Abstract
Mutations in BRCA1 account for the majority of familial aggregations of early onset breast and ovarian cancer (~70%) and about 1/5 of all early onset breast cancer families; in contrast, mutations in BRCA2 account for a smaller proportion of breast/ovarian cancer families and a similar proportion of early onset breast cancer families. BRCA2 has also been shown to be associated with a much more pleiotropic disease spectrum compared to BRCA1. Since the identification of both BRCA1 and BRCA2 investigations into the functions of these genes have revealed that both are associated with the maintenance of genomic integrity via their apparent roles in cellular response to DNA damage, especially their involvement in the process of double strand DNA break repair. This review will focus on the specific roles of both genes and how functional differences may account for the diverse clinical findings observed between families that harbour BRCA1 or BRCA2 mutations.
Keywords: BRCA1, BRCA2, double strand break repair, breast cancer, predisposition
Genomic integrity
The integrity of the genome is crucial for the correct functioning of the individual organism. Any perturbation that results in a loss of genomic integrity is likely to result in disregulation and consequent uncontrolled cellular proliferation. Maintenance of genomic integrity is orchestrated by a number of different but interrelated DNA repair mechanisms that include DNA mismatch repair, nucleotide excision repair, base excision repair and DNA double strand break repair. In relation to human neoplasia all of these mechanisms have been implicated in disease including such illnesses as hereditary non-polyposis colorectal cancer (HNPCC), adenomatous polyposis, xeroderma pigmentosum and the focus of this review, inherited predispositions to breast cancer.
There are essentially two types of genes that have been associated with inherited predispositions to cancer, gatekeeper genes or caretaker genes [1]. Gatekeeper genes such as the adenomatous polyposis coli (APC) gene can be directly associated with disease by virtue of its involvement in the control of cell signalling pathway(s) that are associated with cellular proliferation. Caretaker genes, in contrast, are not involved directly with cellular proliferation but are intimately associated with the maintenance of genomic integrity. Loss of caretaker function will lead to an accelerated accumulation of change within the genome, which has the potential to affect cellular control and hence result in accelerated tumour development [2].
Initially it was considered that errors in genes associated with the maintenance of genomic integrity could barely be tolerated and most resulted in severe symptoms that culminated in the early demise of the affected individual. Such conditions include Nijmegen Breakage Syndrome, Ataxia Telangiectasia, Fanconi Anaemia, Bloom's Syndrome, Werner's Syndrome and Rothmund Thompson Syndrome [3]. All of these conditions are extremely rare and are autosomal recessive diseases that are characterised by an enormous increase in the risk of developing malignancy. In 1993 the first hint that deficient DNA repair processes were more common than originally believed was revealed by the identification of the genetic basis of a relatively common inherited predisposition to colorectal cancer, namely HNPCC, where the underlying defect is in DNA mismatch repair [4]. In 1994 the first gene associated with the genetic basis of early onset breast and/or ovarian cancer was identified (BRCA1), which at the time could not be linked to any known cellular process [5]. Subsequently, BRCA1 has been implicated in the cellular response to DNA double strand break damage [6].
Double strand break (DSB) repair
The mechanisms involved in the repair of DSBs are poorly understood as they are complex and require the accurate coordination of a large variety of proteins that are not only involved in the process of repair but also the recognition of repair, placing the cell cycle into a holding pattern until the repair is complete and transcriptional control.
Double strand breaks in DNA can be caused by a variety of environmental insults but most notably ionising radiation has been considered the most likely cause of such DNA damage. There are essentially three mechanisms of DSB, one involves nonhomologous end joining (NHEJ) repair, another is via homologous recombination repair (HRR) and the third is single strand annealing (SSA). There are differences between the three types of DSB repair in regards to the likelihood of errors. Both NHEJ and SSA are error prone as both mechanisms do not involve a sister chromatid in the repair process whereas HRR is essentially error free since this process relies on the use of the homologous chromosome for repair [7,8].
