Abstract
Studies on environmental changes provide important insights into modes of speciation, into the (adaptive) reoccupation of ecological niches and into species turnover. Against this background, we here examine the history of the gastropod genus Lanistes in the African Rift Lake Malawi, guided by four general evolutionary scenarios, and compare it with patterns reported from other endemic Malawian rift taxa. Based on an integrated approach using a mitochondrial DNA phylogeny and a trait-specific molecular clock in combination with insights from the fossil record and palaeoenvironmental data, we demonstrate that the accumulation of extant molecular diversity in the endemic group did not start before approximately 600 000 years ago from a single lineage. Fossils of the genus from the Malawi Rift, however, are over one million years older. We argue that severe drops in the lake level of Lake Malawi in the Pleistocene offer a potential explanation for this pattern. Our results also challenge previously established phylogenetic relationships within the genus by revealing parallel evolution and providing evidence that the endemic Lanistes species are not restricted to the lake proper but are present throughout the Malawi Rift.
Keywords: ancient lake, endemism, molecular clock, fossil record, environmental change, Lanistes
1. Introduction
The influence of abiotic factors on speciation and extinction events plays a key role in evolutionary biology (e.g. Benton 2009). As such, the importance of environmental changes has been discussed in the context of biodiversity (Sanders 1968; Kondoh 2001; Martens 2002; Kadmon & Benjamini 2006), of rates of morphological evolution (Martens 1994), and of species turnover (Bennett 2004; Peters 2008). In particular, research on faunas of the Great African Lakes has underlined the importance of limnological and geological data for understanding evolutionary processes (e.g. Palacios-Fest et al. 2005; Seehausen et al. 2008). Lake-level changes and hence habitat (in)stability have provided insights into extinction events (Van Damme & Pickford 1995; Verheyen et al. 2003), the (adaptive) reoccupation of ecological niches (e.g. Seehausen 2006), and modes of speciation (e.g. Genner et al. 2007a; Turner 2007).
Two extremes of potential limnological patterns are conceivable for large lacustrine systems. The first is characterized by long-term stability of the abiotic setting. This stability is often assumed to be derived from the remarkable depth of some of the lakes, presumably mitigating effects of environmental changes. The endemic radiations within these lakes have regularly proved to be remarkably old. World-renowned examples are Lake Tanganyika in Central East Africa and Lake Baikal in Siberia (Sherbakov 1999; Kocher 2004; Wilson et al. 2004; Macdonald et al. 2005; Day et al. 2008). The other extreme is that of seemingly long-lived lacustrine systems that, however, were afflicted by catastrophic disturbances, which eliminated the inhabiting biota. The subsequent reconstitution of habitats may have triggered new faunal radiations that are considerably younger than lineages that have experienced long-term stability. East African examples of this second extreme are Lake Victoria that desiccated completely during the Late Pleistocene (Johnson et al. 1996; Stager & Johnson 2008), and the palaeolacustrine systems in the Turkana Basin (Brown & Feibel 1991; Van Bocxlaer et al. 2008).
Intermediates between the two extreme hydrological patterns show neither a pronounced long-term stability nor recent clear-cut limnological disruptions. Such aquatic systems may permit the study of consecutive evolutionary phases in the process of adaptive radiation, thus providing additional insights into speciation processes and how they are affected by the abiotic setting. A candidate for this intermediate type is Lake Malawi (Van Bocxlaer 2005; Seehausen 2006), the southernmost African Great Lake. At present, the lake (maximum depth 706 m; Spigel & Coulter 1996) largely fills the Malawi Rift, which started to develop approximately 8.6 Myr ago (Ebinger et al. 1987, 1989; Delvaux 1995; Ring & Betzler 1995). Owing to its very small catchment area, the water balance of Lake Malawi is strongly influenced by regional and even local climate fluctuations (DeBusk 1998; Barker et al. 2007), and lake-level changes from a few dozen metres to several hundred metres take place on various time scales (Delvaux 1995; Finney et al. 1996; Scholz et al. 2007).
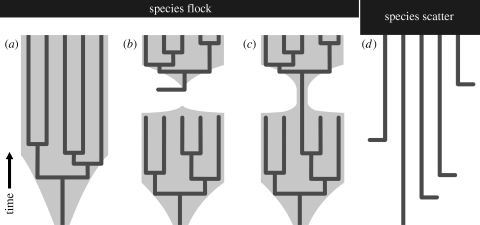
Based on these hydrological settings, four evolutionary scenarios for endemic taxa within Lake Malawi are conceivable (figure 1; note that the scenarios are not mutually exclusive).
An ancient intralacustrine radiation: the endemic group would display a deep branching phylogenetic signal similar to that of some endemic taxa of Lake Tanganyika (corresponding to figure 1a).
Re-colonization: a former endemic taxon in the lake became extinct as a result of environmental changes, for example, a desiccation or a severe lake level drop. Hence, the extant endemic radiation would be comparatively young, consisting of descendants of a founder species that re-colonized the lake (figure 1b).
Following the founder-flush model sensu Schön & Martens (2004) (figure 1c), an endemic radiation might have experienced a strong bottleneck event during an ecological crisis. One or few surviving lineages then proliferated within the lake after the recovery of ecological settings.
Applying the species scatter model of Hauswald et al. (2008) to Lake Malawi would suggest that several species of a taxon colonized the lake independently (figure 1d). These lineages would have evolved via anagenesis into the endemic extant taxa. The phylogenetic analysis would probably reveal a non-monophyletic pattern.
Figure 1.
Evolutionary scenarios relative to the ecological stability of a lacustrine system. The stability is indicated by the width of the grey surface underlying the schematic species trees (broad representing stable; narrow, unstable conditions). See text for details. (a) Ancient radiation; (b) re-colonization; (c) founder flush; (d) species scatter.
The influence of Malawi's lake-level changes on the evolution of its cichlid fauna has already been discussed in the literature (Owen et al. 1990; Sturmbauer et al. 2001; Genner & Turner 2005; Cohen et al. 2007; Turner 2007). However, possibilities of deducing taxonomic assemblages and timing of cichlid fish evolution beyond molecular phylogenetics are limited, since their fossil remains are often restricted to isolated pieces of bone and teeth. Mostly, these remains do not allow taxonomic identification at the generic or specific level (Van Neer 1994). Mollusc shells, by contrast, are preserved relatively well in lacustrine strata and often allow specific identification (Van Damme 1984). Thus, their fossil record may provide an independent temporal and palaeobiological reference for molecular phylogenetic analyses (Michel 1994).
