Abstract
BORIS (CTCFL) is the only known paralog of the versatile regulatory protein CTCF, a multifunctional DNA binding protein that mediates distinct gene regulatory functions involved in cell growth, differentiation, and apoptosis. Unlike CTCF, the expression of BORIS is normally restricted to specific cells in testes (the only cells where CTCF is not expressed), where it may play a role in reprogramming the methylation pattern of male germ line DNA. Frequent amplification of the 20q13.2 region, which contains the BORIS gene, and expression of BORIS transcripts in diverse human tumors and cell lines have led to the hypothesis that aberrant expression of BORIS may play a role in tumorigenesis by interfering with CTCF functions. However, recent studies using more quantitative methods indicate low frequency of BORIS expression in melanoma, ovarian, prostate, and bladder carcinomas. To investigate the relationship between chromosome 20q13 amplification and BORIS mRNA levels within breast cancer cell lines and tissues, we developed a quantitative RT-PCR assay to measure the levels of BORIS mRNA. Endpoint RT-PCR assays were also used to investigate the possible expression of alternatively spliced variants. Using multiple primer sets and controls, we found that neither mature BORIS transcripts nor spliced variants are commonly expressed at detectable levels in malignant breast cells or tissues, although endogenous BORIS transcripts can be induced in MCF-7 cells following 5-aza-2′-deoxycytidine treatment. In conclusion, in most breast cancer cells, endogenous BORIS is unlikely to be expressed at sufficient levels to interfere with CTCF functions. Thus it is improbable that aberrant BORIS expression plays a role in most human breast cancers.
Introduction
BORIS, first described as “Brother Of the Regulator of Imprinted Sites,” or CTCF-like protein (CTCFL; NM_080618), is the sole known paralog of CTCF (CCCTC-binding factor; NM_006565) - a multifunctional DNA binding protein that uses different sets of zinc fingers to mediate distinct functions in regulation of gene expression. These functions include context-dependent promoter repression or activation, creation of modular hormone-responsive gene silencers, and formation of enhancer blocking elements (insulators) (reviewed in [1]–[3]). Recent evidence indicates that CTCF is involved in the global organization of chromatin, and “may be a heritable component of an epigenetic system regulating the interplay between DNA methylation, higher-order chromatin structure, and lineage-specific gene expression” [3]. Unlike CTCF, the expression of BORIS is normally restricted to specific cells in testes (the only cells where CTCF is not expressed), where it may play a role in reprogramming the methylation pattern of male germ line DNA [4].
The genomic organizations of the BORIS and CTCF genes, which are located on chromosomes 20q13.2 and 16q22.1 respectively, suggest that the two genes evolved from a gene duplication event during vertebrate evolution [5]. The amino acid sequences composing the two proteins' eleven zinc finger motifs are nearly identical, but the sequences at the amino- and carboxy- terminal ends diverge markedly. This likely provides the proteins with similar DNA binding specificities/affinities, yet distinct protein functions [4]. In fact, Sun and colleagues have recently demonstrated, using a DNA methylase-deficient cell model, that competition between BORIS and CTCF is a possibility when both proteins are present in equal amounts [6], a situation that may occur in certain cancer cells.
Aberrant expression of BORIS has been proposed to play a role in tumorigenesis [7]. The 20q13.2 region where the BORIS gene is located is commonly amplified in significant percentages of malignancies in a variety of organs, and may harbor one or more oncogenes [8]–[11]. Aberrantly expressed BORIS transcripts have also been reportedly detected in diverse human tumors and tumor-derived cell lines, including nearly all those derived from breast tissues [12]–[21]. Other reports indicate BORIS contributes to the promoter-specific demethylation and derepression of several cancer-testis (CT; a class of genes expressed normally in the testis, but activated in a wide range of tumor types) genes [15], [19], although BORIS expression by itself appears to be insufficient for the induction of CT gene expression [22].
Despite its relationship to CTCF and its location within a commonly amplified genomic region, recent findings in melanoma, ovarian, prostate, and bladder carcinomas [14], [20], [22] appear to controvert a broad tumorigenic role for BORIS. These studies found that BORIS transcript expression was not as frequent in primary melanomas (27%) [14], [20], [22] as originally estimated for melanoma cell lines (90%) [14], [20], [22], and when measured quantitatively, levels in tumors were not statistically different from those in normal prostate, bladder, and ovarian tissues [14], [20], [22]. While initiating a study to investigate the molecular mechanism(s) leading to the aberrant BORIS expression in breast cancers, we have obtained similar discordant results regarding the expression of BORIS. Using sensitive RT-PCR-based assays, employing multiple primer sets, we find that neither mature BORIS transcripts, nor spliced variants, are commonly expressed in malignant breast cells. Thus it is unlikely that aberrant BORIS expression plays a role in most human breast cancers.
