Abstract
In cultured hippocampal neurons and in adult brain, the splicing regulatory protein Sam68 is partially relocated to the somatodendritic domain, and associates with dendritic polysomes. Transfer to the dendrites is activity-dependent. We have investigated the repertoire of neuronal mRNAs to which Sam68 binds in vivo. Using co-immunoprecipitation and micro-array screening techniques, Sam68 was found to associate with a number of plasticity-related mRNA species, including Eef1a1, an activity-responsive mRNA coding for translation elongation factor eEF1A. In cortical neuronal cultures, translation of the Eef1a1 mRNA was strongly induced by neuronal depolarisation, and correlated with enhanced association of Sam68 with polysomal mRNAs. The possible function of Sam68 in Eef1a1 mRNA utilization was studied by expressing a dominant-negative, cytoplasmic Sam68 mutant (GFP-Sam68ΔC) in cultured hippocampal neurons. The level of eEF1A was lower in neurons expressing GFP-Sam68ΔC than in control neurons, supporting the proposal that endogenous Sam68 may contribute to the translational efficiency of the Eef1a1 mRNA. These findings are discussed in the light of the complex, potentially crucial regulation of eEF1A biosynthesis during longterm synaptic change.
Keywords: Adaptor Proteins, Signal Transducing; genetics; metabolism; Animals; Cells, Cultured; DNA-Binding Proteins; genetics; metabolism; Embryo, Mammalian; Gene Expression Regulation; physiology; Hippocampus; cytology; Humans; Immunoprecipitation; methods; Neurons; metabolism; Peptide Elongation Factor 1; biosynthesis; Protein Binding; physiology; Protein Biosynthesis; RNA, Messenger; metabolism; RNA-Binding Proteins; genetics; metabolism; Rats; Receptors, N-Methyl-D-Aspartate; metabolism; Transfection; methods
Keywords: Dendritic mRNA, activity-dependent translation, RNA-binding protein, plasticity
Introduction
The translation of neuronal mRNAs is regulated by their association with specific RNA-binding proteins, of which an increasing variety have been identified (Bramham and Wells 2007; Kiebler and Bassell 2006). In the well-studied cases of CPEB, FMRP and ZBP-1, the proteins maintain the target mRNAs in a dormant state during transport or storage; this inhibitory function is thought to be suppressed by activity-dependent phosphorylation events, e.g. in the vicinity of synapses (Bagni and Greenough 2005; Huang et al. 2002; Huttelmaier et al. 2005). In addition to these predominantly cytoplasmic proteins, recent evidence from non-neuronal systems indicates that nuclear proteins of the splicing machinery may also contribute to the control of translation. For example, SR splicing factors continuously shuttle between nucleus and cytoplasm, where they can bind to mature mRNAs and promote translation (Sanford et al. 2004). Translational efficiency can also be increased by splicing proteins which are deposited onto RNA substrates during the splicing process itself, and which remain bound to the mRNA after nuclear processing and export have been completed (Kunz et al. 2006; Le Hir et al. 2000; Wiegand et al. 2003). Interestingly, a diversity of splicing factors have been found to exist in neuronal dendrites (Glanzer et al. 2005). One of those is Sam68, a ubiquitous RNA-binding protein implicated in signal-dependent splice site selection, and which can also function as an adaptor and substrate for Src family kinases (Lukong and Richard 2003; Matter et al. 2002; Paronetto et al. 2007). In cultured cell lines, Sam68 is a nuclear, non-shuttling protein; however, it is required to enhance the nuclear export and/or translation of certain retroviral mRNAs, indicating a potential interaction with translation-activating factors (McLaren et al. 2004; Modem et al. 2005). Beside viral mRNAs, Sam68 can bind to a select population of mature cellular mRNAs derived from cultured HeLa cells (Itoh et al. 2002); the functional outcome of these binding interactions has not been determined.
We previously reported that Sam68 was among the proteins associated with neuronal mRNAs (Ben Fredj et al. 2004; Grange et al. 2004). In adult cortical and hippocampal tissue, biochemical fractionation and immuno-electron microscopy indicated that Sam68 molecules were associated with dendritic polysomes, suggesting a possible positive role in translational regulation (Grange et al. 2004; Khandjian et al. 2004). In cultured hippocampal neurons, a minor but detectable fraction of endogenous or GFP-tagged Sam68 molecules were found to occur in the soma and dendrites, concentrated in generally immobile, RNA-rich granules. Following prolonged membrane depolarisation by elevated KCl, the proportion of Sam68 in the cytoplasm was strongly increased, due to massive migration of the protein to these somatodendritic sites (Ben Fredj et al. 2004). These results suggest that Sam68 may play a part in the regulation of those mRNAs which become translationally activated in response to electrical activity. To test this hypothesis, we have investigated the identity of mRNA species that were bound to Sam68 in cortical neurons in vivo. We found that one major target was Eef1a1, an mRNA known to be activated during long-term synaptic change in the hippocampus. In cultured neurons, depolarisation enhanced the association of both Sam68 and the Eef1a1 mRNA with polyribosomes, and increased the amount of its product, the eEF1A protein. Expression of a dominant-negative Sam68 mutant confined to the cytoplasm was able to lower the amount of eEF1A, supporting the proposal that Sam68 promotes the translation of neuronal Eef1a1.
Materials and Methods
Immunoprecipitation of Sam68-associated RNA and preparation of amplified RNA probes
Cortical Sam68-RNA complexes were prepared as previously described (Grange et al. 2004) Briefly, the cortex from one adult rat was dissected and lysed at 4°C in polysome dissociation buffer [10 mM Tris-HCl, pH 7.5, 50 mM KCl, 1 mM sodium orthovanadate, 2 mM dithiothreitol (DTT), 100 nM microcystine-LR (Sigma), protease inhibitors (Complete; Roche Molecular)] containing 0.5% Triton X-100, 160 mM sucrose, 30 mM EDTA, pH 8.0 and 400 U/ml RNAsin (Promega, Inc., Madison, WI). After clarification by centrifugation at 82,000g for 20 min at 4°C, the lysate was precleared with protein G–Sepharose beads (GE Healthcare), then immunoprecipitated with affinity-purified anti-Sam68 IgG (AD-1), or with preimmune protein A-purified IgG from the same rabbit. Immune complexes were collected onto protein G –Sepharose beads, washed, and processed for RNA purification using the “RNEasy” kit (Qiagen France, Courtaboeuf). The immunoprecipitated polyA+ RNAs were linearly amplified by cDNA synthesis and T7 polymerase transcription (Gelder et al. 1990), using the MessageAmp™ aRNA Kit (Ambion). According to whether the template RNA originated in the specific immunoprecipitate or the preimmune control, either Cyanine 3™ (Cy3) or Cyanine 5™ (Cy5)-labeled UTP was incorporated during transcript synthesis, as per manufacturer’s instructions. Transcripts amplified from the specific immunoprecipitate and the corresponding control were mixed in equal amounts and used to probe the 15000 cDNAs arrayed on the NIA micro-array (Tanaka et al. 2000). To control for possible biasing by differential incorporation of Cy3 and Cy5, the experiment was repeated with swapping the fluorophores between “specific” and “background” transcripts.
Quantitation of hybridization
After hybridization and washing, digital images of the micro-array at 635 nm and 532 nm were acquired by laser scanning confocal microscopy. Fluorescent spots were delineated and quantitated with GenePix software (Axon Instruments). Raw fluorescence values (F) in each channel were converted into their Log2. After Cy3 labelling of background RNAs, immunoprecipitation of any given mRNA was considered specific when the value in the Cy5 channel exceeded that in the Cy3 channel by at least 4 Log2 units (a 16-fold difference). When background RNAs were labelled with Cy5, the cut-off was brought down to 0.5 Log2 unit (an 1.4-fold difference). This differential cut-off was applied to compensate for the systematic bias in labelling efficiency in favour of Cy5. Note that the requirement for higher Cy3 labelling of specifically immunoprecipitated RNAs is conservative and likely excludes a number of Sam68-associated mRNAs. Z scores were calculated as (Log2F − <Log2F>)/σLog2F, where <Log2F> is the mean and σLog2F is the standard deviation, both calculated over the entire population of fluorescence values in the relevant channel (Mayne et al. 2001). Reproducibility of the overall population of z scores was measured by simple linear regression between experiments. 95% confidence intervals were calculated for the z scores of Table Ia using t statistics (n=3), and individual mRNAs with lower confidence limit zmin < - 1.0 were rejected for inclusion in the Table; more than 80% of these scores had lower limit above zero. Frequencies of GO descriptors were computed from ~4000 functionally described genes of the microarray (available through http://lgsun.grc.nia.nih.gov/cDNA/NIA-CloneSet-GoTermFinder.html).
Table 1.