Initially, evidence suggested that DSBs were primarily rejoined by NHEJ repair in mammalian cells [9]. More recent evidence has, however, indicated that HRR is active in mammalian cells [9] and that both mechanisms are important for the repair of DSBs. Apart from ionising radiation DSBs are frequently created during DNA replication, which are under normal circumstances very efficiently repaired via HRR [10]. Loss of either type of repair process results in an increased frequency of chromosomal rearrangements, which can result in a loss or gain of genetic material, and/or recombination of the genome. The repair of DSBs is a complex process and involves not only the orchestration of a variety of proteins but also of different DNA repair processes. DSB repair can involve MLH1, a protein primarily involved in DNA mismatch repair and XPD, a protein involved in nucleotide excision repair. Furthermore, since the identification of genes associated with rare DNA repair disorders it has become apparent that they are interrelated and that there is considerable overlap in cellular phenotypes, for example Fanconi anaemia (group B and D) is related to deficiencies in BRCA2 [11].
The repair of DSBs requires the formation of a protein complex that involves three different proteins, RAD50, MRE11 and NBS1 that interact with BRCA1.
A breakdown in DSB repair will result in chromosomal rearrangements that include deletions, duplications, inversions, and translocations. Since the genes involved in this process are integrally linked to cell cycle checkpoint control any perturbation in a cell ability to recognise DNA damage may adversely affect its ability to repair DSBs, which can result in adverse chromosomal change.
DNA damage and breast cancer
A small but significant proportion of breast cancer patients come from families where there are other 1st or 2nd degree relatives with disease and represents somewhere in the order of 10% of all breast cancer patients [12]. From this subset approximately two thirds of them can be accounted for by mutations in the breast cancer susceptibility genes BRCA1 or BRCA2. It is now ten years since BRCA1 was identified and during the intervening period much has been learnt about the functions of both BRCA1 and BRCA2 proteins [13]. The most striking finding about the function of the two genes has been the recognition of their roles in either sensing DNA damage or being intimately involved in its repair. BRCA1 and BRCA2 proteins are involved in either the recognition of DNA DSBs or the repair of these lesions, respectively.
BRCA1
BRCA1 appears to be unique to mammals and is likely to be relatively new in evolutionary terms as there are no orthologs found in yeast, drosophila or c. elegans genomes [14].
The function of BRCA1 is complex and involves a series of proteins that have distinct binding sites on BRCA1 (see Fig. 1) and there appears to be specific functional activity localized in different regions of the protein. Evidence of additional proteins binding to BRCA1 is shown in Table 1. The majority of proteins that have been identified which interact with BRCA1 are also associated with some aspect of DNA repair or cell cycle checkpoint control [15]. Initial studies suggest that BRCA1 is involved in at least two types of DSB repair, NHEJ and HRR as well as the control of transcription [16]. A number of different facets to DNA repair have been associated with BRCA1 and these can be broadly categorised into those that are intimately involved in DNA damage sensing [17], DNA damage recognition [18], a contribution to S and G2 phase checkpoint control [19], transcriptional control [20] and X-chromosome inactivation [21]. The following summarises the functions that BRCA1 has been associated with:
Figure 1.
Schematic representation of the various binding sites on BRCA1 protein.
Table 1.
Proteins that bind to BRCA1 or BRCA2
| BRCA1 | BRCA2 |
|---|---|
| ATF1 | P/CAF |
| BARD1 | RAD51 |
| BAP1 | BRAF35 |
| Rb | DSS1 |
| TP53 | BCCIPα |
| RAD50 | EMSY |
| Mre1 | |
| Nbs1 | |
| ZBRK1 | |
| Importinα | |
| BRAP2 | |
| γ-tubulin | |
| STAT1 | |
| P300/CBP | |
| BRCA2 | |
| c-Abl | |
| RNA pol II | |
| HDAC1 | |
| HDAC2 | |
| CTIP | |
| LMO4 | |
| BACH1 | |
| ATM | |
| ATR | |
| c-Myc | |
| Nmi | |
| ERCC2 | |
| CyclinD2 | |
| Swi/Snf complex | |
| P300 | |
| Oestrogen Receptor (ER) |
1. BRCA1 and RAD51 involved in focus formation. Once a DNA break has been recognised one of the first events that can be visualized is the formation of foci around the DSB. Foci formation is mediated by RPA (replication protein A), which is a multifunctional molecule that not only functions in DSB repair but is also required for DNA replication [22]. If cells are irradiated RPA and BRCA1 can be observed collocating together. In BRCA1 deficient cells the number of damage-induced RPA foci is much greater than that compared to their wild-type counterparts [23].
