Summary
The mammalian APOBEC3 family of cytidine deaminases includes several members that possess potent antiretroviral activity. Human APOBEC3F and APOBEC3G are specifically incorporated into human immunodeficiency virus type 1 (HIV-1) progeny virions in the absence of virion infectivity factor (Vif), where they deaminate deoxycytidine to deoxyuridine on the minus strand of nascent reverse transcripts. Editing of the HIV-1 cDNA leads to its degradation or to G to A hypermutation of the integrated provirus [1–8]. Here, we show that APOBEC3 proteins also restrict the activity of a distantly related long terminal repeat (LTR) retrotransposon. When expressed in the yeast Saccharomyces cerevisiae, human APOBEC3C, APOBEC3F, or APOBEC3G or mouse APOBEC3 potently inhibit replication of the Ty1 LTR retrotransposon. APOBEC3G interacts with Ty1 Gag and is packaged into Ty1 virus-like particles (VLPs) by a mechanism that closely resembles the one it uses to enter HIV-1 virions. Expression of APOBEC3G results in a reduced level of Ty1 cDNA integration and G to A editing of integrated Ty1 cDNA. Our findings indicate that APOBEC3G restricts Ty1 and HIV-1 by similar mechanisms and suggest that the APOBEC3 proteins target a substantially broader spectrum of retroelements than previously appreciated.
Results and Discussion
LTR retrotransposons and retroviruses are related families of mobile elements that transpose through an RNA intermediate. They are characterized by the presence of long-terminal repeats (LTRs), orthologous gag and pol genes, and common mechanisms of expression and replication. The major difference between these two classes of retroelements is that retroviral genomes encode an envelope glycoprotein (Env) that promotes the dissemination of viral particles to new host cells. Evidence from several lines of investigation favors the hypothesis that retroviruses evolved from LTR retrotransposons in part by acquisition of an env-like gene (reviewed in [9]). Given the evolutionary relationship between LTR retrotransposons and retroviruses, it seems reasonable to hypothesize that they share susceptibility to mechanisms of innate regulation by eukaryotic host cells [10–12]. However, little evidence for this idea exists.
The human genome encodes seven APOBEC3 genes, which are the products of a recent expansion from a single mammalian APOBEC3 gene [13, 14]. Human APOBEC3B (hA3B), APOBEC3C (hA3C), APOBEC3F (hA3F), APOBEC3G (hA3G), and mouse APOBEC3 (mA3) restrict the replication of distinct classes of primate lentiviruses [2, 5–7, 15]. APOBEC3G is also active against other retroviruses and even the hepadnavirus hepatitis B virus but does not inhibit the abundant LINE-1 non-LTR retrotransposon, which, unlike LTR retroelements, is reverse transcribed in the nucleus [2, 4, 16, 17]. Given these observations, we asked whether the antiretroviral activity of APOBEC3G proteins might extend to LTR retrotransposons. To test this possibility, we focused on the Ty1 LTR retrotransposon in the yeast Saccharomyces cerevisiae. Ty1 belongs to the Ty1/copia clade of LTR retrotransposons, which is distantly related to mammalian retroviruses [18]. The transposition of Ty1, unlike most other LTR retrotransposons, can be detected when the element is located in its normal context in the chromosome and driven from its own promoter. Hence, it is an ideal system for analyzing host-factor regulation.
APOBEC3 open-reading frames, fused to the GAL1 promoter on the pYES2 expression plasmid and to a HA-epitope at the C terminus, were introduced into a yeast strain containing a chromosomal Ty1 element marked by the retrotransposition indicator gene, his3AI. The his3AI gene, which is defective due to an antisense intron within the HIS3 coding sequence, is contained within the 3′UTR of Ty1 in the opposing transcriptional orientation [19]. Therefore, the intron in his3AI can be spliced from the Ty1his3AI transcript. When this spliced RNA is used as a template for retrotransposition, a functional HIS3 gene is recreated in the newly integrated element, allowing cells that sustain a retrotrans-position event to be scored as His+ prototrophs.