| Table Ia. Identity and generic function of major Sam68-associated mRNAs. ID: Genbank identifier of cloned sequence on the NIA 15 K micro-array. Name: name of transcript homologous to the sequence on the array (available through http://lgsun.grc.nia.nih.gov/cDNA/15k.html). Mean z score and its standard deviation (n=3) were calculated for each transcript as described in the text. P values were calculated from z scores, assuming normal distribution of scores. Biological process or component associated with transcripts were identified by reporting Gene Ontology (GO) descriptors used to annotate the transcripts in the NIA database (available through http://lgsun.grc.nia.nih.gov/cDNA/NIA-CloneSet-GoTermFinder.html). Rightmost column: identification of the GO descriptors by their numbering in the Gene Ontology database. | |||||
|---|---|---|---|---|---|
| ID | Name | Mean z ± S.D. | p value | Process or component | GO |
| AW557547 | Translation elongation factor 1 alpha 1, Eef1a1 | 2.66 ± 0.97 | 0.004 | protein biosynthesis | 06412 |
| BG086025 | ATPase, H+ transporting, V1 subunit B, isoform 2, Atp6v1b2 | 2.55 ± 1.07 | 0.005 | proton transport | 15992 |
| BG076172 | Calmodulin-1, Calm1 | 2.36 ± 1.11 | 0.009 | signal transduction | 07165 |
| BG088462 | Zfhx1a, C2-H2 zinc finger protein homeobox 1a | 2.35 ± 1.29 | 0.009 | regulation of transcription | 06355 |
| AW558451 | Reticulon 3, Rtn3 | 2.28 ± 0.60 | 0.011 | protein transport | 15031 |
| BG088310 | Prosaposin | 2.1 ± 0.88 | 0.018 | metabolism | 08152 |
| BG071424 | BRI3 | 2.04 ± 0.28 | 0.021 | integral to membrane | 16021 |
| BG073409 | Stathmin 1, Stmn1 | 1.98 ± 0.48 | 0.024 | signal transduction | 07165 |
| BG065012 | 14-3-3 eta, Ywhah | 1.86 ± 0.82 | 0.031 | signal transduction | 07165 |
| BG075901 | Gamma-aminobutyric acid receptor subunit, Gabra3 | 1.86 ± 0.36 | 0.031 | synaptic transmission | 07268 |
| BG076067 | Inositol hexaphosphate kinase 1 | 1.86 ± 0.50 | 0.032 | metabolism | 08152 |
| BG085717 | 14-3-3 epsilon, Ywhae | 1.85 ± 0.84 | 0.032 | signal transduction | 07165 |
| BG082675 | ADP-ribosylation factor 1, Arf-1 | 1.85 ± 0.90 | 0.032 | signal transduction | 07165 |
| BG075782 | Protein phosphatase 1, catalytic subunit, gamma, Ppp1cc | 1.83 ± 0.76 | 0.034 | signal transduction | 07165 |
| BG075243 | Zinc transporter ZnT-3 | 1.8 ± 0.27 | 0.036 | cation transport | 06812 |
| BG077677 | Beta-actin | 1.78 ± 0.53 | 0.037 | cytoskeleton | 07010 |
| BG065113 | Branched chain aminotransferase, bcat-1 | 1.78 ± 1.51 | 0.038 | metabolism | 08152 |
| BG064914 | Malate dehydrogenase, Mor2 | 1.73 ± 0.61 | 0.042 | metabolism | 08152 |
| BG087365 | Neuroplastin | 1.69 ± 0.86 | 0.046 | surface receptor signal transduction | 07166 |
| BG073370 | Tetratricopeptide repeat domain 3, Ttc3 | 1.69 ± 1.29 | 0.046 | protein binding | 05515 |
| BG074109 | Heat shock protein 1, alpha, Hspca | 1.67 ± 1.15 | 0.047 | chaperone activity | 03754 |
| BG088451 | Tissue inhibitor of metalloproteinase 2, Timp2 | 1.61 ± 0.77 | 0.053 | extracellular matrix | 05578 |
| BG075319 | Mitogen activated protein kinase 3, pp44 MAPK, Erk1 | 1.61 ± 0.57 | 0.054 | signal transduction | 07165 |
| BG074398 | Matrix glycoprotein Sparcl1/hevin | 1.54 ± 0.64 | 0.062 | extracellular matrix | 05578 |
| BG073255 | Tetraspanin TM4-A | 1.5 ± 0.86 | 0.066 | integral to membrane | 16021 |
| BG082737 | Proline-arginine-rich end leucine-rich repeat protein, Prelp | 1.46 ± 0.64 | 0.073 | extracellular matrix | 05578 |
| BG073185 | BTB (POZ) domain containing 4, KBTBD4 | 1.41 ± 1.05 | 0.079 | regulation of transcription | 06355 |
| BG088334 | Vacuolar protein sorting 41 | 1.4 ± 0.05 | 0.081 | protein transport | 15031 |
| BG087861 | Dynein, cytoplasmic, intermediate chain 1, Dncic1 | 1.39 ± 0.39 | 0.082 | microtubule-based movement | 07018 |
| BG085988 | Heterogeneous nuclear ribonucleoprotein H2, Hnrph2 | 1.35 ± 0.91 | 0.089 | mRNA processing | 06397 |
| BG073339 | Splicing factor, arginine/serine-rich 5, Srp40/HRS | 1.31 ± 0.72 | 0.096 | mRNA processing | 06397 |
| AW553322 | G protein-coupled receptor associated sorting protein 1 | 1.27 ± 0.86 | 0.102 | protein transport | 15031 |
| BG073253 | Rab11B | 1.23 ± 1.25 | 0.109 | protein transport | 15031 |
| BG086823 | Myeloid leukemia factor 2, parathymosin | 1.23 ± 0.78 | 0.109 | immune response | 06955 |
| BG072963 | Dynein heavy chain | 1.23 ± 0.21 | 0.110 | microtubule-based movement | 07018 |
| BG086722 | Ubiquitin carboxyl-terminal esterase L1, UCH-L1 | 1.16 ± 0.98 | 0.122 | ubiquitin-dependent protein catabolism | 06511 |
| BG086087 | Fanconi anemia-associated protein of 100 kDa, Faap100 | 1.11 ± 1.31 | 0.134 | response to DNA damage | 06974 |
| BG086010 | Dynein intermediate chain 2, Dync1i2 | 1.11 ± 0.12 | 0.134 | microtubule-based movement | 07018 |
| BG088587 | Beclin 1 (coiled-coil, myosin-like BCL2-interacting protein) | 1.09 ± 0.43 | 0.137 | autophagy | 06914 |
| BG080018 | Nuclear protein Nulp1/bHLH Tcf25 | 0.99 ± 0.72 | 0.161 | regulation of transcription | 06355 |
| BG088909 | Transducin-like enhancer of split 6, TLE6/Grg6 | 0.99 ± 1.01 | 0.162 | regulation of transcription | 06355 |
| BG076556 | Farnesyltransferase beta subunit | 0.98 ± 1.10 | 0.162 | regulation of cell proliferation | 42127 |
| BG069542 | hnRNP-E1, poly(rC)-binding protein 1 | 0.98 ± 1.38 | 0.163 | mRNA processing | 06397 |
| Table Ib Sam68-associated transcripts with documented neurobiological function. | |||||
|---|---|---|---|---|---|
| Transcripts involved in synaptic plasticity | Published evidence | ||||
| Translation elongation factor 1 alpha 1, Eef1a1 | Tsokas et al. 2005, Huang et al. 2005 | ||||
| ATPase, H+ transporting, V1 subunit B2, Atp6v1b2 | Munton et al. 2007 | ||||
| Calmodulin-1, Calm1 | Xia and Storm 2005 | ||||
| 14-3-3 eta, Ywhah | Simsek-Duran et al. 2004 | ||||
| Protein phosphatase 1, catalytic subunit, gamma, Ppp1cc | Hu et al. 2006 | ||||
| Beta-actin | Eom et al. 2003 | ||||
| Mitogen activated protein kinase 3, pp44 MAPK, Erk1 | Thomas and Huganir 2004 | ||||
| Transcripts involved in neurodegeneration | Published evidence | ||||
| 14-3-3 epsilon, Ywhae | Toyo-oka et al. 2003 | ||||
| Tetratricopeptide repeat domain 3, Ttc3 | Berto et al. 2007 | ||||
| Reticulon 3, Rtn3 | He et al. 2004 | ||||
| Prosaposin | Matsuda et al. 2004 | ||||
| Bri3 | Wickham et al. 2005 | ||||
| Ubiquitin carboxyl-terminal esterase L1, Uch-L1 | Liu et al. 2002, Gong et al. 2006 | ||||
| Beclin-1 | Pacheco et al. 2007 | ||||
| Transcript involved in psychiatric disease | Published evidence | ||||
| Gamma-aminobutyric acid receptor, subunit Gabra3 | Massat et al. 2002 | ||||
Polymerase chain reactions
Immunoprecipitated or total RNA was reverse-transcribed with the universal primer GGAATTC(T)17, and the resulting cDNA was amplified by using the gene-specific primer pairs listed in Table II. For classical PCR, after 2 min at 95°C, 30 cycles were performed (15 s at 95°C, 30 s annealing, and 30 s at 72°C for each cycle). For quantitative PCR by real-time fluorometry (Light Cycler, Roche Molecular Systems), DNA was denatured at 95°C for 5 min, then subjected to ~35–40 amplification cycles (15 s at 95°C, 5 s annealing, 15 s at 72°C, all temperature changes at 20°C/s). Annealing took place at 55°C except for Arc (57°C). The relative amounts of specific DNA template were determined by counting the number of cycles needed to reach the inflection point (“crossing point” at which d2[DNA]/dt2=0) of the DNA accumulation curve (typically between 25 and 32), and standardized by relating to a scale generated with known amounts of total cDNA. The specificity of the amplification product was confirmed by checking its melting curve with the Light Cycler, and its size on an agarose gel.
Table II.
PCR primers used in this study.
| Transcript | Primers |
|---|---|
| Arc | Forward: 5′-TGTTGGTAACTGCTCGTGTCTGTAG-3′ |
| Reverse: 5′-TTTGAGGTAAGATGGTGTGGGC-3′ | |
| β-Actin | Forward: 5′-TGGCACCACACTTTCTACAATGAG-3′ |
| Reverse: 5′-AGGCATACAGGGACAACACAGC-3′ | |
| Calm-1 | Forward: 5′-TCTTCAAGTGCCCCAATCCC-3′ |
| Reverse: 5′-TCAAGTCCACAGACACAGCCTACTC-3′ | |
| Eef1a1 | Forward:5′-GGACAGCAAAAATGACCCACC-3′ |
| Reverse: 5′-TTGATGACACCCACAGCAACTG -3′ | |
| Gephyrin | Forward: 5′-AAAGAGGGGAGTGTGTTTTGGC-3′ |
| Reverse: 5′-AAGGCATTGAGTAAGTCATCTGGG-3′ | |
| NR1-1 | Forward: 5′-CGTATCCTAGGCATGGTGTGG-3′ |
| Reverse: 5′-CCCGGTGCTCGTGTCTTTGG-3′ | |
| β-Tubulin | Forward: 5′-CAGACCAGACAACTTCGTTTTCG-3′ |
| Reverse: 5′-GTATCAGACACTTTGGGCGAGG-3′ | |
| Uch-L1 | Forward: 5′-TGCTGCTGCTGTTTCCCCTC-3′ |
| Reverse: 5′-CCATCCTCAAATCCCAGGTTG-3′ |
Immunoprecipitation of RNA-mycSam68 complex from HEK cells
HEK-293 cells were grown in Dulbecco’s modified Minimum Essential Medium, containing 10% fetal calf serum (Invitrogen). The myc-Sam68 expressing plasmid was a kind gift from I. Barlat and B. Tocqué (at the time at Aventis Pharma, Vitry, France). One 6 cm dish of cells was transfected with either myc-Sam68 or with control vector (containing a CMV promoter). 48 h later, cells were lysed in polysome dissociation buffer and, after clarifying by centrifugation, the lysate was immunoprecipitated with 2 μg anti-myc monoclonal antibody (12CA5, Santa Cruz) and protein G-Sepharose beads, as described for brain lysates. Total RNA was isolated and amplified by RT-PCR as decribed above.