Results
Nearly all breast cancer cell lines and tumors lack detectable levels of BORIS mRNA
To investigate the relationship between chromosome 20q13 amplification and BORIS mRNA levels, we developed a quantitative RT-PCR assay (exons 10–11, Figure 1) to measure the levels of BORIS mRNA within breast cell lines having either normal or amplified 20q13 DNA regions [23]. Sensitivity and amplification efficiencies of the BORIS, CTCF, and TBP PCR reactions were determined by amplifying dilutions of plasmids containing the corresponding coding sequences (Table 1). The BORIS qRT-PCR assay remained linear down to 2.86 attograms (2.86−18 g) of plasmid DNA, equivalent to 24 copies of BORIS. Using the same assay, BORIS transcript levels were undetectable in control BJ fibroblasts; but in BJ fibroblasts transduced with CMV-HA-BORIS adenovirus, were over 32,000 times greater than those in testis (Table 1). Altogether, the results of these individual assays and controls established the specificity, sensitivity, and large dynamic range of the BORIS qRT-PCR assay.
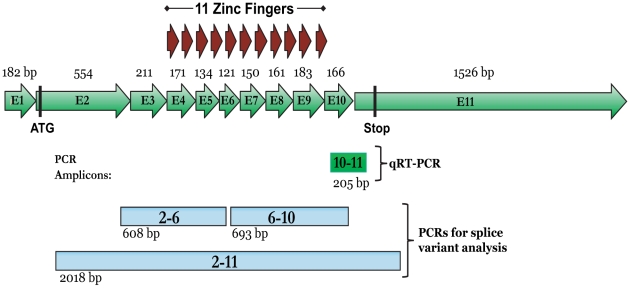
Figure 1. BORIS mRNA and PCR design.
The eleven exons of BORIS mRNA (ENSEMBL# ENST00000243914, CTCFL), and their corresponding sizes (base pairs) are depicted as green arrows. The ATG start codon, area encoding the eleven zinc fingers (red arrows), and TGA stop codon are marked for reference. The areas and sizes of the real-time PCR amplicon (10-11; bright-green box), and end-point PCR amplicons (2–6, 6–10, 2–11; blue boxes) used to detect alternatively spliced variants, are indicated.
Table 1. Detection of BORIS mRNA in breast cell lines, primary tumors, and controls.
| Gene | BORIS | BORIS | BORIS | BORIS | TBP | ||
| Exons | (2–11) | (2–6) | (6–10) | (10–11) | |||
| Data Type | PCR band | PCR band | PCR band | Ct value | Ct value | BORIS (%TBP) | |
| Category | Sample | ||||||
| Cell Lines | MDA-MB-453 | NP | - | - | ND | 26.9 | 0 |
| MDA-MB-231 | - | - | - | ND | 25.72 | 0 | |
| MCF-10A | NP | - | - | ND | 26.52 | 0 | |
| T47D | NP | - | - | ND | 26.79 | 0 | |
| MCF-12A | NP | - | - | ND | 26.76 | 0 | |
| MDA-MB-436 | NP | - | - | 37.71 | 26.17 | 0.033 | |
| MCF-7* | - | - | - | ND | 25.82 | 0 | |
| SUM185PE* | NP | - | - | ND | 26.91 | 0 | |
| UACC812* | NP | - | - | ND | 26.67 | 0 | |
| AU565* | NP | - | - | ND | 26.94 | 0 | |
| BT474* | NP | - | - | ND | 26.38 | 0 | |
| SKBR3* | NP | - | - | ND | 28.5 | 0 | |
| MDA-MB-435† | NP | + | + | 35.11 | 28.16 | 0.809 | |
| Breast | |||||||
| Tumors | T686 | NP | - | - | ND | 28.59 | 0 |
| T688 | NP | - | - | ND | 31.39 | 0 | |
| T689 | NP | - | - | ND | 28.99 | 0 | |
| T691 | NP | - | - | ND | 29.9 | 0 | |
| T693 | NP | - | - | ND | 28.1 | 0 | |
| T694 | NP | - | - | ND | 27.85 | 0 | |
| T695 | NP | - | - | ND | 29.78 | 0 | |
| T-Amb | NP | - | - | ND | 29.95 | 0 | |
| Normal Breast | |||||||
| Tissues | N-Amb | NP | - | - | ND | 29.9 | 0 |
| N697 | NP | - | - | NP | # | 0 | |
| Controls | Water | - | - | - | ND | ND | 0 |
| minus RT | - | - | - | ND | ND | 0 | |
| genomic DNA | - | - | - | ND | ND | 0 | |
| Testes | + | + | + | 27.35 | 24.03 | 9.151 | |
| BORIS plasmids (dilution series) | + | + | + | 18.47-35.34 | NA | NA | |
| BJ Fibroblasts | NP | - | - | ND | 24.07 | 0 | |
| BJ + Adeno-BORIS | NP | + | + | 13.09 | 24.62 | 2.96×105 |