2. BRCA1 is part of a large super protein complex known as BASC and evidence suggests that it could act as a damage sensor. BRCA1 appears to have an increased affinity for branched structures compared to double strand DNA implicated this protein as a damage sensor [24].
3. Phosphorylation of BRCA1 suggests that it acts as a signalling factor between damage sensing and recognition. After DNA damage BRCA1 is rapidly phosphorylated in dividing cells suggesting that it may function downstream of the checkpoint mechanisms that sense and signal DNA damage or problems with DNA replication during S-phase of the cell [25].
4. Routing DSB repair through HRR that are in appropriate phases of the cell cycle [26].
5. Mediates resectioning of broken termini to generate long single stranded tails required for RAD51 assembly. This is mediated by activating Chk1 kinase upon DNA damage [27].
6. During mitosis BRCA1 is clearly associated with the centrosome, and γ-tubulin associates preferentially with the hypophosphorylated form of BRCA1. This suggests that BRCA1 may direct a role in centrosome regulation and chromosome segregation - an important point in relation to the maintenance of genomic integrity and the increase chromosomal errors observed in BRCA1 deficient cell lines [28].
7. Contribution of checkpoints in both S and G2 phases. BRCA1 is implicated in several different checkpoint events. After ionising radiation (IR) BRCA1 deficient cells fail to arrest scheduled DNA synthesis in S-phase [29]. G2 arrest does not occur in the absence of BRCA1 and it must be phosphorylated to initiate this checkpoint [30]. Both these functions of BRCA1 are in response to DSBs. Chromatid decatenation checkpoint control is also deregulated [31].
8. Possible role in down regulating mRNA 3' processing [32].
9. Role in transcriptional control
a) interacting with proteins to remodel chromatin structure [33]
b) transactivating genes through interaction with TP53 [34]
c) interacting with LMO4 repressing BRCA1's transcriptional activation function [35]
d) acting as a co-repressor of transcription [36].
10. X-chromosome inactivation. BRCA1, BARD1 and Xist co-localise on the inactive X-chromosome in female somatic cells and associates with Xist RNA. Perturbations of the relationship between BRCA1 and X-inactivation have the potential to destabilise silencing of the X-chromosome and thereby affect normal gene expression patterns [37].
BRCA2
Similar to BRCA1, BRCA2 is evolutionarily speaking a relative newcomer, as it is only found in the same genomes as BRCA1 [38]. This coupled with a high degree of similarity in disease expression between individuals harbouring changes in BRCA1 and BRCA2 suggests that their functions are either similar or that they have unique and different functional specificity within the overall process [38].
Less is known about the function of BRCA2 than BRCA1. Furthermore, what is known about BRCA2 has been gained from studies on cell lines examining specific processes that can be observed as phenotypic differences compared to control cells.
Does a deficiency in DSB repair orchestrated by BRCA1 or BRCA2 result in an increased sensitivity to ionising radiation? The evidence is as follows:
1. Linked to homologous recombination. BRCA2 directly interacts with RAD51 and they in association with BRCA1 can be identified in nuclear foci of undamaged replicating cells. Defective RAD51 focus formation is observed in cells where BRC repeats are over-expressed, thereby inhibiting the self-association of RAD51 and consequently the formation of RAD51 nucleoprotein filaments on single strand DNA [39].
2. Gene expression of BRCA2 is contingent on which phase of the cell cycle the cell happens to be in. This has lead to difficulties in determining what over-expression of BRCA2 may confer but it appears that it may inhibit the cell cycle [40].
3. Mitotic checkpoint control. Thymic tumours from BRCA21492Tr/Tr mice had defective mitotic checkpoint control defects suggesting that there is a promotion of transformed cells that develop into malignancy [41].
4. Deficiency in the error free repair mechanism of DSBs. BRCA2 mutant cells are phenotypically similar to mutants of the five RAD51 paralogs strongly implicating BRCA2 in DSB repair [42].
5. Reduced resistance to DNA-damaging agents. IR results in an increased (1.5 - 2 fold) rate of cell killing compared to control cells [43].
6. Missegregation of chromosomes. There is an excess of aneuploidy in BRCA2 mutant cells caused by anomalies in the centrosome [44].
7. Reduced levels of HRR. There are gross defects in HRR in BRCA2 mutant cells. HNEJ is not affected [45].
8. Reduced sister chromatid exchange (SCE). SCE is a consequence of HRR events where crossing over occurs between sister chromatids. The rate of SCE is reduced in BRCA2 mutant cells implying a reduction in HRR efficiency [46].