Neuronal cultures and transfections
Primary cultures of rat hippocampal neurons were prepared from fetal (E19) hippocampi and seeded at a density of 6000 cells/cm2 onto poly-L-lysine-coated glass coverslips (Marienfeld, Germany), adhering to the procedure described by Goslin and Banker (Banker et al. 1998), except that the culture medium was Neurobasal with B27 supplement (Invitrogen). Housing and handling of animals complied with the regulations issued by the European Union and INSERM and were approved by the local veterinary authorities. The GFP-Sam68 (WT) and GFP-Sam68ΔC plasmids were kind gifts from S. Richard (McGill University, Montréal, Canada). The mRFP1 plasmid (expressing monomeric red fluorescent protein) was a kind gift from R.Y. Tsien (UCSD, San Diego, CA, USA). Transfection was performed at 9 DIV by calcium phosphate coprecipitation, exactly according to (Goetze et al. 2004). We used a mix of 2.5 μg GFP-Sam68 and 0.5 μg mRFP1 plasmids per coverslip. Expression of the GFPSam68 and GFP-Sam68ΔC plasmid was stable for the next 10 days. Neurons were fixed at 14–19 DIV and processed for immunofluorescent staining.
Primary cortical neurons were prepared from E16 fetal cortices, using essentially the same procedure as for hippocampal neurons. The cortical cells were seeded at a density of 105 cells/cm2 onto poly-L-lysine coated 10-cm dishes (Nunc). Neurons at 9 DIV were treated for 3 h with either depolarizing medium (containing, in mM: KCl, 25; CaCl2, 1.8; MgCl2, 0.8; NaCl, 100; NaHCO3, 26; NaH2PO4, 1; glucose, 30; Hepes-NaOH, 15) or control solution (same except for KCl, 5; NaCl, 120).
Sedimentation of polysomes on sucrose gradients
RNA-protein complexes were fractionated as described by (Krichevsky and Kosik 2001). Briefly, cortical neurons were lysed in low salt buffer [20 mM Tris-HCl (pH 7.5), 10 mM NaCl, 3 mM MgCl2, 1 mM RNasin, 1 mM dithiothreitol, 0.3% Triton X-100, and 0.05 M sucrose] containing protease inhibitors (EDTA-free Complete, Roche Molecular), 1 mM vanadate and 100 nM microcystin LR. Nuclei and the majority of mitochondria were sedimented by centrifugation for 10 min at 10,000 × g at 4°C. The NaCl and MgCl2 concentrations in the cytoplasmic extracts (supernatants) were adjusted to 170 and 13 mM, respectively. Linear sucrose gradients (15%–45% w/w in 25 mM Tris-HCl [pH 7.5], 25 mM NaCl, 5 mM MgCl2) were prepared using an AutoDensi Flow apparatus (Büchner). Cytoplasmic extracts (1.2 ml) were overlaid onto 10.6 ml gradients and centrifuged at 32,000 rpm for 2.5 hr at 4°C in a SW41 rotor. Fractions of 1 ml volume were collected with on-line UV absorbance measurement at 254 nm (GE Healthcare Akta Prime). For Western immunoblotting, the protein content of the fractions was concentrated by trichloroacetic acid precipitation, redissolved in Laemmli buffer, and analysed on 8% polyacrylamide/SDS gels. For RT-PCR, RNA was extracted by treating the relevant fractions with Trizol reagent (Invitrogen), precipitated with ethanol, and subjected to RT-PCR with the same primers as above.
Detection of eEF1A protein increase by immunoblotting
Cortical neurons were treated with control or depolarising medium as above, then lysed in icecold RIPA buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1 % Triton X- 100, 0.5 % Na deoxycholate, 0.1 % SDS) containing protease inhibitors (EDTA-free Complete, Roche Molecular). After clarification by centrifugation (15 min at 12,000 × g), equal protein amounts of each lysate were analysed by 10 % polyacrylamide/SDS gel electrophoresis and immunoblotting, first with anti-eEF1A monoclonal antibody (UBI) then with anti-actin polyclonal antibody (Santa Cruz). Immunoreactive bands were revealed by chemiluminescence (Pierce Femto) and autoluminography. The films were digitized by scanning and the integrated intensity of the bands was determined using Metamorph software (Universal Imaging). Immunoblotting and exposure conditions were calibrated so that the integrated intensity of the bands was in the linear range with respect to protein amount.
Immunofluorescence of eEF1A in hippocampal neurons
Neurons were fixed with 4% PFA containing 4% sucrose (30 min, 4°C); permeabilized with 0.2% Triton X-100; blocked with 10% preimmune goat serum in PBS (25°C, 1 h); and stained with 1 μg/ml anti-eEF1A monoclonal antibody (UBI) in PBS containing 3% goat serum (25°C, 2 h), then with Alexa350™-conjugated secondary antibody (Molecular Probes). After washing, coverslips were mounted with Mowiol and observed on a Zeiss microscope fitted with an epifluorescence setup and a cooled CCD camera (Zeiss Axiocam), or with a confocal laser scanning microscope (Zeiss LSM 510).
For quantitation of eEF1A fluorescence, fields containing transfected neurons were imaged at low magnification (20x). Imaging conditions were identical for all fields. Images were processed with Metamorph (Universal Imaging). For each image, a background image was produced with a low-pass filter and subtracted so as to remove low-frequency fluorescence noise due e.g. to local unevenness in illumination. Next, a lower fluorescence threshold was set, which allowed automatic delineation and analysis of the fluorescent objects (cells), using Integrated Morphometric Analysis. The same threshold was used throughout the entire analysis. The relative eEF1A fluorescence of each transfected neuron i was calculated according to the equation:
where eEF1Atransf.,i is the mean grey level per pixel within the transfected neuron, and <eEF1A>nontransf is the average (over 15 surrounding neurons) of the mean grey level per pixel within non-transfected neurons. These IFtransf.,i values were averaged over the entire set of transfected neurons, to yield the values of Fig. 4E.
Fig. 4.
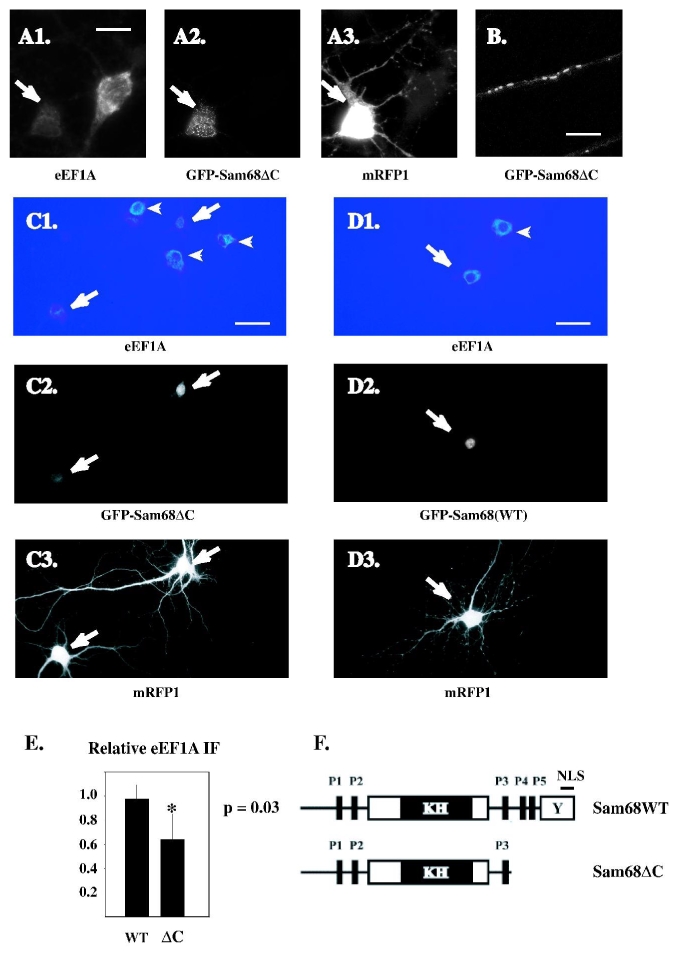
Effect of dominant-negative Sam68 on eEF1A expression in cultured hippocampal neurons.
A-C: The GFP-Sam68 (WT) and GFP-Sam68ΔC proteins were expressed in hippocampal neurons, together with monomeric Red Fluorescent Protein (mRFP) as a morphological marker. Five days later (at 14 DIV), the neurons were fixed and stained by immunofluorescence with anti-eEF1A antibody; mRFP, GFP-Sam68 and endogenous eEF1A were simultaneously visualized by triple-colour fluorescence microscopy.
A1-A3: close-up view of the three fluorescent markers in a GFP-Sam68 ΔC-transfected neuron and a nearby, untransfected neuron (wide-field microscopy). A1: Note the lack of eEF1A immunostaining in the transfected cell (arrow), compared to the neighbouring cell. Scalebar: 13 μm. B: clustered distribution of the GFP-Sam68ΔC protein within the dendrite of a hippocampal neuron (single plane of a confocal z series). Scalebar: 4.7 μm.
C1-C3: representative field showing GFP-Sam68ΔC transfected neurons and neighbouring untransfected cells at low magnification. Scalebar: 65 μm. C1: the relative intensity of eEF1A immunofluorescence is represented by pseudocolour coding. Perinuclear staining is brighter in untransfected neurons (arrowheads) than in transfected (arrows). C3: normal neuronal morphology of the transfected cells.
D1-D3: same as C1-C3 with GFP-Sam68WT transfected neuron.
E: the eEF1A immunofluorescence was measured (as mean grey level per pixel) in transfected neurons and in surrounding, nontransfected neurons of the same field. The ratio between the fluorescence of transfected neurons and the average fluorescence of surrounding nontransfected cells was then calculated for each transfected neuron. The histogram bars show the average (+/− S.D.) of these ratios for neurons transfected with GFP-Sam68ΔC and with GFP-Sam68WT. Neurons expressing GFP-Sam68ΔC had a ratio lower than 1, indicating a decrease in endogenous eEF1A. Neurons expressing GFP-Sam68WT had a ratio close to 1. The difference between GFP-Sam68ΔC and GFP-Sam68WT was significant (p=0.03, two-sided Mann-Whitney U test) (n=3 experiments).
F: schematic depiction of the wild-type and C-terminally deleted, dominant-negative Sam68 proteins. The major protein domains are shown as boxes. KH: core RNA-binding module (hnRNP-K homology domain). P1 to P5: proline-rich motifs. Y: tyrosine-rich region. NLS: nuclear localisation signal.