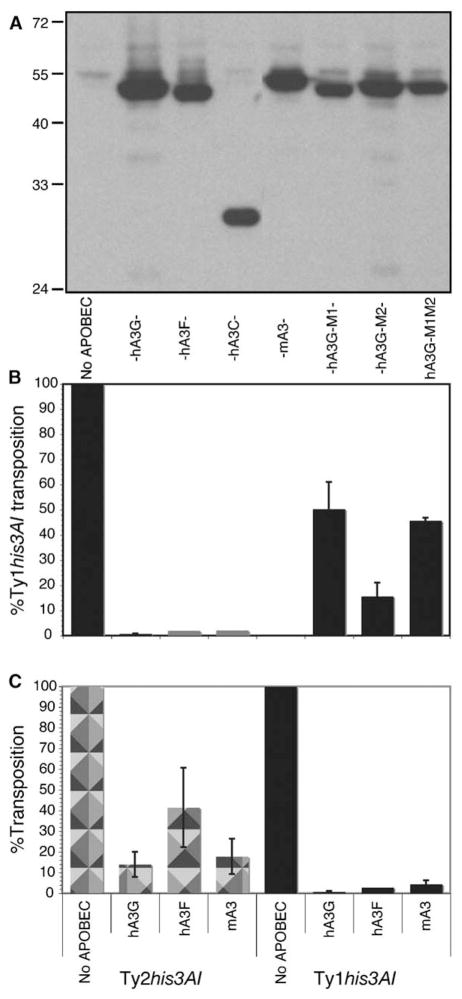
The frequency of His+ prototroph formation was measured in strain JC3212 expressing hA3C, hA3F, hA3G, or mA3 (Figure 1A). Remarkably, expression of any of the APOBEC3 proteins reduced Ty1his3AI transposition to 2% or less of the activity in the strain lacking APOBEC (Figure 1B). Transposition was not inhibited when the GAL1 promoter was repressed by growth in glucose (data not shown). Mutant hA3G proteins harboring substitutions predicted to disrupt one or both of the two zinc binding catalytic sites (hA3G-M1: S95A, P96A, C97A; hA3G-M2: S286A, P287A, C288A; hA3G-M1M2: S95A, P96A, C97A, S286A, P287A, C288A) eliminated most of the inhibitory activity of hA3G, suggesting that the deaminase activity of hA3G is critical for inhibition of retrotransposition. The residual inhibitory activity of these hA3G mutants may be due to a deaminase-independent function of hA3G [20] or to an indirect effect of expressing high levels of the hA3G protein on a modulator of Ty1 transposition.
Figure 1. Human and Mouse APOBEC3 Proteins Specifically Inhibit Ty1 Retrotransposition.
(A) Western blot analysis of whole cell lysates from strain JC3212 induced for expression of the HA-tagged APOBEC3 protein indicated, or no APOBEC3, with anti-HA antibodies.
(B) Relative frequency of Ty1his3AI transposition in three transformants harboring pYES2 (no APOBEC) or pYES2 with the APOBEC3 allele indicated in strain JC3212. The percentage of Ty1his3AI transposition is the average frequency of transposition in APOBEC3 plasmid transformants divided by the average frequency of Ty1his3AI transposition in pYES2 transformants.
(C) Similar to (B) except that strain JC297 containing Ty1his3AI, or strain JC560 harboring Ty2his3AI, was used in the analysis. Error bars, ± standard error.
The ability of hA3F and hA3G to restrict HIV-1 replication is correlated with the specific incorporation of these proteins into HIV-1 virions [1, 6, 8]. If APOBEC3 proteins are specifically incorporated into Ty1 VLPs, then they might have disparate effects on transposition of other LTR retrotransposons in yeast, depending on their relative levels of VLP incorporation. To test this hypothesis, we examined the ability of APOBEC3 proteins to block transposition of Ty2 elements, which are more than 90% homologous to Ty1 but show a significant level of divergence within gag [21, 22]. Derivatives of pYES2 containing various APOBEC3-HA alleles were introduced into yeast strains harboring either a Ty1his3AI element (strain JC297) or a Ty2his3AI element (strain JC560). Similar to the results observed with strain JC3212 (Figure 1B), expression of hA3G, hA3F, or mA3 reduced Ty1his3AI transposition to between 0.8% and 4.5% of the activity in strain JC297 lacking APOBEC (Figure 1C). In contrast, each of the APOBEC3 proteins reduced Ty2his3AI transposition only to 14% to 42% of the activity in strain JC560 lacking APOBEC. Because Ty2 RNA and transposition are both lower than those of Ty1 [23], these data argue against a passive interaction between hA3G and Ty cDNA or RNA; rather, they raise the possibility that APOBEC3 proteins are preferentially incorporated into Ty1 VLPs.