In the present study, we aim to take advantage of this reference. Guided by the scenarios outlined above, we examine the evolutionary history of the gastropod genus Lanistes Montfort, 1810 in Lake Malawi. Based on extensive sampling from the lake itself and other parts of Eastern and Southern Africa, we establish a molecular phylogeny using fragments of the mitochondrial cytochrome oxidase c subunit I gene (COI) and of the large ribosomal subunit RNA (LSU rRNA). These inferences are framed along with findings from Malawi's Lanistes fossil record as well as with recently reported palaeohydrological data. Finally, we compare and discuss effects of environmental change on the evolution of lacustrine taxa within the Malawi Rift and relate them to examples from other Great African lakes.
2. Material and methods
(a). Sampling design
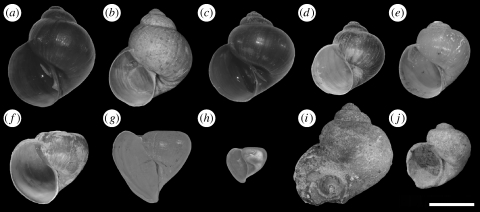
Three endemic species of Lanistes (Ampullariidae) are described from within the lake (figure 2): Lanistes solidus Smith, 1877, Lanistes nasutus (Mandahl-Barth 1972) and Lanistes nyassanus Dohrn, 1865. Additionally, two non-endemic species are reported from swamps and lagoons nearby the lake: Lanistes ovum Troschel, 1845 and Lanistes ellipticus Martens, 1866 (Mandahl-Barth 1972; Louda & McKaye 1982; Louda et al. 1984; Berthold 1990a,b).
Figure 2.
Shells of endemic Lanistes species of the Malawi Rift and two more widely distributed species: (a) Lanistes ovum and (c) Lanistes ellipticus, both from their type locality at Tete, Mozambique (specimens 9740 and 9741 in table 1 in the electronic supplementary material). Specimens (b) and (d) from Malawi were originally also determined as L. ovum and L. ellipticus by G. Mandahl-Barth (HLMD 9891-1-4 and MRAC 796867). Specimens (e–g) are Lanistes solidus (MRAC 796869), Lanistes nyassanus (6586 in table 1 in the electronic supplementary material) and Lanistes nasutus (NHM, Eccles collection). Specimen (h) is the juvenile L. nasutus (7412 in table 1 in the electronic supplementary material) that was analysed in the course of this study. Specimens (i) and (j) are Pliocene fossils from Unit 3A of the Chiwondo Beds that morphologically resemble L. ellipticus (D. Clark collection RUP-P-2104) and L. solidus (A. Gorthner collection HLMD-CF-451).
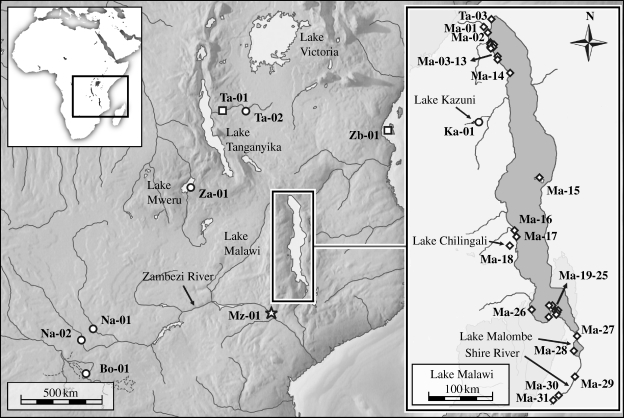
We have collected specimens of these taxa at 31 localities in Lake Malawi and its vicinity (Republic of Malawi) in 2006, 2007 and 2008 (figure 3). Despite intensive search, we have found only a single specimen presumably belonging to the deep-water species L. nasutus (figure 2h). Although juveniles of this species may be difficult to distinguish from juvenile L. nyassanus, our specimen has the widely open umbilicus typical for L. nasutus (in contrast to the narrow umbilicus of juvenile and adult L. nyassanus; Martens 1897, p. 168). Material was also obtained from the Shire River, the only outlet of Lake Malawi at its southern end, from Lake Malombe and the satellite lake Chilingali. Additionally, Lanistes species from Botswana, Burkina Faso, Mozambique, Namibia, Tanzania, Zambia and Zanzibar were included in the analyses (locality details are provided in table 1 in the electronic supplementary material).
Figure 3.
Map of central East Africa showing the sampling points for the present study. Most specimens were collected within the Malawi Rift (enlarged map section to the right). Sampling points for specimens of the Malawi group are marked with diamonds; circles indicate sampling points for specimens of the ovum-complex (classification is a result of the present study; figure 4). The shared type locality of L. ovum and L. ellipticus is marked with a star symbol. Squares indicate sampling points for other taxa. Locality details are given in table 1 in the electronic supplementary material.
Identification was carried out based on shell characteristics as described in the original literature. Shell and tissue vouchers have been deposited at the Natural History Museum, London (BMNH) and the zoological collection of Berlin Museum of Natural History (ZMB; table 1 in the electronic supplementary material).
(b). DNA isolation and sequencing
DNA was isolated using a CTAB protocol (Wilke et al. 2006). We amplified a fragment of the COI gene using the universal COI primer pair LCO1490 and HCO2198 of Folmer et al. (1994) and optionally the reverse primer COI-BR as described in Colgan et al. (2007). In addition, we amplified a fragment of the more conservative LSU rRNA from specimens with unique COI haplotypes using the universal primer pair 16Sar-L and 16Sbr-H of Palumbi et al. (1991; see §2c below and table 1 in the electronic supplementary material). PCR conditions were as given in Schultheiß et al. (2008). Sequences (forward and reverse) were determined using a LI-COR DNA sequencer Long ReadIR 4200 (Lincoln, NE) and a Thermo Sequenase fluorescent labelled primer cycle sequencing kit (Amersham Pharmacia Biotech, Piscataway, NJ). The protein-coding COI gene was aligned unambiguously using the default settings in ClustalW, v. 1.4 (Thompson et al. 1994) as implemented in BioEdit v. 7.0.5.3 (Hall 1999). Alignment of the LSU rRNA was carried out using a heads or tails approach (Landan & Graur 2007). Since the first base pairs of the forward and reverse sequences were difficult to read, we uniformly cut off the first and last few base pairs, leaving a 641 bp-long completely overlapping fragment for the COI gene and a 536 bp-long fragment (after alignment) for the LSU rRNA. All sequences are available from NCBI GenBank (see table 1 in the electronic supplementary material).