Immunoprecipitation of Sam68-NMDA receptor complexes
Rat cortical synaptic membranes were prepared by differential centrifugation as described by Grange et al. (2004), and resuspended in ice-cold buffer containing 4mM Hepes (pH 7.4), 1mM preboiled sodium orthovanadate, 1mM dithiothreitol, 100 nM microcystin (Sigma), and a mix of protease inhibitors (“Complete”, Roche). An aliquot of suspension (530 μl, 4.5 μg/μl total protein) was solubilized by adding 1:9 vol. of detergent solution (10% sodium deoxycholate in 0.5M Tris, pH 9.0) and gently agitating for 20 min. Triton X-100 (0.1% final concn.) was then added, and the extract was dialyzed overnight against immunoprecipitation buffer (10 mM Tris-HCl, pH 7.4, 100 mM NaCl, 0.1 mM sodium orthovanadate, 5 mM EDTA, 1 mM dithiothreitol, 0.1 % Triton X-100, 1 nM microcystin, and protease inhibitors). The dialyzed extract was centrifuged for 30 min at 105g in a TL-100 tabletop ultracentrifuge (Beckman), and the supernatant (300 μg protein diluted to 500 μl buffer per IP) was used for immunoprecipitation for 3 h with 10 μg affinity-purified anti-Sam68 polyclonal antibody, or preimmune IgG purified from the same rabbit, or with anti-NR1 monoclonal antibody (Pharmingen). Immunoprecipitates were collected onto protein G-Sepharose beads and analysed by SDS-PAGE (8% polyacrylamide gel) and Western immunoblotting. Antibodies used for immunoblotting were purchased from UBI (anti-PSD95 monoclonal antibody) and Chemicon (anti-GluR2/3 rabbit polyclonal antibody). The bands were revealed by chemiluminescence (Pierce Femto).
Results
Identification of Eef1a1 in the repertoire of Sam68-associated mRNAs
Following an approach initiated by Tenenbaum et al. (2000), we decided to use a combination of co-immunoprecipitation and array screening to probe the repertoire of Sam68-associated mRNAs in cortical neurons. Sam68-mRNA complexes were immunoprecipitated from a rat cortical cytoplasmic extract with an anti-Sam68 antibody, as previously reported (Ben Fredj et al. 2004). A parallel immunoprecipitation was performed with preimmune antibody. The polyA+ RNAs associated with the Sam68 and control immunoprecipitates were linearly amplified by cDNA synthesis and T7 polymerase-mediated transcription, as initially described by Eberwine and coworkers (Gelder et al. 1990), and the amplified transcripts were fluorescently labeled with Cy3 or Cy5, respectively. The two transcript populations were then mixed and hybridized with the ~15,000 cDNAs of the NIA “15K” clone set arrayed onto a glass slide (Tanaka et al. 2000), and for each cDNA the fluorescence values at both wavelengths were measured and compared (Fig. 1A, B). A given mRNA was considered to be specifically immunoprecipitated when the fluorescence level was consistently higher in the channel corresponding to the immunospecific than in the background precipitation. To compare mRNA abundance measurements across several experiments, normalized measures of the relative abundance of each mRNA in the immunoprecipitated population were calculated by converting the fluorescence values into z scores (Lopez de Silanes et al. 2004; Mayne et al. 2001; see Materials and Methods). Duplicate assays of the same RNA population showed that z scores were highly reproducible through the entire set of ~15,000 genes, except for a limited number of low abundance mRNAs, indicating that abundance measurements were not grossly perturbed by fluctuations in the hybridization and detection procedure (Fig. 1C). When distinct experiments with different brain samples were compared, fluctuations in individual z scores were larger, likely due to random differences in the hydrolysis or dissociation rates of individual mRNAs during immunoprecipitation. The correlation between samples remained sufficient to ensure that the species scoring highest (z > 1.5) in one trial retained a 85% probability of scoring positive (z > 0) in a second trial (Fig. 1D). To obtain reliable identification of the most abundant mRNAs in the immunoprecipitate, z scores were measured in three independent experiments, mRNAs were ranked by decreasing mean z score, and some species with unacceptably large standard deviations were excluded. We also discarded a few arrayed sequences for which no cognate mRNAs had been identified. Table Ia shows those specifically immunoprecipitated mRNA species for which the mean relative abundance was highest after applying this screen.
Fig. 1.
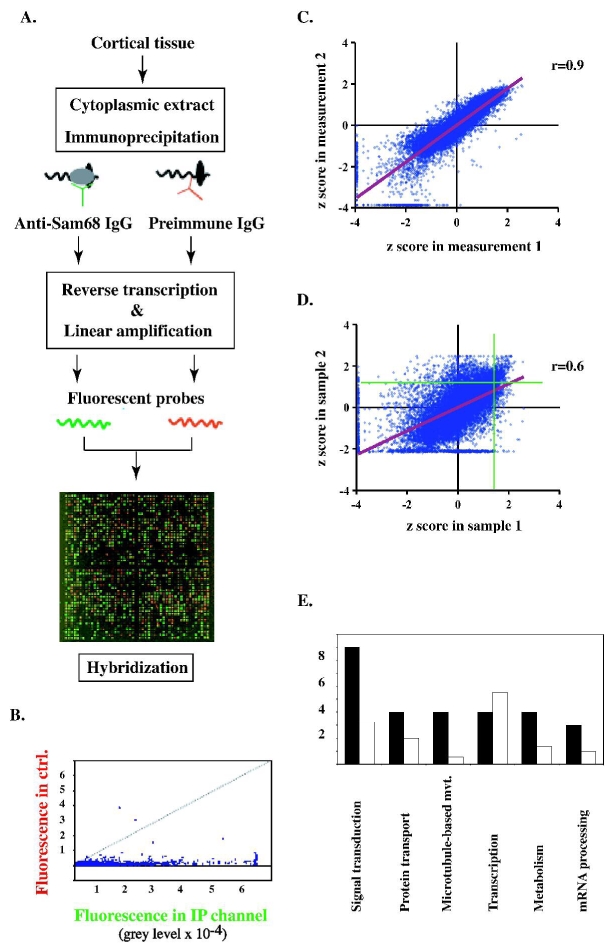
A. Scheme of the RNA-protein coimmunoprecipitation procedure, and image of a representative hybridization experiment. Green channel: Cy5-labelled probes obtained by amplification of polyA+ mRNA immunoprecipitated with the anti-Sam68 antibody. Red channel: Cy3-labelled probes obtained after immunoprecipitation with control (preimmune) antibody. B. Distribution of paired fluorescence values obtained in A. X-axis: specifically immunoprecipitated transcripts. Y-axis: adventitious transcripts. A large number of transcripts are more abundant in the specific immunoprecipitate than in the control. C. Within-sample reproducibility of z score measurements. The results of two different hybridization experiments performed with the same sample are compared. The regression line is shown in red. D. Between-sample reproducibility of z scores. The results of hybridization experiments performed on two different brain extracts are compared. The regression line is shown in red. Note that mRNAs with highest values in either sample (to the right of the vertical green line and above the horizontal green line) are mostly confined to the top right quadrant. E. Functional profile of the 43 mRNAs displaying the highest mean z scores in the immunoprecipitates (see Table Ia). For each transcript, a generic function was registered (identified by Gene Ontology descriptors of the mRNA in the NIH database; see Table Ia). Black barss: number of occurrences of the indicated functional descriptors in the 43 mRNA sample. Empty bars: expected number of occurrences of the descriptors, based on their frequencies among mRNAs of the micro-array. The difference is highly significant (p<0.0001, chi-square test), indicating enrichment for specific functional categories.
The dual-channel procedure was then used to compare the relative abundances of mRNA species in the immunoprecipitated vs. total polyA+ populations. The total population of polyA+ RNAs present in the input extract (before immunoprecipitation) was amplified and the resulting Cy5-labeled transcripts were mixed with an aliquot of Cy3-labeled transcripts derived from the Sam68 immunoprecipitate. After hybridization to the array, z scores were calculated for each channel. The species of Table Ia scored 0.6 to 1.0 units higher in the immunoprecipitated than in the total population, consistent with their enrichment.
There was a discernible pattern in the biological functions of these mRNAs (Fig. 1E). When assessed by Gene Ontology annotation for generic cellular process or component, more than 20% of the mRNAs (9 transcripts) were involved in intracellular or receptor-associated signal transduction, as they coded for e.g. receptor subunits, protein kinases, protein phosphatases, and associated regulatory factors. Another 20% of the mRNAs coded for proteins involved in protein transport, either as regulators of vesicular traffic, or of microtubule-dependent motion. These proportions are significantly higher than what would be expected based on the overall distribution of biological functions among the genes represented on the micro-array (Fig. 1E). By comparison, transcription regulators appeared to be under-represented among the major Sam68-associated mRNAs. Assessment of the litterature further indicates that some of the proteins of Table I contribute to exocytic and endosomal traffic (Arf-1, BRI-3, vacuolar ATPase) or to cytoskeletal regulation (stathmin) event-though this is not apparent from their GO function label. All of these processes are heavily involved in the local response to stimuli which affect neuronal homeostasis and plasticity. Indeed, more than one third of the genes of Table Ia code for proteins directly involved in synaptic plasticity, connectivity development, or neurodegenerative disease (Table Ib).
The β-actin mRNA, which was detected among the most abundant species in our screen, had formerly been detected by RT-PCR in immunoprecipitates of brain Sam68 (Ben Fredj et al. 2004), and was known as a specific target for recombinant Sam68 (Itoh et al. 2002). This provided a positive control for the immunoprecipitation procedure. The Gephyrin mRNA provided a negative control, as it was found neither by RT-PCR, nor in the micro-array analysis (data not shown and (Grange et al. 2004)).
The most enriched mRNA, Eef1a1, coded for the ubiquitous isoform of translation elongation factor eEF1A, an important player in both protein synthesis and long-term synaptic change (see Discussion). The z score value of the Eef1a1 fluorescence indicates that the representation of Eef1a1 among immunoprecipitated mRNAs is significantly higher than average (p=0.004).
RT-PCR with gene-specific primer pairs (see Table II) was used to confirm the specific coimmunoprecipitation of a range of mRNAs detected in the micro-array analysis, either among the more enriched species listed in Table Ia, or among mRNAs with lower relative abundance. In addition to β-actin, confirmed species included Eef1a1, Calmodulin I and Uch-L1 (Fig. 2). Interestingly, the Arc/Arg 3.1 mRNA was present among the less abundant mRNAs in the micro-array analysis, and could also be detected by RT-PCR (see Fig. 2A). Since both Arc and Eef1a1 are dendritic, activity-responsive mRNAs, this raises the possibility that upon exit to the somatodendritic compartment Sam68 may play a role in activity-induced translation of neuronal mRNAs.
