SUMMARY
Chitinases have been implicated to be of importance in the life cycle development and transmission of a variety of parasitic organisms. Using a molecular approach, we identified and characterized the structure of a single copy LmexCht1-chitinase gene from the primitive trypanosomatid pathogen of humans, Leishmania mexicana. The LmexCht1 encodes an ~50 kDa protein, with well-conserved substrate-binding and catalytic domains characteristic of members of the Chitinase-18 protein family. Further, we showed that LmexCht1 mRNA is constitutively expressed by both the insect vector (i.e. promastigote) and mammalian (i.e. amastigote) life cycle developmental forms of this protozoan parasite. Interestingly, however, amastigotes were found to secrete/release ~ >2-4 fold higher levels of chitinase activity during their growth in vitro than promastigotes. Moreover, a homologous episomal-expression system was devised and used to express an epitope–tagged LmexCht1 chimeric construct in these parasites. Expression of the LmexCht1 chimera was verified in these transfectants by RT-PCR, Western blots and indirect immunofluorescence analyses. Further, results of coupled-immunoprecipitation/ enzyme activity experiments demonstrated that the LmexCht1 chimeric protein was secreted/released by these transfected L. mexicana parasites and that it possessed functional chitinase enzyme activity. Such transfectants were also evaluated for their infectivity both in human macrophages in vitro and in two different strains of mice. Results of those experiments demonstrated that the LmexCht1 transfectants survived significantly better in human macrophages and also produced significantly larger lesions in mice than control parasites. Taken together, our results indicate that the LmexCht1-chimera afforded a definitive survival advantage to the parasite within these mammalian hosts. Thus, the LmexCht1 could potentially represent a new virulence determinant in the mammalian phase of this important human pathogen
Keywords: Leishmania, human parasite, gene structure, infectivity, macrophage, mouse infections, trypanosomatid, kinetoplastid protozoan, chitinase
INTRODUCTION
Leishmania sp. are a group of trypanosomatid protozoan pathogens which are transmitted to their mammalian hosts via the bite of infected sand fly vectors. In humans, these parasites cause a broad spectrum of diseases ranging from mild cutaneous ulcerations to severe and typically fatal visceral disease (1). All Leishmania have a life cycle that includes two major parasite developmental stages: 1) extracellular, flagellated promastigote forms that reside and multiply within the alimentary tract of their sand fly vector hosts and 2) obligate intracellular, amastigote forms which reside and multiply within the phago-lysosomal system of infected mammalian macrophages (2). Parasite proteins which mediate the survival, growth and development of Leishmania within these diverse host environments generally remain to be elucidated. However, one such molecule thought to be involved in these processes is a parasite-derived chitinase. The putative substrate of this enzyme, chitin, is a ubiquitous structural polysaccharide found in arthropods that may create barriers to parasite development in such hosts (3). In that regard, based on light and electron microscopic observations, previously it has been hypothesized that a Leishmania chitinase might function to facilitate both parasite infection and survival within their sand fly vector hosts (4,5). Further, it was suggested that a parasite chitinase might also aid in the transmission of these organisms to their mammalian hosts (5).
Despite its apparent relevance, to date little direct evidence exists per se concerning the role(s) of such a chitinase in the developmental biology of Leishmania parasites in either their sand fly vector or mammalian hosts. However, previously we used a heterologous probe to indicate that promastigote forms of Leishmania mexicana (i.e. a stage present in its insect vector) contained a putative chitinase-like sequence within their genome (6). It is important to note that this parasite is the major etiologic agent of human cutaneous leishmaniasis in the New World (7). Further, experimental studies of L. mexicana have been greatly facilitated because culture systems exist for generating the various life cycle developmental forms of this parasite in vitro (8-10). Moreover, several mouse model systems have also been established to study the infectivity and pathogenicity of L. mexicana in vivo (8,11-13). In light of the above, in the current report we used a molecular approach to identify, characterize and examine the expression of the LmexCht1-chitinase gene throughout the various developmental life cycle stages of this human pathogen. In addition, parasites transfected with an LmexCht1- chimeric construct were evaluated in both human macrophages and in mice to determine whether this gene may play a role(s) in the survival and virulence of this parasite within its mammalian host.
EXPERIMENTAL PROCEDURES
Reagents
All chemicals used, unless specified, were of analytical grade and purchased from Sigma-Aldrich Chemical Co. Enzymes used for molecular studies were obtained from Roche Molecular Biochemicals; DNA and RNA molecular mass standards were from Invitrogen, Inc. and protein molecular mass standards were purchased from Amersham Biosciences.
Parasites and culture conditions
Promastigote developmental forms of the M379 strain of Leishmania mexicana (WHO designation: MNYC/BZ/62/M379) were grown at 26°C in Medium M199 (Invitrogen Inc.), supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS, Gemini Bio-Products, Woodland, CA) essentially as described previously (14). Amastigote forms of the L. mexicana M379 strain were maintained in BALB/c mice as previously described (8). Lesion amastigotes were isolated from infected mice and cultured axenically in vitro at 32°C; procyclic promastigote cultures were established in vitro by transformation of lesion amastigotes at 26°C; and metacyclic promastigotes were generated from procyclic promastigotes all as described previously (8-10). For isolation of nucleic acids and proteins, parasite cultures were harvested at ~mid-log phase (~1-2 × 107 cells ml−1) by centrifugation at 2100 × g for 15 min. at 4°C (15). The resulting cell pellets were washed twice in ice-cold phosphate buffered saline (PBS, 10mM sodium phosphate, 145 mM NaCl, pH 7.4) by centrifugation as above and finally resuspended in the appropriate buffers.
To assess whether wild-type L. mexicana parasites released/secreted any measurable chitinolytic activity during their growth in vitro, promastigotes and axenic amastigotes were grown to ~1 × 107 cells ml−1 as above, harvested by centrifugation and their cell-free culture supernatants were processed as previously described (16). Aliquots of such culture supernatants were used for both enzyme assays and immunoprecipitation experiments. Prior to use in assays, parasite culture supernatants and unused medium controls were neutralized to ~pH 7.0 using 1M Tris-HCl buffer, pH8.0.
Enzyme assays
Cell-free culture supernatants from both wild-type L. mexicana promastigotes and axenic amastigotes (above) were analyzed for their exochitinase (which releases dimers of N-acetylglucosamine from the non-reducing ends of chitin) and endochitinase (which hydrolyzes bonds within the chitin polymer) activities (17). For these assays, the fluorogenic substrates: 4-methylumbelliferyl β-D-N, N’-diacetylchitiobioside (4MU-chitobiose) and 4-methylumbelliferyl β-D-N,N’,N”-triacetylchitiotrioside (4MU-chitotriose) were used to measure exo- and endo-chitinase activities, respectively (6). The concentration of the fluorescent 4-methylumbelliferone (4MU) product generated from these substrates was determined using 4MU standards. Fluorescence was measured with a Turner Digital Filter Fluorometer (Model 112; Turner Designs, Sunnyvale, CA) using a 360 nm excitation filter and a 450 nm emission filter. In these experiments, fresh, unused, complete parasite growth (culture) medium was used as a background control. Values obtained from such controls were subtracted from those generated with various test samples. The results of these enzyme assays are expressed as pmoles of 4MU product generated h−1 ml−1 of parasite culture supernatant using either 4MU-chitobiose or 4MU-chitotriose as substrates. All samples were assayed in triplicate and these assays were repeated on multiple samples.
In parallel, such parasite culture supernatants were also subjected to immunoprecipitation reactions using an anti-LdCht1- peptide antiserum or pre-immune serum from the same rabbit (i.e., normal rabbit serum, NRS) in a protein A-based assay as described previously (6,18). The immunoprecipitated-Protein-A bound complexes resulting from such reactions were subsequently assayed for their exochitinase and endochitinase activities as described above. Results of these immunoprecipitation experiments were normalized by subtracting the values obtained with NRS from those obtained with the anti-LdCht1- peptide antiserum. All samples were assayed in triplicate and these assays were repeated on three independently generated samples.
Nomenclature
The designations used in this report for genes, proteins and plasmids follow the nomenclature for Trypanosoma and Leishmania as outlined by Clayton et al (19).
Oligonucleotide primers, PCR and probe preparation
Oligonucleotide primers: PCR-Fwd and PCR-Rev, respectively were designed to portions of Region II (i.e. a putative catalytic site) and Region III (i.e. a putative substrate binding site) of the Leishmania donovani Chitinase 1 gene (18). Previously, these regions were shown to be relatively well conserved among chitinase genes from various sources (18). These primers (PCR-Fwd : 5′-GACGGCATCGACTTCAACTGGGAGTA-3′ and PCR-Rev: 5′-GTACGCCATCAAGTGCACATAGTCGAG-3′) were synthesized by ß-cyanoethylphosphoramidite chemistry using an Expedite™ nucleic acid synthesis system (PE Applied Biosystems) and used in PCR amplifications with L. mexicana genomic (g) DNA as a template. It was possible to use gDNA as template since trypanosomatid protozoans generally do not possess introns within the coding region of their open reading frames (ORF) (20,21). After an initial “hot start” at 94°C for 2 min, the conditions used for amplification were: 94°C for 15 sec, 55°C for 30 sec, 72°C for 30 sec (35 cycles) and a final step at: 72°C for 5 min. The resulting 270-bp amplified-product was cloned into the pCR®2.1-TOPO vector (Invitrogen) and the resulting plasmid (Lmex-PCR270) was subjected to nucleotide sequencing. Analyses of the sequence data obtained from the Lmex-PCR270 clone showed that it had high sequence identity with the L. donovani Cht1 gene (18). Subsequently this cloned PCR fragment was labeled with digoxigenin-dUTP using the PCR Dig Labeling Kit according to manufacturer’s instructions (Roche). The resulting digoxigenin-labeled probe (Lmex-DIG270) was used to screen an L. mexicana cosmid library.
Generation and screening of an L. mexicana Cosmid Library
Genomic DNA was isolated from 109 mid-log phase L. mexicana M379 promastigotes using a Gnome DNA Isolation Kit (Bio101, Carlsbad, CA). Such gDNA was used to construct an L. mexicana cosmid library. To that end, following restriction endonuclease digestion, gDNA was ligated into the SuperCos I vector, phage-packaged and adsorbed onto host E. coli, XL-1 Blue MR cells according to manufacturer’s instructions (Stratagene). Subsequently, ~3000 colonies from this cosmid library were screened using the Lmex-DIG270 probe under high stringency hybridization and washing conditions (i.e. 0.1X SSC; 0.1% SDS at 65°C). Amongst the several positive clones identified from such screening, one (Lmex-Cos1) was chosen for further analyses. DNA was isolated from this Lmex-Cos1 cosmid clone using a plasmid purification kit (Qiagen). Such DNA was subsequently subjected to nt sequencing.
Nucleotide sequencing and Analyses
DNA was sequenced using the fluorescent di-deoxy chain terminator cycle sequencing method (22) at the Johns Hopkins University DNA Analysis Facility (Baltimore, MD). Sequence data obtained from both strands were analyzed using the Genetic Computer Group (GCG) software package (23) running on an N.I.H. Unix System and Sequencher™3.0 software (Gene Codes Corp., Ann Arbor, MI). Further, such sequences were also subjected to BLAST-N and BLAST-P analyses using the NCBI BLAST-link (http://www.ncbi.nlm.nih.gov/BLAST/). Signal sequence and protease cleavage sites were predicted using the SignalP link available at the world wide ExPASy (Expert Protein Analysis System) proteomics server of the Swiss Institute of Bioinformatics (SIB) available at: http://www.expasy.ch/tools. Protein domain analysis was done using the Worldwide Web based Simple Modular Architecture Research Tool (http://smart.embl-heidelberg.de) available via EMBL-EBI, European Bioinformatics Institute (at http://www.ebi.ac.uk). Protein multiple sequence alignments were done using the ClustalW program (24) using a MacVector 7.0 software package (GCG, Madison,WI).
Isolation of genomic DNA and Southern Blot Analysis
L. mexicana gDNA was prepared from log phase promastigote cells using the GNome DNA isolation kit as described above. For Southern blot analyses, such DNA was digested with several restriction endonucleases and separated in 1% agarose gels, transferred to positively charged Nylon membranes (Roche) and cross-linked to the membranes by UV irradiation using a Stratalinker® 2400 (Stratagene). Subsequently, blots were hybridized under high stringency using a second digoxigenin-labeled DNA probe (i.e. LmexCht1-DIG1151) which corresponded to a major portion of the L. mexicana chitinase open reading frame (i.e. nt 1– nt 1151). After washing these blots at high stringency (0.1X SSC, 0.1% SDS at 65°C), the hybridized fragments were visualized using an anti-digoxigenin-alkaline phosphatase conjugated antibody in conjunction with a chemiluminescent reagent (CSPD) according to manufacturer’s instructions (Roche). Images were captured from such blots using BIOMAX™-MR X-ray film (Kodak). DNA was also isolated from the Lmex-Cos1 cosmid clone and digested with several different restriction endonucleases. Subsequently, such preparations were subjected to Southern blot analysis as above.
Isolation of RNA and Northern Blot Analysis
Total RNA was isolated from exponential and stationary phase promastigote and axenic amastigote cultures of L. mexicana, as well as from the lesion-derived amastigotes using TRIzol® according to the manufacturer’s instructions (Invitrogen). Aliquots of 5 μg of total RNA were separated in 1.2% agarose gels using the glyoxal method (25), transferred onto nylon membranes, and cross-linked by UV irradiation as above. Blots were hybridized under high stringency conditions using a DIG-labeled probe (LmexCht1-DIG270). Subsequent to high stringency washing, such blots were subjected to immunological detection using anti-digoxigenin antibody conjugated to alkaline phosphatase and developed using the CSPD chemiluminescent reagent as described above.