(c). Phylogenetic analyses
After preliminary analyses, we reduced our original COI dataset of 90 sequences to a dataset with 42 unique haplotypes (see table 1 in the electronic supplementary material). We chose two species of Pila Röding, 1798 as out-group, since the genus has recently be shown to be the sister taxon of Lanistes (Jørgensen et al. 2008). COI and LSU rRNA sequences of Pila ovata Olivier, 1804, Pila conica Gray, 1828 and of seven Lanistes specimens were obtained from NCBI GenBank (see table 1 in the electronic supplementary material). Prior to the phylogenetic analyses, we used the program PhyML v. 3.0 (Guindon & Gascuel 2003) to determine the optimal model of DNA sequence evolution for each fragment. Hereby, the general time reversible (GTR) model with invariable sites and gamma distribution was selected for both fragments. The subsequent test for substitution saturation within our datasets using the entropy-based index in DAMBE (Xia et al. 2003) revealed no substantial saturation. For phylogenetic reconstruction, we then conducted a partitioned Bayesian analysis using the program MrBayes v. 3.1 (Huelsenbeck & Ronquist 2001) with the combined COI/LSU rRNA dataset comprising 1177 bp. As an alternative to Bayesian inference, a partitioned maximum-likelihood analysis was carried out with the combined dataset using RAxML v. 7.0.4 (Stamatakis et al. 2008) with 1000 bootstrap replicates. Again, the GTR+I+Γ model was used with the parameters separately estimated for each fragment.
(d). Molecular clock estimation
Molecular clock estimations are among the most debated issues in phylogenetic reconstructions (Ayala 1997; Ho & Larson 2006; Pulquério & Nichols 2007). We have aimed at mitigating common problems associated with the molecular clock approach by: (i) testing for substantial nucleotide saturation under the respective model of sequence evolution (Wilke 2003, 2004); (ii) testing for deviant lineages and/or rate heterogeneity in our dataset (Sanderson 1997; Huelsenbeck et al. 2000; Kishino et al. 2001; Welch & Bromham 2005); and (iii) inferring error rates for node depth variations (Hillis et al. 1996; Wilke 2004).
First, we carried out a Bayesian analysis using the Lanistes COI dataset without enforcing the molecular clock. In the second step, we repeated this analysis but enforced the molecular clock by imposing a uniform substitution rate over all branches. Subsequent calculations were carried out with the program R (R Development Core Team 2007), the add-on package APE (Paradis et al. 2004) and our own set of R-functions (available upon request). We tested the applicability of the strict molecular clock by performing a likelihood-ratio test (Felsenstein 1988) for the single best Bayesian inference trees of both approaches. Since the log-likelihood ratio (−2 log Λ) did not exceed 28.12 at 40 degrees of freedom (critical Χ2 value 55.76 at p=0.05), the strict clock model was accepted. In order to date the age of the endemic Lanistes species of Lake Malawi, the node depth for its most recent common ancestor was calculated in the 1000 best trees of the Bayesian posterior for the two independent runs.
In the absence of sufficient internal calibration points for Lanistes, we applied the trait-specific COI Protostomia clock rate of Wilke et al. (in press). Under the GTR+I+Γ model, the average substitution rate equals 1.76±0.34% per million years. This trait-specific clock rate has shown to be robust in a wide range of invertebrate taxa with similar biological and life-history characteristics (i.e. dioeceous tropical or subtropical taxa with a generation time of 1 year and a body size of approximately 2–50 mm). In order to provide conservative error estimations, we calculated the total error using a propagation of uncertainty approach and, thus accounted for the variation of the applied clock rate as well as the variation of node depths. In addition, a Bayesian molecular clock analysis of the COI fragment was conducted using the software BEAST v. 1.4.8 (Drummond & Rambaut 2007) under a strict clock model with the same clock rate (Monte-Carlo Markov chain length 10 000 000 generations).
3. Results
(a). Phylogenetic analyses
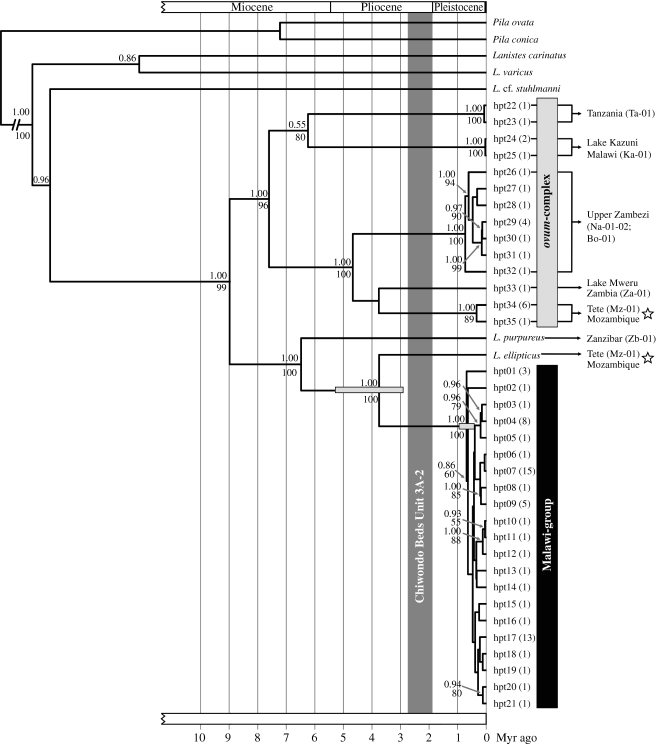
Since the phylogenetic trees based on the combined COI/LSU rRNA fragments generated in MrBayes and RAxML show a highly similar topology to the COI tree inferred for molecular clock analyses using BEAST, we here only provide the latter phylogeny with the respective support values from the other analyses (figure 4). The monophyly of the genus Lanistes in the two-fragment analysis is well supported (Bayesian posterior probability (BPP) of 1.00 and a bootstrap support (BS) of 100). The phylogenies reveal two similarly well supported major groups: one contains the monophyletic group of all specimens collected in the Malawi Rift (BPP 1.00; BS 100; henceforth referred to as the Malawi group), L. ellipticus from the type locality as sister taxon (BPP 1.00; BS 100; locality Mz-01 in figure 3) and L. purpureus (BPP 1.00; BS 100). The second major group contains specimens of L. ovum from the type locality (haplotypes 34 and 35 in figure 4; locality Mz-01 in figure 3), specimens from Lake Kazuni (hpt24 and hpt25 in figure 4; locality Ka-01 in figure 3) and specimens from Tanzania, Zambia, Namibia, Botswana and Mozambique. This major group is henceforth referred to as the ovum-complex (BPP 1.00; BS 96). Specimens neither from Lake Malawi nor from any other sampled locality within the Malawi Rift (including Lake Malombe, Lake Chilingali and the Shire River) cluster with specimens of L. ovum or L. ellipticus from their common type locality. None of the phylogenetic analyses resolve the species relationships within the Malawi group. Basal to both major groups is the split to L. cf. stuhlmanni from Tanzania (BPP 0.96) and the clade containing Lanistes varicus from Burkina Faso and Lanistes carinatus from Uganda (BPP 0.86).
Figure 4.