Three endpoint RT-PCR assays, spanning different intron splice junctions, were designed to detect mature transcripts and alternatively spliced variants of BORIS mRNA (2–11, 2–6, 6–10; Figure 1). These RT-PCR reactions were performed using RNA derived from cell lines, breast tumors, normal breast tissue, or several different positive and negative controls (described in text). Cell lines with evidence of 20q13.2 amplification [23] are indicated by an asterisk (*). The detection of BORIS mRNA using the different end-point PCR assays is indicated by a “+”, whereas the lack of a product is indicated by a “-”. NP = not performed. A quantitative real-time RT-PCR (qRT-PCR) assay, spanning intron 10 (10–11), was used to quantify BORIS mRNA levels. The median threshold cycle (Ct; the PCR cycle for which products are detected above baseline), of triplicate reactions is reported for both BORIS (10–11) and TBP. BORIS mRNA was not detected (ND) in most cell lines and all tumors. Quantitative results were normalized and are expressed as a percentage of TBP expression (% TBP). For the N697 sample, only TBP end-point PCR was performed and it was positive (#). MDA-MB-435 is considered to be melanoma-derived (†).
Using this assay, we found evidence of BORIS expression in one breast cancer-derived cell line, MDA-MB-436, at a level (0.033% TBP) near the detection limit of 40 cycles (Figure 2A). A 24-fold higher level of BORIS (0.809% TBP) was detected, however, in the melanoma-derived MDA-MB-435 cell line. No evidence of BORIS expression was found in any of the remaining breast cell lines. Notably, neither MDA-MB-436 nor MDA-MB-435 exhibit 20q13 amplification (Table 1). Testis RNA was used as both a positive control and reference of physiological levels of BORIS in all experiments, and BORIS mRNA levels in testis were 272-fold higher than levels in the MDA-MB-436 cells. The successful amplification in all samples of TBP transcripts, used as internal controls, confirmed both the integrity and successful reverse transcription of the RNA (Table 1).
Figure 2. BORIS and CTCF mRNA levels in cell lines.
Levels of (A) BORIS and (B) CTCF mRNA were measured by real time PCR in cell lines and testis (provided as a physiological control), and are reported relative to the levels of internal reference transcript TBP. BORIS mRNA was detectable in the breast cell line, MDAMB436, and in the melanoma-derived cell line, MDAMB435, but not in the remaining breast-derived cell lines. CTCF mRNA levels were near or above the levels of TBP mRNA in all cell lines. Testis expressed the lowest CTCF level, near the level of BORIS in this sample (note the differences in scale between the graphs).
In addition to RT-PCR assays, we attempted to detect BORIS protein by immunoblot analyses; however, all of the commercially available antibodies that we tested were unable to detect BORIS protein in MCF-7, MDA-MB-231, or MDA-MB-435 cell lines infected with adenoviral BORIS expression vectors (data not shown). Virally mediated BORIS mRNA expression was confirmed in all three infected cell lines (7.1, 5.1, and 1.1% of the level of TBP transcripts, respectively).
To investigate the prevalence and levels of BORIS expression within breast tumors, samples of eight unrelated tissue specimens were analyzed by qRT-PCR. All tumors were high grade (grade III), invasive ductal carcinomas. BORIS transcripts were not detected in any breast tumor samples (Table 1). In addition, two non-malignant breast samples were also analyzed, and did not express detectable levels of BORIS transcripts (Table 1).