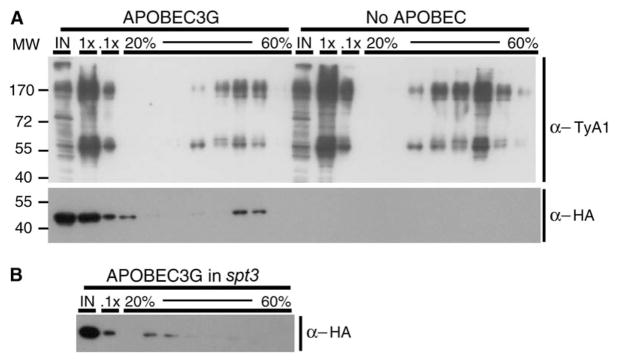
To test this idea, we determined whether hA3G cosediments with Ty1 VLPs in sucrose gradients. Cell lysate prepared from a strain expressing hA3G-HA was fractionated on two consecutive sucrose gradients. The p58 and p54 Ty1 Gag proteins and the p190 Ty1 Gag-Pol fusion protein were detected in four fractions collected near the bottom of the second gradient by Western blot analysis with anti-VLP antisera (Figure 2A). Human A3G cosedimented with Ty1 VLPs in the two densest fractions of the four containing Ty1 VLPs. In contrast, hA3G was not present in these dense fractions when expressed in an spt3 strain, which lacks Ty1 VLPs because of the absence of a factor, Spt3, required for Ty1 transcription (Figure 2B). Therefore, the inhibition of Ty1 transposition by hA3G may reflect the packaging of hA3G into Ty1 VLPs.
Figure 2. Human APOBEC3G Cosediments with Ty1 VLPs.
(A) Western blot analysis of sucrose gradient fractions of cell lysates from strain JC3212 expressing APOBEC3G or no APOBEC with rabbit antiserum specific for Ty1 Gag (top) or anti-HA antibodies (bottom). For each strain, 10 ug of whole cell lysate (IN), 7 μl (1×) and 0.7 μl (0.1×) of the 45% fraction of the first sucrose gradient and 11.5 μl of the first and then every third fraction of the second 20% to 60% gradient (20%–60%) were analyzed.
(B) Similar to (A) except an spt3 strain expressing APOBEC3G was analyzed with anti-HA antibodies.
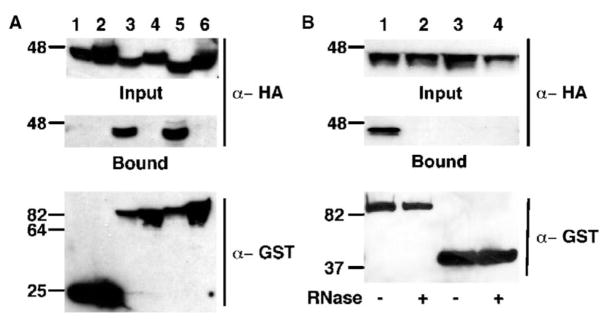
Packaging of hA3G into HIV-1 virions is mediated by a specific interaction between hA3G and the HIV-1 Gag protein [24–27]. We used glutathione-S-transferase (GST)-based coprecipitation experiments to determine whether hA3G interacts with Ty1 Gag (Figure 3A). Human 293T cells that expressed a HA epitope-tagged form of either hA3G (Figure 3A, top, lanes 1, 3, and 5) or β-arrestin (a cytoplasmic human protein that serves as a negative control; lanes 2, 4, and 6) and coexpressed nonfused GST (bottom, lanes 1 and 2), HIV-1 GST-Gag (lanes 3 and 4), or Ty1 GST-Gag (lanes 5 and 6) were lysed, and the fraction of each lysate that bound to glutathione-Sepharose beads was purified. Western analysis of the bound fractions with anti-HA antibodies revealed that hA3G was strongly associated with HIV-1 GST-Gag (middle, lane 3) and Ty1 GST-Gag (lane 5), but not with non-fused GST (lane 1). In contrast, HA-tagged β-arrestin was not coprecipitated by the GST-Gag fusion proteins (middle, lanes 2, 4, and 6). We have also observed coimmunoprecipitation of hA3G with Ty1 Gag in yeast (data not shown).