Fig. 2.
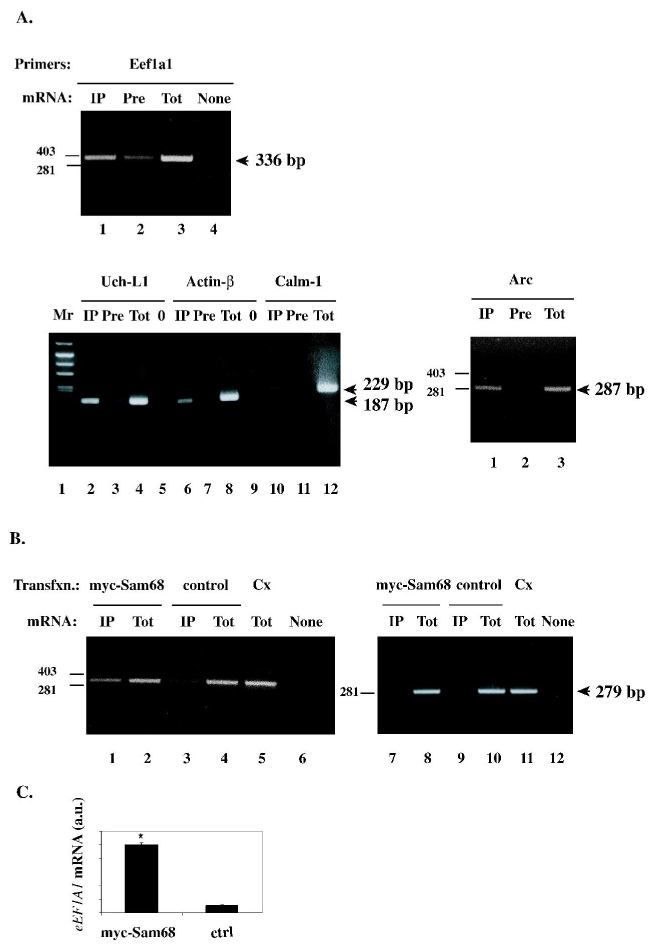
A. Validation of specific micro-array hybridization results by RT-PCR. Cortical tissue extract was subjected to immunoprecipitation with anti-Sam68 (“IP”) or preimmune (“Pre”) IgG as shown; total mRNA was purified from the immunoprecipitates, and subjected to reverse transcription and amplification by PCR, using the indicated gene-specific primers (Table II). The PCR products were visualized by fluorescent staining with ethidium bromide. The expected sizes of PCR products are indicated (Eef1a1: 336 bp; Uch-L1: 187 bp; β-actin: 178 bp; Calm-1: 229 bp; Arc: 287 bp ). Positive controls for the amplification reactions were obtained by performing PCR with total mRNA (“Tot”) directly derived from the input extract. No amplification took place without cDNA (“None” or “0”).
B. Reconstitution of the association between Sam68 and the Eef1a1 mRNA in cultured cells. HEK 293 cells were transfected with a myc-tagged Sam68 expression construct, or with control vector, as indicated. Cytoplasmic extracts were prepared and immunoprecipitated with anti-myc antibody, and subjected to RT-PCR with Eef1a1 (lanes 1, 3) or with β-tubulin (lanes 7,9) primers. As positive controls for amplification, RT-PCR products were generated with total mRNA from the transfected cells (lanes 2, 4, 8, 10) or from rat cortex (lane 5, 11). Lanes 6, 12: negative control for PCR.
C. The amounts of endogenous Eef1a1 mRNA in anti-myc immunoprecipitates from the indicated transfections were determined by quantitative (“real-time”) PCR (n=3 for each transfection).a.u., arbitrary units.
As Eef1a1 was the most enriched of Sam68-associated mRNAs in our assay, and since its translational regulation by synaptic inputs is well-documented (see below), we decided to investigate further its relationship with Sam68. To reconstitute the interaction between Eef1a1 mRNA and Sam68 in a heterologous system, myc-tagged Sam68 or a control plasmid were transiently expressed in HEK 293 cells; cytoplasmic extracts prepared from the transfected cells were immunoprecipitated with anti-myc antibody, and the immmunoprecipitates were probed by RT-PCR using the Eef1a1-specific primer pair. Endogenous Eef1a1 transcripts were detected in anti-myc immunoprecipitates from myc-Sam68-expressing cells but not from control cells (Fig. 2C, lanes 1–6). This result was reproduced by quantitative RT-PCR (Fig. 2C). Thus, the Eef1a1 mRNA associates with bona fide, overexpressed Sam68. By contrast, the β-tubulin mRNA was absent from myc-Sam68 immunoprecipitates (Fig. 2B, lanes 7–12), supporting the mRNA sequence specificity of binding.
Identification of Eef1a1 as a translationally activated mRNA in depolarized neurons
The Eef1a1 mRNA contains a 5′ oligopyrimidine tract and belongs to the general group of signal-responsive mRNAs encoding components of the translation apparatus (Petroulakis and Wang 2002). Moreover, translation of Eef1a1 is stimulated during synaptic change ((Tsokas et al. 2005). To determine whether Eef1a1 might belong to the population of translationally activated mRNAs in our system of in vitro depolarised neurons, primary cultures of cortical neurons were subjected to control treatement or prolonged depolarisation, lysed under nondenaturing conditions, and the post-nuclear supernatants were fractionated on sucrose density gradients, as described by (Krichevsky and Kosik 2001). Fractions were analysed for their RNA content by continuous monitoring of UV absorbance during collection. Fractions corresponding to the free mRNPs, 80S monosomes, polyribosomes, and RNA granules (migrating down to the bottom of the gradient) were analysed by RT-PCR with Eef1a1- specific primers. In non-stimulated neurons, the Eef1a1 mRNA was associated to the same extent with free mRNPs and with polysomes. Upon depolarisation, Eef1a1 became nearly undetectable in the free mRNP fraction, and strongly accumulated in the polysome fraction (Fig. 3A). These data indicate that Eef1a1 mRNA translation is activated by depolarisation. Consistent with this result, depolarisation of cultured cortical neurons induced a significant increase in the total level of eEF1A protein, as detected by Western immunoblotting (Fig. 3B).
Fig. 3.
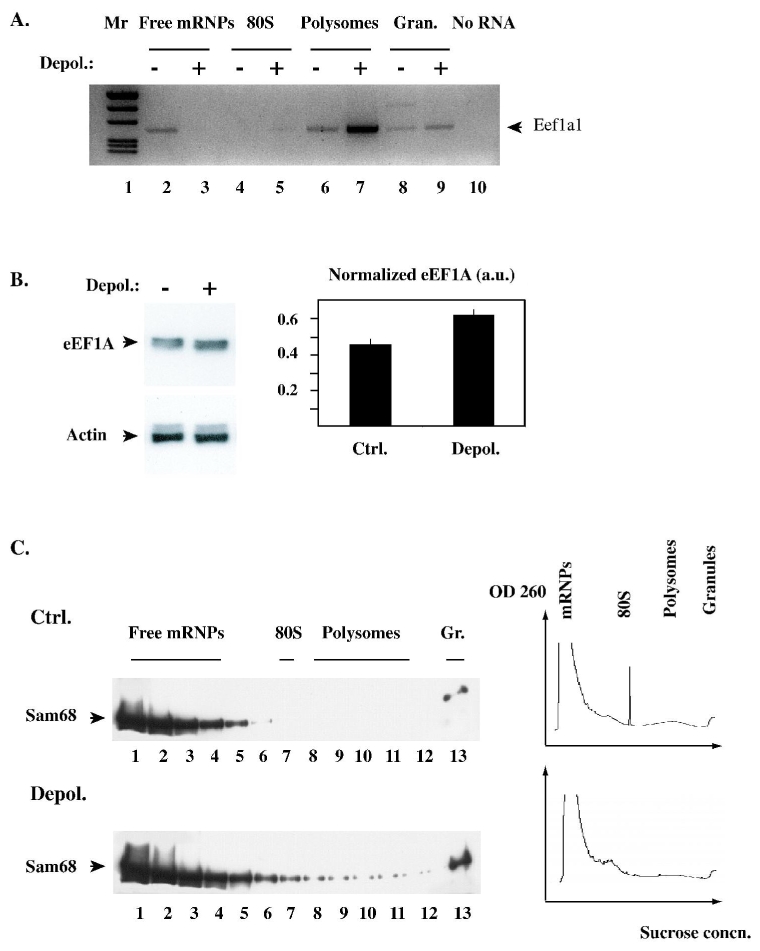
A. Depolarisation induces an increase in Eef1a1 mRNA translation. Cultured cortical neurons were incubated for 6 h in 5mM or 25 mM KCl, then cytoplasmic extracts were prepared and equal protein amounts were subjected to fractionation on sucrose density gradients. For each gradient, representative fractions corresponding to translationally inactive mRNPs, 80S monosomes, polysomes, and RNA granules were analysed by RT-PCR with specific primers to amplify the Eef1a1 mRNA. The PCR products were analysed side by side on an agarose gel (“−” and “+” lanes: gradient fractions from control and depolarised neurons; respectively) and visualized by fluorescent staining with ethidium bromide. Greyscale was inverted to improve contrast.
B. Depolarisation increases the amount of eEF1A protein. Cortical neurons were treated as in A, then lysed, and equal amounts of total protein were analysed by SDS-polyacrylamide gel electrophoresis and immunoblotting. Left: the same blot was probed first with anti-eEF1A monoclonal antibody, then with anti-actin antibody. Right: the relative amount of eEF1A in each lane was measured by densitometric scanning, and normalized to the amount of actin in the same lane to correct for gel loading error. The histogram shows the average (+/− S.D.) of normalized eEF1A amount in each condition.
C. Depolarisation enhances the association of Sam68 with polysomal RNA and RNA granules. Cytoplasmic extracts from control and depolarised neurons were prepared and fractionated as in A. The fractions were analysed by immunoblotting with anti-Sam68 antibody (left). The distribution of RNA along the gradient was determined by continuous monitoring of UV absorbance at 254 nm during collection of the fractions (right). The various types of ribonucleoproteic assemblies are indicated.