An alternatively spliced variant of BORIS, missing exon 7, is present in testis, but is absent in breast cancer cell lines and tumors
To investigate the possibility that alternatively spliced forms of BORIS, undetectable by the above qRT-PCR assay, were being expressed in breast cell lines and tissues, we developed three endpoint RT-PCR assays (Figure 1). The primers were designed to detect all published BORIS transcripts, including those generated through the use of alternative promoters [24]. Using these assays and the above-described controls, we found, in agreement with the qRT-PCR data, that BORIS transcripts were detectable in MDA-MB-435, testis, and BORIS-transduced fibroblasts. BORIS expression was not detectable in any of the remaining breast cell lines or tissues (Table 1). A splice variant was present, however, in testis, as indicated by two bands in both the Exon 2/11 (data not shown) and Exon 6/10 PCR reactions (Figure 3); these two variants were roughly equal in abundance. Variants were not detected using the Exon 2/6 primers (Figure 3). The Exon 6/10 products were isolated from agarose, cloned into the pGEM® T-Easy T/A vector (Promega, Madison, WI), and analyzed by DNA sequencing. The sequence of the larger product corresponded to the expected full-length BORIS sequence located between the assay's PCR primers. The smaller product was identical, except that it lacked 150 bases found in the larger product, in accordance with its estimated size determined from the agarose gel. The missing sequence was identified to be the entire seventh exon of BORIS.
Figure 3. Expression of an alternatively spliced variant of BORIS in testis.
BORIS specific primers, designed to amplify the region of BORIS between exons 2–6 (lanes 1–3) and exons 6–10 (lanes 5–7), were used to screen for possible alternatively spliced variants of BORIS. The expected sizes of these two products, based on the published sequence of mature BORIS mRNA, are 608 bp and 693 bp, respectively. These full-length products were present when testis cDNA (lane 1 & 5) and a plasmid containing a full length BORIS cDNA (lane 3 & 7) were used as PCR templates. In testis, an additional band (543 bp) was also amplified by the exon 6–10 PCR primers (lane 5). PCR products were not present in the testis (-RT) negative control reactions (lanes 2 & 6).
BORIS expression is detected in MCF-7 cells following 5-aza-2′-deoxycytidine treatment
Treatment of cells with the demethylating agent, 5-aza-2′-deoxycytidine (5-aza-dC), has been previously demonstrated to result in transcriptional activation and elevated levels of BORIS mRNA [14], [15], [19], [20]. To determine if similar effects occur in breast cells (as well as to test our assay's ability to detect endogenous BORIS transcripts), we evaluated BORIS mRNA levels by qRT-PCR in MCF-7 cells following exposure to 10 µM 5-aza-dC (72 hr exposure, followed by 24 hrs in growth medium without 5-aza-dC). This treatment of MCF-7 cells resulted in an increase in BORIS transcripts from undetectable levels to levels roughly equivalent to those in MDA-MB-435 melanoma cells (1.02 vs. 0.81%TBP), about 10% of the levels present in testis (Figure 4).
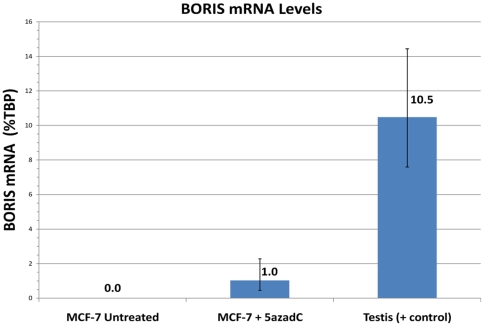
Figure 4. Endogenous BORIS mRNA expression is induced by 5-aza-2′-deoxycytidine.
BORIS mRNA levels in MCF-7 cells were determined by qRT-PCR (as in Figure 2). Endogenous BORIS mRNA was detectable (1.02% TBP) only after treatment with the demethylating agent, 5-aza-2′-deoxycytidine (5-aza-dC) and at a level approximately one tenth of that in testis (1.02% vs. 10.47% TBP).