Mapping of the 5′ Spliced-leader acceptor site
As indicated above, trypanosomatids generally do not possess introns within their ORFs. However, the pre-mRNAs in these organisms are joined to a 39 nt conserved spliced-leader at their 5′-end by trans-splicing to generate mature, translatable mRNAs (26,27). To identify the 5′-splice acceptor site in the LmexCht1 gene, RT-PCR analysis was performed essentially as described previously (18). For these reactions, cDNA was generated from total RNAs isolated from both L. mexicana M379 promastigotes and axenic amastigotes. Such cDNAs were used as template in PCR with a forward primer (i.e. SpliceFwd) based on the L. mexicana spliced leader sequence [i.e. nt 5 to nt 31 of the 39nt SL sequence] (28): 5′-AACGCTATATAAGTATCAGTTTCTGTA-3′ and a reverse primer (i.e. ORF-RT/Rev) based on a portion of the 5′-end (i.e. nt 138 to nt 157) of the LmexCht1 ORF: 5′-CAGTTATCGAGGTGTTGTGC-3′. The resulting PCR amplified products obtained from these reactions (i.e. using cDNAs from both promastigote and axenic amastigotes) were cloned into the pCR®2.1-TOPO plasmid vector (Invitrogen), sequenced and analyzed. Sequence data obtained from these two parasite developmental forms were compared.
Generation of an epitope tagged expression construct
In this report, the pKSNEO leishmanial vector (29) was used to episomally-express an L. mexicana gene construct. This vector has been used previously to express a variety of homologous and heterologous genes in both promastigotes and axenic amastigotes in several different species of Leishmania parasites (30-33). The pKSNEO vector contains 3′-UTR regulatory elements from an L. donovani amastigote-specific (i.e. A2) gene (34) which can favor some “differential” up-expression (albeit not completely parasite stage-specific) in amastigote forms of these organisms (31). This leishmanial vector was used to express a construct encoding an LmexCht1-hemagglutinin-tagged (-HA) chimeric protein in L. mexicana parasites. To that end, a construct was designed that contained the complete open reading frame of the L. mexicana Cht1 gene (including its 5′-end encoding the signal peptide) joined, at its 3′-end, with a sequence encoding a nine amino acid epitope of the influenza virus hemagglutinin (Roche). This construct was generated by PCR using the Lmex-Cos1 cosmid as template. The forward primer used in these reactions was: 5′-GATACTAGTATGGTGCAGAGGAGCTCACTT-3′ (containing an Spe I restriction endonuclease site shown in bold); and the reverse primer was: 5′-CAAACTAGTTCACGCGTAGTCCGGCACGTCGTACGGGTATAGATCGCGGTC-3′ (containing an Spe I restriction endonuclease site shown in bold; stop codon in bold italics; and a hemagglutinin (-HA) epitope tag [underlined sequence]). The resulting amplified product was gel purified and cloned into the pCR2.1-TOPO vector (Invitrogen) to generate a pCR2.1∷LmexCht1-HA plasmid. The insert was excised from the latter plasmid using Spe I restriction endonuclease. Subsequently, the excised fragment was ligated into the pKSNEO (Spe I-linearized) plasmid to generate the pKSNEO∷LmexCht1-HA plasmid construct. The orientation of LmexCht1-HA in pKSNEO was verified using restriction endo-nuclease analysis. Further, the sequence of the pKSNEO∷LmexCht1-HA construct was verified by nt sequencing.
Transfection of plasmids into Leishmania mexicana promastigotes
Log phase L. mexicana promastigotes were transfected with either the pKSNEO control plasmid or the pKSNEO∷ LmexCht1-HA construct by electroporation using methods derived from Debrabant et al. (32). To that end, L. mexicana promastigote cells were harvested, washed two times with 1X PBS by centrifugation (2100 × g, 10 min at 4°C) and resuspended in ice-cold electroporation buffer (21 mM HEPES, 137 mM NaCl, 5mM KCl, 0.7 mM Na2HPO4, 6mM D-glucose, pH 7.0) at 108 cells ml−1. A 500μl aliquot of such cell suspension was added to a 2-mm gap electroporation cuvette (BTX® Harvard Apparatus, Holliston, MA) to which 20 μl of purified plasmid DNA (at 1 μg μl−1) in sterile 10 mM Tris, 2 mM EDTA, pH 8.0 (Quality Biological, Inc., Gaithersburg, MD) was added. The cells were electroporated using a single pulse (conditions: 475 V, 800 microfarads and 13 ohms) in a BTX ECM-600 electroporation system (BTX® Harvard Apparatus). Electroporated cells were incubated on ice for 10 min, transferred into 5 ml of complete culture medium (M199 containing 10% [v/v] FBS) as described above and incubated at 26°C for 24 h. Following this, the transfected cells were harvested by centrifugation as above and resuspended in the same culture medium containing 10 μg ml−1 Geneticin® (G418, Invitrogen). Subsequently, these transfectants were selected for their growth in increasing concentrations (up to 200 μg ml−1) of G418 over a period of several weeks. Promastigote cultures of such transfectants were routinely maintained and grown at 26°C in complete growth medium containing 200 μg ml−1 of G418. For some experiments, such transfected promastigotes were allowed to transform and grow under axenic amastigote growth conditions (i.e. in pH 5.5 medium at 32°C) (35) in the presence of G418. Growth kinetics of both pKSNEO control and pKSNEO∷LmexCht1-HA transfected parasites were monitored at regular intervals during the time course of their growth in vitro. To that end, aliquots of such cultures were diluted appropriately with an isotonic buffer (ISOTON-II, Beckman-Coulter Particle Characterization, Miami, FL) and counted using a Model Z1 Coulter Counter (Beckman-Coulter) essentially as described by Debrabant et al. (35).
RT-PCR Analysis in L. mexicana transfectants
Total RNA was isolated, as described above, from both pKSNEO control and pKSNEO∷LmexCht1-HA transfected promastigote and axenically grown amastigote developmental forms of the parasite. Prior to use in RT-PCR, RNA samples were treated with RNase-free DNase I (Stratagene) to eliminate DNA contamination. cDNA was generated from these RNA samples and used in PCR amplification reactions with the High Fidelity PCR Master® kit (Roche) as per manufacturers instructions. A gene-specific primer pair (NeoFwd and NeoRev) was designed to amplify a portion of the message (i.e. ΔNEO, 324 bp) transcribed from the neomycin phosphotransferase (NEO) gene present in the pKSNEO-vector backbone. The sequence of NeoFwd was: 5′-GATTGAACAAGATGGATTGCACGCAGG-3′, and of NeoRev: 5′-AGGAGCAAGGTGAGATGACAGGAGAT-3′. A second primer pair (Cht1-Fwd and HA-Rev) was designed to amplify a portion of the message (i.e. ΔCht1HA, 979 bp) transcribed from the LmexCht1-HA chimeric gene construct present in the pKSNEO∷LmexCht1-HA plasmid. The sequence of Cht1-Fwd was: 5′-TTTGCGGACCTCGTTGGGGACACGGTG-3′ and of HA-Rev: 5′-CGCGTAGTCCGGCACGTCGTACGGGTA-3′. The conditions used for these PCR amplifications were: 94°C for 15 sec, 50°C for 30 sec, 72°C for 1 min (35 cycles) and 72°C for 5 min. Amplified products were separated by agarose gel electrophoresis, stained with ethidium bromide and imaged using an Eagle Eye-II ® video system (Stratagene) equipped with a UV trans-illuminator.
Western blot detection of the LmexCht1∷HA protein in transfected cells
Both pKSNEO control and pKSNEO∷LmexCht1-HA transfected L. mexicana promastigotes and axenically grown amastigotes were harvested from log-phase cultures and washed three times in 1X PBS by centrifugation as above. Such cell pellets were solubilized in SDS-PAGE sample buffer (36) and protein concentrations were determined using the bicinchoninic acid method (Micro BCA, Pierce Biotechnology, Inc., Rockford, IL). Aliquots of these cell lysates (15 μg/lane) were separated by SDS-PAGE (10 %, pre-cast, Tris-Glycine polyacrylamide, Novex® gels, Invitrogen) and the proteins were trans-blotted onto nylon (polyvinylidene diflouride, PVDF) membranes (Immobilon™P, Millipore Corp., Bedford, MA) as previously described (32). Such membranes were, blocked with 5 % (w/v) non-fat, powdered milk in a buffer containing 0.05% (v/v) Tween-20, 20 mM Tris-HCl, 150 mM NaCl, pH 7.4 (37), probed with an anti-HA monoclonal antibody (Covance Research Products, Inc., Berkeley, CA) or an appropriately matched purified mouse IgG1, κ control immunoglobulin (Sigma-Aldrich) followed by a goat anti-mouse horse radish peroxidase (HRP)-conjugated secondary antibody (Amersham Biosciences). Immuno-detection was carried out using the ECL Western Blot Kit reagents (Amersham Biosciences) and images were captured from such blots using BIOMAX™-MR X-ray film (Kodak).
Immunofluorescence microscopy of transfected parasites
Prior to use, the wells (5 mm diam.) of Teflon-coated, glass microscope slides (Cel-Line/Erie Scientific Co. Portsmouth, NH) were treated for 30 min at room temperature (RT) with an aqueous solution (10 mg ml−1 [w/v]) of poly-L-lysine hydrobromide (Sigma-Aldrich), rinsed with deionized water, air dried and stored at RT. For experiments, mid-log (exponential)-phase cultures of both pKSNEO control and pKSNEO∷LmexCht1-HA transfected L. mexicana promastigotes and axenically grown amastigotes were harvested and washed three times in PBS by centrifugation as above. Cells were fixed in suspension with 4% (w/v) paraformaldehyde (Sigma) in PBS for 20 min on ice, washed three times in PBS by centrifugation at ~10, 000 × g for 30 sec in an Eppendorf Microcentrifuge (Model 5414 D, Brinkman Instruments Inc., Westbury, NY). The final cell pellets (~ 2 × 107 cells) were resuspended in 200 μl of PBS. Aliquots (20μl ea.) of such cell suspensions were applied for 15-20 min to wells of the microscope slides above, drops aspirated and the wells allowed to air dry (all at RT). These cells were permeabilized with absolute methanol at −20°C for 5 min, rinsed in PBS and incubated (blocked) at RT for 60 min in PBS containing 5% (W/V) bovine serum albumin (BSA, Sigma). Subsequently, cells were reacted for 1 hr at RT with an anti-HA monoclonal antibody (Covance Research Products), or an isotype-matched control immunoglobulin (Sigma-Aldrich), appropriately diluted in PBS containing 1% (w/v) BSA (Sigma). Following three washes in PBS, cells were reacted for 1 hr at RT with a fluorescein isothiocyanate (FITC)-conjugated, goat anti-mouse IgG (H+L) secondary antibody (Sigma-Aldrich) diluted in PBS containing 1% (w/v) BSA. Subsequently, cells were washed three times with PBS and mounted in Vectashield® Mounting medium (Vector Laboratories, Inc., Burlingame, CA). Images were captured using a Zeiss Axioplan Microscope (Carl Zeiss, Inc., Thornwood, NY) equipped with epifluorescence, a cooled CCD camera (Photometrics, Tucson, AZ) and appropriate FITC excitation /barrier filters. All captured images were processed using Adobe Photoshop 5.5 (Adobe Systems Inc., San Jose, CA).
Detection of secreted/released LmexCht1∷HA in culture supernatants of transfected parasites
To ascertain whether transfected parasites secreted/released the LmexCht1∷HA chimeric protein during their growth in vitro, culture supernatants of both pKSNEO control and pKSNEO∷LmexCht1-HA transfected L. mexicana promastigotes and axenic amastigotes were harvested, neutralized to ~pH 7 and subjected to immunoprecipitation with an anti-LdCht1 antibody or NRS as above. The resulting immunoprecipitated complexes were separated in SDS-PAGE, (4-20 %, Tris-Glycine polyacrylamide gels, Invitrogen), trans-blotted onto PVDF membranes and subjected to Western Blot analysis. These blots were probed with an anti-HA-biotin conjugated monoclonal antibody (Covance Research Products) and subsequently reacted with a streptavidin-HRP conjugate (Jackson ImmunoResearch, West Grove, PA). Such blots were developed using the ECL Western Blot Kit reagents and images were captured using BIOMAX™-MR X-ray film (Kodak) as described above.
Measurement of secreted/released LmexCht1∷HA chitinase activity
In initial experiments, we observed that the anti-HA antibody cross-reacted with a number of serum components present in the complete–FBS containing, parasite growth/culture media. Further, we found that no detectable chitinase activity could be immunoprecipitated with this anti-HA antibody from culture supernatants of pKSNEO∷LmexCht1-HA transfectants grown in such FBS-containing media. Thus, an alternative protocol was devised in order to measure the chitinase activity of the secreted/released LmexCht1∷HA chimeric protein. To that end, we set-up short-term “release assays” similar to those previously described by Bates et al. (38) to assay leishmanial secretory acid phosphatase activity. For such assays, mid-log-phase cultures (~1.5 × 107 cells ml−1) of both pKSNEO control and pKSNEO∷LmexCht1-HA transfected promastigotes and axenic amastigotes were grown as above and subsequently harvested by centrifugation at 6000 × g for 10 min at 4°C. The cell pellets were washed three times by resuspension in ice-cold PBS and centrifugation as above. Subsequently, promastigotes were resuspended to ~ 5×107 cells ml−1 in a non-nutrient buffer (10mM HEPES, 145mM NaCl, containing 1mM glucose, pH 7.0) and incubated on a platform rocker at 26°C for 4 h. Axenic amastigotes were treated similarly except in a buffer containing: 10mM MES, 145mM NaCl, 1mM glucose, pH 5.5 and incubated at 37°C. In agreement with previous reports (16,38), following such incubation, these cell suspensions remained >99% viable as ascertained by phase contrast microscopy. Subsequent to incubation, these cell suspensions were centrifuged at 6000 × g for 15 min at 4°C and the supernatants were carefully harvested by aspiration. To ensure the complete removal of cells, such supernatants were recentrifuged at high speed as previously described by Shakarian et al.(16). The resulting cell-free supernatants were neutralized to ~pH 7.0 using 1M Tris-HCl buffer, pH8.0 as above. Aliquots of these release assay samples were used to measure chitinase enzyme activity using 4MU-substrates as above. Neutralized, unused release assay incubation buffers served as controls in these assays. Further, such neutralized release assay samples were also used in immunoprecipitation/enzyme activity experiments, below.