Depolarisation-inducible association of Sam68 with polysomes
We previously reported that Sam68 was associated with polyribosomes in cytoplasmic extracts derived from adult rat cortical tissue (Grange et al. 2004). To confirm that a similar association occurred in cultured cortical neurons, and to determine whether the interaction could be regulated by depolarisation, extracts were prepared and fractionated exactly as in Fig. 3A. Fractions were then analysed by Western immunoblotting with anti-Sam68 antibody. In extracts from control, quiescent neurons, Sam68 mostly comigrated with free mRNPs; a minor fraction sedimented in the RNA granule fraction (Fig. 3C, top). Following depolarisation, Sam68 accumulated in fractions containing polysomes and granules, even though the total amount of RNA in those fractions was slightly lower than in the control condition (Fig. 3, bottom). This result is consistent with the depolarisation-induced accumulation of Sam68 fluorescence in RNA-enriched dendritic clusters, which we have previously reported (Ben Fredj et al. 2004). Thus, in depolarised neurons, the increase in Eef1a1 translation correlates with the loading of Sam68 onto translated mRNAs.
Effect of dominant-negative Sam68 on neuronal eEF1A expression
The association of Sam68 to actively translated Eef1a1 mRNA suggests that Sam68 might regulate the synthesis of eEF1A. To investigate this point, we assayed the level of eEF1A immunoreactivity in cultured hippocampal neurons that expressed GFP-tagged derivatives of either wild-type Sam68 or Sam68ΔC, a C-terminally truncated Sam68 mutant. The latter mutant retains a significant affinity for specific RNA sequences, but is deprived from tyrosine-rich and proline-rich motifs involved in protein-protein interactions with SH2 and SH3-containing ligands, and also from a C-terminal (nonclassical) nuclear localisation signal (Fig. 4F). In cultured cell lines, Sam68ΔC is constitutively cytoplasmic and displays transdominant-negative activity towards endogenous Sam68, when coexpressed with retroviral (HIV-1) mRNAs that require Sam68 for efficient utilization (Reddy et al. 1999; Soros et al. 2001). Sam68ΔC is specifically recruited to these viral mRNAs once they have reached the cytoplasm, and appears to block their normal interaction with the translation apparatus (Soros et al. 2001). We reasoned that expression of Sam68ΔC in neurons should similarly allow us to probe the contribution of endogenous Sam68 to the translational activity of target neuronal mRNAs, as distinct from its role in nuclear mRNA processing. In transfected neurons, the GFP-Sam68ΔC protein was mostly excluded from the neuronal nucleus and concentrated in clusters within the cell body; it was also detectable in dendritic shafts (Fig. 4, A2, B). The relative expression level of endogenous eEF1A in transfected and non-transfected neurons was evaluated by immunofluorescent staining with an anti-eEF1A monoclonal antibody. Among the neurons of a field, those expressing GFP-Sam68ΔC typically had a lower level of eEF1A immunoreactivity than surrounding, nontransfected neurons (Fig. 4, A1, C1). This was not the case in neurons that had been transfected with GFP-tagged wild-type Sam68 (Fig. 4, D1). Neurons expressing GFP-Sam68ΔC maintained a normal morphology (Fig. 4, A1, C3), indicating that the decrease in eEF1A did not arise from some general toxicity of the construct. The effect of GFP-Sam68ΔC expression on endogenous eEF1A immunofluorescence was quantitated by using as internal reference value the average immunofluorescence of nontransfected neurons surrounding the transfected cells (see Materials and Methods). This quantitation procedure indicated that GFP-Sam68ΔC reproducibly lowered the level of eEF1A immunofluorescence, whereas GFP-Sam68 did not (Fig. 4E). As a control, GFP-Sam68ΔC did not affect the abundance of coexpressed mRFP. Since GFP-Sam68ΔC functions in the cytoplasm, and presumably competes with endogenous Sam68 for binding sites in mRNA-protein complexes, this result suggests that endogenous Sam68 positively regulates the translation of eEF1A-encoding mRNAs.
Possible post-synaptic anchoring by Sam68
In non-neuronal cells, Sam68 is known to bind to membrane-associated signalling proteins (Lukong and Richard 2003). In neurons, given its association with dendritic mRNAs that become actively translated in response to local synaptic stimuli (Tsokas et al. 2005), we wondered whether Sam68 might physically interact with synaptic receptors. In synaptic membranes prepared from rat cortex, both the NMDA receptor and the post-synaptic density protein PSD95 could be co-immunoprecipitated with Sam68 (Fig. 5, lanes 1–6). By contrast, AMPA receptors were essentially absent from these immunoprecipitates (lanes 7–9), consistent with the notion that Sam68 was specifically recruited by the NMDA receptor complex. We could also detect Sam68 in NMDA receptor immunoprecipitates (lanes 10–11). Thus Sam68 might conceivably provide a platform for docking specific neuronal mRNAs and/or for recruiting specific translation factors at subsynaptic sites.
Fig. 5.
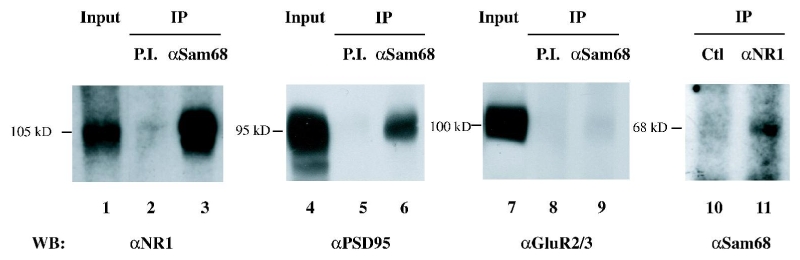
Binding of Sam68 to the post-synaptic NMDA receptor complex. Purified synaptic membranes were solubilized under non-denaturing conditions, and subjected to immunoprecipitation with the indicated antibodies. Lanes 1, 4, 7: aliquot (10%) of input lysate. After detecting PSD-95, the blot of lanes 4–6 was stripped and reprobed to detect GluR2/3 subunits of AMPA receptors, shown in lanes 7–9. Lane 10: control immunoprecipitation with a mouse monoclonal IgG, class-matched to the anti-NR1 antibody.
Discussion
The present sudy indicates that brain Sam68 is associated with a wide but selective range of cytoplasmic mRNAs, among which transcripts encoding long-term plasticity related proteins are prominent. Sam68 might collectively regulate the synthesis of an entire group of such proteins, in an activity-dependent fashion. The set of Sam68-associated mRNAs we found in brain differs from those reported in non-neuronal cell lines or lineages (Itoh et al. 2002; Paronetto et al. 2006; Tremblay and Richard 2006). Extensive comparison of the different sets is difficult, since the screening methods were different. However, two of the mRNAs, β-actin and dynein, were found in both the study by (Itoh et al. 2002) and our screen, providing a reliable positive control. Taken together, the various screens suggest that mRNA recognition by Sam68 is highly dependent on cellular context.
One of most enriched among the Sam68-associated cortical transcripts was the Eef1a1 mRNA. Although Eef1a1 is ubiquitous, the Sam68-Eef1a1 mRNA interaction was apparently not detected in the previous studies. This may be due to the sensitivity of the micro-array sampling procedure; alternatively, the interaction may be favoured by neuron-specific factors. However, it was possible to reconstitute the Sam68-Eef1a1 mRNA interaction in transfected HEK cells by using overexpressed, myc-tagged Sam68. The interaction is phylogenetically conserved since HEK 293 cells are of human origin; this is consistent with the >90% identity of human and rat Eef1a1 nucleotide sequences, including in the 3′ untranslated region. Interestingly, based on free energy minimization, the entire 3′UTR of the Eef1a1 mRNA is predicted to fold into a single, long hairpin displaying at its extremity a U-rich terminal loop (Fig. 6). This is reminiscent of the U6 motif-containing stem-loop structure that mediates recognition of the β-actin mRNA by Sam68 in vitro (Itoh et al. 2002). However, our experiments do not rule out the possibility that the Sam68 – Eef1a1 mRNA association may be mediated by a third molecule. Further studies will be required to establish whether the 3′ UTR structure forms a direct binding site for Sam68 on the Eef1a1 mRNA.
Fig. 6.

The 3′ UTR sequences of human EEF1A1 and rat Eef1a1 mRNAs have been aligned using Clustal W (v. 1.83). The first nucleotide of each sequence is that immediately following the stop codon, and the last nucleotide corresponds to the major polyadenylation site. Stars indicate identical residues. Using the Rnaalifold programme of the Vienna RNA package (Ivo L Hofacker, http://www.tbi.univie.ac.at/~ivo/RNA/), the predicted folding of the aligned sequences displays a large stem structure with a U-rich terminal loop (overlined).
In dissociated cortical neurons, chemically induced depolarisation is known to generate bursts of action potentials and to recruit some of the pathways that underlie synaptic plasticity. While causing the translational repression of multiple mRNA species, depolarisation activates the translation of a select group of transcripts (Krichevsky and Kosik 2001). Our data indicate that Eef1a1 belongs to this group. Eef1a1 encodes the ubiquitous isoform of translation elongation factor eEF1A, which transports amino-acyl-tRNA to the A site of the ribosome during protein synthesis. In addition, eEF1A also anchors mRNAs to the actin cytoskeleton, and can bundle actin filaments according to a unique spatial arrangement (Liu et al. 2002a). Local provision of eEF1A may play a role in both biosynthetic and structural aspects of synaptic plasticity, and perhaps establish a link between these two facets of long-term synaptic change. In Aplysia, synthesis of the eEF1A homologue is instrumental to maintenance of synapse-specific long-term facilitation (Giustetto et al. 2003). In the rodent hippocampus, translation of the Eef1a1 mRNA is regulated by synaptic signals in complex ways, which appear to differ between preparations. In hippocampal slices from young rats, Eef1a1 translation has been shown to be rapidly induced during tetanus-induced LTP of CA3- CA1 synapses, subject to activation of the rapamycin-sensitive mTOR/S6 Kinase pathway (Tsokas et al. 2005). This finding is consistent with the fact that the Eef1a1 mRNA contains a 5′ oligopyrimidine tract, which is a signature for mTOR-inducible mRNAs. Interestingly, in adult rat hippocampus in vivo, LTP of dentate granule synapses induced redistribution rather than neosynthesis of eEF1A protein. In contrast, long-term depression elicited by pharmacological stimulation of metabotropic glutamate receptors (mGluRs) was accompanied by a large increase in eEF1A protein, without change in Eef1a1 mRNA level (Huang et al. 2005). Thus, the induction of eEF1A synthesis seems to be exquisitely sensitive to the spatio-temporal and cell-specific details of synaptic signalling, suggesting that multiple effectors regulate the translation of Eef1a1 in neurons. One of these effectors may be the RNA-binding protein FMRP, since in vitro and in cell lines, FMRP has been reported to bind and repress the Eef1a1 mRNA (Sung et al. 2003); furthermore it is tempting to speculate that removal of this putative inhibition underlies the effect of mGluRs on Eef1a1 translation (Bear et al. 2004). The present data suggest that Sam68 is also part of the regulatory machinery that controls neuronal Eef1a1 translation, with an action distinct from that of FMRP. Since depolarisation increased the recruitment of Sam68 to polysomal mRNAs, it is likely that Eef1a1 transcripts bound to Sam68 were engaged in translation. The finding that the dominant-negative GFP-Sam68ΔC, but not GFP-Sam68(WT) inhibited eEF1A protein accumulation suggests that endogenous Sam68 may be required for the translational activation of Eef1a1 mRNA. Since the dominant-negative effect of GFP-Sam68ΔC was already detected in unstimulated neurons, the low amount of Sam68 existing in the cytoplasm prior to depolarisation may contribute to generating the proportion of actively translated Eef1a1 mRNA seen under basal conditions. In line with this possibility, Sam68 has been reported to interact with RNA helicase A and with the mRNA export factor Tap (Reddy et al. 2000), two proteins known to promote translation of specific viral and cellular mRNAs (Hartman et al. 2006; Li et al. 2006). Kinases recruited by the C terminus of Sam68 might also be required to phosphorylate and release mRNA-bound translation inhibitory factors, similar to the case of hnRNP-K phosphoylation by Src (Ostareck-Lederer et al. 2002). GFP-Sam68 ΔC, which is defective for certain protein-protein interactions, may displace the endogenous Sam68 and associated translation factors out of the target mRNA; indeed, in the case of viral mRNAs, GFP-Sam68ΔC functionally antagonizes RNA helicase A and Tap (Reddy et al. 2000). Upon depolarisation, the largely enhanced export of Sam68 to the cytoplasm may cooperate with the action of signalling cascades such as the mTOR pathway for translational stimulation of activity-responsive mRNAs exemplified by Eef1a1. A similar scheme may apply to certain non-neuronal systems; in the case of meiotic male gametes, Paronetto et al. (2006) recently confirmed the occurrence of signal-induced transfer of Sam68 to the cytoplasm and its binding to polysomes.