CTCF mRNA is abundant in breast cancer cell lines
Aberrant BORIS expression is currently hypothesized to contribute to tumorigenesis by competing with CTCF for DNA binding sites. To evaluate the feasibility of this hypothesis, we measured CTCF mRNA levels in the breast tumor-derived cell lines (Figure 2B). CTCF transcripts were detected within all cell lines (and the testis control). Levels of CTCF mRNA ranged from 75–581% of TBP levels in the cell lines. Notably, testis exhibited the lowest level of CTCF transcripts, nearly equivalent to the level of BORIS mRNA in this sample (23.3 vs. 9.2% of TBP).
Discussion
In the course of investigating the relationship between chromosome 20q13 amplification and its effects on BORIS expression, we have found only a single cell line among 12 breast cancer cell lines and 8 tumors to express detectable levels of BORIS mRNA. We therefore conclude that the presence of 20q13 amplification is insufficient to derepress BORIS transcription, and that aberrant BORIS expression in breast cells is not a prevalent factor facilitating or maintaining their malignant character.
These results were unexpected, given claims of BORIS expression in a large number of tumors and its absence in associated somatic tissues [7], [19], [21]. To investigate this discordance, we re-evaluated both the specificity and sensitivity of our qRT-PCR assay using several controls. Specificity has been demonstrated by cloning and sequencing PCR products, by detecting BORIS transcripts in cells transduced with a BORIS-encoding adenovirus, and by the lack of PCR products in all negative controls (-RT, water, genomic DNA). Sensitivity of the assay has been demonstrated by amplifying log dilutions of a BORIS-containing plasmid. In addition, we were able to successfully detect endogenous BORIS mRNA in testis, two (breast- and melanoma-derived) cell lines, as well as in MCF-7 cells treated with 5-aza-dC.
We also performed PCR assays designed to detect possible splice variants that, if expressed in place of full-length BORIS, would lead to false-negative results. No such variants were determined to be present in any of the breast tissues or cell lines. However, a splice variant was detected in the testis, and it was expressed at approximately the same level as the full-length BORIS mRNA. This variant, which lacks exon 7 (encoding part of the zinc finger region), may have an important function in the testis, and its presence warrants further investigation.
Our results are inconsistent with those of Vatolin and colleagues, who reportedly detected expression of BORIS by endpoint RT-PCR in 7/8 breast cell lines and 11/12 breast tumors using PCR primers that span exons 3–10 [19]. Five of these eight cell lines are common to this study: T47D, MDA-MB-231, MDA-MB-453, MCF-7, and MDA-MB-435. Notably, the region amplified in our exon 6–10 PCR reactions was flanked by the same reverse primer sequence used by Vatolin et al. [19]. Therefore, the results of these two assays should have corresponded. Furthermore, we did not detect BORIS transcripts in any of the eight primary breast tumors we investigated, all of which were grade III tumors. Histopathological information was not provided in the original publication by Vatolin et.al., precluding a comparison of the tumor stage/grade in the two studies. In a more recent publication, this group reported that both BORIS mRNA and protein were detectable in multiple invasive breast carcinomas, although again stage and grade were not specified [7], [19], [21].
Overall, we did not detect BORIS mRNA in 6 of the 7 breast cell lines determined to be positive by Vatolin et.al. [19]. However, in accordance with their results, we did demonstrate BORIS mRNA expression in MDA-MB-435 cells and lack of expression in MCF-7 cells. The later report from this group [21] that MCF-7 cells express BORIS protein is in apparent conflict with this result. Our finding that 5-aza-dC treatment resulted in the activation of BORIS expression in MCF-7 cells is also consistent with previous findings. Of the two cell lines that we did find to be BORIS-positive, MDA-MB-435 cells expressed the highest levels of BORIS transcripts. However, although previously believed to be a breast cell line, MDA-MB-435 is currently regarded as a derivative of the malignant melanoma cell line M14 [25], [26]. This is notable due to the frequent expression of CT genes in melanomas; e.g., MAGEA1 (reviewed in [27]). Thus, the melanoma origin of MDA-MB-435 may explain why BORIS expression was found in this cell line.