Figure 3. Ty1 Gag Binds APOBEC3G Specifically.
(A) Western blot analysis of whole cell lysates from human 293T cells cotransfected with pK-GST (lanes 1 and 2), pK/HIVGAG-GST (lanes 3 and 4), or pK/TYA-GST (lanes 5 and 6) and with pK-h3G-HA (lanes 1, 3, and 5) or pK-βARR-HA (lanes 2, 4, and 6) by using anti-HA antibodies (top, input) or anti-GST antibodies (bottom). The fraction of each lysate that bound to glutathione-Sepharose beads was subjected to Western analysis with anti-HA antibodies (middle, bound).
(B) 293T cells were cotransfected with phA3G-HA together with pK/TY1GAG-GST (lanes 1 and 2) or pK/MS2-GST and pDM128/4XMS2 (lanes 3 and 4). Binding assays were performed as described in (A) except that the cell lysate was incubated at 37°C for 10 min in the presence (lanes 2 and 4) or absence (lanes 1 and 3) of 50 μg of RNase A, prior to addition of the glutathione-Sepharose beads.
Incorporation of APOBEC3G into HIV-1 virion particles is mediated by the nucleocapsid domain of HIV-1 Gag, one of whose functions is to bind and recruit the HIV-1 RNA genome into virion particles [24–27]. Indeed, binding of APOBEC3G by HIV-1 Gag also requires the presence of RNA, although nonspecific, cellular RNAs function as well as viral RNA [24, 25]. It has therefore been proposed that APOBEC3G exploits the essential RNA packaging step to gain entry into retroviral particles [25].
To test whether RNA also plays a role in Ty1 Gag binding by APOBEC3G, we asked whether RNase treatment would block Ty1 Gag binding by APOBEC3G, as previously reported for HIV-1 Gag [24, 25]. Indeed, as shown in Figure 3B, lanes 1 and 2, RNase treatment entirely prevented this interaction. As a control for the specificity of this interaction, a fusion protein consisting of GST linked to the bacteriophage MS2 coat protein, a well-characterized RNA binding protein active in human cells (27), was tested for interaction with APOBEC3G in transfected cells. As shown in Figure 3B, lanes 3 and 4, no interaction was observed. Of note, these 293T cells were also transfected with an expression vector, pDM128/4XMS2, that we have previously shown expresses high levels of an mRNA bearing four tandem high-affinity MS2 target sites [28]. So the lack of any interaction between APOBEC3G and GST-MS2 noted in Figure 3B is unlikely to be due to the absence of RNAs able to interact with the GST-MS2 protein. Taken together, these findings demonstrate that hA3G forms a specific ribonucleoprotein complex with Ty1 Gag leading to the packaging of hA3G into Ty1 VLPs.
Because hA3G induces C to T hypermutation on the minus strand of newly synthesized HIV-1Δvif cDNA, we asked whether hA3G also induces C to T editing in Ty1HIS3 cDNA. Because Ty1 cDNA is in very low abundance [29], we analyzed integrated Ty1HIS3 cDNA from His+ prototrophs to obtain independent cDNA sequences. A fragment of Ty1HIS3 cDNA was amplified with one primer that recognizes the splice junction of HIS3 and one primer that hybridizes in Ty1 pol, and the Ty1 pol region was sequenced. Eleven C to T substitutions and one G to A substitution in 12,227 nucleotides sequenced were detected on the minus strand of Ty1HIS3 cDNA in strains expressing hA3G. Between one and six C to T substitutions were found on five of seventeen cDNA templates. In contrast, zero substitutions in 14,559 nucleotides sequenced from 21 Ty1HIS3 templates were detected in strains lacking hA3G. These findings support the hypothesis that hA3G deaminates C to U residues on the minus strand of Ty1 reverse transcripts, although the level of editing appears to be lower than that seen on HIV-1 cDNA [2–5].