To specially determine whether the LmexCht1∷HA chimeric protein possessed chitinase activity, neutralized release assay samples from of both pKSNEO control and pKSNEO∷LmexCht1-HA transfected promastigotes and axenic amastigotes were subjected to immunoprecipitations with an anti-HA immuno-affinity bead matrix (Covance Research Products). In these experiments, aliquots of release assay supernatants were reacted with the anti-HA conjugated beads on a platform rocker for 4 h at 4°C. Subsequently, these beads were pelleted by centrifugation, washed three times with 1X PBS and the bound immunoprecipitates analyzed for their chitinase activity using 4MU-chitotriose/-chitobiose as substrates (6). All samples were assayed in triplicate and such assays were repeated using at least two independently generated samples. Results of these assays are expressed as pmoles of 4MU-product generated hour −1 ml−1 of parasite release assay supernatant. Neutralized, unused release assay incubation buffers served as controls in these assays.
Infections of human macrophages in vitro
Elutriated human peripheral-blood monocytes were obtained from the National Institutes of Health Blood Bank (Bethesda, MD). These monocytes were resuspended to ~3.6 × 105 cells ml−1 in RPMI-1640 medium containing 25 mM HEPES (Invitrogen), 10% (v/v) heat-inactivated fetal bovine serum (HyClone, Logan, UT), 2 mM L-glutamine (Invitrogen), 100 IU ml−1 penicillin, 50 μg ml−1 streptomycin sulfate, 5 μg ml−1 gentamycin (Sigma-Aldrich) and 20 ng ml–1 recombinant human macrophage colony stimulating factor (M-CSF, PeproTech Inc., Rocky Hill, NJ). The monocytes were plated in 0.5 ml volumes each in, Lab-Tek®, 8 chamber, tissue culture slides (Nalge Nunc) and incubated for 7 days at 37°C in a humidified atmosphere containing 5% CO2 in air to facilitate their differentiation into macrophages. Stationary phase L. mexicana M379 promastigotes transfected with either the control pKSNEO or the pKSNEO∷LmexCht1-HA plasmids were used to infect these monocyte-derived macrophages. To that end, promastigotes from stationary-phase cultures were harvested and washed three times by centrifugation (1,500 × g for 10 min at 4°C) with complete macrophage culture medium and finally resuspended in the same medium (pre-warmed to 32°C). Parasites were incubated with the monocyte-derived macrophages at a ratio of 10 parasites per host cell at 32°C in a humidified atmosphere containing 5% CO2 in air for 5 hr. Subsequently, non-adherent (i.e. free, extracellular) parasites were removed by aspiration and repeated washing with pre-warmed RPMI medium as above. Following this, one set of such macrophage cultures (T5hr) was immediately processed for light microscopy: i.e. culture medium was removed, the slides were air-dried for 5 min at RT, fixed by immersion in absolute methanol for 10 min and finally stained using the Diff-Quick Stain set (Dade Behring, Inc., Newark, DE). A second set of parasite-infected macrophage cultures was incubated for an additional 72 hr in the RPMI medium at 32°C in 5% CO2 in air, as above. After 72 hr, this set of macrophage cultures (T72 hr) was also fixed, stained and processed for microscopy as described above. Subsequently, triplicate chambers of both the T5hr and T72 hr parasite-infected macrophage cultures were examined by light microscopy. For analysis, a minimum of 300 macrophages was counted from each chamber. Values obtained in these experiments with the pKSNEO control and pKSNEO∷ LmexCht1-HA transfected parasites are expressed as: (1) percentage of macrophages that were infected by these parasites and (2) the total number of intracellular parasites (amastigotes) within 100 macrophages.
In vivo infections in mice
L. mexicana amastigotes were isolated from infected BALB/c mice and allowed to transform into promastigotes at 26°C in vitro, as described above. The promastigotes (i.e. in their first in vitro passage after transformation) were transfected with either the pKSNEO (control) or pKSNEO∷LmexCht1-HA plasmids, as described above. Transfected promastigotes were selected for growth in increasing concentrations of G418 up to 200 μg ml−1. Susceptible BALB/c mice (Charles River UK, Ltd) were infected (into the right foot) with 106 late stationary-phase promastigotes of these transfectants. Subsequently, amastigotes were isolated from the resulting mouse lesions and used to generate metacyclic promastigotes in vitro as previously described by Bates and Tetley (9), except that 200 μg ml−1 G418 was included in the culture medium. These metacyclic promastigotes were used for mouse infectivity experiments. To that end, age (9-10 weeks) and weight (18-20g)-matched female BALB/c and CBA/Ca mice (Charles River UK Ltd.), 8 per group, were each infected with 500 metacyclic promastigotes of either L. mexicana pKSNEO (control) or pKSNEO∷ LmexCht1-HA transfectants. These parasites were injected into the dorsal surface of the right hind foot. The course of infection in such mice was followed by measuring the swelling of the right foot relative to the uninfected left foot over a period of 15 weeks. At the end of these experiments, the mice were humanely sacrificed, and the total parasite burden in each infected foot was determined by homogenization in culture medium, followed by direct counting using a Neubauer hemocytometer counting chamber (Fischer Scientific Ltd, UK) and phase-contrast light microscopy (8). All procedures involving animals were performed in accordance with UK Government (Home Office) and EC regulations.
RESULTS
Analysis of the L. mexicana chitinase gene
Genes encoding chitinolytic enzymes have been reported from a wide variety of plant, animal and microbial sources (39,40). However, despite their diverse origins, many chitinases possess regions of amino acid sequence homology within their functional domains. These domains include the substrate binding and the active/catalytic sites of these enzymes (39). Based on these observations, we used a PCR-based approach to identify a chitinase gene from the human pathogen Leishmania mexicana. To that end, oligonucleotide primers were designed corresponding to portions of Region II (i.e. a putative catalytic site) and Region III (i.e. a putative substrate binding site) of a chitinase (LdCht1) from a closely related organism (18). These primers were used in PCR amplifications with L. mexicana gDNA as a template. The resulting 270 bp product obtained from such amplification reactions was gel purified, cloned (Lmex-PCR270) and subsequently sequenced. Such sequences were subjected to BLAST-N and BLAST-P analyses. Results of these analyses showed that the Lmex-PCR270 clone contained an open reading frame (ORF), which showed both high nt and deduced-amino acid sequence identity (92% and 82%, respectively) to a portion of the LdCht1 gene of Leishmania donovani (18). These results suggested that we had amplified a portion of a gene encoding a L. mexicana chitinase homolog.
The L mex-PCR270 fragment above, was labeled with digoxigenin-dUTP. The resulting probe (Lmex-DIG270) was used to screen an L. mexicana gDNA cosmid library by hybridization. Following several rounds of screening with this probe, a positive cosmid clone (Lmex-Cos1) was selected for further analysis. Results of nt-sequence analyses revealed that the Lmex-Cos1 clone contained a complete ORF (LmexCht1) of 1374 bp. The composition of this ORF is GC-rich (~62%) and the third base position of the codons used show a strong bias (~71%) towards G or C residues. These observations are consistent with the overall GC content of the Leishmania genome (41). Further, the LmexCht1 ORF encodes a polypeptide of 457 amino acids with a calculated molecular mass of 50,350.88 Da (Fig. 1A).
FIG. 1. The sequence and structure of the L. mexicana chitinase.
A. The deduced amino acid sequence of the L. mexicana chitinase 1 (LmexCht1) gene. The underlined sequence delineates a putative 28 aa signal peptide (Met1 – Ser28). The light gray boxes (marked: I [Leu123-Ala139] and III [Leu249-Gly264]) indicate the two putative substrate binding sites and the open box (marked: II [Arg150- Thr179]) indicate the putative catalytic/active site of this enzyme. The bold-italicized residues within the open box (Leu167- Glu175) represent the signature sequence of the Chitinase-18 Protein Family. The two potential N-linked glycosylation sites (Asn48 and Asn384) are marked with an asterisk and the single potential O-linked glycosylation site (Thr52) is indicated by (τ). The dashed-underlined aa sequences (designated as Pep1-Pep4) represent regions with high levels of conservation to antigenic peptide epitopes in the L. donovani Cht1 deduced protein. B. Comparison of the conserved functional domains of the L. mexicana Cht1 (L. mex.) and L. donovani Cht1 (L.d.) chitinases. The conserved substrate binding sites (i.e. Regions I and III) and the conserved catalytic/active site (i.e. Region II) are indicated. The divergent aa residues between these two proteins are shown in bold-face.
Comparison of the deduced-amino acid sequence encoded by the LmexCht1 ORF to all available, non-redundant data bases using BLAST-P, showed that it has homology to a variety of chitinases from diverse sources. Amongst those, the LmexCht1 deduced-protein showed the highest level of amino acid sequence identity (i.e. 83%) with the LdCht1 chitinase of L. donovani (GenBank™ AF009354, GI:3169726) (data not shown). Further, analysis of the LmexCht1 deduced-protein using the Pfam database (via Pfam server: http://pfam.wustl.edu) indicated that it belongs to the Glycohydrolase-18 family of proteins. Moreover, such analyses showed that the LmexCht1 deduced-protein also contained a signature sequence characteristic of the Chitinase-18 family (i.e. spanning aa residues 167-175: LDGIDFNWE) (Fig. 1A). In addition, the LmexCht1 deduced-protein was also found to possess the three functional domains that are characteristic of chitinases in general (i.e. Regions I and III, substrate binding sites and Region II, the catalytic/active site) (39). Taken together, these observations indicate that the LmexCht1 ORF encodes a L. mexicana chitinase.
Based on the von Heijine algorithm (42,43), the hydrophobic, N-terminal, 28 amino acids of the LmexCht1 deduced protein constitute a putative signal peptide (Fig. 1A). Cleavage at this site, presumably in the endoplasmic reticulum of this parasite, would result in a mature protein with Ala29 as its N-terminal amino acid residue. Such cleavage would result in a mature protein consisting of 429 amino acids with a calculated molecular mass of 47,468.48 Da and a predicted pI of 6.26. The conserved putative functional domains/regions of the LmexCht1 protein are shown in Fig. 1A. These include: Regions I and III (i.e. the two conserved putative substrate binding sites) which span residue Leu123-Ala139 and Leu249- Gly264, respectively, and Region II (i.e. the putative catalytic/active site of the enzyme) which spans residues Arg150- Thr179. A comparison of these LmexCht1 functional domains with those of its closest homolog (i.e. the LdCht1 chitinase) (18) showed that in Regions I, II and III, they share ~88, 90 and 94% amino acid sequence identity, respectively (Fig. 1B).
Previously, four antigenic peptide epitopes (i.e. Pep1-Pep4) were identified in the LdCht1 deduced protein aa sequence and used to generate a rabbit anti- LdCht1 peptide antibody (6). Results of comparative sequence alignments with the latter showed that regions corresponding to Pep1 [Val221-Val239]; Pep2 [Lys309-Arg325]; Pep3 [Arg325-Ala342] and Pep4 [Asn430-Pro449] were also conserved in the LmexCht1 deduced protein (Fig. 1A) and within these regions, they shared ~90, 100, 100 and 80% amino acid sequence identity, respectively.
The LmexCht1 deduced protein was analyzed using: NetNGlyc, NetOGlyc and NetPhos web-based tools (available at http://www.cbs.dtu.dk/services) to identify amino acid residues that are potential sites for post-translational modifications of this protein. Such analyses showed that the LmexCht1 possesses two potential sites for N-linked glycosylation at Asn48 and Asn384 and one potential site for O-linked glycosylation at Thr52 (Fig. 1A). In addition, the LmexCht1 deduced protein also contained multiple potential sites for phosphorylation by several different mechanisms (e.g. casein kinase II, protein kinase-C, etc.).
As predicted by the Kyte-Doolitle algorithm (44) the LmexCht1 deduced protein is hydrophilic in nature. Further, no apparent hydrophobic transmembrane domains or predicted glycosyl-inositol phosphate (GPI-) anchor signature sequences (45) were present in the C-terminus of this protein. Similarly, no KDEL- or analogous endoplasmic reticulum (ER) retention sequences (46) or other intracellular organelle specific-targeting sequences were identified in the LmexCht1 deduced protein. Based on its overall hydrophilicity, the presence of an N-terminal signal peptide and the absence of both membrane anchor and ER-retention motifs, suggest that the LmexCht1 is a soluble/released protein.