9. Reduced efficiency of gene targeting but less than the 20-fold difference in BRCA1 mutant cells [47].
10. Reduced numbers of RAD51 foci after IR in BRCA2 deficient cells. For HRR to occur one of the first events is the formation of RAD51 foci. This implies that BRCA2 is critical for the efficient formation of this initial step in HRR [48].
Given the plethora of binding partners and functions and the limited disease profiles observed in women with mutations in BRCA1 why is there such specificity in disease expression?
Why do mutations in BRCA1 and BRCA2 result in such a specific disease pattern?
Mutations in BRCA1 and BRCA2 essentially predispose to an increase in breast and/or ovarian cancer although BRCA2 changes confer an increased risk of a variety of other cancers, their relative frequency in comparison to breast or ovarian cancer is somewhat low. Given that the function of both genes is associated with the maintenance of genomic integrity and that this is a fundamental process that occurs in all cells it is surprising to observe such disease specificity. There are a number of possibilities that could account for the observed disease spectrum. During breast development at the time of puberty and again during pregnancy rapid clonal proliferation occurs that is retained within the breast lobule [49-51]. Clonal expansion of any somatic cell runs the risk of amplifying genetic errors that may be harboured within the original set of cells. In this way it can be envisaged that women who harbour BRCA1 or BRCA2 germline mutations run the risk of amplifying a "second hit" and thereby alter the likelihood of developing disease [52]. Since both BRCA1 and BRCA2 are associated with genomic integrity why is there not an increased frequency of other malignancies in persons harbouring these changes? There are many possibilities with respect to the answer of this question and it should be emphasised that at this point in time there is insufficient knowledge about the functions of both BRCA1 and BRCA2. Several different notions have been put forward but none have been unequivocally supported by experimental evidence. Evidence from one source has indicated that BRCA1 can function as an inhibitor of oestrogen receptor signalling [53] suggesting that in cells deficient in this gene a loss of repression of oestrogen mediated mammary or ovarian epithelial proliferation occurs, thereby contributing to mammary and ovarian carcinogenesis [54]. Other more complex functions of BRCA1 or BRCA2 may also be tissue specific, not necessarily as a result of some intrinsic factor associated with only the breast or ovary but rather as a result of the growth and differentiation pathways of these organs [26,55,56].
BRCA1 and BRCA2 and sporadic breast cancer
One of the most surprising results that became rapidly apparent after the identification of BRCA1 and BRCA2 was the absence of involvement of these genes in breast cancer patients who had no family history of disease (hereafter referred to as sporadic). Molecular analysis of sporadic breast tumours revealed that BRCA1 or BRCA2 did not behave as typical tumour suppressor genes by virtue of the fact that there appears to be an almost complete absence of 2ndBRCA1 or BRCA2 allele loss. This evidence implies that either haploinsufficiency is enough to initiate disease or that BRCA1 or BRCA2 are not involved in sporadic cases of breast cancer. Currently, this view for BRCA1 remains intact whereas evidence has now emerged implicating a novel gene product that interacts with BRCA2 to suppress its transactivational activity. The gene in question is termed EMSY and when overexpressed potentially mimics the effects of BRCA2 inactivation [57]. Whether BRCA1 will be implicated in sporadic breast and/or ovarian cancer remains to be determined.
Conclusions
There remains much to discover with respect to the function of BRCA1 and BRCA2, notwithstanding new mechanisms of disease are being discovered which will eventuate in a better understanding not only of inherited forms of breast cancer but also of sporadic breast cancer. A thorough understanding of the events that underlie this devastating disease will result in better outcomes for those women unfortunate enough to develop this malignancy.
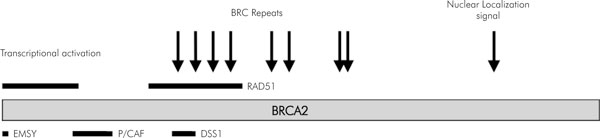
Figure 2.
Schematic representation of the various binding sites on BRCA2 protein.