Strict clock Bayesian phylogeny of Lanistes spp. inferred from the mitochondrial COI gene. The topology is congruent with the combined phylogeny inferred from COI and LSU rRNA with maximum-likelihood and Bayesian methods. Bayesian posterior probabilities (BPP) and bootstrap support values (BS) of the combined analyses are given next to the respective nodes (only values above 0.5 BPP and 50 BS are shown). The tree was rooted with sequences of Pila ovata and Pila conica. Grey node bars indicate the 95% confidence intervals of the two age estimations. Locality information is provided next to haplotypes in the phylogenetic tree (abbreviations correspond to localities in figure 3). Numbers in parentheses next to the tree tips indicate the numbers of specimens sharing the respective haplotype. Star symbols show the phylogenetic position of specimens of L. ovum and L. ellipticus from their type locality. The vertical grey bar indicates the age of Unit 3A-2 of the Chiwondo Beds in Malawi from where Lanistes fossils are known (see specimens (i) and (j) in figure 2). Detailed locality information is provided in table 1 in the electronic supplementary material.
(b). Molecular clock estimation
The depths of the nodes for the start of accumulation of extant molecular diversity in the Malawi group of the best 1000 MrBayes trees yielded a mean depth of 1.18±0.24% sequence divergence. By applying the trait-specific COI Protostomia clock, we calculated the maximum age of the start of the molecular diversification to be 640 000 years (95% confidence interval: 310 000–970 000 years). Age estimations with BEAST yielded an age of 680 000 years (95% CI: 380 000–910 000 years). The split between the Malawi group and its apparent sister taxon L. ellipticus was calculated to be 3.74 million years old (95% CI: 2.9–5.3 million years; figure 4).
4. Discussion
(a). Endemism and parallel evolution of Lanistes in the Malawi Rift
Previous authors have argued that Lake Malawi's shoreline forms a strict boundary between endemic species in the lake proper (L. nasutus, L. nyassanus and L. solidus), and non-endemic species inhabiting ponds and marshes surrounding the lake (L. ovum and L. ellipticus) (Crowley et al. 1964; Mandahl-Barth 1972; Berthold 1990a,b). Our analysis strongly indicates that this is not the case. None of the specimens collected throughout the Malawi Rift cluster with L. ellipticus or L. ovum from their type locality (figure 4). We therefore conclude that these two species are not present in the rift. Instead, our findings suggest that two morphologically similar, yet endemic species arose in the rift (see specimens a–d in figure 2 for a comparison of shell morphology). These two species are part of the Malawi group and henceforth referred to as Lanistes sp. (ovum-like) and Lanistes sp. (ellipticus-like). Thus, we reinterpret the paraphyly suggested by Jørgensen et al. (2008) for the endemic Lanistes species of Lake Malawi as a result of parallel evolution or plesiomorphy in shell morphology and stress the need for a molecular based taxonomic revision of the genus.
Whereas these findings indicate that the shoreline does not separate Lanistes endemics from non-endemics, we have found such a potential faunal transition for the genus approximately 50 km west of the lake—outside of the Malawi Rift. In Lake Kazuni (figure 3; Ka-01), we found only specimens that cluster in the ovum-complex (figure 4; hpt24 and hpt25). Although Lake Kazuni connects to Lake Malawi via the South Rukuru River, L. ovum has apparently not entered the Malawi Rift nor has it been found in sympatry with species from the Malawi group. By contrast, we have not observed such a faunal transition in the Shire River although it was sampled 140 km downstream from Lake Malawi. All specimens collected from there cluster within the Malawi group. This indicates that the Malawi group consists of species endemic to the entire Malawi Rift rather than being endemic to the lake proper.
(b). Evolutionary history of the endemic Lanistes species in Lake Malawi
The occurrence of Lanistes species in the lake in Pliocene times was reported from Unit 3A-2 of the Chiwondo fossil beds in Malawi (2.7–1.8 Myr ago; see fossil specimens in figure 2; Schrenk et al. 1995; Kullmer 2008). However, our results show that the accumulation of extant molecular diversity within the recent Lanistes group started approximately 600 000 years ago (figure 4). Moreover, it is likely that the group is even younger because of an inherent bias in the molecular clock estimation due to ancestral polymorphism (see Edwards & Beerli 2000; for a discussion of ancestral polymorphism in trait-specific clocks see also Wilke et al. (in press)). In the light of this young phylogenetic age, the Malawi group may have lacked sufficient time for complete lineage sorting, which is also indicated by numerous low support values in figure 4. Hence, the apparent paraphyly may not be an indication of the underlying species relationships (see Knowles & Carstens 2007). Note that without nuclear markers, we also cannot exclude that hybridization contributes to this paraphyly. Thus, a detailed interspecific analysis of the Malawi group needs to be based on faster evolving, nuclear markers such as microsatellites or amplified fragment length polymorphism.
The comparative young age of the group argues against an ancient radiation of the endemic Lanistes species (scenario (a) in figure 1) and against the species scatter model (scenario (d)). In fact, the presence of Lanistes in the lake in Pliocene times in combination with the Pleistocene onset of molecular diversification is in concordance with evolutionary scenarios (b) and (c) (figure 1). A potential trigger for this recent onset of molecular diversification is an increase in ecological opportunity, for example, through increased heterogeneity of the lacustrine habitat enabling ecological segregation (sensu Schluter 2000). Such segregation (e.g. different habitat preferences) was reported for the endemic Lanistes species in Lake Malawi (Louda & McKaye 1982; Louda et al. 1984). Thus, we conclude that appropriate limnological conditions for the evolution of the extant Lanistes radiation have (re-)emerged only very recently; for example, after recovery from a severe lake level low stand. The existence of such low stands in the history of Lake Malawi was reported on the basis of deep drilling core data and linked to phases of pronounced tropical African aridity (Cohen et al. 2007; Scholz et al. 2007). The authors describe two events 135 000 and 75 000 years ago when Malawi's lake level dropped at least 600 and 350 metres, respectively. Delvaux (1995) reported another severe Pleistocene low stand 1.6–1.0 Myr ago. The validity of this assumption, however, was recently questioned (Cohen et al. 2007).
The question arises as to whether part of the Pliocene Lanistes fauna in the Malawi Rift has survived such lake-level changes up to present times. Although it is not possible to answer the question unambiguously with the present data, a recent colonization (scenario (b), figure 1) would require a source area in the vicinity of Lake Malawi to have existed during arid phases. However, as Cohen et al. (2007) pointed out, an aridity that would provoke lake level drops in Lake Malawi by more than 600 metres would convert the surrounding watershed into a semi-desert. Considering the low probability of small paludal and lacustrine habitats persisting in a semi-desert landscape, the founder-flush model (scenario (c), figure 1) appears to be the more likely scenario. This suggestion is also supported by the occurrence of Lanistes shells in the deep drilling core during the low stand 135 000 years ago (A. Cohen 2008, personal communication).