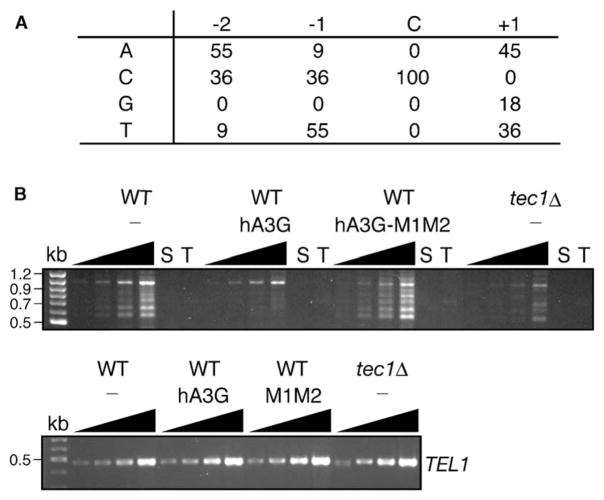
APOBEC3G preferentially recognizes the target sequence CCCA and does not effectively edit C residues flanked by 5′ G residues. (The edited C residue is underlined.) The C residue at position −1 is the most highly favored, but T at position −1 is recognized at a low frequency [7]. Analysis of nucleotides adjacent to the edited C residues in Ty1 indicated that APOBEC3G has a relaxed target specificity in yeast (Figure 4A). At position −1, T or C is preferred, and at position −2, either A or C is preferred. We note that G residues were not detected at either position. This somewhat relaxed specificity may reflect the fact that Ty1 transposition assays are conducted at 20°C, which is significantly lower than the temperature of 37°C at which HIV-1 replication is assayed. This reduced temperature may stabilize the binding of hA3G to less favorable consensus sequences.
Figure 4. Human APOBEC3G Edits C Residues in Ty1 cDNA and Inhibits Integration of Total Ty1 cDNA Upstream of tRNAgly Genes.
(A) Sequence context for eleven independent dC deaminations on the minus strand of Ty1 cDNA. The fraction of events (expressed as a percentage) in which a particular base (A, C, G, or T) was found at the −2, −1, or +1 position relative to the deaminated C residue is indicated.
(B) Top, PCR analysis of genomic DNA from a wild-type (WT) strain expressing no APOBEC protein (−) or the APOBEC3 protein indicated, or from a tec1Δ strain lacking APOBEC, with one primer that hybridizes in the Ty1 pol domain (T) and one that hybridizes to 16 tRNAgly genes (S). PCR reactions were terminated after 29, 31, 33, or 35 cycles (black triangles). Control PCR reactions contained one primer (T or S). Bottom, PCR reactions of the TEL1 gene terminated after 17, 19, 21, or 23 cycles, as a control for equivalent template concentrations.