We have determined and described both the specificity and sensitivity of our assays, and cannot account for these discrepant results other than to suggest that the levels described by Vatolin et.al. may have been lower than our threshold of detection. It is possible that the high number of PCR cycles used by Vatolin et al. (40–45 cycles) led to the detection of faint BORIS expression, possibly from subpopulations of cells. Based on their endpoint RT-PCR data, however, BORIS and CTCF transcripts appeared to be expressed at roughly equivalent levels in the breast cell lines. This contrasts with our qRT-PCR data in which we saw robust CTCF expression but undetectable BORIS levels in most cell lines and tissues. Although one other study has reported that BORIS protein is expressed in MDA-MB-231 cells [7], [19], [21], we were unable to detect BORIS mRNA in this cell line. In addition, using the same commercial antibody, we were unable to confirm the expression of BORIS protein in MDA-MB-231 cells. We cannot account for these discrepant findings, other than to suggest that variations in the cell line might be responsible. Nevertheless, our general conclusion is supported by other studies that have used serial analysis of gene expression (SAGE) to investigate aberrant gene expression in breast tumors and cell lines [28], [29]. These SAGE studies did not report the expression of BORIS mRNA in the samples analyzed. Notably, the study by Yao et al. [28] was specifically designed to identify differentially expressed genes encoded within commonly amplified chromosomal regions, including the 20q13 region where the BORIS gene is located.
Aberrant BORIS expression has been proposed to facilitate cell transformation by competing for CTCF binding sites, leading to disruption of insulator boundaries and abnormal gene expression [7]. For a competition model to be feasible in breast cancer, levels of BORIS and CTCF mRNA and/or protein would have to be comparable. We found however, that while CTCF mRNA was quite abundant in breast cells, BORIS mRNA was generally below our detection limit. Even in the MDA-MB-468 breast cell line, in which BORIS transcripts were detectable, levels of CTCF mRNA were, on average, at least 9000-fold higher than those of BORIS mRNA. At these disparate levels, it is unlikely that differences in protein stability would make sufficient BORIS protein available to compete with CTCF at DNA binding sites. Thus, aberrant BORIS expression is unlikely to compete with CTCF, even in the rare breast cancer cell line in which it is expressed.
Materials and Methods
Tissue samples
Frozen breast tissue specimens (seven malignant, and one reduction mammoplasty sample) were provided by the Cooperative Human Tissue Network. The tumor specimens were grade III (Scarf-Bloom-Richardson) invasive ductal carcinomas. Individual samples of RNA isolated from non-malignant and malignant breast, as well as testis, were purchased from Ambion (Austin, TX).
Cell culture
Normal BJ fibroblasts and the breast cancer-derived cell lines: MDA-MB-453; MDA-MB-231; T47D; MDA-MB-436; MCF-7; UACC-812; AU-565; BT474; and SK-BR-3; the non-malignant derived cell lines: MCF-10A and MCF-12A; as well as the MDA-MB-435 cell line, which is currently regarded to be a derivative of the M14 malignant melanoma cell line (and will be referred to as such herein) [25], [26], were obtained from the American Type Culture Collection. Another breast cancer cell line, SUM185PE, was provided by Dr. Joe Gray (LBNL). Cells were propagated and subcultured using conditions indicated by the supplier.
BORIS adenovirus
To create the recombinant adenovirus, a 3483 bp NheI/NotI DNA fragment, containing the BORIS coding region, was subcloned from pBIG2i-HA BORIS (generously provided by V. Lobanenkov, NIH) [15]. This fragment was ligated into an NheI/Not I linearized pShuttle vector (Adeno-X™ Expression System, Clontech). Generation of recombinant adenovirus and virus particles were completed using the manufacturer's protocol. This adenovirus contains the coding sequence of HA-tagged BORIS, verified by DNA sequencing, under control of the CMV promoter. Adenovirus titers were determined by immunocytochemical detection of the adenovirus hexon protein in infected HEK293 cells (Adeno-X™ Rapid Titer, Clontech). This virus was used to infect BJ fibroblasts using 10 infection-forming units per cell. Functional activity of the BORIS adenovirus was confirmed by qRT-PCR using RNA isolated from transduced cells.
RNA isolation and reverse transcription
Total RNA was isolated using silica-based spin-column extraction kits (RNeasy mini kit, Qiagen) using the manufacturer's protocol. Total RNA was treated with DNA-free DNAse I (Ambion) to reduce contaminating DNA. RNA integrity was evaluated by agarose gel electrophoresis using Gel Star (Cambrex) nucleic acid gel stain. Complimentary DNA (cDNA) was synthesized from 2 µg of total RNA by random-decamer primed reverse transcription using the Retroscript reverse transcription kit (Ambion) and the manufacturer's standard protocol. Negative (-RT) controls contained RNase-free water substituted for reverse transcriptase.