(c). Comparison with other Malawian taxa
We note a remarkable lack of studies on the evolution of Malawi's endemic invertebrates. Besides morphological work on molluscs (e.g. Berthold 1990b), copepods (e.g. Fryer 1959) and ostracods (e.g. Martens 2003), we are aware of only one molecular study examining evolutionary patterns in more detail (Genner et al. 2007b). This study on the thiarid snail genus Melanoides Olivier, 1804 reveals a substantially different pattern than the one we have found for Lanistes. First, members of the genus colonized Lake Malawi independently at least two times. Second, Lanistes and Melanoides from Lake Malawi might also differ in their geographical origin. Genner et al. (2007b) suggested a possible origin of the Melanoides polymorpha complex west of the rift, e.g. in a shared watershed of the Congo–Zambezi. The Lanistes endemics from the Malawi Rift, by contrast, are only distantly related to the specimens from the Upper Zambezi but closely to L. ellipticus from the Lower Zambezi (figure 3: Na-01/02, Bo-01; figure 4). Further work with a wider geographical sampling coverage is needed to assess whether the Lanistes endemics of the Malawi Rift do indeed have their origin in the Lower Zambezi.
The mitochondrial genetic architecture of the Lanistes group resembles the pattern that was found for Malawi cichlids by Sturmbauer et al. (2001). These authors suggested that the observed presence of shared mitochondrial haplotypes between geographically distant lake populations, in combination with the high degree of haplotype diversity within populations, indicate a rapid range extension in the aftermath of a severe lake level low stand in the Pleistocene, and subsequent speciation. Lake-level changes on a smaller scale, influencing persistence and isolation of peripheral habitats, have recently been shown to promote peripatric speciation in the Malawi Rift (Genner et al. 2007a; see also Martens (2003) for a discussion on ostracod diversity). Peripatric speciation may also have triggered the evolution of Lanistes sp. (ellipticus-like) and Lanistes sp. (ovum-like) in the swamps and lagoons surrounding the lake.
(d). Environmental change in African Rift lakes and the evolution of lacustrine taxa—concluding remarks
Basin morphology and physical settings of the Great African lakes have been shown to crucially influence evolution of their endemic faunas: whereas the shallow Lake Victoria desiccated completely in the Pleistocene (Stager & Johnson 2008), severe lake level drops of Lake Tanganyika resulted in three separate basins, providing opportunity for allopatric speciation (Scholz & Rosendahl 1988; Verheyen et al. 1996; Cohen et al. 1997; Marijnissen et al. 2006). In Lake Malawi, on the other hand, basin separation did not occur and severe lake level drops may rather have promoted a breakdown of species barriers and hybridization (see above; Sturmbauer et al. 2001; Seehausen 2006). Our study also indicates that morphological adaptation recurred in the Malawi Rift not only spatially (parallel evolution of Malawian and Zambezian species) but also over time: fossil specimens of the Chiwondo Beds resemble morphologically the extant endemic L. solidus (see specimens (e) and (j) in figure 2), pointing towards consecutive adaptations to comparable selection pressures (e.g. wave action and predation; for a more detailed discussion on the morphological adaptations of the endemic Lanistes species see Louda & McKaye 1982; Louda et al. 1984).
Whereas the influence of environmental changes as an evolutionary driving force is often discussed on large temporal and spatial scales (Barnosky 2001; Benton 2009), its effect within shorter time frames and isolated habitats appears to be crucial as well (Owen et al. 1990; Genner et al. 2007a). In concordance with previous studies on Malawi's cichlid fish fauna, the present work corroborates the lake to have been a lacustrine system with intermediate to low ecosystem stability in its recent past.
Acknowledgements
We thank Aslak Jørgensen, Ellinor Michel, Frank Riedel, Andrew S. Cohen and Vincenzo Mercurio for kindly providing materials. Andrew S. Cohen, Dirk Van Damme, Martin Genner, Ellinor Michel, David Reid, Andrew Smith, Jonathan Todd and two anonymous reviewers are gratefully acknowledged for valuable comments on previous versions of the manuscript. We thank Friedemann Schrenk, Thies Geertz, Diederike Geelhoed, Harrison Simfukwe, Patrick Msoyhaya and Dany Simbeye for support in the field and logistic help in Malawi and Mozambique, and Silvia Nachtigall for assistance in the laboratory. This study was funded by the DFG project SCHR352/9-1, the SYNTHESYS grant GB-TAF3455 (RS), the Bijzonder Onderzoeksfonds (BOF) of Ghent University and the Flanders Research Foundation (FWO-Vlaanderen) (both B.V.B). B.V.B is Aspirant of the FWO. Fieldwork was, in part, funded by DFG project RI809/20-1 (F. Riedel).
References
- Ayala F.J.1997Vagaries of the molecular clock. Proc. Natl Acad. Sci. USA 94, 7776–7783 (doi:10.1073/pnas.94.15.7776) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker P.A., Leng M.J., Gasse F., Huang Y.2007Century-to-millennial scale climatic variability in Lake Malawi revealed by isotope records. Earth Planet. Sci. Lett 261, 93–103 (doi:10.1016/j.epsl.2007.06.010) [Google Scholar]
- Barnosky A.D.2001Distinguishing the effects of the Red Queen and Court Jester on Miocene mammal evolution in the northern Rocky Mountains. J. Vert. Paleontol 21, 172–185 (doi:10.1671/0272-4634(2001)021[0172:DTEOTR]2.0.CO;2) [Google Scholar]
- Bennett K.D.2004Continuing the debate on the role of Quaternary environmental change for macroevolution. Phil. Trans. R. Soc. Lond. B 359, 295–303 (doi:10.1098/rstb.2003.1395) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton M.J.2009The Red Queen and the Court Jester: species diversity and the role of biotic and abiotic factors through time. Science 323, 728–732 (doi:10.1126/science.1157719) [DOI] [PubMed] [Google Scholar]