Southern blot analysis of LmexCht1
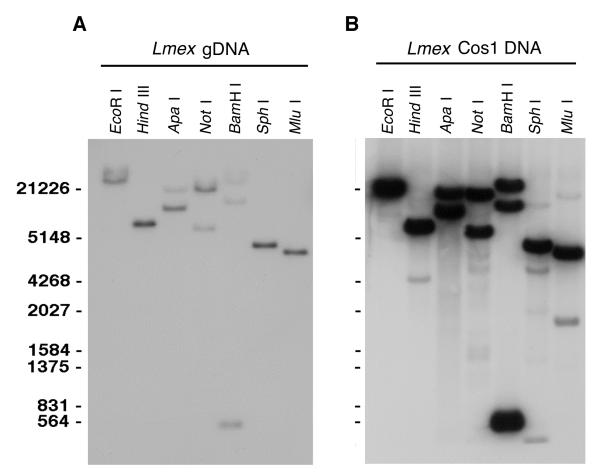
To examine the genomic organization and copy number of the LmexCht1 gene, L. mexicana gDNA was digested with various restriction endonucleases, separated by gel electrophoresis and blotted onto nylon membranes. Such membranes were subjected to Southern hybridization using the digoxigenin-labeled LmexCht1-DIG1151 probe (i.e. corresponding to nt 1 to nt 1151of the L. mexicana chitinase 1 ORF) under high stringency conditions. Enzymes that did not cut within the LmexCht1ORF (e. g. Eco RI, Hind III, Sph I and Mlu I) gave a single band of hybridization with the LmexCht1-DIG1151 probe (Fig. 2A). In contrast, gDNA digested with enzymes which cut once within the LmexCht1ORF (e. g. Apa I and Not I) gave two bands of hybridization; while digestion with Bam HI (which cuts twice within the ORF) gave three bands of hybridization with this DIG-labeled probe (Fig. 2A). Taken together, these results suggest that the LmexCht1 gene is present as a single copy within the diploid genome of this organism.
FIG. 2. Southern Blot analyses of the L. mexicana chitinase 1 gene locus.
A. Southern hybridization of L. mexicana gDNA with the LmexCht1-DIG1151 probe. gDNA (5 μg) was digested with various individual restriction endonucleases ( EcoRI, Hind III, Apa I, Not I, BamH I, Sph I and Mlu I, as indicated), separated in 1% agarose gel, transferred to a nylon membrane and hybridized with the digoxigenin-labeled LmexCht1-DIG1151probe (i.e. corresponding to nt 1 - nt 1151 of the LmexCht1-ORF). DNA standards (in bp) are shown on the left. B. Southern hybridization of the Lmex-Cos1 cosmid DNA with the LmexCht1-DIG1151 probe. Cosmid DNA (1 μg) was digested with the same restriction endonucleases as shown in Panel A and subjected to Southern hybridization with the LmexCht1-DIG1151 probe. DNA standards (in bp) are shown on the left as in Panel A.
Cosmid DNA from the Lmex-Cos1 clone was also digested with the various restriction endonucleases above and subjected to Southern blot analysis with the LmexCht1-DIG1151 probe. Results obtained in these hybridizations were essentially identical to those obtained with L. mexicana gDNA (Fig. 2B). These observations indicate that the Lmex-Cos1 cosmid clone reflects the genomic locus containing a single copy of the Cht1 gene of L. mexicana.
Expression of LmexCht1 mRNA in various L. mexicana developmental stages
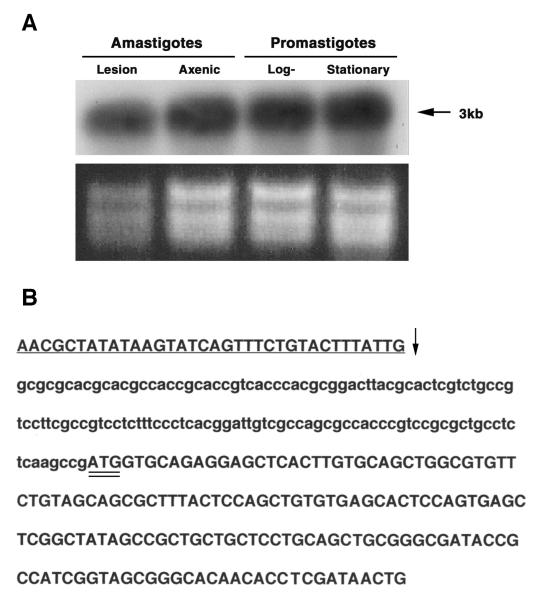
Northern blot analyses were carried out to examine the expression of LmexCht1 mRNA in various life cycle developmental stages of L. mexicana. To that end, total RNA was isolated from in vitro grown procyclic (log-phase) promastigotes, metacyclic (stationary-phase) promastigotes, axenic amastigotes as well as, from tissue-(lesion) derived amastigotes isolated from BALB/c mice. Equivalent amounts (5 μg per lane) of RNA from these various L. mexicana developmental forms were separated by agarose gel electrophoresis and stained with ethidium bromide (Fig. 3A, lower panel). RNA from such gels was transferred to nylon membranes and hybridized with the LmexCht1-DIG270 probe at high stringency. Results of these analyses showed that this gene-specific probe hybridized to a single ~3.0 kb-mRNA transcript in each of the parasite life-cycle developmental stages examined (Fig. 3A, top panel). Further, the intensity of the hybridization signal appeared to be comparable among the RNA samples obtained from both the insect vector forms (i.e. procyclic and metacyclic promastigotes) and the mammalian developmental forms (i.e. in vitro-grown axenic amastigotes and mouse lesion-derived amastigotes) of this parasite. These Northern blot results indicated that LmexCht1 mRNA is constitutively transcribed throughout the developmental life cycle of L. mexicana.
FIG. 3. Northern analyses and mapping of the spliced-leader acceptor site.
A. Northern blot analysis of LmexCht1 mRNA transcripts present in various L. mexicana parasite developmental forms. Top Panel. Total RNA (5 μg) isolated from L. mexicana mouse lesion amastigotes (i.e. in vivo-derived), axenic amastigotes (i.e. in vitro-grown), and log (procyclic)- and stationary (metacyclic)-phase promastigotes was separated in an agarose gel, transferred onto a nylon membrane and hybridized with the LmexCht1-DIG 270 probe (i.e. corresponding to nt 502– nt 771 of the LmexCht1-ORF). Arrow indicates the position of the ~3 kb LmexCht1 mRNA transcript in these samples. Bottom Panel. Ethidium bromide strained agarose gel (used in panel A) showing the ribosomal RNA in each of the total RNA samples used above. B. Mapping of the LmexCht1 5′-spliced-leader acceptor site. Nucleotide sequence of the RT-PCR product obtained with LmexCht1 mRNA amplified with an L. mexicana-spliced leader (forward: SpliceFwd) oligonucleotide primer and an internal LmexCht1 (reverse: ORF-RT/Rev) primer. The 5′-untranslated region of the LmexCht1 gene is shown in lower case letters. The LmexCht1 open reading frame is shown in Upper case letters, a portion of the conserved spliced leader sequence (i.e. forward primer) is shown in underlined Caps and the arrow (↓) marks the position of the spliced-leader acceptor site. The ATG-start codon of the LmexCht1 open reading frame is double-underlined.
Mapping of 5′ splice site
A unique feature of trypanosomatid parasites is that all of their mature, translatable mRNAs are capped at the 5′-end with a conserved 39 nt, “spliced-leader” (SL), sequence (26,27). Such capping involves the addition (i.e. via trans-splicing) of a 39 nt spliced-leader to a specific “splice addition/acceptor site” (i.e. a consensus motif: Py AG) in the 5′-non-coding region of the parasite’s pre-mRNAs. To identify the 5′-splice acceptor site in LmexCht1, RT-PCR analyses was performed using total RNA isolated from both L. mexicana promastigotes and axenic amastigotes. cDNAs generated from such RNAs were used as template in PCR amplifications with a 5′-forward primer (i.e. SpliceFwd, above) corresponding to a portion of the L. mexicana spliced leader sequence (i.e. nt 5 to nt 31 of the 39 nt SL sequence) and a reverse primer (i.e. ORF-RT/Rev, above) corresponding to an internal sequence in the LmexCht1 ORF (i.e. nt 138 to nt 157). The nt sequence of the 316 bp product obtained in these reactions using cDNA from L. mexicana promastigotes is shown in Fig. 3B. Analyses of these results indicated that the start codon (i.e. the first ATG) of the LmexCht1 ORF, was preceded by 124 nt of 5′-UTR (Fig. 3B). Further, these results showed that the spliced leader addition/acceptor-site mapped to −125 nt from the ATG start codon of the LmexCht1 ORF. It is important to note that identical results were obtained in these assays using cDNA generated from total RNA isolated from L. mexicana axenic amastigotes. In addition to defining the 5′UTR and spliced-leader addition/acceptor site, these results also verified that both parasite developmental forms synthesized mature, translatable mRNAs for the LmexCht1 gene.
Measurement of chitinase activity secreted/released by L. mexicana promastigotes and axenic amastigotes
To determine whether wild-type L. mexicana secreted/released any measurable chitinolytic activity during their growth in vitro, aliquots of cell-free culture media supernatants from both parasite developmental forms (i.e. promastigotes and axenic amastigotes) were assayed for chitinase activity using 4MU-chitobiose and 4MU-chitotriose as substrates. Fresh, unused, complete, FBS-containing parasite growth/culture media were used as controls in these assays. Values obtained from these controls were subtracted from those generated with the various experimental samples. Such controls were essential since the FBS used in these parasite growth media possesses some apparent endogenous chitinolytic activity. Results of these enzyme assays showed that both parasite developmental forms of L. mexicana secreted/released significant amounts of both exochitinase and endochitinase activity into their culture media supernatants (Table 1). Interestingly, axenic amastigotes of these parasites appeared to secrete/release considerably more chitinase activity into their growth medium (i.e. ~2.5 fold and ~4.5 fold higher levels of exo- and endo-chitinase activity, respectively) than promastigotes. Further, it is also of interest to note that the culture supernatants from both promastigotes and axenic amastigotes showed higher activities with 4MU-chitotriose (i.e. an endochitinase substrate) than with 4MU-chitobiose (i.e. an exochitinase substrate).
TABLE 1.
Soluble -released chitinase activity in L. mexicana parasite culture supernatants.
| Developmental Stage | Total Activity | Immunoprecipitated Activityc | ||
|---|---|---|---|---|
|
| ||||
| (pmoles of 4MU released ml−1 h−1) | ||||
| Exochitinase activitya |
Endochitinase activityb |
Exochitinase activitya |
Endochitinase activityb |
|
| Promastigotes | 1081 ± 102 | 1375 ± 142 | 497 ± 55 | 632 ± 73 |
| Amastigotes | 2440 ± 289 | 6207 ± 583 | 1122 ± 148 | 2731 ± 310 |
Activity assayed using 4MU-chitibiose as substrate.
Activity assayed using 4MU-chitotriose as substrate.
Activity immunoprecipitated using an anti-LdCht1-peptide antibody in a protein A-Sepharose 4B/CL bead-based assay.
Immunoprecipitated complexes were assayed for exochitinase and endochitinase activity using the appropriate substrate. The results were normalized by subtracting values obtained with NRS (i.e. preimmune serum from same rabbit). The data shown reflect the mean results of triplicate assays for each sample from three separate experiments.
Previously, a rabbit anti-LdCht1-peptide serum was generated against four antigenic peptide epitopes (Pep1-Pep4) from the deduced aa sequence of the L. donovani chitinase (6). Since the LmexCht1 deduced aa sequence (cf. Fig. 1A) showed a very high level of conservation over these four peptides (i.e. >92.5% identity, overall) with the latter, the anti-LdCht1-peptide antibody and NRS controls were tested in immunoprecipitation reactions with cell-free culture supernatants obtained from L. mexicana promastigotes and axenic amastigotes. Such immunoprecipitates were assayed for both their exochitinase and endochitinase activities using 4MU-chitobiose and 4MU-chitotriose as substrates, respectively. Results of these assays showed that both promastigotes and axenic amastigotes of L. mexicana secreted/released chitinase activities which were immunoprecipitated by the anti-LdCht1-peptide serum (Table 1). Further, as observed in our direct enzyme assays, such immunoprecipitates showed higher activites with 4MU-chitotriose than with 4MU-chitobiose. In these assays, the anti-LdCht1-peptide serum immunoprecipitated ~ 45% of the detectable chitinase activity released by these two L. mexicana parasite developmental forms. These values are similar to those obtained with this antibody in immunoprecipitation assays with culture supernatants derived from the homologous (L. donovani) parasite system (Dwyer, unpublished observations).
Taken together, results of these enzyme assays demonstrated that both L. mexicana promastigotes and axenic amastigotes secrete/release chitinolytic activity into their culture media supernatants during their growth in vitro. Further, axenic amastigotes appeared to secrete higher levels of such enzyme activity than promastigote developmental forms of this parasite. In addition, results of our immunoprecipitation experiments suggested that the four antigenic peptide epitopes (i.e. Pep1-Pep4) present in the LmexCht1 deduced protein (cf. Fig. 1A) must be structurally expressed in the native enzyme secreted by these parasites during their growth in vitro. Further, these results also indicate that these L. mexicana peptide epitopes must share a high level of structural conservation with those present in the L. donovani chitinase.