Acknowledgments
Work supported by INSERM; Université Joseph Fourier; Ministère de l’Enseignement Supérieur et de la Recherche; Association pour la Recherche contre le Cancer (ARC); Fondation pour la Recherche Médicale.
We thank Michel Volovitch and Alain Prochiantz for their kind cooperation in the use of micro-array analysis; Stéphane Richard and Roger Y. Tsien for the generous gift of the GFP-Sam68 ΔC and mRFP1 plasmids, respectively; Isabelle Barlat and Bruno Toqué for the kind gift of myc-Sam68 plasmid; and Michael Kiebler for discussions and reagents.
References
- Bagni C, Greenough WT. From mRNP trafficking to spine dysmorphogenesis: the roots of fragile X syndrome. Nat Rev Neurosci. 2005;6(5):376–387. doi: 10.1038/nrn1667. [DOI] [PubMed] [Google Scholar]
- Banker G, Asmussen H, Goslin K. In: Rat Hippocampal Neurons in Low-Density Culture. Banker G, Goslin K, editors. Cambridge, Massachusetts: MIT Press; 1998. p. 666. [Google Scholar]
- Bear MF, Huber KM, Warren ST. The mGluR theory of fragile X mental retardation. Trends in Neurosciences. 2004;27(7):370–377. doi: 10.1016/j.tins.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Ben Fredj N, Grange J, Sadoul R, Richard S, Goldberg Y, Boyer V. Depolarization-induced translocation of the RNA-binding protein Sam68 to the dendrites of hippocampal neurons. J Cell Sci. 2004;117(Pt 7):1079–1090. doi: 10.1242/jcs.00927. [DOI] [PubMed] [Google Scholar]
- Berto G, Camera P, Fusco C, Imarisio S, Ambrogio C, Chiarle R, Silengo L, Di Cunto F. The Down syndrome critical region protein TTC3 inhibits neuronal differentiation via RhoA and Citron kinase. J Cell Sci. 2007;120(Pt 11):1859–1867. doi: 10.1242/jcs.000703. [DOI] [PubMed] [Google Scholar]
- Bramham CR, Wells DG. Dendritic mRNA: transport, translation and function. Nat Rev Neurosci. 2007;8(10):776–789. doi: 10.1038/nrn2150. [DOI] [PubMed] [Google Scholar]
- Cole TB, Wenzel HJ, Kafer KE, Schwartzkroin PA, Palmiter RD. Elimination of zinc from synaptic vesicles in the intact mouse brain by disruption of the ZnT3 gene. Proc Natl Acad Sci U S A. 1999;96(4):1716–1721. doi: 10.1073/pnas.96.4.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Scala F, Dupuis L, Gaiddon C, De Tapia M, Jokic N, Gonzalez de Aguilar JL, Raul JS, Ludes B, Loeffler JP. Tissue specificity and regulation of the N-terminal diversity of reticulon 3. Biochem J. 2005;385(Pt 1):125–134. doi: 10.1042/BJ20040458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eom T, Antar L, Singer R, Bassell GJ. Localization of a beta-actin messenger ribonucleoprotein complex with zipcode-binding protein modulates the density of dendritic filopodia and filopodial synapses. J Neurosci. 2003;3(32):10433–44. doi: 10.1523/JNEUROSCI.23-32-10433.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelder R, von Zastrow M, Yool A, Dement W, Barchas J, Eberwine J. Amplified RNA Synthesized from Limited Quantities of Heterogeneous cDNA. PNAS. 1990;87(5):1663–1667. doi: 10.1073/pnas.87.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giustetto M, Hegde AN, Si K, Casadio A, Inokuchi K, Pei W, Kandel ER, Schwartz JH. Axonal transport of eukaryotic translation elongation factor 1a mRNA couples transcription in the nucleus to long-term facilitation at the synapse. PNAS. 2003;100(23):13680–13685. doi: 10.1073/pnas.1835674100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glanzer J, Miyashiro KY, Sul J-Y, Barrett L, Belt B, Haydon P, Eberwine J. RNA splicing capability of live neuronal dendrites. PNAS. 2005;102(46):16859–16864. doi: 10.1073/pnas.0503783102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetze B, Grunewald B, Baldassa S, Kiebler M. Chemically controlled formation of a DNA/calcium phosphate coprecipitate: application for transfection of mature hippocampal neurons. J Neurobiol. 2004;60(4):517–525. doi: 10.1002/neu.20073. [DOI] [PubMed] [Google Scholar]
- Gong B, Cao Z, Zheng P, Vitolo OV, Liu S, Staniszewski A, Moolman D, Zhang H, Shelanski M, Arancio O. Ubiquitin hydrolase Uch-L1 rescues beta-amyloid-induced decreases in synaptic function and contextual memory. Cell. 2006;126(4):775–788. doi: 10.1016/j.cell.2006.06.046. [DOI] [PubMed] [Google Scholar]
- Grange J, Boyer V, Fabian-Fine R, Fredj NB, Sadoul R, Goldberg Y. Somatodendritic localization and mRNA association of the splicing regulatory protein Sam68 in the hippocampus and cortex. J Neurosci Res. 2004;75(5):654–666. doi: 10.1002/jnr.20003. [DOI] [PubMed] [Google Scholar]
- Hartman TR, Qian S, Bolinger C, Fernandez S, Schoenberg DR, Boris-Lawrie K. RNA helicase A is necessary for translation of selected messenger RNAs. Nat Struct Mol Biol. 2006;13(6):509–516. doi: 10.1038/nsmb1092. [DOI] [PubMed] [Google Scholar]
- He W, Lu Y, Qahwash I, Hu XY, Chang A, Yan R. Reticulon family members modulate BACE1 activity and amyloid-beta peptide generation. Nat Med. 2004;10(9):959–965. doi: 10.1038/nm1088. [DOI] [PubMed] [Google Scholar]
- Hu XD, Huang Q, Roadcap DW, Shenolikar SS, Xia H. Actin-associated neurabin-protein phosphatase-1 complex regulates hippocampal plasticity. Journal of Neurochemistry. 2006;98(6):1841–1851. doi: 10.1111/j.1471-4159.2006.04070.x. [DOI] [PubMed] [Google Scholar]
- Huang F, Chotiner JK, Steward O. The mRNA for Elongation Factor 1α Is Localized in Dendrites and Translated in Response to Treatments That Induce Long-Term Depression. J Neurosci. 2005;25(31):7199–7209. doi: 10.1523/JNEUROSCI.1779-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y-S, Jung M-Y, Sarkissian M, Richter JD. N-methyl-D-aspartate receptor signaling results in Aurora kinase-catalyzed CPEB phosphorylation and αCaMKII mRNA polyadenylation at synapses. EMBO J. 2002;21(9):2139–2148. doi: 10.1093/emboj/21.9.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttelmaier S, Zenklusen D, Lederer M, Dictenberg J, Lorenz M, Meng X, Bassell GJ, Condeelis J, Singer RH. Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature. 2005;438(7067):512–515. doi: 10.1038/nature04115. [DOI] [PubMed] [Google Scholar]
- Itoh M, Haga I, Li Q-H, Fujisawa J-i. Identification of cellular mRNA targets for RNA-binding protein Sam68. Nucl Acids Res. 2002;30(24):5452–5464. doi: 10.1093/nar/gkf673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandjian EW, Huot M-E, Tremblay S, Davidovic L, Mazroui R, Bardoni B. Biochemical evidence for the association of fragile X mental retardation protein with brain polyribosomal ribonucleoparticles. PNAS. 2004;101(36):13357–13362. doi: 10.1073/pnas.0405398101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiebler MA, Bassell GJ. Neuronal RNA granules: movers and makers. Neuron. 2006;51(6):685–690. doi: 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- Krichevsky AM, Kosik KS. Neuronal RNA granules: a link between RNA localization and stimulation-dependent translation. Neuron. 2001;32(4):683–696. doi: 10.1016/s0896-6273(01)00508-6. [DOI] [PubMed] [Google Scholar]
- Kunz JB, Neu-Yilik G, Hentze MW, Kulozik AE, Gehring NH. Functions of hUpf3a and hUpf3b in nonsense-mediated mRNA decay and translation. RNA. 2006;12(6):1015–1022. doi: 10.1261/rna.12506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. Embo J. 2000;19(24):6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Bor Y-c, Misawa Y, Xue Y, Rekosh D, Hammarskjold M-L. An intron with a constitutive transport element is retained in a Tap messenger RNA. Nature. 2006;443(7108):234–237. doi: 10.1038/nature05107. [DOI] [PubMed] [Google Scholar]
- Liu G, Grant WM, Persky D, Latham VM, Jr, Singer RH, Condeelis J. Interactions of Elongation Factor 1alpha with F-Actin and beta -Actin mRNA: Implications for Anchoring mRNA in Cell Protrusions. Mol Biol Cell. 2002a;13(2):579–592. doi: 10.1091/mbc.01-03-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Fallon L, Lashuel HA, Liu Z, Lansbury PT., Jr The UCH-L1 gene encodes two opposing enzymatic activities that affect alpha-synuclein degradation and Parkinson’s disease susceptibility. Cell. 2002b;111(2):209–218. doi: 10.1016/s0092-8674(02)01012-7. [DOI] [PubMed] [Google Scholar]