- Berthold T.Intralacustrine speciation and the evolution of shell sculpture in gastropods of ancient lakes: application of Günther's niche concept. Abh. Naturwiss. Ver. Hamburg 31/32, 1990a85–118 [Google Scholar]
- Berthold T.Phylogenetic relationships, adaptations and biogeographic origin of the Ampullariidae (Mollusca, Gastropoda) endemic to Lake Malawi. Abh. Naturwiss. Ver. Hamburg 31/32, 1990b47–84 [Google Scholar]
- Brown F. H., Feibel C. S.1991Stratigraphy, depositional environments and paleogeography of the Koobi Fora Formation. In Koobi Fora Research Project, stratigraphy, artiodactyls and paleoenvironments, vol. 3 (ed. Harris J. M.), pp. 1–30 Oxford, UK: Clarendon Press [Google Scholar]
- Cohen A.S., Lezzar K.E., Tiercelin J.J., Soreghan M.1997New palaeogeographic and lake-level reconstructions of Lake Tanganyika: implications for tectonic, climatic and biological evolution in a rift lake. Basin Res 9, 107–132 (doi:10.1046/j.1365-2117.1997.00038.x) [Google Scholar]
- Cohen A.S., et al. 2007Ecological consequences of early Late Pleistocene megadroughts in tropical Africa. Proc. Natl Acad. Sci. USA 104, 16 422–16 427 (doi:10.1073/pnas.0703873104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colgan D.J., Ponder W.F., Beacham E., Macaranas J.2007Molecular phylogenetics of Caenogastropoda (Gastropoda: Mollusca). Mol. Phylogen. Evol 42, 717–737 (doi:10.1016/j.ympev.2006.10.009) [DOI] [PubMed] [Google Scholar]
- Crowley T.E., Pain T., Woodward F.R.1964A monographic review of the Mollusca of Lake Nyasa. Ann. Mus. r. Afr. Centr. Tervuren 8° Sci. Zool 131, 1–58 [Google Scholar]
- Day J.J., Cotton J.A., Barraclough T.G.2008Tempo and mode of diversification of Lake Tanganyika cichlid fishes. PLoS ONE 3, e1730.(doi:10.1371/journal.pone.0001730) [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBusk G.H.1998A 37,500-year pollen record from Lake Malaŵi and implications for the biogeography of afromontane forests. J. Biogeogr 25, 479–500 (doi:10.1046/j.1365-2699.1998.2530479.x) [Google Scholar]
- Delvaux D.1995Age of Lake Malawi (Nyasa) and water level fluctuations. Mus. R. Afr. Centr. Tervuren (Belg.) Dept. Geol. Min. Rapp. Ann 1995–1996, 99–108 [Google Scholar]
- Drummond A.J., Rambaut A.2007BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol 7, 214.(doi:10.1186/1471-2148-7-214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebinger C.J., Rosendahl B.R., Reynolds D.J.1987Tectonic model of the Malawi rift, Africa. Tectonophysics 141, 215–235 (doi:10.1016/0040-1951(87)90187-9) [Google Scholar]
- Ebinger C.J., Deino A.L., Drake R.E., Tesha A.L.1989Chronology of volcanism and rift basin propagation: Rungwe Volcanic Province, East Africa. J. Geophys. Res 94, 15 785–15 803 (doi:10.1029/JB094iB11p15785) [Google Scholar]
- Edwards S.V., Beerli P.2000Perspective: gene divergence, population divergence, and the variance in coalescence time in phylogeographic studies. Evolution 54, 1839–1854 (doi:10.1111/j.0014-3820.2000.tb01231.x) [DOI] [PubMed] [Google Scholar]
- Felsenstein J.1988Phylogenies from molecular sequences: inference and reliability. Annu. Rev. Genet 22, 521–565 (doi:10.1146/annurev.ge.22.120188.002513) [DOI] [PubMed] [Google Scholar]
- Finney B.P., Scholz C.A., Johnson T.C., Trumbore S., Southon J.1996Late Quaternary lake-level changes of Lake Malawi. In The limnology, climatology and paleoclimatology of the East African Lakes (eds. Johnson T.C., Odada E.O.), pp. 495–508 Amsterdam, UK: Gordon and Breach Scientific Publishers [Google Scholar]
- Folmer O., Black M., Hoeh W.R., Lutz R., Vrijenhoek R.1994DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol 3, 294–299 [PubMed] [Google Scholar]
- Fryer G.1959The trophic interrelationships and ecology of some littoral communities of Lake Nyasa with especial reference to the fishes, and a discussion of the evolution of a group of rock-frequenting Cichlidae. Proc. Zool. Soc. Lond 132, 153–281 [Google Scholar]
- Genner M.J., Turner G.F.2005The mbuna cichlids of Lake Malawi: a model for rapid speciation and adaptive radiation. Fish Fish 6, 1–34 (doi:10.1111/j.1467-2679.2005.00173.x) [Google Scholar]
- Genner M.J., Nichols P., Carvalho G.R., Robinson R.L., Shaw P.W., Smith A., Turner G.F.Evolution of a cichlid fish in a Lake Malawi satellite lake. Proc. R. Soc. B 274, 2007a2249–2257 (doi:10.1098/rspb.2007.0619) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genner M.J., Todd J.A., Michel E., Erpenbeck D., Jimoh A., Joyce D.A., Piechocki A., Pointier J.P.Amassing diversity in an ancient lake: evolution of a morphologically diverse parthenogenetic gastropod assemblage in Lake Malawi. Mol. Ecol 16, 2007b517–530 (doi:10.1111/j.1365-294X.2006.03171.x) [DOI] [PubMed] [Google Scholar]
- Guindon S., Gascuel O.2003A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol 52, 696–704 (doi:10.1080/10635150390235520) [DOI] [PubMed] [Google Scholar]
- Hall T.A.1999BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser 41, 95–98 [Google Scholar]
- Hauswald A.-K., Albrecht C., Wilke T.2008Testing two contrasting evolutionary patterns in ancient lakes: species flock vs. species scatter in valvatid gastropods of Lake Ohrid. Hydrobiologia 615, 169–179 (doi:10.1007/s10750-008-9556-0) [Google Scholar]
- Hillis D.M., Mable B.K., Moritz C.1996Applications of molecular systematics. In Molecular systematics (eds. Hillis D.M., Moritz C., Mable B.K.), pp. 515–543 Sunderland, MA: Sinauer Associates, Inc [Google Scholar]
- Ho S.Y.W., Larson G.2006Molecular clocks: when times are a-changin. Trends Genet 22, 79–83 (doi:10.1016/j.tig.2005.11.006) [DOI] [PubMed] [Google Scholar]
- Huelsenbeck J.P., Ronquist F.2001MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755 (doi:10.1093/bioinformatics/17.8.754) [DOI] [PubMed] [Google Scholar]