Homologous episomal expression of the LmexCht1 gene in L. mexicana promastigotes and axenic amastigotes
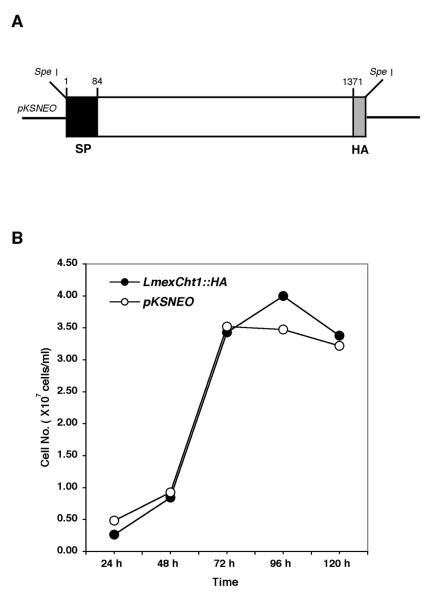
A homologous episomal expression system was devised to further examine in detail the role of the LmexCht1 protein in the developmental cycle of L. mexicana. To that end, a chimeric construct was generated containing the complete ORF of LmexCht1 fused, in frame, at its 3′-end with a sequence encoding a hemagglutinin (HA) epitope. Following ligation into the pKSNEO leishmanial expression vector, this construct (LmexCht1∷HA) was used to transfect L. mexicana promastigotes (Fig. 4A). In parallel, promastigotes were transfected with the pKSNEO vector alone and these served as controls in all transfection experiments. Following electroporation, both the LmexCht1∷HA and pKSNEO control transfectants were selected for their growth in increasing concentrations of G418 (i.e. up to 200 μg ml−1) over a period of several weeks. Subsequent to such drug selection, the growth kinetics of these transfectants were compared. To that end, quadruplicate cultures of both LmexCht1∷HA and pKSNEO control transfectants were initiated at 1-2 × 106 cells ml−1 (i.e. from stock cultures in their exponential phase of growth) in complete growth medium containing 200 μg ml−1 of G418. Aliquots were taken from such cultures at 24 hr intervals over a period of 5 days, diluted appropriately and counted using a Coulter Counter as described above. Results of these assays showed that promastigotes transfected with either the LmexCht1∷HA chimeric construct or the control plasmid (pKSNEO) had virtually identical growth kinetics over the time course of these experiments (Fig. 4B). Further, non-transfected (“wild-type”) L. mexicana promastigotes (grown in complete medium lacking G418) displayed growth kinetics identical to those obtained with the transfectants, above (data not shown). Taken together, these observations indicate that these episomal-transfections did not overtly alter the characteristic growth kinetics of the parental L. mexicana M379 promastigote cell line.
FIG. 4. Episomal expression of the LmexCht1∷HA chimera in L. mexicana promastigotes.
A. Map of the LmexCht1∷HA chimeric construct. A schematic representation showing the complete open reading frame (i.e. nt 1 – nt 1371, minus the terminal TAG stop codon) of the LmexCht1 gene fused at its 3′-end with a 27-nt sequence encoding the hemagglutinin (HA) epitope (light gray box). The black box at the 5′-end represents nt-1 to nt-84 encoding the putative 28 aa signal peptide (SP) of the LmexCht1 protein. The thick black line represents the pKSNEO plasmid (leishmanial) expression vector, and the Spe I restriction endonuclease sites used for cloning are shown. B. Growth kinetics of L. mexicana transfectants in vitro. The growth kinetics of promastigotes transfected with either LmexCht1∷HA (●) or pKSNEO control (○) constructs were monitored for their growth in vitro over a period of five days. Quadruplicate cultures were initiated at ~1-2 × 106 cells ml−1 and aliquots taken at various time points for cell counting. Values shown represent the mean of three separate determinations for each culture.
L. mexicana promastigotes transfected with either the LmexCht1∷HA or the pKSNEO control plasmid were placed under conditions (i.e. 32 °C, pH 5.5) to allow them to transform into, and grow as, axenic amastigotes in vitro. Both promastigotes and axenic amastigotes of these L. mexicana transfectants were subsequently analyzed for their expression of the LmexCht1∷HA gene and chimeric protein using RT-PCR, Western Blots and immuno-fluorescence microscopy.
Detection of LmexCht1∷HA and NEO control mRNAs in L. mexicana transfectants
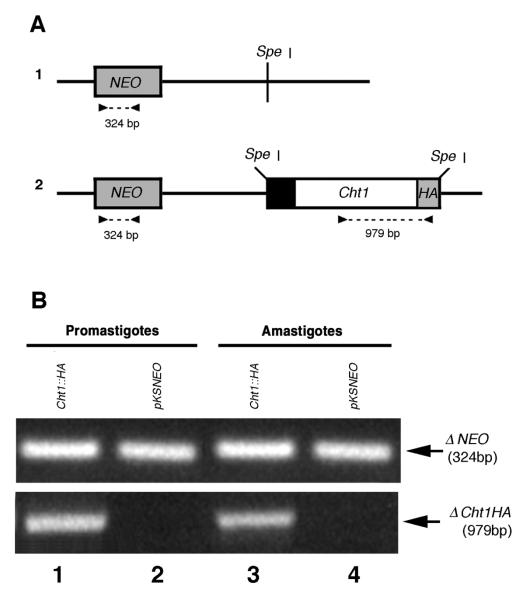
RT-PCR analyses were performed to detect the mRNA transcripts produced by the LmexCht1∷HA and pKSNEO (control) episomes in transfected cells. For these assays, total RNA was isolated from both promastigote and axenic amastigotes of pKSNEO control and LmexCht1∷HA transfectants. cDNAs generated from such RNAs were subsequently used as templates in PCR amplifications with two different primer pairs: 1) NeoFwd and NeoRev, designed to amplify a portion (i.e. 324 bp) of the NEO gene present in the pKSNEO vector backbone and 2) Cht1-Fwd and HA-Rev designed to amplify a portion (i.e. 979 bp) of the episomally expressed LmexCht1∷HA chimeric construct (Fig 5A, 1 and 2).
FIG. 5. RT-PCR analysis of episomally expressed NEO and LmexCht1∷HA mRNAs.
A. Schematic representations of the pKSNEO control plasmid and the LmexCht1∷HA construct. 1. The pKSNEO plasmid: the dark gray box represents the nt sequence encoding neomycin phosphotransferase (NEO), the black line denotes the pKSNEO plasmid vector backbone and Spe I indicates the restriction site used for insert cloning. 2. The LmexCht1∷HA construct: the black and white boxes represent nt-1 to nt-84 encoding the putative signal peptide and nt 85 - nt 1371 of the LmexCht1 ORF, respectively. The light gray box represents the nt sequence encoding the hemagglutinin (HA) epitope fused in-frame with the LmexCht1 ORF followed by a terminal TGA stop codon. The Spe I restriction sites used for cloning this insert are indicated. The dark gray box and black lines represent NEO and the vector backbone as in 1, above. Arrowheads show the positions of the primers used for amplification, the dashed lines and numbers denote the predicted size (in bp) of the PCR products. B. A portion of an ethidium bromide stained agarose gel showing the amplification products obtained in RT-PCR. RNA isolated from L. mexicana promastigotes and (axenic) amastigotes transfected with either the pKSNEO control plasmid (lanes 2 and 4) or the LmexCht1∷HA (Cht1∷HA) construct (lanes 1 and 3) was reverse transcribed using oligo(dT). Aliquots of the resulting cDNAs were subjected to PCR amplification using the primer pairs shown above. Top panel: shows the resulting 324 bp product (← ΔNEO) amplified from the neomycin phosphotransferase gene present in the pKSNEO plasmid backbone of all transfectants. Bottom panel: the 979 bp product (← ΔCht1HA) amplified from LmexCht1∷HA transfectants.
In PCR amplifications with the NeoFwd and NeoRev primers, a ~324 bp product was obtained using cDNA generated from both promastigotes and axenic amastigotes of LmexCht1∷HA (Fig. 5B, Top Panel, lanes 1 and 3) as well as from pKSNEO (control) transfectants (Fig. 5B, Top Panel, lanes 2 and 4). These results demonstrated that mRNA for the NEO gene was effectively transcribed at approximately equivalent levels by both promastigotes and axenic amastigotes transfected with either the LmexCht1∷HA or the pKSNEO (control) plasmids. Moreover, these observations were verified by Northern blot analyses using total RNA from these cells and a NEO-specific probe (data not shown).
Further, in PCR amplifications with the Cht1-Fwd and HA-Rev primers, a single ~979 bp product was obtained using cDNA synthesized from LmexCht1∷HA promastigote RNA (Fig. 5B, Bottom Panel, lane 1). An identical result was obtained in parallel reactions with these primers and cDNA generated from axenic amastigotes of LmexCht1∷HA transfectants (Fig. 5B, Bottom Panel, lane 3). These results indicated that both promastigote and axenic amastigote developmental forms of the LmexCht1∷HA transfectants effectively transcribed approximately equivalent amounts of mRNA transcripts for the LmexCht1∷HA chimeric gene. Moreover, these observations were verified by Northern blot analyses using total RNA from these cells and a Cht-specific probe (data not shown). No reaction products were obtained with the Cht1-Fwd and HA-Rev primers using cDNA synthesized from either promastigotes or axenic amastigotes of pKSNEO (control) transfectants (Fig. 5B, Bottom Panel, lanes 2 and 4, respectively).
The results obtained above were confirmed using several independently generated RNA-cDNA preparations. In control reactions, in which cDNAs were not generated prior to PCR, no amplified products were obtained (data not shown). Moreover, in control reactions in which the forward or reverse primers were omitted from the reaction mixtures, no amplified products were obtained (data not shown).
Expression of the LmexCht1∷HA chimeric protein in L. mexicana transfectants
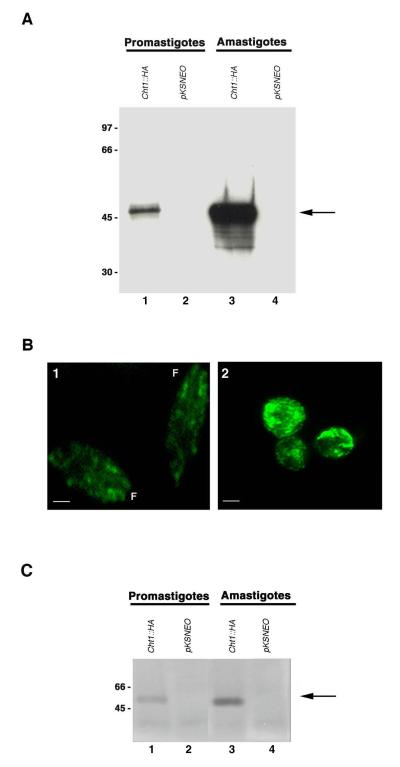
Having demonstrated that both L. mexicana promastigotes and axenic amastigotes synthesized mRNA transcripts from the LmexCht1∷HA episomal construct, it was of interest to determine whether such transcripts were actually translated into the chimeric protein by these two parasite developmental stages. To address this, lysates from promastigotes and axenic amastigotes of pKSNEO (control) and LmexCht1∷HA transfectants were subjected to SDS-PAGE and reacted in Western blots with a mouse anti-HA monoclonal antibody or with an appropriately matched purified mouse control immunoglobulin. In such blots, the anti-HA antibody reacted with a single ~50 kDa Cht1∷HA chimeric-protein present in lysates from promastigotes of the LmexCht1∷HA transfected parasites (Fig. 6A, lane 1). The anti-HA antibody appeared to react even more strongly with a similarly sized (i.e. ~50 kDa) Cht1∷HA chimeric-protein present in lysates from axenic amastigotes of the LmexCht1∷HA transfectants (Fig. 6A, lane 3). In the latter sample, this antibody also reacted to a much lesser extent with several proteins of lower apparent molecular mass (~35-40kDa) which presumably reflect proteolytic degradation fragments of the mature ~50 kDa Cht1∷HA chimeric-protein. The anti-HA antibody showed no reactivity with lysates of either control, pKSNEO transfected promastigotes or axenic amastigotes (Fig. 6A, lanes 2 and 4, respectively). Similarly, the isotype-matched purified control mouse immunoglobulin showed no reactivity with any of the samples tested in these assays (data not shown). Taken together, the results of these Western blot experiments demonstrated that the LmexCht1∷HA chimeric-gene construct was readily transcribed and translated into an ~50 kDa chimeric protein by both promastigotes and axenic amastigotes of these transfectants. Further, our observations indicated that such expression was significantly enhanced in the axenic amastigote form of these transfected parasites. This pattern of differential up-expression may reflect an inherent property of amastigotes per se, as shown in Table1, but may also be facilitated by the A2 gene-regulatory sequences (30,31,34) present in the backbone of the pKSNEO expression vector.
FIG. 6. Episomal expression of the LmexCht1∷HA chimeric protein in L. mexicana transfectants.
A. Western blot of whole cell lysates of L. mexicana promastigotes and (axenic) amastigotes transfected with either the LmexCht1∷HA construct (Cht1∷HA) (lanes 1 and 3) or the pKSNEO control plasmid (lanes 2 and 4), probed with a mouse anti-HA monoclonal antibody. Molecular mass standards (in kDa) are shown on the left. Arrow denotes the ~50 kDa LmexCht1∷HA chimeric protein. B. Indirect immunofluorescence images of LmexCht1∷HA transfected promastigotes (panel 1) and axenic amastigotes (panel 2) probed with a primary mouse anti-HA monoclonal antibody and a FITC-labeled goat anti-mouse secondary antibody. F-denotes the anterior, flagellar end of promastigotes. Bar represents 2μm. C. Immunoprecipitates from culture supernatants of L. mexicana promastigotes and (axenic) amastigotes transfected with either the LmexCht1∷HA construct (Cht1∷HA) (lanes 1 and 3) or the pKSNEO control plasmid (lanes 2 and 4) were obtained using a rabbit anti-LdCht1-peptide antibody. Such immune complexes were separated in SDS-PAGE, transblotted onto PVDF membranes and probed with a mouse anti-HA monoclonal antibody. Molecular mass standards (in kDa) are shown on the left. Arrow denotes the ~50 kDa LmexCht1∷HA chimeric protein.
Immunofluorescence Microscopy
The LmexCht1∷HA and pKSNEO tranfectants of L. mexicana were examined by indirect immunofluorescence microscopy to visualize the cellular distribution of the expressed Cht1∷HA chimeric protein. For these experiments, both transfected promastigotes and axenic amastigotes were fixed, permeabilized, reacted with anti-HA mouse monoclonal antibody, followed by a goat anti-mouse FITC-conjugated secondary antibody and examined using an epi-fluorescence microscope. Results of such observations revealed that the LmexCht1∷HA transfected promastigotes displayed only very low levels of intra-cellular immuno-fluorescence with the anti-HA antibody (Fig. 6B, panel 1). In contrast, LmexCht1∷HA transfected axenic amastigotes showed very bright, punctate intracellular fluorescence (Fig. 6B, panel 2). No fluorescent signal was detected in either promastigotes or axenic amastigotes of pKSNEO (control) transfectants treated with the anti-HA monoclonal antibody (data not shown). Similarly, none of the cell samples tested showed any reactivity with the control isotype-matched mouse immunoglobulin used in these assays (data not shown). Results of these immunofluorescence assays demonstrated that both LmexCht1∷HA transfected promastigotes and axenic amastigotes synthesized and expressed the LmexCht1∷HA chimeric protein. Further, our immunofluorescence observations are in agreement with the Western blotting data described above, indicating that the LmexCht1∷HA chimeric protein appeared to be up-expressed in the axenic amastigote form of these transfectants.
