | 2.30E-05 | 1.19 (1.10-1.30) | 1.43 (1.21-1.68) |
| rs657152 (G, T) | Stage 1 Cohorts | 0.380|0.437 | 1463|1408 | 18.05 | 2.15E-05 | 1.26 (1.13-1.40) | 1.59 (1.28-1.97) |
| 9q34 | Stage 1 All | 0.380|0.430 | 1806|1773 | 18.13 | 2.06E-05 | 1.23 (1.12-1.35) | 1.51 (1.25-1.83) |
| 135129086 | Stage 2 | 0.374|0.404 | 1791|1729 | 7.24 | 7.13E-03 | 1.14 (1.04-1.26) | 1.30 (1.07-1.58) |
| ABO | Stage 1 + 2 | 0.377|0.417 | 3597|3502 | 24.29 | 8.28E-07 | 1.19 (1.11-1.27) | 1.41 (1.23-1.61) |
| rs630014 (C, T) | Stage 1 Cohorts | 0.475|0.421 | 1463|1408 | 18.04 | 2.16E-05 | 0.80 (0.72-0.88) | 0.63 (0.51-0.78) |
| 9q34 | Stage 1 All | 0.473|0.427 | 1805|1773 | 16.74 | 4.28E-05 | 0.82 (0.75-0.90) | 0.67 (0.56-0.81) |
| 135139543 | Stage 2 | 0.479|0.441 | 2196|2118 | 11.83 | 5.84E-04 | 0.86 (0.79-0.94) | 0.74 (0.63-0.88) |
| ABO | Stage 1 + 2 | 0.477|0.435 | 4001|3891 | 27.49 | 1.58E-07 | 0.85 (0.79-0.90) | 0.71 (0.63-0.81) |
| rs167020 (G, A) | Stage 1 Cohorts | 0.250|0.313 | 1462|1408 | 27.28 | 1.76E-07 | 1.37 (1.22-1.54) | 1.88 (1.48-2.38) |
| 7q36 | Stage 1 All | 0.259|0.307 | 1805|1773 | 20.06 | 7.52E-06 | 1.27 (1.15-1.41) | 1.62 (1.31-2.00) |
| 155312494 | Stage 2 | 0.278|0.294 | 1802|1 734 | 2.39 | 1.22E-01 | 1.09 (0.98-1.20) | 1.18 (0.96-1.45) |
| SHH | Stage 1 + 2 | 0.269|0.301 | 3607|3507 | 18.12 | 2.07E-05 | 1.17 (1.09-1.26) | 1.38 (1.19-1.60) |
| rs172310 (C, A) | Stage 1 Cohorts | 0.272|0.336 | 1454|1399 | 27.02 | 2.01E-07 | 1.36 (1.21-1.53) | 1.85 (1.47-2.34) |
| 7q36 | Stage 1 All | 0.282|0.329 | 1796|1763 | 17.43 | 2.98E-05 | 1.25 (1.12-1.38) | 1.56 (1.26-1.92) |
| 155308388 | Stage 2 | 0.305|0.323 | 1768|1699 | 2.80 | 9.45E-02 | 1.09 (0.99-1.21) | 1.19 (0.97-1.46) |
| SHH | Stage 1 + 2 | 0.293|0.326 | 3564|3462 | 17.04 | 3.66E-05 | 1.17 (1.08-1.25) | 1.36 (1.17-1.57) |
| rs288746 (T, C) | Stage 1 Cohorts | 0.109|0.144 | 1458|1403 | 14.57 | 1.35E-04 | 1.37 (1.16-1.61) | 1.87 (1.36-2.59) |
| 7q36 | Stage 1 All | 0.114|0.138 | 1800|1768 | 8.08 | 4.48E-03 | 1.23 (1.07-1.42) | 1.52 (1.14-2.02) |
| 155299433 | Stage 2 | 0.116|0.128 | 1805|1735 | 2.59 | 1.08E-01 | 1.12 (0.97-1.30) | 1.26 (0.95-1.68) |
| SHH | Stage 1 + 2 | 0.115|0.133 | 3605|3503 | 10.14 | 1.45E-03 | 1.18 (1.07-1.30) | 1.39 (1.13-1.70) |
| rs8028529 (T, C) | Stage 1 Cohorts | 0.198|0.255 | 1457|1404 | 25.92 | 3.55E-07 | 1.38 (1.22-1.56) | 1.91 (1.49-2.45) |
| 15q14 | Stage 1 All | 0.202|0.249 | 1800|1768 | 23.13 | 1.51E-06 | 1.31 (1.17-1.47) | 1.72 (1.38-2.15) |
| 34441889 | Stage 2 | 0.231|0.229 | 1800|1736 | 0.02 | 8.92E-01 | 0.99 (0.89-1.11) | 0.98 (0.79-1.23) |
| none | Stage 1 + 2 | 0.217|0.239 | 3600|3504 | 11.12 | 8.53E-04 | 1.14 (1.06-1.24) | 1.31 (1.12-1.53) |