It is possible that the low level of editing we observed in Ty1 cDNAs after expression of hA3G is a consequence of having selected for a functional HIS3 gene, which may have enriched for cDNAs that escaped editing. However, Ty1 cDNA extracted from sucrose-gradient-purified Ty1 VLPs from strains expressing hA3G also had a similarly low but significant level of C to T transitions on the minus strand (data not shown). A second possibility is that hypermutated Ty1 cDNA might be targeted for degradation, as has been observed for HIV-1, and therefore would be unavailable for integration. Alternatively, editing in the primer binding site or U3 region of the LTR, which has also been observed in HIV-1 [30], might interfere with plus strand cDNA synthesis or integration, respectively. To determine if expression of APOBEC3G blocks Ty1 cDNA integration or a step prior to it, we measured the level of de novo integration into regions upstream of tRNA genes, which are preferred sites of Ty1 integration, in the presence or absence of hA3G. Strains were grown at 20°C to allow unselected Ty1 transposition events to occur. Genomic DNA was prepared, and total Ty1 element integration into the upstream region of any of the 16-tRNAgly genes was detected by PCR (Figure 4B). As anticipated, the strain lacking hA3G yielded a ladder of PCR products between 0.5 kb and 1.2 kb (representing Ty1 insertions ~100 to 800 bp upstream of one or more tRNAgly genes [31]). The appearance of this ladder was dependent on Ty1 transcription, which is reduced in a tec1Δ mutant. Expression of hA3G sharply reduced the appearance of Ty1 integration bands. Importantly, amino-acid substitutions in the catalytic domains of hA3G restored Ty1 integration upstream of tRNAgly genes. These findings indicate that the cytidine deaminase activity of hA3G interferes with cDNA integration or a step prior to it, such as cDNA synthesis or stability. Therefore, we hypothesize that heavily deaminated Ty1 cDNA may be rapidly degraded in vivo.
The HIV-1 protein Vif suppresses the antiviral activity of hA3G by targeting it for ubiquitination and degradation [32–34]. Therefore, we determined whether HIV-1Vif could restore Ty1 retrotransposition when hA3G was expressed. Although Vif was expressed at high levels, hA3G inhibited Ty1his3AI transposition equally in its presence or absence (data not shown). In human cells, a Vif mutant that interacts with hA3G, but not the Cul5-elongin B/C E3-ubiquitin ligase, does not induce degradation of hA3G. Hence, our results may be explained by the fact that the yeast homologs of Cul5 and elongins B and C are not highly conserved. It is possible, however, that the function of Vif can be reconstituted in yeast by introduction of one or more of these human cofactors.
In summary, we have shown that human and mouse APOBEC3 proteins can inhibit not only exogenous retroviruses such as HIV-1 but also the yeast Ty1 retrotransposon, despite the great evolutionary distance between these retroelements. Moreover, Ty1 Gag, like HIV-1 Gag, interacts specifically with APOBEC3G, and both interactions are RNA dependent (Figure 3). Interestingly, the nucleocapsid domain of HIV-1 Gag, which mediates its interaction with hA3G [24, 25, 27], is highly divergent from the nucleocapsid domain of Ty1 Gag. Thus, APOBEC3 proteins may recognize a conserved structural intermediate involved in RNA packaging into both HIV-1 virion particles and Ty1 VLPs, rather than a conserved sequence. Overall, these data argue that Ty1 and retroviral Gag proteins have not only conserved functions critical for their replication, such as genomic RNA packaging, but also their susceptibility to the APOBEC3 family of innate resistance factors. This result supports the idea that yeast cell factors that repress Ty1 transposition may also be candidates for innate regulators of exogenous retroviral infection in mammals.
Although the ancient Ty1/copia elements have yet to be identified in mammalian genomes, the more recent superfamily of Ty3/gypsy LTR retrotransposons as well as endogenous retroviruses constitute a significant fraction of typical mammal genomes [35, 36]. Moreover, steady retrotransposition of these elements over time is likely to be more deleterious to genome integrity than sporadic instances of retroviral infection. Our findings, taken together with a recent study demonstrating that hA3G inhibits retrotransposition of endogenous retroviruses in mice [37], suggest that APOBEC3 proteins can target a surprisingly broad range of LTR retroelements. Hence, mammalian APOBEC3 proteins may have evolved originally to restrict the transposition of LTR retrotransposons in these genomes, with inhibition of retroviral infection either being fortuitous or appearing later in evolution.
Supplementary Material
Acknowledgments
We thank Marlene Belfort for critical manuscript review and the Wadsworth Center Molecular Genetics Core Facility for DNA sequencing. This work was supported by National Institutes of Health grants GM52072 to M.J.C. and AI57099 to B.R.C.
Footnotes
Supplemental Data
Supplemental Experimental Procedures are available with this article online at http://www.current-biology.com/cgi/content/full/15/7/661/DC1/.