Detection of the secreted/released LmexCht1-HA chimeric protein in culture supernatants of transfected parasites
As shown in Table 1 above, the anti-LdCht1-peptide antibody readily immunoprecipitated the endogenous chitinase activity secreted/released by wild-type L. mexicana parasites during their growth in vitro. In light of those observations, experiments were carried out to determine whether with this antibody could also immunoprecipitate the LmexCht1∷HA chimeric protein produced by transfected L. mexicana parasites. For these experiments, culture supernatants of both pKSNEO control and LmexCht1-HA transfected L. mexicana promastigotes and axenic amastigotes were reacted with the anti-LdCht1-peptide antibody in a Protein A Sepharose bead-based assay. The resulting immunoprecipitated complexes were solubilized, separated in SDS-PAGE, transblotted onto PVDF membranes and reacted in Western blots with a mouse anti-HA monoclonal antibody or with an appropriately matched purified mouse control immunoglobulin. In such blots, the anti-HA antibody reacted with a single ~50 kDa protein which was immunoprecipitated by the anti-LdCht1-peptide antibody from culture supernatants of both promastigotes and axenic amastigotes of LmexCht1∷HA transfected parasites (Fig. 6C, lanes 1 and 3, respectively). Interestingly, immunoprecipitates obtained from axenic amastigotes appeared to react more strongly with the anti-HA antibody compared to promastigotes (cf. Fig. 6C, lanes 3 and 1, respectively). These observations indicated that LmexCht1∷HA transfected axenic amastigotes appeared to secrete/release higher levels of the LmexCht1∷HA-chimeric protein than promastigotes. In these coupled-assays, the anti-HA antibody showed no reactivity with immunoprecipitates from either pKSNEO control transfected promastigotes or axenic amastigotes (Fig. 6C, lanes 2 and 4, respectively). Similarly, the isotype-matched purified control mouse immunoglobulin showed no reactivity with any of the samples tested in these assays (data not shown). No reactivity was observed in these blots with any samples immunoprecipitated with NRS (data not shown).
Results of these combined immunoprecipitation and Western-blotting experiments demonstrated that the complete (full length) LmexCht1∷HA chimeric protein was synthesized and secreted/released by both transfected L. mexicana promastigotes and axenic amastigotes during their growth in vitro. Further, they indicated that the secreted LmexCht1∷HA chimeric protein possessed the structurally conserved antigenic peptide epitopes (i.e. Pep1 to Pep4, Fig. 1A) recognized by the rabbit anti-LdCht1-peptide antibody.
Chitinase activity of the secreted/released LmexCht1∷HA chimeric protein
Experiments were designed to use a mouse anti-HA monoclonal antibody to specifically immunoprecipitate and measure the chitinase activity of the secreted/released LmexCht1∷HA chimeric protein. However, in preliminary experiments, we found that no detectable chitinase activity could be immunoprecipitated with this antibody from the FBS-containing culture media supernatants of LmexCht1∷HA transfectants. Therefore, short-term “release assays”(38) were set-up to measure such activity. In these assays, transfected cells were grown in complete media, harvested, washed and resuspended in an appropriately buffered, balanced salt solution which maintained both parasite viability and secretion but not de novo biosynthesis. Cell-free supernatants from such “release assays” were: 1) analyzed directly for their “total-released” chitinase activity using 4MU-substrates and 2) reacted with a mouse anti-HA monoclonal antibody to specifically immunoprecipitate the secreted /released LmexCht1∷HA chimeric protein which were subsequently analyzed for their chitinase activity as above.
Results of direct enzyme assays using 4MU-chitotriose showed that both LmexCht1∷HA transfected promastigotes and axenic amastigotes released significantly higher levels of total chitinase activity into their supernatants during incubation in vitro than similarly transfected pKSNEO control parasites (Table 2). Further, results of our coupled anti-HA immunoprecipitation/4MU-chitotriose enzyme activity assays demonstrated that the LmexCht1∷HA chimeric protein secreted/released by both transfected L. mexicana promastigotes and axenic amastigotes, did in fact, possess chitinase activity (Table 2). Under these conditions, the mouse anti-HA monoclonal antibody immunoprecipitated ~13% of the total activity released by LmexCht1∷HA transfected promastigotes and axenic amastigotes in these in vitro release assays. The relatively low amount of enzyme activity immunoprecipitated in these assays might be due to this antibody’s affinity for the HA-epitope under our conditions and/or the steric availability/accessibility of the HA-epitope in the LmexCht1∷HA chimeric protein. No chitinase activity was detected in immunoprecipitates obtained from pKSNEO control transfected parasites. Overall, values very similar to, albeit somewhat lower than, those measured in these assays with 4MU-chitotriose were also obtained using 4MU-chitobiose as substrate (data not shown).
TABLE 2.
Chitinase activity released by L. mexicana transfectants during short-term release assays* in vitro.
| Developmental Stage /transfected with |
Total Chitinase Activitya | Immunoprecipitated Activityb | ||
|---|---|---|---|---|
|
| ||||
| (pmoles of 4MU released ml−1 h−1) | ||||
| LmexCht1∷HA | pKSNEO | LmexCht1∷HA | pKSNEO | |
| Promastigotes | 900 ± 123 | 64 ± 10 | 114 ± 18 | - |
| Amastigotes | 1500 ± 173 | 369 ± 45 | 183 ± 26 | - |
Parasites were harvested at mid log phase (~1.5 × 107 ml−1), washed and incubated at 5 × 107 cells ml−1 in an appropriate balanced buffered solution for 4 h and cell-free culture supernatants were assayed for activity.
Activity assayed using 4MU-chitotriose as substrate.
Activity immunoprecipitated using a mouse anti-HA monoclonal antibody Affinity Matrix (Covance). Bound-immunoprecipitates were assayed for chitinase activity using 4MU-chitotriose as substrate.
The cumulative results of these experiments demonstrated that the LmexCht1∷HA chimeric protein secreted/released by the LmexCht1∷HA transfected parasites actually possessed functional chitinase (i.e. exo- and endo-) activity.
Survival of LmexCht1∷HA transfected parasites in human macrophages
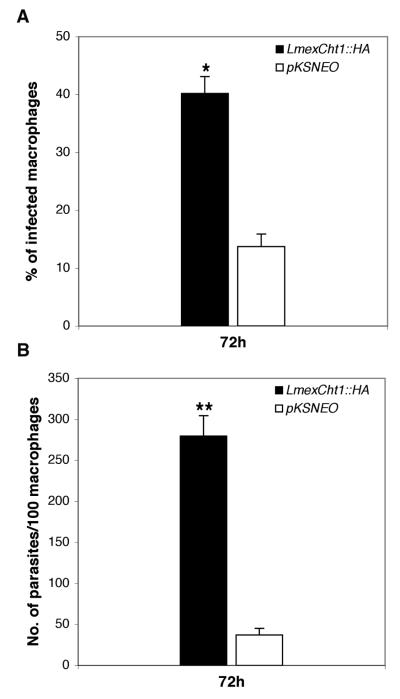
Experiments were designed to assess the viability/survival of LmexCht1∷HA transfected parasites within human macrophages in vitro. To that end, human peripheral blood-derived monocytes were differentiated into macrophages in vitro. Two parallel sets of macrophages were exposed to stationary phase cultures of either LmexCht1∷HA or pKSNEO (control) transfected promastigotes for 5 h and then washed extensively to remove extracellular parasites. Subsequently, one set of these cultures was immediately fixed and stained for light microscopy. The second set of infected macrophages was incubated for an additional 72 h prior to processing as above. These preparations were examined by light microscopy and scored for: 1) the percentage of macrophages infected with parasites and 2) the number of parasites per hundred macrophages (i.e. parasite burden/load). Results of these analyses showed that after 5 h of contact, approximately equivalent numbers of human macrophages (~50-55%) were infected with either the LmexCht1∷HA or the pKSNEO control transfectants (data not shown). However, after 72 hr of incubation, there was a significant difference (p < 0.01) between the percentage of macrophages infected with the LmexCht1∷HA compared to the pKSNEO control parasites (>40% vs. ~12%, respectively) (Fig. 7A). These infected macrophages were also scored for their parasite burden/load after 5 h and 72h post-infection. Results of such analyses showed that after 5 h of contact, these human macrophages contained comparable numbers of phagocytosed pKSNEO and LmexCht1∷HA transfected parasites (data not shown). In contrast, after 72 h, macrophages infected with the LmexCht1∷HA transfectants possessed a significantly higher parasite burden/load ( ~7 times more, p < 0.001) than those infected with the pKSNEO control parasites (Fig. 7B). Taken together, these observations indicated that the LmexCht1∷HA transfected parasites survived significantly better than the pKSNEO control transfectants within human monocyte-derived macrophages in vitro.
FIG. 7. Survival of L. mexicana transfectants in human macrophages in vitro.
Human monocyte-derived macrophages were incubated with stationary phase L. mexicana promastigotes, transfected with either the LmexCht1∷HA chimeric construct or the pKSNEO control plasmid at a parasite to host cell ratio of 10:1. After 5h of incubation, extracellular (free) parasites were removed by aspiration. Following this, one set of cultures was immediately fixed for light microscopy and a second set was incubated for an additional 72 h prior to fixation. Subsequently, both sets of cultures were stained and examined by light microscopy. A. Percentage of macrophages infected by L. mexicana parasites transfected with either the LmexCht1∷HA chimeric construct (∎) or the pKSNEO control plasmid (□) after 72 h post-infection, respectively. B. Total number of intracellular parasites (i.e. amastigotes) per 100 macrophages after 72h post-infection with either LmexCht1∷HA (∎) or the pKSNEO (□) control transfectants. The data shown are from a single experiment but are typical of those obtained from two separate and independent macrophage infection experiments. The values given represent the mean ± SD obtained from triplicate samples analyzed at each time point. Statistically significant differences between experimental and control groups are as indicated (* p < 0.01 and ** p < 0.001).
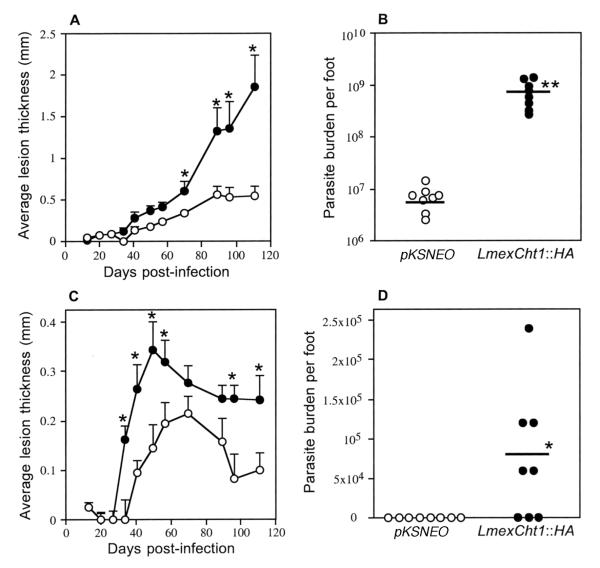
Infectivity of L. mexicana Cht1∷HA transfectants in Mice
In light of our in vitro results with human macrophages, experiments were set up to test the infectivity of the LmexCht1∷HA and pKSNEO control transfected parasites in mice. For these experiments, age and weight matched female BALB/c (a highly susceptible “non-healing” strain) and CBA/Ca (a more resistant “healing” strain) mice were injected in the dorsal surface of the right hind foot with metacyclic promastigotes of either LmexCht1∷HA or pKSNEO control transfectants. The course of infection in these animals was monitored by measuring the footpad-swelling relative to the uninfected left foot over a period of 15 weeks. In these experiments, BALB/c mice infected with the LmexCht1∷HA or pKSNEO controls showed negligible changes in foot pad lesion thickness during the initial 4 weeks of infection (Fig. 8A). However, by ten weeks post infection, mice infected with the LmexCht1∷HA parasites had significantly larger lesions (p < 0.05) than those infected with the pKSNEO control parasites. The difference in lesion size between these two groups of infected BALB/c mice became even more pronounced over the remaining course of the experiment (Fig. 8A). In that regard, after 15 weeks post infection, BALB/c mice infected with the LmexCht1∷HA parasites had lesions which were ≥ 3.6 times larger than those infected with the pKSNEO control parasites. After 15 weeks post infection, mice were sacrificed and the parasite burden in their lesions was determined. Results of these assays showed that lesions obtained from LmexCht1∷HA infected BALB/c mice had a significantly higher (p <0.0005) mean parasite burden than those infected with the pKSNEO control parasites (i.e. ≥ 7 × 108 versus 7 × 106 parasites per infected foot, respectively) (Fig. 8B).
FIG. 8. Infectivity of L. mexicana transfectants in mice.