| rs4130461 (G, T) | Stage 1 Cohorts | 0.224|0.273 | 1463|1408 | 18.71 | 1.53E-05 | 1.31 (1.16-1.47) | 1.70 (1.34-2.17) |
| 15q14 | Stage 1 All | 0.231|0.272 | 1806|1773 | 16.64 | 4.52E-05 | 1.25 (1.12-1.39) | 1.56 (1.26-1.94) |
| 34439130 | Stage 2 | 0.256|0.250 | 1802|1736 | 0.39 | 5.32E-01 | 0.97 (0.87-1.08) | 0.93 (0.75-1.16) |
| none | Stage 1 + 2 | 0.243|0.261 | 3608|3509 | 6.15 | 1.32E-02 | 1.10 (1.02-1.19) | 1.21 (1.04-1.41) |
| rs4459505 (G, A) | Stage 1 Cohorts | 0.177|0.218 | 1455|1402 | 15.51 | 8.21E-05 | 1.30 (1.14-1.49) | 1.70 (1.30-2.21) |
| 15q14 | Stage 1 All | 0.178|0.214 | 1796|1765 | 14.92 | 1.12E-04 | 1.26 (1.12-1.42) | 1.59 (1.26-2.01) |
| 34443314 | Stage 2 | 0.196|0.198 | 1803|1737 | 0.08 | 7.81E-01 | 1.02 (0.90-1.14) | 1.03 (0.82-1.31) |
| none | Stage 1 + 2 | 0.187|0.206 | 3599|3502 | 8.52 | 3.51E-03 | 1.13 (1.04-1.23) | 1.28 (1.08-1.51) |
NCBI dbSNP identifier.
Major allele, minor allele.
Chromosome and NCBI Human genome Build 36 location.
Gene neighborhood within 20 kb upstream and 10 kb downstream of SNP.
Stage 1 is the initial GWAS and stage 2 the replication.
Minor allele frequency in control and case participants.
Controls, cases.
1 d.f. score test.
Estimate assuming multiplicative odds model OR, odds ratio; Het, heterozygous; Hom, homozygous for minor allele. CI, 95% confidence interval.
In a combined analysis of individuals of European background24 the strongest association with pancreatic cancer below the threshold for genome-wide significance25 was observed for a locus on chromosome 9q34, located within the first intron of ABO, a well-described blood group gene, marked by rs505922 (P=5.37 × 10−8; trend model; heterozygous odds ratio [ORHet] of 1.20; 95% CI 1.12-1.28 and homozygous odds ratio [ORHom] of 1.44; 95% CI 1.26-1.63). A comparable result was observed when all ethnic groups were included in the first stage (P=2.61 × 10−8; Supplemental Table 3). In the case-control replication set, we genotyped a second SNP, rs687621 (r2=1 with rs505922 in HapMap CEU and r2=0.91 in Stage 2 control individuals), located 12 kb centromeric in intron 2; the results provided confirmation of the locus (P=1.57×10−4 in the second-stage case-control studies only). In the combined analysis, a comparably strong signal was observed for rs630014 (P=1.58×10−7; ORHet 0.85, ORHom 0.71), which resides within 500 bp of rs505922 and is in linkage disequilibrium (r2=0.52 in HapMap CEU and 0.40 in PanScan GWAS European control individuals). After adjusting for rs505922, none of the remaining SNPs in ABO were significant at the P<0.01 level. The SNPs reside in a haplotype block that encompasses the proximal promoter and introns 1 and 2 (Figure 2).
Figure 2. Association and linkage disequilibrium plot of the 9q34 locus.