References
- 1.Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 2.Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 3.Zhang H, Yang B, Pomerantz RJ, Zhang C, Arunachalam SC, Gao L. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature. 2003;424:94–98. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 5.Mariani R, Chen D, Schrofelbauer B, Navarro F, Konig R, Bollman B, Munk C, Nymark-McMahon H, Landau NR. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell. 2003;114:21–31. doi: 10.1016/s0092-8674(03)00515-4. [DOI] [PubMed] [Google Scholar]
- 6.Wiegand HL, Doehle BP, Bogerd HP, Cullen BR. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 2004;23:2451–2458. doi: 10.1038/sj.emboj.7600246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bishop KN, Holmes RK, Sheehy AM, Davidson NO, Cho SJ, Malim MH. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr Biol. 2004;14:1392–1396. doi: 10.1016/j.cub.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 8.Zheng YH, Irwin D, Kurosu T, Tokunaga K, Sata T, Peterlin BM. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J Virol. 2004;78:6073–6076. doi: 10.1128/JVI.78.11.6073-6076.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim FJ, Battini JL, Manel N, Sitbon M. Emergence of vertebrate retroviruses and envelope capture. Virology. 2004;318:183–191. doi: 10.1016/j.virol.2003.09.026. [DOI] [PubMed] [Google Scholar]
- 10.Aye M, Irwin B, Beliakova-Bethell N, Chen E, Garrus J, Sandmeyer S. Host factors that affect Ty3 retrotransposition in Saccharomyces cerevisiae. Genetics. 2004;168:1159–1176. doi: 10.1534/genetics.104.028126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bolton EC, Mildvan AS, Boeke JD. Inhibition of reverse transcription in vivo by elevated manganese ion concentration. Mol Cell. 2002;9:879–889. doi: 10.1016/s1097-2765(02)00495-1. [DOI] [PubMed] [Google Scholar]
- 12.Scholes DT, Banerjee M, Bowen B, Curcio MJ. Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics. 2001;159:1449–1465. doi: 10.1093/genetics/159.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jarmuz A, Chester A, Bayliss J, Gisbourne J, Dunham I, Scott J, Navaratnam N. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics. 2002;79:285–296. doi: 10.1006/geno.2002.6718. [DOI] [PubMed] [Google Scholar]
- 14.Conticello SG, Thomas CJ, Petersen-Mahrt SK, Neuberger MS. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol Biol Evol. 2005;22:367–377. doi: 10.1093/molbev/msi026. [DOI] [PubMed] [Google Scholar]
- 15.Yu Q, Chen D, Konig R, Mariani R, Unutmaz D, Landau NR. APOBEC3B and APOBEC3C are potent inhibitors of simian immunodeficiency virus replication. J Biol Chem. 2004;279:53379–53386. doi: 10.1074/jbc.M408802200. [DOI] [PubMed] [Google Scholar]
- 16.Turelli P, Mangeat B, Jost S, Vianin S, Trono D. Inhibition of hepatitis B virus replication by APOBEC3G. Science. 2004;303:1829. doi: 10.1126/science.1092066. [DOI] [PubMed] [Google Scholar]
- 17.Turelli P, Vianin S, Trono D. The innate antiretroviral factor APOBEC3G does not affect human LINE-1 retrotransposition in a cell culture assay. J Biol Chem. 2004;279:43371–43373. doi: 10.1074/jbc.C400334200. [DOI] [PubMed] [Google Scholar]
- 18.Eickbush TH, Malik HS. Origins and evolution of retrotransposons. In: Craig N, Craigie R, Gellert M, Lambowitz A, editors. J Biol Chem, DNA Mobile. Washington, D.C: American Society for Microbiology; 2002. pp. 1111–1144. [Google Scholar]
- 19.Curcio MJ, Garfinkel DJ. Single-step selection for Ty1 element retrotransposition. Proc Natl Acad Sci USA. 1991;88:936–940. doi: 10.1073/pnas.88.3.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newman EN, Holmes RK, Craig HM, Klein KC, Lingappa JR, Malim MH, Sheehy AM. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr Biol. 2005;15:166–170. doi: 10.1016/j.cub.2004.12.068. [DOI] [PubMed] [Google Scholar]