Highly susceptible BALB/c (Panel A) and more resistant CBA/Ca (Panel C) mice (8 animals per group) were injected with 500 metacyclic promastigotes of either LmexCht1∷HA (●) or the pKSNEO control (○) transfectants into the dorsal surface of the right hind foot. Lesion development was monitored weekly by measuring the difference in thickness between the infected and the uninfected foot. The data shown are from a single experiment but are typical of those obtained from two separate and independent mouse infection experiments. The values given represent the mean ± SE obtained from each group of infected mice. Asterisk (*) above error bars represent statistically significant differences (p < 0.05) between experimental and control groups. After 15 weeks post-infection, the parasite burden in each individual BALB/c (Panel B) and CBA/Ca (Panel D) mouse foot-lesion was determined following homogenization and direct microscopic counting of released parasites (amastigotes). Horizontal bars represent the mean parasite burden determined for each group (n= 8) of infected animals. Statistically significant differences between experimental and control groups are indicated (* p < 0.05 and ** p < 0.0005).
In parallel experiments, the infectivity of LmexCht1∷HA and pKSNEO control transfectants was also tested in resistant CBA/Ca mice. By 5 weeks post-infection, animals infected with LmexCht1∷HA transfected parasites showed significant lesion development (p < 0.05) compared to those infected with the pKSNEO controls (Fig. 8C). The difference in lesion size between these two groups of CBA/Ca mice was most pronounced by 7 weeks post-infection (p.i.). At this point, mice infected with LmexCht1∷HA transfected parasites had lesions which were significantly larger (i.e. ~2.7 times; p < 0.05) than those produced by the pKSNEO controls. It is of interest to note that peak lesion development occurred at 7 weeks p.i. in mice infected with the LmexCht1∷HA transfectants, three weeks earlier than those infected with the pKSNEO control transfectants (Fig. 8C). Subsequent to these time points, the lesion size decreased in both groups of infected animals over the next several weeks. However, by 12 weeks post-infection, the lesion size stabilized in mice infected with the LmexCht1∷HA transfectants and, the majority of these mice were unable to heal their lesions over the remaining course of the experiment (Fig. 8C). At the end of 15 weeks, mice were sacrificed and the parasite burden in their lesions was determined. Results of these assays showed that lesions from LmexCht1∷HA infected CBA/Ca mice contained a mean burden of ~ 7.5 × 104 parasites each (Fig. 8D). In contrast, no parasites were detectable in samples obtained from mice infected with the pKSNEO control transfectants.
Taken together, results of these experiments demonstrated that LmexCht1∷HA transfected parasites were significantly more infectious for both BALB/c and CBA/Ca mice than the pKSNEO control transfectants. Further, these data also showed that the larger lesions produced by the LmexCht1∷HA transfectants in both strains of mice were not merely the result of exacerbated local pathology but rather they reflected the total parasite burden present in these lesions.
DISCUSSION
Leishmania mexicana is an important protozoan pathogen of humans throughout Central and South America and is the major causative agent of human cutaneous leishmaniasis in the New World (7). Studies of this organism have been greatly facilitated because culture systems exist for generating its various different life cycle developmental forms in vitro (8-10). In addition, several mouse model-infection systems have also been established to study the pathogenicity of this parasite in vivo (8,11-13). Considering the speculated importance of “chitinase” to parasite survival and transmission (4,5), to date, little evidence exists per se concerning its role(s) in the developmental biology of Leishmania sp. In that regard, it is of relevance to point-out that, previously, we used a heterologous probe to show that in vitro-grown L. mexicana promastigotes (i.e. a parasite developmental form present in its insect vector) contained a putative chitinase-like sequence within their genome (6). Thus, in light of the fore going, the current study was carried out to isolate, identify and characterize the L. mexicana chitinase gene throughout the developmental life cycle of this human pathogen.
Using a PCR based approach, we identified a single copy, 1374 bp ORF (LmexCht1) in L. mexicana parasites. The LmexCht1 encodes a deduced protein of 457 aa with a calculated molecular mass of 50,350.88 Da which has homology to known chitinases. Structural analysis of the LmexCht1 deduced protein showed that it possessed three conserved functional domains/regions (i.e. two putative substrate binding sites and a catalytic/active site) characteristic of chitinases in general (39). Further, within the active site region, the LmexCht1 deduced protein contained a nine aa consensus sequence characteristic of the catalytic domain of the chitinase-18 family of glycosyl-hydrolase enzymes (47). This includes, a conserved terminal glutamic acid residue that has been shown to be critical for the catalytic function of these proteins (48-50). Some members of this family include chitinases from prokaryotes [e.g. Bacillus circulans (51) and Serratia marcescens (51,52)], fungi [e.g. Rhizopus oligosporus (53), and Streptomyces lividans (54)], nematodes [e.g. Brugia malayi (55)], insects [e.g. Manduca sexta (56)] and plants [e.g. Glycine max (57) and tobacco Nicotiana tabacum (58)]. Virtually all of these chitinase-18 family chitinases are soluble-released /secretory proteins which function extracellularly. In that regard, results of our computer analyses suggested that the LmexCht1 deduced protein: 1) had an overall hydrophilic composition, 2) possessed a putative N-terminal signal peptide and 3) lacked both GPI- and transmembrane-anchor motifs. These predictions suggest that the LmexCht1 has properties typical of a soluble-released/secretory protein. In support of those predictions, results of our enzyme analyses showed that L. mexicana parasites secrete/release chitinase activity into their culture (media) supernatants during their growth in vitro. Further, in addition to chitinases per se, the chitinase-18 protein family also includes other members with interesting properties e.g.: 1) hevamine, a rubber tree protein with both chitinase and lysozyme-like activities (59); 2) secreted endo-beta-N-acetyl-glucosaminidases of Flavobacterium (e.g. Endo F), and Streptomyces (e.g. Endo H) which hydrolyze the glycosidic bond between the core N-acetylglucosamine residues of asparagine-linked high-mannose oligosaccharides (60,61); 3) a mammalian lysosomal enzyme, di-N-acetyl-chitobiase, which is a involved in the degradation of asparagine-linked glycoproteins (62); 4) human cartilage glycoprotein, Gp-39, a chitin-binding lectin which is associated with tissue remodeling and is expressed by activated macrophages (63); 5) a 50 kDa acidic mammalian chitinase (AMCase) (33) of lung epithelial cells and macrophages which is highly up-expressed and is implicated to mediate pathogenic inflammatory responses in asthma (64) and 6) a lysosomal chitotriosidase of activated human macrophages (65) which is secreted and accumulates at very high levels in the plasma of patients with Gaucher disease (66), to a modest level in several other lysosomal storage diseases e.g., Niemann Pick- and Krabbe-disease (66,67) and to a lesser extent in certain infectious parasitic diseases [e.g. malarial infections caused by Plasmodium falciparum (68) and visceral leishmaniasis (69)].
In trypanosomatid parasites, all mature, translatable mRNAs are capped at their 5′-end with a conserved 39-nt spliced-leader sequence (26,27). Results of our RT-PCR analysis using spliced-leader and gene-specific primers, demonstrated that both promastigotes and axenic amastigotes of L. mexicana synthesized mature (5′-capped) transcripts of the LmexCht1 gene. In parallel, results of Northern blot analyses showed that each of the L. mexicana developmental forms examined in this study (i.e. procyclic and metacyclic promastigotes, in vitro-grown axenic amastigotes and mouse lesion-derived amastigotes) constitutively transcribed an ~3kb mRNA transcript of the LmexCht1 gene. Taken together, these results indicate that this gene is actively transcribed throughout the developmental life cycle of the L. mexicana parasite. To determine whether such mRNAs were translated into a functional product, a series of enzyme activity assays were performed. Results of such assays showed that both L. mexicana promastigotes and axenic amastigotes secreted/released considerable levels of chitinase activity into their culture media supernatants during their growth in vitro. It is of significance to point out that these assays also demonstrated that axenic amastigotes appeared to secrete substantially higher levels of such chitinase activity than promastigote developmental forms of this parasite. Further, results of our coupled-immunoprecipitation/enzyme activity assays using an anti-LdCht1-peptide antibody, indicated that the native LmexCht1-chitinase shared some structurally conserved antigenic peptide-epitopes with a chitinase from closely related parasite. To examine the role(s) of the LmexCht1 protein in L. mexicana parasites, an episomal expression system was devised using the pKSNEO leishmanial expression vector (29). In these experiments, parasites were transfected with either a pKSNEO chimeric construct containing the LmexCht1 ORF fused at its 3′-end with a hemagglutinin (HA) epitope sequence or the pKSNEO vector alone. Results of RT-PCR analyses showed that episomally-transfected promastigote and axenic amastigote forms of the parasite both made mRNA transcripts from the LmexCht1∷HA chimeric construct. Further, the cumulative results of our Western blot and indirect immunofluorescence assays showed that the chimeric-gene construct was translated into a (~50 kDa) LmexCht1∷HA chimeric protein by both developmental forms of this parasite. In addition, immunoprecipitations of parasite culture (media) supernatants with a rabbit anti-LdCht1-peptide antibody followed by Western blot analysis using a mouse anti-HA monoclonal antibody demonstrated that the full-length LmexCht1∷HA chimeric protein was in fact secreted/released by both LmexCht1∷HA transfected promastigotes and axenic amastigotes during their growth in vitro. Moreover, results of our coupled anti-HA immunoprecipitations/ enzyme activity experiments using “release assay” supernatants from such transfected parasites demonstrated that the LmexCht1∷HA chimeric protein which they secreted/released had functional chitinase activity.
The results of our endogenous enzyme activity assays in conjunction with the cumulative results obtained from the LmexCht1∷HA transfection experiments showed that the LmexCht1 chitinase is expressed by both the insect (promastigote) and the mammalian (amastigote) developmental forms of this parasite. Interestingly, however, expression of the LmexCht1 chitinase appeared to be quantitatively enhanced in axenic amastigotes of L. mexicana. Taken together, these observations suggest that this protein has a hitherto unsuspected role(s) in the mammalian phase of the L. mexicana life cycle.
To test this concept, the infectivity and survival of the LmexCht1∷HA and pKSNEO (vector control) transfectants was assessed in human monocyte-derived macrophages in vitro. Results of those assays showed that the macrophages infected with the LmexCht1∷HA parasites had ~7 times higher parasite burdens/loads than those infected with the pKSNEO control parasites. In addition, the infectivity of these transfectants was also tested in vivo using two different strains of mice. Results of those experiments showed that LmexCht1∷HA transfected parasites produced significantly larger lesions in susceptible BALB/c mice than the pKSNEO control transfectants. Similar results were also obtained with these transfectants using more resistant CBA/Ca mice. Further, results of quantitative analyses showed that the larger lesions produced by the LmexCht1∷HA transfectants in both strains of mice reflected the higher total parasite burdens present in these lesions. Taken together, the results of these in vitro and in vivo infection experiments demonstrate that the LmexCht1∷HA chimera afforded a significant survival advantage to the transfected parasite both within infected human and mouse macrophages. The mechanisms underlying this survival advantage remain to be elucidated experimentally, and a variety of possibilities exist given the wide range of biological activities ascribed to members of the chitinase-18 protein family. However, it is interesting to speculate that the over-expression of a chitinase-18 family member (i.e. LmexCht1) by L. mexicana might alter the endogenous homeostatic/regulatory mechanisms of infected macrophages (i.e. cidal or stasis) to this obligate intracellular parasite/pathogen. Thus, we hypothesize that the parasite chitinase could potentially represent a new virulence determinant in the pathology of this human disease.
Previously, it was postulated that chitinase activity released by Leishmania would facilitate parasite escape from the chitinous peritrophic matrix which surrounds the infected blood meal within the sand fly vector midgut (4,70). Through this process, the parasite could gain access to the lumen of the midgut and thus, establish an infection within its sand fly vector host. Further, it has been proposed that damage to the chitin-covered stomodeal valve at the junction of the sand fly mid- and foregut is a mechanism to aid parasite transmission (5). These hypotheses suggest that the leishmanial chitinase may also play essential roles in parasite survival within sand flies and ultimately for their transmission to mammalian hosts. To date, the role of the parasite chitinase in these processes has not been addressed directly. However, in light of the current study, such in situ experiments in sand flies are now possible using L. mexicana parasites expressing various chimeric constructs of the LmexCht1 gene.
Acknowledgments
Drs. Manju Joshi and Alison Shakarian were supported by Postdoctoral Intramural Research Training Award Fellowships from the NIAID, NIH. Dr. Mat Yamage was a Visiting Fellow supported by a postdoctoral fellowship from the Fogarty International Center and NIAID, NIH. Dr. Matthew Rogers and parts of this work carried out in the UK were supported by grant 064945 from The Wellcome Trust, UK, and Dr. Saeed Al-Harthi was supported by a fellowship from Umm Al-Qura University, Saudi Arabia. We thank Ms. Sue Moriyasu for her technical assistance in the initial phases of this study; Dr. Greg Matlashewski (Institute of Parasitology, McGill University) for providing the pKSNEO leishmanial expression vector; Drs. Alain Debrabant and Angamuthu Selvapandiyan (Center for Biologics Evaluation and Research, FDA) for help with the human macrophage infection experiments and Duane Bartley of The Johns Hopkins University DNA Analysis Facility for assistance with nucleotide sequencing.