References
- Kinzler KW, Vogelstein B. Cancer-susceptibility genes. Gatekeepers and caretakers. Nature. 1997;386:761–762. doi: 10.1038/386761a0. [DOI] [PubMed] [Google Scholar]
- Levitt NC, Hickson ID. Caretaker tumour suppressor genes that defend genome integrity. Trends in Mol Med. 2002;8:179–186. doi: 10.1016/S1471-4914(02)02298-0. [DOI] [PubMed] [Google Scholar]
- Thompson LH, Schild D. Recombination DNA repair and human disease. Mutat Res. 2002;509:49–78. doi: 10.1016/s0027-5107(02)00224-5. [DOI] [PubMed] [Google Scholar]
- Peltomaki P, Aaltonen LA, Sistonen P, Pylkkanen L, Mecklin JP, Jarvinen H, Green JS, Jass JR, Weber JL, Leach FS, Petersen GM, Hamilton SR, de la Chapelle A, Vogelstein B. Genetic mapping of a locus predisposing to human colorectal cancer. Science. 1993;260:810–812. doi: 10.1126/science.8484120. [DOI] [PubMed] [Google Scholar]
- Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W, Bell R, Rosenthal J, Hussey Ch, Tran T, McClure M, Frye Ch, Hattier T, Phelps R, Haugen-Strano A, Katcher H, Yakumo K, Gholami Z, Shaffer D, Stone S, Bayer S, Wray Ch, Bogden R, Dayananth P, Ward J, Tonin P, Narod S, Bristow PK, Norris FH, Helvering L, Morrison P, Rosteck P, Lai M, Barrett JC, Lewis C, Neuhausen S, Cannon-Albright L, Goldgar D, Wiseman R, Kamb A, Skolnick MH. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- Scully R, Chen J, Plug A, Xiao Y, Weaver D, Feunteun J, Ashley T, Livingston DM. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell. 1997;88(2):265–275. doi: 10.1016/S0092-8674(00)81847-4. [DOI] [PubMed] [Google Scholar]
- Bosch M van den, Lohman PH, Pastink A. DNA double-strand break repair by homologous recombination. Biol Chem. 2002;383:873–892. doi: 10.1515/BC.2002.095. [DOI] [PubMed] [Google Scholar]
- Khanna KK, Jackson SP. DNA double strand breaks: signalling, repair and the cancer connection. Nat Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- Thompson LH, Schild D. Homologous recombination repair of DNA ensures mammalian chromosome stability. Mutat Res. 2001;477:131–153. doi: 10.1016/s0027-5107(01)00115-4. [DOI] [PubMed] [Google Scholar]
- Liu N, Lammerdin JE, Tebbs RS, Schild D, Tucker JD, Shen MR, Brookman KW, Siciliano MJ, Walter CA, Fan W, Narayana LS, Zhou ZQ, Adamson AW, Sorenson KJ, Chen DJ, Kones NJ, Thompson LH. XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA crosslinks and other damage. Mol Cell. 1998;1:783–793. doi: 10.1016/S1097-2765(00)80078-7. [DOI] [PubMed] [Google Scholar]
- Howlett NG, Taniguchi T, Olson S, Cox B, Waisfisz Q, de Die-Smulders C, Persky N, Grompe M, Joenje H, Pals G, Ikeda H, Fox EA, D'Andrea AD. Biallelic inactivation of BRCA2 in Fanconi Anemia. Science. 2002;297:606–609. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- Easton DF, Bishop DT, Ford D, Crockford GP. The Breast Cancer Linkage Consortium. Genetic linkage analysis in familial breast and ovarian cancer: results from 214 families. Am J Hum Genet. 1993;52:678–701. [PMC free article] [PubMed] [Google Scholar]
- Powell SN, Kachnic LA. Roles of BRCA1 and BRCA2 in homologous recombination, DNA replication fidelity and the cellular response to ionizing radiation. Oncogene. 2003;22:5784–5791. doi: 10.1038/sj.onc.1206678. [DOI] [PubMed] [Google Scholar]
- Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 2002;108:171–182. doi: 10.1016/S0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- Dasika GK, Lin SC, Zhao S, Sung P, Tomkinson A, Lee EY. DNA damage-induced cell cycle checkpoints and DNA strand break repair in development and tumorigenesis. Oncogene. 1999;18:7883–7899. doi: 10.1038/sj.onc.1203283. [DOI] [PubMed] [Google Scholar]