Quantitative real-time PCR
BORIS and CTCF mRNA levels were measured in breast tissue specimens and cell lines using the Platinum® Quantitative PCR Supermix (Invitrogen). The sequences of forward primers, reverse primers, and Taqman probes for the systems are described in Table 2. Both transcripts were amplified in parallel, along with a stably expressed reference gene, TBP (TATA box binding protein; NM_003194), in triplicate reactions, from equal amounts of cDNA (1 µl of the reverse transcription reaction). The qRT-PCR BORIS primers amplify a 205 base pair (bp) sequence spanning the splice junction between exons 10 and 11, flanking a 4734 bp intron. CTCF mRNA levels were measured by SYBR Green qRT-PCR (SYBR Greener™, Invitrogen), using the same conditions and controls described for the BORIS qRT-PCR assay. The CTCF primers amplify a 135 bp sequence element and flank a 3818 bp intron. The final concentrations were 300 nM of each forward and reverse primer, and 100 nM of probe, when used. PCR amplification was performed using the Biorad MyiQ Single-Color Real-Time PCR Detection System using the following standard amplification protocol: 50°C×2 min, 95°C×2 min, 40 cycles: 95°C×15 sec, 60°C×60 sec. Testis cDNA and plasmid DNA containing the BORIS coding sequence were used as positive controls, as was cDNA from fibroblasts transduced with the BORIS-containing adenovirus. Water and –RT reactions were both used as negative controls to detect possible amplification from contaminating DNA. The identities of the PCR products were confirmed by gel analyses and DNA sequencing. Amplification efficiencies, determined by amplifying log dilutions of plasmids containing the corresponding coding sequences, were determined to be near 100%. Relative levels of BORIS and CTCF transcripts were calculated using the delta Ct method and normalized to those of the TBP reference transcript using the formula: %TBP = 2−(Ct (BORIS)−Ct (TBP))×100%. Standard deviations of triplicate reactions were used to propagate error using the square root of the sum of squares method.
Table 2. Quantitative RT-PCR primer and probe sequences.
| BORIS, Exon 10/11 (qPCR) | For | 5′- ACCTGCACAGACATTCGGAGAAGT |
| Rev | 5′- AACTGTTCTCCCTTCGTGGTGGAA | |
| Probe | 5′- 6FAM-TTCCCTTTCCTGAAGCAGCCGACTTTGC-BHQ | |
| BORIS, Exon 2/11 | For | 5′- TGTGCAGAGAGAAAGACCATCGGA |
| Rev | 5′- GCAGTGAACATGCAACCTGACTCT | |
| BORIS, Exon 2/6 | For | 5′- TGGTGGCCAGTGAAGACAGTAAGT |
| Rev | 5′- GGATCGGACATGGCGCTTCAATTT | |
| BORIS, Exon 6/10 | For | 5′- CTTTCAGTGTTGCCAGTGCAGCTA |
| Rev | 5′- TTCTGACCCTTTGTGGCTTCCTTC | |
| CTCF | For | 5′- TCGTCGTTACAAACACACCCACGA |
| Rev | 5′- CTGCACAAACTGCACTGAAACGGA | |
| TBP | For | 5′- CACGAACCACGGCACTGATT |
| Rev | 5′- TTTTCTTGCTGCCAGTCTGGAC | |
| Probe | 5′- 6FAM-TGTGCACAGGAGCCAAGAGTGAAGA-BHQ |
Splice variant screening by conventional PCR
To detect the possible presence of alternatively spliced BORIS transcripts, three end-point PCR reactions were developed (Figure 1). Primer pairs were designed to amplify regions of BORIS mRNA between: 1) exons 2 and 11, 2) exons 2–6, and 3) exons 6–10. Reactions were performed using SYBR® GreenER PCR reagent (Invitrogen) and the Biorad PCR Detection System. The amplification parameters for these PCR assays were: 1) 50°C×2 min, 95°C×8.5 min, followed by 45 cycles 95°C×30 sec, 58°C×30 sec, 72°×2.5 min; 2) and 3) 50°C×2 min, 95°C×8.5 min, followed by 45 cycles 95°C×15 sec, 60°C×30 sec, 72°×1.5 min. Reaction products were analyzed as described above.