- Huelsenbeck J.P., Larget B., Swofford D.2000A compound Poisson process for relaxing the molecular clock. Genetics 154, 1879–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson T.C., Scholz C.A., Talbot M.R., Kelts K., Ricketts R.D., Ngobi G., Beuning K., Ssemmanda I., McGill J.W.1996Late Pleistocene desiccation of Lake Victoria and rapid evolution of cichlid fishes. Science 273, 1091–1093 (doi:10.1126/science.273.5278.1091) [DOI] [PubMed] [Google Scholar]
- Jørgensen A., Kristensen T.K., Madsen H.2008A molecular phylogeny of apple snails (Gastropoda, Caenogastropoda, Ampullariidae) with an emphasis on African species. Zool. Scr 37, 245–252 (doi:10.1111/j.1463-6409.2007.00322.x) [Google Scholar]
- Kadmon R., Benjamini Y.2006Effects of productivity and disturbance on species richness: a neutral model. Am. Nat 167, 939–946 (doi:10.1086/504602) [DOI] [PubMed] [Google Scholar]
- Kishino H., Thorne J.L., Bruno W.J.2001Performance of a divergence time estimation method under a probabilistic model of rate evolution. Mol. Biol. Evol 18, 352–361 [DOI] [PubMed] [Google Scholar]
- Knowles L.L., Carstens B.C.2007Delimiting species without monophyletic gene trees. Syst. Biol 56, 887–895 (doi:10.1080/10635150701701091) [DOI] [PubMed] [Google Scholar]
- Kocher T.D.2004Adaptive evolution and explosive speciation: the cichlid fish model. Nat. Rev. Genet 5, 288–298 (doi:10.1038/nrg1316) [DOI] [PubMed] [Google Scholar]
- Kondoh M.2001Unifying the relationships of species richness to productivity and disturbance. Proc. R. Soc. Lond. B 268, 269–271 (doi:10.1098/rspb.2000.1384) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullmer O.2008The fossil suidae from the Plio-Pleistocene Chiwondo Beds of northern Malawi, Africa. J. Vert. Paleontol 28, 208–216 (doi:10.1671/0272-4634(2008)28[208:TFSFTP]2.0.CO;2) [Google Scholar]
- Landan G., Graur D.2007Heads or tails: a simple reliability check for multiple sequence alignments. Mol. Biol. Evol 24, 1380–1383 (doi:10.1093/molbev/msm060) [DOI] [PubMed] [Google Scholar]
- Louda S.M., McKaye K.R.1982Diurnal movements in populations of the prosobranch Lanistes nyassanus at Cape Maclear, Lake Malawi, Africa. Malacologia 23, 13–21 [Google Scholar]
- Louda S.M., McKaye K.R., Kocher T.D., Stackhouse C.J.1984Activity, dispersion, and size of Lanistes nyassanus and L. solidus (Gastropoda, Ampullariidae) over the depth gradient at Cape Maclear, Lake Malawi, Africa. Veliger 26, 145–152 [Google Scholar]
- Macdonald K.S., Yampolsky L., Duffy J.E.2005Molecular and morphological evolution of the amphipod radiation of Lake Baikal. Mol. Phylogen. Evol 35, 323–343 (doi:10.1016/j.ympev.2005.01.013) [DOI] [PubMed] [Google Scholar]
- Mandahl-Barth G.1972The freshwater Mollusca of Lake Malawi. Rev. Zool. Bot. Afr 86, 257–289 [Google Scholar]
- Marijnissen S.A.E., Michel E., Daniels S.R., Erpenbeck D., Menken S.B.J., Schram F.R.2006Molecular evidence for recent divergence of Lake Tanganyika endemic crabs (Decapoda: Platythelphusidae). Mol. Phylogen. Evol 40, 628–634 (doi:10.1016/j.ympev.2006.03.025) [DOI] [PubMed] [Google Scholar]
- Martens E. V.1897Beschalte Weichthiere Ost-Afrikas. In Deutsch-Ost-Afrika, vol. 4 (ed. Stuhlmann F.), pp. 1–308 Berlin, Germany: Dietrich Reimer [Google Scholar]
- Martens K.1994Speciation in ancient lakes: 40 years after Brooks. Arch. Hydrobiol. Beih 44, 75–96 [Google Scholar]
- Martens K.2002Redundancy and ecosystem stability in the fluctuating environments of long-lived lakes. In The East African Great Lakes: limnology, palaeolimnology and biodiversity, vol. 12 (ed. Odada E. O., Olago D. O.), pp. 309–319 Dordrecht, The Netherlands: Academic Publishers [Google Scholar]
- Martens K.2003On the evolution of Gomphocythere (Crustacea, Ostracoda) in Lake Nyassa/Malawi (East Africa), with the description of 5 new species. Hydrobiologia 497, 121–144 (doi:10.1023/A:1025417822555) [Google Scholar]
- Michel E.1994Why snails radiate: a review of gastropod evolution in long-lived lakes, both recent and fossil. Arch. Hydrobiol. Beih 44, 285–317 [Google Scholar]
- Owen R.B., Crossley R., Johnson T.C., Tweddle D., Kornfield I., Davison S., Eccles D.H., Engstrom D.E.1990Major low levels of Lake Malawi and their implications for speciation rates in cichlid fishes. Proc. R. Soc. Lond. B 240, 519–553 (doi:10.1098/rspb.1990.0052) [Google Scholar]
- Palacios-Fest M.R., Alin S.R., Cohen A.S., Tanner B., Heuser H.2005Paleolimnological investigations of anthropogenic environmental change in Lake Tanganyika. IV. Lacustrine paleoecology. J. Paleolimn 34, 51–71 (doi:10.1007/s10933-005-2397-1) [Google Scholar]
- Palumbi S., Martin A., Romano S., Mcmillian W.O., Stice L., Grabowski G.1991The simple fool's guide to PCR Honolulu, HI: University of Hawaii [Google Scholar]
- Paradis E., Claude J., Strimmer K.2004APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290 (doi:10.1093/bioinformatics/btg412) [DOI] [PubMed] [Google Scholar]
- Peters S.E.2008Environmental determinants of extinction selectivity in the fossil record. Nature 454, 626–629 (doi:10.1038/nature07032) [DOI] [PubMed] [Google Scholar]
- Pulquério M.J.F., Nichols R.A.2007Dates from the molecular clock: how wrong can we be?. Trends Ecol. Evol 22, 180–184 (doi:10.1016/j.tree.2006.11.013) [DOI] [PubMed] [Google Scholar]
- R Development Core Team 2007R: a language and environment for statistical computing Vienna, Austria: R Foundation for Statistical Computing [Google Scholar]
- Ring U., Betzler C.1995Geology of the Malawi Rift: kinematic and tectonosedimentary background to the Chiwondo Beds, Northern Malawi. J. Hum. Evol 28, 7–21 (doi:10.1006/jhev.1995.1003) [Google Scholar]
- Sanders H.L.1968Marine benthic diversity: a comparative study. Am. Nat 102, 243–282 (doi:10.1086/282541) [Google Scholar]
- Sanderson M.J.1997A nonparametric approach to estimating divergence times in the absence of rate constancy. Mol. Biol. Evol 14, 1218–1231 [Google Scholar]
- Schluter D.The ecology of adaptive radiation. In Oxford series in ecology and evolution 2000Oxford, UK: Oxford University Press [Google Scholar]
- Scholz C.A., Rosendahl B.R.1988Low lake stands in lakes Malawi and Tanganyika, East Africa, delineated with multifold seismic data. Science 240, 1645–1648 (doi:10.1126/science.240.4859.1645) [DOI] [PubMed] [Google Scholar]