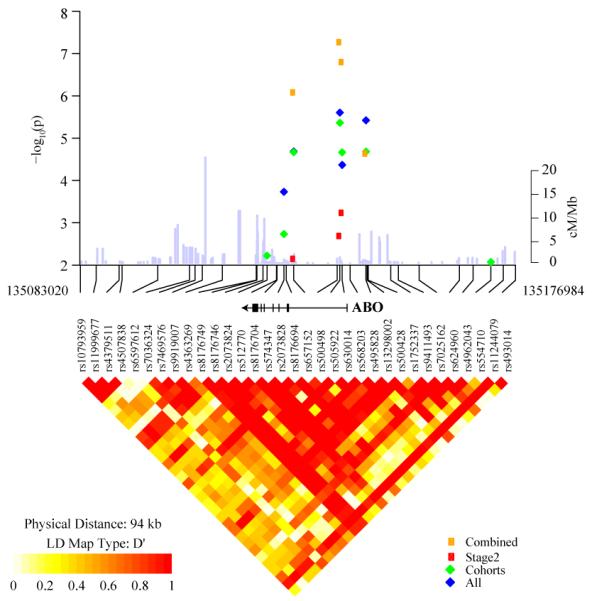
Association results are shown for all GWAS studies (blue diamonds), GWAS cohorts (green diamonds), replication studies (red circles) and all studies combined (yellow circles). Overlaid on the association panel is a plot of estimated recombination rates (cM/Mb) across the region from HapMap Phase II. The LD plot shows a region of chromosome 9 marked by SNPs, rs505922 and rs630014 (r2=0.52 in HapMap CEU and 0.40 in PanScan European control individuals) and bounded by SNPs between chr9; 135,083,020-135,176,984 (NCBI Genome build 36). LD is depicted for SNPs with MAF > 5% within PanScan. Controls of European background (n=1,799 unrelated individuals). Note that rs505922 and rs630014 are located in the first intron of the ABO gene, shown above the LD plot. Only SNPs genotyped in both the GWAS and “Fast Track” replication are shown.
Blood groups were first described by Karl Landsteiner in 1900 but the structure of the ABO antigens and their biosynthesis remained elusive until after 1950. The ABO gene encodes a glycosyltransferase that catalyzes the transfer of carbohydrates to the H antigen, forming the antigenic structure of the ABO blood groups. The proteins encoded by the A and B alleles of the ABO gene differ minimally in amino acid sequence but catalyze the transfer of different carbohydrates (N-acetylgalactosamine or galactose) onto the H antigen to form the A or B antigens. Individuals with the O blood group do not produce either the A or B antigens due to a single base deletion.
Our findings are notable because multiple studies, mainly from the 1950s and 1960s reported an association between ABO blood type and gastrointestinal cancers, strongest for gastric cancer but also for pancreatic cancer26,27. The protective allele (T) for rs505922 is in complete linkage disequilibrium (LD) (r2=1.0) with the O allele of the ABO locus, consistent with earlier reports showing increased risk of gastric and pancreatic cancer for individuals of the A and B blood groups. It is plausible that the single base deletion that generates the O blood group underlies the association signal but further mapping and laboratory work is required to determine which variant(s) account for the observed association.
Genetic variation in the first intron of the ABO gene has also been associated with circulating levels of serum tumor necrosis factor alpha (TNF-alpha) levels28 circulating soluble intracellular adhesion molecule 1 (sICAM-1)29 and plasma levels of alkaline phosphatase30. Although higher TNF-alpha levels are associated with the common allele of rs505922, protective for pancreatic cancer in our study, the data concerning the relationship between blood groups and TNF-alpha levels are inconsistent28. Furthermore, this region could be important for regulating circulating sICAM-1 levels as rs507666 and rs505922 (located 170 bps apart) were recently reported to be associated with circulating ICAM-1 levels29. Also, SNPs in the ABO locus, including rs657152, have been associated with plasma levels of liver derived alkaline phosphatase30. Lastly, altered ABO antigen expression has been observed in primary and metastatic pancreatic cancer as compared to normal pancreatic tissues31.
For rapidly fatal conditions, case-control studies are prone to distortion because they disproportionately include survivors. For variants unrelated to survival, case-control data are suitable for discovery and replication of risk-related markers. However, for variants related to survival, case-control studies yield biased estimates of the association with pancreatic cancer risk. ABO variants appear unrelated to survival and show strong and similar signals in both cohort and case-control data.
We observed an association at the genome-wide level (P=1.76 × 10−7) with SHH among cohorts that was not replicated in follow-up in case-control studies (P=0.12), raising three possibilities: the cohort finding is due to chance, SHH is related to both survival and to risk or that the SNPs failed to replicate because of chance (Table 1). Because there is substantial evidence that SHH plays a role in pancreatic carcinogenesis, further work is required to investigate this region32.