- 21.Kim JM, Vanguri S, Boeke JD, Gabriel A, Voytas DF. Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 1998;8:464–478. doi: 10.1101/gr.8.5.464. [DOI] [PubMed] [Google Scholar]
- 22.Jordan IK, McDonald JF. Tempo and mode of Ty element evolution in Saccharomyces cerevisiae. Genetics. 1999;151:1341–1351. doi: 10.1093/genetics/151.4.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Curcio MJ, Hedge AM, Boeke JD, Garfinkel DJ. Ty RNA levels determine the spectrum of retrotransposition events that activate gene expression in Saccharomyces cerevisiae. Mol Gen Genet. 1990;220:213–221. doi: 10.1007/BF00260484. [DOI] [PubMed] [Google Scholar]
- 24.Schafer A, Bogerd HP, Cullen BR. Specific packaging of APOBEC3G into HIV-1 virions is mediated by the nucleocapsid domain of the gag polyprotein precursor. Virology. 2004;328:163–168. doi: 10.1016/j.virol.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 25.Zennou V, Perez-Caballero D, Gottlinger H, Bieniasz PD. APOBEC3G incorporation into human immunodeficiency virus type 1 particles. J Virol. 2004;78:12058–12061. doi: 10.1128/JVI.78.21.12058-12061.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alce TM, Popik W. APOBEC3G is incorporated into virus-like particles by a direct interaction with HIV-1 Gag nucleocapsid protein. J Biol Chem. 2004;279:34083–34086. doi: 10.1074/jbc.C400235200. [DOI] [PubMed] [Google Scholar]
- 27.Cen S, Guo F, Niu M, Saadatmand J, Deflassieux J, Kleiman L. The interaction between HIV-1 Gag and APOBEC3G. J Biol Chem. 2004;279:33177–33184. doi: 10.1074/jbc.M402062200. [DOI] [PubMed] [Google Scholar]
- 28.Coburn GA, Wiegand HL, Kang Y, Ho DN, Georgiadis MM, Cullen BR. Using viral species specificity to define a critical protein/RNA interaction surface. Genes Dev. 2001;15:1194–1205. doi: 10.1101/gad.888201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conte D, Barber E, Banerjee M, Garfinkel DJ, Curcio MJ. Posttranslational regulation of Ty1 retrotransposition by mitogen-activated protein kinase Fus3. Mol Cell Biol. 1998;18:2502–2513. doi: 10.1128/mcb.18.5.2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu Q, Konig R, Pillai S, Chiles K, Kearney M, Palmer S, Richman D, Coffin JM, Landau NR. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat Struct Mol Biol. 2004;11:435–442. doi: 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
- 31.Bachman N, Eby Y, Boeke JD. Local definition of Ty1 target preference by long terminal repeats and clustered tRNA genes. Genome Res. 2004;14:1232–1247. doi: 10.1101/gr.2052904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marin M, Rose KM, Kozak SL, Kabat D. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat Med. 2003;9:1398–1403. doi: 10.1038/nm946. [DOI] [PubMed] [Google Scholar]
- 33.Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science. 2003;302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- 34.Mehle A, Strack B, Ancuta P, Zhang C, McPike M, Gabuzda D. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J Biol Chem. 2004;279:7792–7798. doi: 10.1074/jbc.M313093200. [DOI] [PubMed] [Google Scholar]
- 35.Butler M, Goodwin T, Simpson M, Singh M, Poulter R. Vertebrate LTR retrotransposons of the Tf1/sushi group. J Mol Evol. 2001;52:260–274. doi: 10.1007/s002390010154. [DOI] [PubMed] [Google Scholar]
- 36.McCarthy EM, McDonald JF. Long terminal repeat retrotransposons of Mus musculus. Genome Biol. 2004;5:R14. doi: 10.1186/gb-2004-5-3-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Esnault C, Heidmann O, Delebecque F, Dewannieux M, Ribet D, Hance AJ, Heidmann T, Schwartz O. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature. 2005;433:430–433. doi: 10.1038/nature03238. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.