- Scholz C.A., et al. 2007East African megadroughts between 135 and 75 thousand years ago and bearing on early-modern human origins. Proc. Natl Acad. Sci. USA 104, 16 416–16 421 (doi:10.1073/pnas.0703874104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schön I., Martens K.2004Adaptive, pre-adaptive and non-adaptive components of radiations in ancient lakes: a review. Org. Divers. Evol 4, 137–156 (doi:10.1016/j.ode.2004.03.001) [Google Scholar]
- Schrenk F., Bromage T.G., Gorthner A., Sandrock O.1995Paleoecology of the Malawi Rift: vertebrate and invertebrate faunal contexts of the Chiwondo Beds, northern Malawi. J. Hum. Evol 28, 59–70 (doi:10.1006/jhev.1995.1006) [Google Scholar]
- Schultheiß R., Albrecht C., Bößneck U., Wilke T.2008The neglected side of speciation in ancient lakes: phylogeography of an inconspicuous mollusc taxon in lakes Ohrid and Prespa. Hydrobiologia 615, 141–156 (doi:10.1007/s10750-008-9553-3) [Google Scholar]
- Seehausen O.2006African cichlid fish: a model system in adaptive radiation research. Proc. R. Soc. B 273, 1987–1998 (doi:10.1098/rspb.2006.3539) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seehausen O., Takimoto G., Roy D., Jokela J.2008Speciation reversal and biodiversity dynamics with hybridization in changing environments. Mol. Ecol 17, 30–44 (doi:10.1111/j.1365-294X.2007.03529.x) [DOI] [PubMed] [Google Scholar]
- Sherbakov D.Y.1999Molecular phylogenetic studies on the origin of biodiversity in Lake Baikal. Trends Ecol. Evol 14, 92–95 (doi:10.1016/S0169-5347(98)01543-2) [DOI] [PubMed] [Google Scholar]
- Spigel R.H., Coulter G.W.Comparison of hydrology and physical limnology of the East African Great Lakes: Tanganyika, Malawi, Victoria, Kivu and Turkana (with references to some North American great lakes). In The limnology, climatology and paleoclimatology of the East African Lakes Johnson T.C., Odada E.O.1996. pp. 103–140 Eds. Amsterdam, UK: Gordon and Breach Scientific Publishers [Google Scholar]
- Stager J.C., Johnson T.2008The late Pleistocene desiccation of Lake Victoria and the origin of its endemic biota. Hydrobiologia 596, 5–16 (doi:10.1007/s10750-007-9158-2) [Google Scholar]
- Stamatakis A., Hoover P., Rougemont J.2008A rapid bootstrap algorithm for the RAxML Web Servers. Syst. Biol 57, 758–771 (doi:10.1080/10635150802429642) [DOI] [PubMed] [Google Scholar]
- Sturmbauer C., Baric S., Salzburger W., Ruber L., Verheyen E.2001Lake level fluctuations synchronize genetic divergences of cichlid fishes in African lakes. Mol. Biol. Evol 18, 144–154 [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J.1994ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22, 4673–4680 (doi:10.1093/nar/22.22.4673) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner G.F.2007Adaptive radiation of cichlid fish. Curr. Biol 17, R827–R831 (doi:10.1016/j.cub.2007.07.026) [DOI] [PubMed] [Google Scholar]
- Van Bocxlaer B.2005Changes in the malacofauna of Lake Malawi since mid-Holocene times. Geol. Belg 8, 124 [Google Scholar]
- Van Bocxlaer B., Van Damme D., Feibel C.S.2008Gradual versus punctuated equilibrium evolution in the Turkana basin molluscs: evolutionary events or biological invasions?. Evolution 62, 511–520 (doi:10.1111/j.1558-5646.2007.00296.x) [DOI] [PubMed] [Google Scholar]
- Van Damme D.The freshwater Mollusca of northern Africa. Distribution, biogeography and palaeoecology. In Developments in hydrobiology 1984Dordrecht, The Netherlands: Dr W. Junk Publishers [Google Scholar]
- Van Damme D., Pickford M.1995The late Cenozoic Ampullariidae (Mollusca, Gastropoda) of the Albertine Rift Valley (Uganda-Zaire). Hydrobiologia 316, 1–32 (doi:10.1007/BF00019372) [Google Scholar]
- Van Neer W.1994Cenozoic fish fossils from the Albertine Rift Valley in Uganda. In Geology and palaeobiology of the Albertine Rift Valley, Uganda-Zaire, vol. IIPalaeobiology Palaeobiology (eds Senut B., Pickford M.), pp. 89–127 Milwaukee, WI: CIFEG Occasional Publications [Google Scholar]
- Verheyen E., Ruber L., Snoeks J., Meyer A.1996Mitochondrial phylogeography of rock-dwelling cichlid fishes reveals evolutionary influence of historical lake level fluctuations of Lake Tanganyika, Africa. Phil. Trans. R. Soc. Lond. B 351, 797–805 (doi:10.1098/rstb.1996.0074) [DOI] [PubMed] [Google Scholar]
- Verheyen E., Salzburger W., Snoeks J., Meyer A.2003Origin of the superflock of cichlid fishes from Lake Victoria, East Africa. Science 300, 325–329 (doi:10.1126/science.1080699) [DOI] [PubMed] [Google Scholar]
- Welch J.J., Bromham L.2005Molecular dating when rates vary. Trends Ecol. Evol 20, 320–327 (doi:10.1016/j.tree.2005.02.007) [DOI] [PubMed] [Google Scholar]
- Wilke T.2003Salenthydrobia gen. nov (Rissooidea: Hydrobiidae): a potential relict of the Messinian salinity crisis. Zool. J. Linn. Soc 137, 319–336 (doi:10.1046/j.1096-3642.2003.00049.x) [Google Scholar]
- Wilke T.2004How dependable is a non-local molecular clock? A reply to Hausdorf et al. (2003). Mol. Phylogen. Evol 30, 835–840 (doi:10.1016/j.ympev.2003.08.008) [DOI] [PubMed] [Google Scholar]
- Wilke T., Davis G.M., Qiu D.C., Spear R.C.2006Extreme mitochondrial sequence diversity in the intermediate schistosomiasis host Oncomelania hupensis robertsoni: another case of ancestral polymorphism?. Malacologia 48, 143–157 [Google Scholar]
- Wilke T., Schultheiß R., Albrecht C.In press As time goes by: a simple fool's guide to molecular clock approaches in invertebrates. Am. Malacol. Bull. [Google Scholar]
- Wilson A.B., Glaubrecht M., Meyer A.2004Ancient lakes as evolutionary reservoirs: evidence from the thalassoid gastropods of Lake Tanganyika. Proc. R. Soc. Lond. B 271, 529–536 (doi:10.1098/rspb.2003.2624) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X., Xie Z., Salemi M., Chen L., Wang Y.2003An index of substitution saturation and its application. Mol. Phylogen. Evol 26, 1–7 (doi:10.1016/S1055-7903(02)00326-3) [DOI] [PubMed] [Google Scholar]