Pancreatic cancer is among the deadliest cancers with mortality rates approaching incidence rates1. Given there are few known risk factors, improved diagnostics and a finer understanding of the molecular pathogenesis are urgently needed. Our findings have identified the contribution of genetic variation in the ABO locus of 9q34 to pancreatic carcinogenesis, a finding that supports an epidemiologic observation first made half a century ago and recently confirmed33. We are currently conducting a GWAS in the eight studies of stage 2 in this study and anticipate that this will bring the identification of additional loci associated to pancreatic cancer. The discovery of additional genetic risk variants for this highly lethal cancer could contribute to novel risk stratification and improvements in prevention, early detection and therapeutic approaches to pancreatic cancer.
Supplementary Material
Acknowledgements
The authors gratefully acknowledge the energy and contribution of our late colleague, Robert Welch. Additional acknowledgements are in the Supplemental Note.
References
- 1.Jemal A, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Bray F, Pisani P, PArkin DM. IARC CancerBase. No 5. IARCPress; Lyon: 2004. GLOBOCAN 2002: Cancer Incidence, Mortality and Prevalence Worldwide. [Google Scholar]
- 3.Anderson KE,MT, Silverman D. Cancer of the pancreas. In: Schottenfeld D, Fraumeni JF Jr., editors. Cancer Epidemiology and Prevention. Oxford University Press; New York: 2006. [Google Scholar]
- 4.Lowenfels AB, et al. Hereditary pancreatitis and the risk of pancreatic cancer. International Hereditary Pancreatitis Study Group. J Natl Cancer Inst. 1997;89:442–6. doi: 10.1093/jnci/89.6.442. [DOI] [PubMed] [Google Scholar]
- 5.Castleman B. Case records of the Massachusetts General Hospital. N Engl J Medicine. 1972;286:1353–1359. doi: 10.1056/NEJM197206222862510. [DOI] [PubMed] [Google Scholar]
- 6.Klein AP, et al. Prospective risk of pancreatic cancer in familial pancreatic cancer kindreds. Cancer Res. 2004;64:2634–8. doi: 10.1158/0008-5472.can-03-3823. [DOI] [PubMed] [Google Scholar]
- 7.Calle EE, et al. The American Cancer Society Cancer Prevention Study II Nutrition Cohort: rationale, study design, and baseline characteristics. Cancer. 2002;94:2490–501. doi: 10.1002/cncr.101970. [DOI] [PubMed] [Google Scholar]
- 8.The alpha-tocopherol, beta-carotene lung cancer prevention study: design, methods, participant characteristics, and compliance. The ATBC Cancer Prevention Study Group. Ann Epidemiol. 1994;4:1–10. doi: 10.1016/1047-2797(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 9.Riboli E, et al. European Prospective Investigation into Cancer and Nutrition (EPIC): study populations and data collection. Public Health Nutr. 2002;5:1113–24. doi: 10.1079/PHN2002394. [DOI] [PubMed] [Google Scholar]
- 10.Gallicchio L, et al. Single nucleotide polymorphisms in inflammation-related genes and mortality in a community-based cohort in Washington County, Maryland. Am J Epidemiol. 2008;167:807–13. doi: 10.1093/aje/kwm378. [DOI] [PubMed] [Google Scholar]
- 11.Wolpin BM, et al. Circulating insulin-like growth factor binding protein-1 and the risk of pancreatic cancer. Cancer Res. 2007;67:7923–8. doi: 10.1158/0008-5472.CAN-07-0373. [DOI] [PubMed] [Google Scholar]
- 12.Zeleniuch-Jacquotte A, et al. Postmenopausal levels of sex hormones and risk of breast carcinoma in situ: results of a prospective study. Int J Cancer. 2005;114:323–7. doi: 10.1002/ijc.20694. [DOI] [PubMed] [Google Scholar]
- 13.Hayes RB, et al. Methods for etiologic and early marker investigations in the PLCO trial. Mutat Res. 2005;592:147–54. doi: 10.1016/j.mrfmmm.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 14.Xu WH, et al. Joint effect of cigarette smoking and alcohol consumption on mortality. Prev Med. 2007;45:313–9. doi: 10.1016/j.ypmed.2007.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng W, et al. The Shanghai Women’s Health Study: rationale, study design, and baseline characteristics. Am J Epidemiol. 2005;162:1123–31. doi: 10.1093/aje/kwi322. [DOI] [PubMed] [Google Scholar]
- 16.Anderson GL, et al. Implementation of the Women’s Health Initiative study design. Ann Epidemiol. 2003;13:S5–17. doi: 10.1016/s1047-2797(03)00043-7. [DOI] [PubMed] [Google Scholar]
- 17.Rexrode KM, Lee IM, Cook NR, Hennekens CH, Buring JE. Baseline characteristics of participants in the Women’s Health Study. J Womens Health Gend Based Med. 2000;9:19–27. doi: 10.1089/152460900318911. [DOI] [PubMed] [Google Scholar]