- Bernstein C, Bernstein H, Payne CM, Garewal H. DNA repair, pro-apoptotic pathway, dual role proteins in five major DNA repair pathways: fail-safe protection against carcinogenesis. Mutat Res. 2002;511:145–178. doi: 10.1016/S1383-5742(02)00009-1. [DOI] [PubMed] [Google Scholar]
- Paull TT, Cortez D, Bowers B, Elledge SJ, Gellert M. Direct DNA binding by BRCA1. Proc Natl Acad Sci USA. 2001;98:6086–6091. doi: 10.1073/pnas.111125998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman AR, Ford JM. BRCA1 induces DNA damage recognition factors and enhances nucleotide excision repair. Nat Genet. 2002;32:180–184. doi: 10.1038/ng953. [DOI] [PubMed] [Google Scholar]
- Xu B, Kim S-T, Kastan MB. Involvement of Brca1 in S-Phase and G2-Phase. Checkpoints after Ionizing Irradiation. Mol Cell Biol. 2001;21:3445–3450. doi: 10.1128/MCB.21.10.3445-3450.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochar DA, Wang L, Beniya H, Kinev A, Xue Y, Lane WS, Wang W, Kashanchi F, Shiekhattar R. BRCA1 is associated with a human SWI/SNF-related complex: linking chromatin remodeling to breast cancer. Cell. 2000;102:257–265. doi: 10.1016/S0092-8674(00)00030-1. [DOI] [PubMed] [Google Scholar]
- Ganesan S, Silver DP, Greenberg RA, Avni D, Drapkin R, Miron A, Mok SC, Randrianarison V, Brodie S, Salstrom J, Rasmussen TP, Marrese C, Marahrens Y, Deng C-X, Feunteun J, Livingston DM. BRCA1 supports XIST RNA concentration on the inactive X chromosome. Cell. 2002;111:393–405. doi: 10.1016/S0092-8674(02)01052-8. [DOI] [PubMed] [Google Scholar]
- Walter J, Newport J. Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Mol Cell. 2000;5:617–627. doi: 10.1016/S1097-2765(00)80241-5. [DOI] [PubMed] [Google Scholar]
- Choudhary SK, Li R. BRCA1 modulates ionizing radiation-induced nuclear focus formation by the replication protein A p34 subunit. J Cell Biochem. 2002;84:666–674. doi: 10.1002/jcb.10081. [DOI] [PubMed] [Google Scholar]
- Wang Y, Cortez D, Yazdi P, Neff N, Elledge SJ, Qin J. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 2000;14(8):927–939. doi: 10.1101/gad.827000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D, Wang Y, Qin J, Elledge SJ. Requirement of ATM-dependent phosphorylation of BRCA1 in the DNA damage response to double stranded breaks. Science. 1999;286:1162–1166. doi: 10.1126/science.286.5442.1162. [DOI] [PubMed] [Google Scholar]
- Xu X, Weaver Z, Linke SP, Li C, Gotay J, Wang XW, Harris CC, Ried T, Deng CX. Centrosome amplification and a defective G2-M cell cycle checkpoint induce genetic instability in BRCA1 exon 11 isoform-deficient cells. Mol Cell. 1999;3:389–395. doi: 10.1016/S1097-2765(00)80466-9. [DOI] [PubMed] [Google Scholar]
- Yarden RI, Pardo-Reoyo S, Sgagias M, Cowan KH, Brody LC. BRCA1 regulates the G2/M checkpoint by activating Chk1 kinase upon DNA damage. Nat Genet. 2002;30:285–289. doi: 10.1038/ng837. [DOI] [PubMed] [Google Scholar]
- Hsu LC, Doan TP, White RL. Identification of a gamma-tubulin-binding domain in BRCA1. Cancer Res. 2001;61:7713–7718. [PubMed] [Google Scholar]
- Somasundaram K, Zhang H, Zeng YX, Houvras Y, Peng Y, Zhang H, Wu GS, Licht JD, Weber BL, El-Deiry WS. Arrest of the cell cycle by the tumour-suppressor BRCA1 requires the CDK-inhibitor p21WAF1/CiP1. Nature. 1997;389:187–190. doi: 10.1038/38291. [DOI] [PubMed] [Google Scholar]