Abbreviations
- aa
amino acid
- Ab
antibody
- bp
base pair
- DIG
digoxigenin
- FBS
fetal bovine serum
- gDNA
genomic DNA
- HA
hemagglutinin
- LmexCht1
gene encoding the chitinase of Leishmania mexicana
- 4MU
4-methylumbelliferone
- nt
nucleotide
- oligo
oligodeoxy-ribonucleotide
- ORF
open reading frame
- PBS
phosphate buffered saline
- PCR
polymerase chain reaction
- RT
reverse transcription
- SDS-PAGE
sodium dodecyl-sulfate polyacrylamide gel electrophoresis
- SP
signal peptide
Footnotes
Note: The nucleotide sequence data reported in this paper has been submitted to the GenBank™ under the accession number: AY572789
REFERENCES
- 1.UNDP/WorldBank/WorldHealthOrganization . Tropical Disease Resaerch: Progress 1997-1998: Fourteenth Programme Report of UNDP/World Bank/WHO Special Programme for Research and Training in Tropical Diseases. World Health Organization; Geneva, Switzerland: 1999. [Google Scholar]
- 2.Handman E. Clin Microbiol Rev. 2001;14:229–243. doi: 10.1128/CMR.14.2.229-243.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sacks D, Kamhawi S. Annu Rev Microbiol. 2001;55:453–483. doi: 10.1146/annurev.micro.55.1.453. [DOI] [PubMed] [Google Scholar]
- 4.Schlein Y, Jacobson RL, Shlomai J. Proc R Soc Lond B Biol Sci. 1991;245:121–126. doi: 10.1098/rspb.1991.0097. [DOI] [PubMed] [Google Scholar]
- 5.Schlein Y, Jacobson RL, Messer G. Proc Natl Acad Sci U S A. 1992;89:9944–9948. doi: 10.1073/pnas.89.20.9944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shakarian AM, Dwyer DM. Exp Parasitol. 2000;94:238–242. doi: 10.1006/expr.2000.4493. [DOI] [PubMed] [Google Scholar]
- 7.Grimaldi G, Jr., Tesh RB. Clin Microbiol Rev. 1993;6:230–250. doi: 10.1128/cmr.6.3.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bates PA, Robertson CD, Tetley L, Coombs GH. Parasitology. 1992;105(Pt 2):193–202. doi: 10.1017/s0031182000074102. [DOI] [PubMed] [Google Scholar]
- 9.Bates PA, Tetley L. Exp Parasitol. 1993;76:412–423. doi: 10.1006/expr.1993.1050. [DOI] [PubMed] [Google Scholar]
- 10.Bates PA. Parasitology. 1994;108(Pt 1):1–9. doi: 10.1017/s0031182000078458. [DOI] [PubMed] [Google Scholar]
- 11.Alexander J, Coombs GH, Mottram JC. J Immunol. 1998;161:6794–6801. [PubMed] [Google Scholar]
- 12.Buxbaum LU, Denise H, Coombs GH, Alexander J, Mottram JC, Scott P. J Immunol. 2003;171:3711–3717. doi: 10.4049/jimmunol.171.7.3711. [DOI] [PubMed] [Google Scholar]
- 13.Aguilar Torrentera F, Laman JD, Van Meurs M, Adorini L, Muraille E, Carlier Y. Infect Immun. 2002;70:5075–5080. doi: 10.1128/IAI.70.9.5075-5080.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sopwith WF, Debrabant A, Yamage M, Dwyer DM, Bates PA. Int J Parasitol. 2002;32:449–459. doi: 10.1016/s0020-7519(01)00372-1. [DOI] [PubMed] [Google Scholar]
- 15.Debrabant A, Bastien P, Dwyer DM. Mol Cell Biochem. 2001;220:109–116. doi: 10.1023/a:1010809420104. [DOI] [PubMed] [Google Scholar]
- 16.Shakarian AM, Joshi MB, Yamage M, Ellis SL, Debrabant A, Dwyer DM. Mol Cell Biochem. 2003;245:31–41. doi: 10.1023/a:1022851914014. [DOI] [PubMed] [Google Scholar]
- 17.McCreath KJ, Gooday GW. Journal of Microbiological Methods. 1992;14:229–237. [Google Scholar]
- 18.Shakarian AM, Dwyer DM. Gene. 1998;208:315–322. doi: 10.1016/s0378-1119(98)00011-0. [DOI] [PubMed] [Google Scholar]
- 19.Clayton C, Adams M, Almeida R, Baltz T, Barrett M, Bastien P, Belli S, Beverley S, Biteau N, Blackwell J, Blaineau C, Boshart M, Bringaud F, Cross G, Cruz A, Degrave W, Donelson J, El-Sayed N, Fu G, Ersfeld K, Gibson W, Gull K, Ivens A, Kelly J, Vanhamme L, et al. Mol Biochem Parasitol. 1998;97:221–224. doi: 10.1016/s0166-6851(98)00115-7. [DOI] [PubMed] [Google Scholar]
- 20.Schneider A, McNally KP, Agabian N. J Biol Chem. 1993;268:21868–21874. [PubMed] [Google Scholar]
- 21.Fong D, Lee B. Mol Biochem Parasitol. 1988;31:97–106. doi: 10.1016/0166-6851(88)90149-1. [DOI] [PubMed] [Google Scholar]
- 22.McCombie WR, Heiner C, Kelley JM, Fitzgerald MG, Gocayne JD. DNA Seq. 1992;2:289–296. doi: 10.3109/10425179209030961. [DOI] [PubMed] [Google Scholar]
- 23.Devereux J, Haeberli P, Smithies O. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson JD, Higgins DG, Gibson TJ. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning. A laboratory manual. 2nd Edition Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1990. [Google Scholar]
- 26.Borst P. Annu Rev Biochem. 1986;55:701–732. doi: 10.1146/annurev.bi.55.070186.003413. [DOI] [PubMed] [Google Scholar]
- 27.Agabian N. Cell. 1990;61:1157–1160. doi: 10.1016/0092-8674(90)90674-4. [DOI] [PubMed] [Google Scholar]
- 28.Fernandes O, Murthy VK, Kurath U, Degrave WM, Campbell DA. Mol Biochem Parasitol. 1994;66:261–271. doi: 10.1016/0166-6851(94)90153-8. [DOI] [PubMed] [Google Scholar]
- 29.Zhang WW, Charest H, Ghedin E, Matlashewski G. Mol Biochem Parasitol. 1996;78:79–90. doi: 10.1016/s0166-6851(96)02612-6. [DOI] [PubMed] [Google Scholar]
- 30.Charest H, Zhang WW, Matlashewski G. J Biol Chem. 1996;271:17081–17090. doi: 10.1074/jbc.271.29.17081. [DOI] [PubMed] [Google Scholar]
- 31.Ghedin E, Charest H, Zhang WW, Debrabant A, Dwyer D, Matlashewski G. J Biol Chem. 1998;273:22997–23003. doi: 10.1074/jbc.273.36.22997. [DOI] [PubMed] [Google Scholar]
- 32.Debrabant A, Ghedin E, Dwyer DM. J Biol Chem. 2000;275:16366–16372. doi: 10.1074/jbc.M908725199. [DOI] [PubMed] [Google Scholar]
- 33.Zhang WW, Mendez S, Ghosh A, Myler P, Ivens A, Clos J, Sacks DL, Matlashewski G. J Biol Chem. 2003;278:35508–35515. doi: 10.1074/jbc.M305030200. [DOI] [PubMed] [Google Scholar]
- 34.Charest H, Matlashewski G. Mol Cell Biol. 1994;14:2975–2984. doi: 10.1128/mcb.14.5.2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Debrabant A, Joshi MB, Pimenta PFP, Dwyer DM. Int J Parasitol. 2004;34:205–217. doi: 10.1016/j.ijpara.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 36.Laemmli UK. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 37.Shakarian AM, Joshi MB, Ghedin E, Dwyer DM. J Biol Chem. 2002;277:17994–18001. doi: 10.1074/jbc.M200114200. [DOI] [PubMed] [Google Scholar]
- 38.Bates PA, Hermes I, Dwyer DM. Exp Parasitol. 1989;68:335–346. doi: 10.1016/0014-4894(89)90115-x. [DOI] [PubMed] [Google Scholar]
- 39.Flach J, Pilet PE, Jolles P. Experientia. 1992;48:701–716. doi: 10.1007/BF02124285. [DOI] [PubMed] [Google Scholar]
- 40.Boot RG, Blommaart EF, Swart E, Ghauharali-van der Vlugt K, Bijl N, Moe C, Place A, Aerts JM. J Biol Chem. 2001;276:6770–6778. doi: 10.1074/jbc.M009886200. [DOI] [PubMed] [Google Scholar]
- 41.Stiles JK, Hicock PI, Shah PH, Meade JC. Ann Trop Med Parasitol. 1999;93:781–807. doi: 10.1080/00034989957781. [DOI] [PubMed] [Google Scholar]
- 42.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 43.von Heijne G. J Mol Biol. 1985;184:99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]
- 44.Kyte J, Doolittle RF. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 45.Gerber LD, Kodukula K, Udenfriend S. J Biol Chem. 1992;267:12168–12173. [PubMed] [Google Scholar]
- 46.Teasdale RD, Jackson MR. Annu Rev Cell Dev Biol. 1996;12:27–54. doi: 10.1146/annurev.cellbio.12.1.27. [DOI] [PubMed] [Google Scholar]
- 47.Henrissat B. Biochem J. 1991;280(Pt 2):309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watanabe T, Kobori K, Miyashita K, Fujii T, Sakai H, Uchida M, Tanaka H. J Biol Chem. 1993;268:18567–18572. [PubMed] [Google Scholar]
- 49.Perrakis A, Tews I, Dauter Z, Oppenheim AB, Chet I, Wilson KS, Vorgias CE. Structure. 1994;2:1169–1180. doi: 10.1016/s0969-2126(94)00119-7. [DOI] [PubMed] [Google Scholar]
- 50.Terwisscha van Scheltinga AC, Kalk KH, Beintema JJ, Dijkstra BW. Structure. 1994;2:1181–1189. doi: 10.1016/s0969-2126(94)00120-0. [DOI] [PubMed] [Google Scholar]
- 51.Watanabe T, Suzuki K, Oyanagi W, Ohnishi K, Tanaka H. J Biol Chem. 1990;265:15659–15665. [PubMed] [Google Scholar]
- 52.Harpster MH, Dunsmuir P. Nucleic Acids Res. 1989;17:5395. doi: 10.1093/nar/17.13.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yanai K, Takaya N, Kojima N, Horiuchi H, Ohta A, Takagi M. J Bacteriol. 1992;174:7398–7406. doi: 10.1128/jb.174.22.7398-7406.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fujii T, Miyashita K. J Gen Microbiol. 1993;139(Pt 4):677–686. doi: 10.1099/00221287-139-4-677. [DOI] [PubMed] [Google Scholar]
- 55.Fuhrman JA, Lane WS, Smith RF, Piessens WF, Perler FB. Proc Natl Acad Sci U S A. 1992;89:1548–1552. doi: 10.1073/pnas.89.5.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kramer KJ, Corpuz L, Choi HK, Muthukrishnan S. Insect Biochem Mol Biol. 1993;23:691–701. doi: 10.1016/0965-1748(93)90043-r. [DOI] [PubMed] [Google Scholar]
- 57.Watanabe A, Nong VH, Zhang D, Arahira M, Yeboah NA, Udaka K, Fukazawa C. Biosci Biotechnol Biochem. 1999;63:251–256. doi: 10.1271/bbb.63.251. [DOI] [PubMed] [Google Scholar]
- 58.Heitz T, Segond S, Kauffmann S, Geoffroy P, Prasad V, Brunner F, Fritig B, Legrand M. Mol Gen Genet. 1994;245:246–254. doi: 10.1007/BF00283273. [DOI] [PubMed] [Google Scholar]
- 59.Bokma E, van Koningsveld GA, Jeronimus-Stratingh M, Beintema JJ. FEBS Lett. 1997;411:161–163. doi: 10.1016/s0014-5793(97)00682-0. [DOI] [PubMed] [Google Scholar]
- 60.Rao V, Guan C, Van Roey P. Structure. 1995;3:449–457. doi: 10.1016/s0969-2126(01)00178-2. [DOI] [PubMed] [Google Scholar]
- 61.Van Roey P, Rao V, Plummer TH, Jr., Tarentino AL. Biochemistry. 1994;33:13989–13996. doi: 10.1021/bi00251a005. [DOI] [PubMed] [Google Scholar]
- 62.Fisher KJ, Aronson NN., Jr. J Biol Chem. 1992;267:19607–19616. [PubMed] [Google Scholar]
- 63.Boot RG, van Achterberg TA, van Aken BE, Renkema GH, Jacobs MJ, Aerts JM, de Vries CJ. Arterioscler Thromb Vasc Biol. 1999;19:687–694. doi: 10.1161/01.atv.19.3.687. [DOI] [PubMed] [Google Scholar]
- 64.Zhu Z, Zheng T, Homer RJ, Kim YK, Chen NY, Cohn L, Hamid Q, Elias JA. Science. 2004;304:1678–1682. doi: 10.1126/science.1095336. [DOI] [PubMed] [Google Scholar]
- 65.Boot RG, Renkema GH, Strijland A, van Zonneveld AJ, Aerts JM. J Biol Chem. 1995;270:26252–26256. doi: 10.1074/jbc.270.44.26252. [DOI] [PubMed] [Google Scholar]
- 66.Hollak CE, van Weely S, van Oers MH, Aerts JM. J Clin Invest. 1994;93:1288–1292. doi: 10.1172/JCI117084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guo Y, He W, Boer AM, Wevers RA, de Bruijn AM, Groener JE, Hollak CE, Aerts JM, Galjaard H, van Diggelen OP. J Inherit Metab Dis. 1995;18:717–722. doi: 10.1007/BF02436762. [DOI] [PubMed] [Google Scholar]
- 68.Barone R, Simpore J, Malaguarnera L, Pignatelli S, Musumeci S. Clin Chim Acta. 2003;331:79–85. doi: 10.1016/s0009-8981(03)00089-5. [DOI] [PubMed] [Google Scholar]
- 69.Aguilera B, Ghauharali-van der Vlugt K, Helmond MT, Out JM, Donker-Koopman WE, Groener JE, Boot RG, Renkema GH, van der Marel GA, van Boom JH, Overkleeft HS, Aerts JM. J Biol Chem. 2003;278:40911–40916. doi: 10.1074/jbc.M301804200. [DOI] [PubMed] [Google Scholar]
- 70.Pimenta PF, Modi GB, Pereira ST, Shahabuddin M, Sacks DL. Parasitology. 1997;115(Pt 4):359–369. doi: 10.1017/s0031182097001510. [DOI] [PubMed] [Google Scholar]