- 18.McWilliams RR, et al. Polymorphisms in DNA repair genes, smoking, and pancreatic adenocarcinoma risk. Cancer Res. 2008;68:4928–35. doi: 10.1158/0008-5472.CAN-07-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eppel A, Cotterchio M, Gallinger S. Allergies are associated with reduced pancreas cancer risk: A population-based case-control study in Ontario, Canada. Int J Cancer. 2007;121:2241–5. doi: 10.1002/ijc.22884. [DOI] [PubMed] [Google Scholar]
- 20.Duell EJ, et al. Detecting pathway-based gene-gene and gene-environment interactions in pancreatic cancer. Cancer Epidemiol Biomarkers Prev. 2008;17:1470–9. doi: 10.1158/1055-9965.EPI-07-2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hassan MM, et al. Risk factors for pancreatic cancer: case-control study. Am J Gastroenterol. 2007;102:2696–707. doi: 10.1111/j.1572-0241.2007.01510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olson SH, et al. Allergies, variants in IL-4 and IL-4R alpha genes, and risk of pancreatic cancer. Cancer Detect Prev. 2007;31:345–51. doi: 10.1016/j.cdp.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 23.Risch HA. Etiology of pancreatic cancer, with a hypothesis concerning the role of N-nitroso compounds and excess gastric acidity. J Natl Cancer Inst. 2003;95:948–60. doi: 10.1093/jnci/95.13.948. [DOI] [PubMed] [Google Scholar]
- 24.Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38:209–13. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- 25.Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aird I, Bentall HH, Roberts JA. A relationship between cancer of stomach and the ABO blood groups. Br Med J. 1953;1:799–801. doi: 10.1136/bmj.1.4814.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marcus DM. The ABO and Lewis blood-group system. Immunochemistry, genetics and relation to human disease. N Engl J Med. 1969;280:994–1006. doi: 10.1056/NEJM196905012801806. [DOI] [PubMed] [Google Scholar]
- 28.Melzer D, et al. A genome-wide association study identifies protein quantitative trait loci (pQTLs) PLoS Genet. 2008;4:e1000072. doi: 10.1371/journal.pgen.1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pare G, et al. Novel association of ABO histo-blood group antigen with soluble ICAM-1: results of a genome-wide association study of 6,578 women. PLoS Genet. 2008;4:e1000118. doi: 10.1371/journal.pgen.1000118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan X, et al. Population-based genome-wide association studies reveal six loci influencing plasma levels of liver enzymes. Am J Hum Genet. 2008;83:520–8. doi: 10.1016/j.ajhg.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Itzkowitz SH, et al. Cancer-associated alterations of blood group antigen expression in the human pancreas. J Natl Cancer Inst. 1987;79:425–34. [PubMed] [Google Scholar]
- 32.Berman DM, et al. Widespread requirement for Hedgehog ligand stimulation in growth of digestive tract tumours. Nature. 2003;425:846–51. doi: 10.1038/nature01972. [DOI] [PubMed] [Google Scholar]
- 33.Wolpin BM, et al. ABO blood group and the risk of pancreatic cancer. J Natl Cancer Inst. 2009;101:424–31. doi: 10.1093/jnci/djp020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wigginton JE, Cutler DJ, Abecasis GR. A note on exact tests of Hardy-Weinberg equilibrium. Am J Hum Genet. 2005;76:887–93. doi: 10.1086/429864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–59. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frazer KA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–61. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas G, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40:310–5. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 38.Hunter DJ, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–4. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu K, et al. Population substructure and control selection in genome-wide association studies. PLoS ONE. 2008;3:e2551. doi: 10.1371/journal.pone.0002551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 41.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun L, Wilder K, McPeek MS. Enhanced pedigree error detection. Hum Hered. 2002;54:99–110. doi: 10.1159/000067666. [DOI] [PubMed] [Google Scholar]
- 43.Lettre G, Lange C, Hirschhorn JN. Genetic model testing and statistical power in population-based association studies of quantitative traits. Genet Epidemiol. 2007;31:358–62. doi: 10.1002/gepi.20217. [DOI] [PubMed] [Google Scholar]
- 44.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–58. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.