- Mullan PB, Quinn JE, Gilmore PM, McWilliams S, Andrews H, Gervin C, McCabe N, McKenna S, White P, Song YH, Maheswaran S, Liu E, Haber DA, Johnston PG, Harkin DP. BRCA1 and GADD45 mediated G2/M cell cycle arrest in response to antimicrotubule agents. Oncogene. 2001;20:6123–31. doi: 10.1038/sj.onc.1204712. [DOI] [PubMed] [Google Scholar]
- Deming PB, Cistulli CA, Zhao H, Graves PR, Piwnica-Worms H, Paules RS, Downes CS, Kaufmann WK. The human decatenation checkpoint. Proc Natl Acad Sci USA. 2001;98:12044–12049. doi: 10.1073/pnas.221430898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiman FE, Manley JL. The BARD1-CstF-50 interaction links mRNA 3' end formation to DNA damage and tumor suppression. Cell. 2001;104:743–753. doi: 10.1016/S0092-8674(01)00270-7. [DOI] [PubMed] [Google Scholar]
- Hu Y-F, Hao Z, Li R. Chromatin remodeling and activation of DNA replication by the BRCT-domain of BRCA1. Genes Dev. 1999;13:637–642. doi: 10.1101/gad.13.6.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongusaha PP, Ouchi T, Kim KT, Nytko E, Kwak JC, Duda RB, Deng CX, Lee SW. BRCA1 shifts p53-mediated cellular outcomes towards irreversible growth arrest. Oncogene. 2003;22:3749–3758. doi: 10.1038/sj.onc.1206439. [DOI] [PubMed] [Google Scholar]
- Sum EY, Peng B, Yu X, Chen J, Byrne J, Lindeman GJ, Visvader JE. The LIM domain protein LMO4 interacts with the cofactor CtIP and the tumor suppressor BRCA1 and inhibits BRCA1 activity. J Biol Chem. 2002;277:7849–7856. doi: 10.1074/jbc.M110603200. [DOI] [PubMed] [Google Scholar]
- Zheng L, Pan H, Li S, Flesken-Nikitin A, Chen PL, Boyer TG, Lee WH. Sequence-specific transcriptional corepressor function for BRCA1 through a novel zinc finger protein, ZBRK1. Mol Cell. 2000;6:757–768. doi: 10.1016/S1097-2765(00)00075-7. [DOI] [PubMed] [Google Scholar]
- Ganesan S, Silver DP, Greenberg RA, Avni D, Drapkin R, Miron A, Mok SC, Randrianarison V, Brodie S, Salstrom J, Rasmussen TP, Marrese C, Marahrens Y, Deng C-X, Feunteun J, Livingston DM. BRCA1 supports XIST RNA concentration on the inactive X chromosome. Cell. 2002;111:393–405. doi: 10.1016/S0092-8674(02)01052-8. [DOI] [PubMed] [Google Scholar]
- Scully R. Role of BRCA gene dysfunction in breast and ovarian cancer predisposition. Breast Cancer Res. 2000;2:324–330. doi: 10.1186/bcr76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies AA, Masson JY, McIlwraith MJ, Stasiak AZ, Stasiak A, Venkitaraman AR, West SC. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol Cell. 2001;7:273–282. doi: 10.1016/S1097-2765(01)00175-7. [DOI] [PubMed] [Google Scholar]
- Wang S-C, Shao R, Pao AY, Zhang S, Hung M-C, Su L-K. Inhibition of cancer cell growth by BRCA2. Cancer Res. 2002;62:1311–1314. [PubMed] [Google Scholar]
- Vaughn JP, Cirisano FD, Huper G, Berchuck A, Futreal PA, Marks JR, Iglehart JD. Cell cycle control of BRCA2. Cancer Res. 1996;56:4590–4594. [PubMed] [Google Scholar]
- Orelli BJ, Bishop DK. BRCA2 and homologous recombination. Breast Cancer Res. 2001;3:294–298. doi: 10.1186/bcr310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraakman-van der Zwet M, Overkamp WJ, van Lange RE, Essers J, van Duijn-Goedhart A, Wiggers I, Swaminathan S, van Buul PP, Errami A, Tan RT, Jaspers NG, Sharan SK, Kanaar R, Zdzienicka MZ. Brca2 (XRCC11) deficiency results in radioresistant DNA synthesis and a higher frequency of spontaneous deletions. Mol Cell Biol. 2002;22(2):669–679. doi: 10.1128/MCB.22.2.669-679.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tutt A, Gabriel A, Bertwistle D, Connor F, Paterson H, Peacock J, Ross G, Ashworth A. Absence of Brca2 causes genome instability by chromosome breakage and loss associated with centrosome amplification. Curr Biol. 1999;9:1107–1110. doi: 10.1016/S0960-9822(99)80479-5. [DOI] [PubMed] [Google Scholar]