- Lopez de Silanes I, Zhan M, Lal A, Yang X, Gorospe M. Identification of a target RNA motif for RNA-binding protein HuR. PNAS. 2004;101(9):2987–2992. doi: 10.1073/pnas.0306453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukong KE, Richard S. Sam68, the KH domain-containing superSTAR. Biochimica et Biophysica Acta (BBA) - Reviews on Cancer. 2003;1653(2):73–86. doi: 10.1016/j.bbcan.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Massat I, Souery D, Del-Favero J, Oruc L, Noethen MM, Blackwood D, Thomson M, Muir W, Papadimitriou GN, Dikeos DG, Kaneva R, Serretti A, Lilli R, Smeraldi E, Jakovljevic M, Folnegovic V, Rietschel M, Milanova V, Valente F, Van Broeckhoven C, Mendlewicz J. Excess of allele1 for alpha3 subunit GABA receptor gene (GABRA3) in bipolar patients: a multicentric association study. Mol Psychiatry. 2002;7(2):201–207. doi: 10.1038/sj.mp.4000953. [DOI] [PubMed] [Google Scholar]
- Matsuda J, Kido M, Tadano-Aritomi K, Ishizuka I, Tominaga K, Toida K, Takeda E, Suzuki K, Kuroda Y. Mutation in saposin D domain of sphingolipid activator protein gene causes urinary system defects and cerebellar Purkinje cell degeneration with accumulation of hydroxy fatty acid-containing ceramide in mouse. Hum Mol Genet. 2004;13(21):2709–2723. doi: 10.1093/hmg/ddh281. [DOI] [PubMed] [Google Scholar]
- Matter N, Herrlich P, Konig H. Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature. 2002;420(6916):691–695. doi: 10.1038/nature01153. [DOI] [PubMed] [Google Scholar]
- Mayne M, Cheadle C, Soldan SS, Cermelli C, Yamano Y, Akhyani N, Nagel JE, Taub DD, Becker KG, Jacobson S. Gene expression profile of herpesvirus-infected T cells obtained using immunomicroarrays: induction of proinflammatory mechanisms. J Virol. 2001;75(23):11641–11650. doi: 10.1128/JVI.75.23.11641-11650.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren M, Asai K, Cochrane A. A novel function for Sam68: Enhancement of HIV-1 RNA 3′ end processing. RNA. 2004;10(7):1119–1129. doi: 10.1261/rna.5263904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modem S, Badri KR, Holland TC, Reddy TR. Sam68 is absolutely required for Rev function and HIV-1 production. Nucl Acids Res. 2005;33(3):873–879. doi: 10.1093/nar/gki231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munton RP, Tweedie-Cullen R, Livingstone-Zatchej M, Weinandy F, Waidelich M, Longo D, Gehrig P, Potthast F, Rutishauser D, Gerrits B, Panse C, Schlapbach R, Mansuy IM. Qualitative and quantitative analyses of protein phosphorylation in naive and stimulated mouse synaptosomal preparations. Mol Cell Proteomics. 2007;6(2):283–293. doi: 10.1074/mcp.M600046-MCP200. [DOI] [PubMed] [Google Scholar]
- Ostareck-Lederer A, Ostareck DH, Cans C, Neubauer G, Bomsztyk K, Superti-Furga G, Hentze MW. c-Src-Mediated Phosphorylation of hnRNP K Drives Translational Activation of Specifically Silenced mRNAs. Mol Cell Biol. 2002;22(13):4535–4543. doi: 10.1128/MCB.22.13.4535-4543.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco CD, Kunkel R, Lieberman AP. Autophagy in Niemann-Pick C disease is dependent upon Beclin-1 and responsive to lipid trafficking defects. Hum Mol Genet. 2007;16(12):1495–1503. doi: 10.1093/hmg/ddm100. [DOI] [PubMed] [Google Scholar]
- Paronetto MP, Achsel T, Massiello A, Chalfant CE, Sette C. The RNA-binding protein Sam68 modulates the alternative splicing of Bcl-x. J Cell Biol. 2007;176(7):929–939. doi: 10.1083/jcb.200701005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paronetto MP, Zalfa F, Botti F, Geremia R, Bagni C, Sette C. The nuclear RNA-binding protein Sam68 translocates to the cytoplasm and associates with the polysomes in mouse spermatocytes. Mol Biol Cell. 2006;17(1):14–24. doi: 10.1091/mbc.E05-06-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroulakis E, Wang E. Nerve Growth Factor Specifically Stimulates Translation of Eukaryotic Elongation Factor 1A-1 (eEF1A-1) mRNA by Recruitment to Polyribosomes in PC12 Cells. J Biol Chem. 2002;277(21):18718–18727. doi: 10.1074/jbc.M111782200. [DOI] [PubMed] [Google Scholar]
- Reddy TR, Tang H, Xu W, Wong-Staal F. Sam68, RNA helicase A and Tap cooperate in the post-transcriptional regulation of human immunodeficiency virus and type D retroviral mRNA. Oncogene. 2000;19(32):3570–3575. doi: 10.1038/sj.onc.1203676. [DOI] [PubMed] [Google Scholar]
- Reddy TR, Xu W, Mau JK, Goodwin CD, Suhasini M, Tang H, Frimpong K, Rose DW, Wong-Staal F. Inhibition of HIV replication by dominant negative mutants of Sam68, a functional homolog of HIV-1 Rev. Nat Med. 1999;5(6):635–642. doi: 10.1038/9479. [DOI] [PubMed] [Google Scholar]
- Sanford JR, Gray NK, Beckmann K, Caceres JF. A novel role for shuttling SR proteins in mRNA translation. Genes Dev. 2004;18(7):755–768. doi: 10.1101/gad.286404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek-Duran F, Linden DJ, Lonart G. Adapter protein 14-3-3 is required for a presynaptic form of LTP in the cerebellum. Nat Neurosci. 2004;7(12):1296–1298. doi: 10.1038/nn1348. [DOI] [PubMed] [Google Scholar]
- Smalla KH, Matthies H, Langnase K, Shabir S, Bockers TM, Wyneken U, Staak S, Krug M, Beesley PW, Gundelfinger ED. The synaptic glycoprotein neuroplastin is involved in long-term potentiation at hippocampal CA1 synapses. Proc Natl Acad Sci U S A. 2000;97(8):4327–4332. doi: 10.1073/pnas.080389297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soros VB, Carvajal HV, Richard S, Cochrane AW. Inhibition of Human Immunodeficiency Virus Type 1 Rev Function by a Dominant-Negative Mutant of Sam68 through Sequestration of Unspliced RNA at Perinuclear Bundles. J Virol. 2001;75(17):8203–8215. doi: 10.1128/JVI.75.17.8203-8215.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung YJ, Dolzhanskaya N, Nolin SL, Brown T, Currie JR, Denman RB. The fragile X mental retardation protein FMRP binds elongation factor 1A mRNA and negatively regulates its translation in vivo. J Biol Chem. 2003;278(18):15669–15678. doi: 10.1074/jbc.M211117200. [DOI] [PubMed] [Google Scholar]
- Tanaka TS, Jaradat SA, Lim MK, Kargul GJ, Wang X, Grahovac MJ, Pantano S, Sano Y, Piao Y, Nagaraja R, Doi H, Wood WH, III, Becker KG, Ko MSH. Genome-wide expression profiling of mid-gestation placenta and embryo using a 15,000 mouse developmental cDNA microarray. PNAS. 2000;97(16):9127–9132. doi: 10.1073/pnas.97.16.9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenenbaum SA, Carson CC, Lager PJ, Keene JD. Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proc Natl Acad Sci USA. 2000;97(26):14085–14090. doi: 10.1073/pnas.97.26.14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas GM, Huganir RL. MAPK cascade signalling and synaptic plasticity. Nat Rev Neurosci. 2004;5(3):173–183. doi: 10.1038/nrn1346. [DOI] [PubMed] [Google Scholar]
- Toyo-oka K, Shionoya A, Gambello MJ, Cardoso C, Leventer R, Ward HL, Ayala R, Tsai L-H, Dobyns W, Ledbetter D, Hirotsune S, Wynshaw-Boris A. 14-3-3[epsi] is important for neuronal migration by binding to NUDEL: a molecular explanation for Miller-Dieker syndrome. Nat Genet. 2003;34(3):274–285. doi: 10.1038/ng1169. [DOI] [PubMed] [Google Scholar]
- Tremblay GA, Richard S. mRNAs associated with the Sam68 RNA binding protein. RNA Biol. 2006;3(2):1–4. doi: 10.4161/rna.3.2.3204. [DOI] [PubMed] [Google Scholar]
- Tsokas P, Grace EA, Chan P, Ma T, Sealfon SC, Iyengar R, Landau EM, Blitzer RD. Local Protein Synthesis Mediates a Rapid Increase in Dendritic Elongation Factor 1A after Induction of Late Long-Term Potentiation. J Neurosci. 2005;25(24):5833–5843. doi: 10.1523/JNEUROSCI.0599-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham L, Benjannet S, Marcinkiewicz E, Chretien M, Seidah NG. Beta-amyloid protein converting enzyme 1 and brain-specific type II membrane protein BRI3: binding partners processed by furin. J Neurochem. 2005;92(1):93–102. doi: 10.1111/j.1471-4159.2004.02840.x. [DOI] [PubMed] [Google Scholar]
- Wiegand HL, Lu S, Cullen BR. Exon junction complexes mediate the enhancing effect of splicing on mRNA expression. PNAS. 2003;100(20):11327–11332. doi: 10.1073/pnas.1934877100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z, Storm DR. The role of calmodulin as a signal integrator for synaptic plasticity. Nat Rev Neurosci. 2005;6(4):267–276. doi: 10.1038/nrn1647. [DOI] [PubMed] [Google Scholar]