- Xia F, Taghian DG, DeFrank JS, Zeng Z-C, Willers H, Iliakis G, Powell SN. Deficiency of human BRCA2 leads to impaired homologous recombination but maintains normal nonhomologous end joining. Proc Natl Acad Sci. 2001;98:8644–8649. doi: 10.1073/pnas.151253498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tutt A, Bertwistle D, Valentine J, Gabriel A, Swift S, Ross G, Griffin C, Thacker J, Ashworth A. Mutation in Brca2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. EMBO J. 2001;20:4704–4716. doi: 10.1093/emboj/20.17.4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell. 2001;7:263–272. doi: 10.1016/S1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- Chen C-F, Chen P-L, Zhong Q, Sharp ZD, Lee W-H. Expression of BRC repeats in breast cancer cells disrupts the BRCA2-Rad51 complex and leads to radiation hypersensitivity and loss of G2/M checkpoint control. J Biol Chem. 1999;274:32931–32935. doi: 10.1074/jbc.274.46.32931. [DOI] [PubMed] [Google Scholar]
- Kordon EC, Smith GH. An entire functional mammary gland may comprise the progeny of a single cell. Development. 1998;125:1921–1930. doi: 10.1242/dev.125.10.1921. [DOI] [PubMed] [Google Scholar]
- Chapke G, Smith GH. Mammary epithelial stem cells: our current understanding. J Mammary Gland Biol Neoplasia. 1999;4:35–52. doi: 10.1023/A:1018752519356. [DOI] [PubMed] [Google Scholar]
- Tsai YC, Lu Y, Nichols PW, Zlotnikov G, Jones PA, Smith HS. Contiguous patches of normal human mammary epithelium derived from a single stem cell. Cancer Res. 1996;56:402–404. [PubMed] [Google Scholar]
- Scully R, Puget N. BRCA1 and BRCA2 in hereditary breast cancer. Biochimie. 2002;84:95–102. doi: 10.1016/S0300-9084(01)01359-1. [DOI] [PubMed] [Google Scholar]
- Fan S, Wang J, Yuan R, Ma Y, Meng Q, Erdos MR, Pestell RG, Yuan F, Auborn KJ, Goldberg ID, Rosen EM. BRCA1 inhibition of estrogen receptor signaling in transfected cells. Science. 1999;284:1354–1356. doi: 10.1126/science.284.5418.1354. [DOI] [PubMed] [Google Scholar]
- Ferguson AT, Davidson NE. Regulation of estrogen receptor alpha function in breast cancer. Crit Rev Oncog. 1997;8:29–46. doi: 10.1615/critrevoncog.v8.i1.20. [DOI] [PubMed] [Google Scholar]
- Scully R, Livingston DM. In search of the tumour-suppressor functions of BRCA1 and BRCA2. Nature. 2000;408:429–432. doi: 10.1038/35044000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett LM, McAllister KA, Malphurs J, Ward T, Collins NK, Seely JC, Gowen LC, Koller BH, Davis BJ, Wiseman RW. Mice heterozygous for a Brca1 or Brca2 mutation display distinct mammary gland and ovarian phenotypes in response to diethylstilbestrol. Cancer Res. 2000;60:3461–3469. [PubMed] [Google Scholar]
- Hughes-Davies L, Huntsman D, Ruas M, Fuks F, Bye J, Chin S-F, Milner J, Brown LA, Hsu F, Gilks B, Nielsen T, Schulzer M, chia S, Ragaz J, Cahn A, Linger L, Ozdag H, Cattaneo E, Joradnova ES, Schuuring E, Yu DS, Venkitaraman A, Ponder B, Doherty A, Aparicio S, Bentley D, Theillet C, Ponting CP, Caldas C, Kouzarides T. EMSY links the BRCA2 pathway to sporadic breast and ovarian cancer. Cell. 2003;115:523–535. doi: 10.1016/S0092-8674(03)00930-9. [DOI] [PubMed] [Google Scholar]