Immunoblot analysis
BORIS protein levels were analyzed in MCF-7, MDA-MB-231, and MDA-MB-435 cell lines. To serve as positive controls, each of these cell lines was infected with the BORIS adenovirus, and the resulting expression of BORIS mRNA was confirmed by qRT-PCR. Parallel cell cultures were lysed in RIPA buffer (50 mM Tris•HCl pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% Na Deoxycholate, 0.1% SDS) containing protease (Protease Inhibitor Cocktail Set III EDTA-free, Calbiochem) and phosphatase (Phosphatase Inhibitor Cocktail Set II, Calbiochem) inhibitors, both diluted 1∶100. The lysates were incubated on ice for 15 minutes, then centrifuged 24,000×g for 30 min at 4°C. Twenty-five micrograms of each lysate were loaded per well onto 4–12% gradient polyacrylamide gels (NuSep). After electrophoresis, the proteins were transferred to nitrocellulose, and probed with antibodies using standard conditions. Three anti-Boris antibodies were tested: Abcam (Ab18337, lots 123956 and 469726, 1∶2500 dilution), Rockland (600-401-907, lot 21606, 1∶1000 dilution), and Sigma (HPA001472, lot A08951, 4 ug/ml final concentration). The blots were probed with anti-β-actin antibodies (Sigma, A1978, lot 118K4827) to control for loading differences. Signals were imaged using an Odyssey infrared imaging system (Licor Biosciences).
Acknowledgments
We thank Dr. V. Lobanenkov for valuable discussions and reagents.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was funded by grants from the California Breast Cancer Research Program (9WB-0193), the Department of Defense Breast Cancer Research Program (W81XWH-04-1-0283), and by the Office of Health and Environmental Research, Health Effects Division, United States Department of Energy (contract no. DE-AC02-05CH11231). W.C. Hines and A. Bazarov received support from the Komen Foundation (fellowship PDF0707408 & grant BCTR0707231) and the Flight Attendant Medical Research Institute (032122). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ohlsson R, Renkawitz R, Lobanenkov V. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 2001;17:520–527. doi: 10.1016/s0168-9525(01)02366-6. [DOI] [PubMed] [Google Scholar]
- 2.Filippova GN. Genetics and epigenetics of the multifunctional protein CTCF. Curr Top Dev Biol. 2008;80:337–360. doi: 10.1016/S0070-2153(07)80009-3. [DOI] [PubMed] [Google Scholar]
- 3.Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loukinov DI, Pugacheva E, Vatolin S, Pack SD, Moon H, et al. BORIS, a novel male germ-line-specific protein associated with epigenetic reprogramming events, shares the same 11-zinc-finger domain with CTCF, the insulator protein involved in reading imprinting marks in the soma. Proc Natl Acad Sci U S A. 2002;99:6806–6811. doi: 10.1073/pnas.092123699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hore TA, Deakin JE, Marshall Graves JA. The evolution of epigenetic regulators CTCF and BORIS/CTCFL in amniotes. PLoS Genet. 2008;4:e1000169. doi: 10.1371/journal.pgen.1000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun L, Huang L, Nguyen P, Bisht KS, Bar-Sela G, et al. DNA methyltransferase 1 and 3B activate BAG-1 expression via recruitment of CTCFL/BORIS and modulation of promoter histone methylation. Cancer Res. 2008;68:2726–2735. doi: 10.1158/0008-5472.CAN-07-6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klenova EM, Morse HC, 3rd, Ohlsson R, Lobanenkov VV. The novel BORIS + CTCF gene family is uniquely involved in the epigenetics of normal biology and cancer. Semin Cancer Biol. 2002;12:399–414. doi: 10.1016/s1044-579x(02)00060-3. [DOI] [PubMed] [Google Scholar]
- 8.Tanner MM, Tirkkonen M, Kallioniemi A, Holli K, Collins C, et al. Amplification of chromosomal region 20q13 in invasive breast cancer: prognostic implications. Clin Cancer Res. 1995;1:1455–1461. [PubMed] [Google Scholar]
- 9.Kallioniemi A, Kallioniemi OP, Piper J, Tanner M, Stokke T, et al. Detection and mapping of amplified DNA sequences in breast cancer by comparative genomic hybridization. Proc Natl Acad Sci U S A. 1994;91:2156–2160. doi: 10.1073/pnas.91.6.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanner MM, Grenman S, Koul A, Johannsson O, Meltzer P, et al. Frequent amplification of chromosomal region 20q12-q13 in ovarian cancer. Clin Cancer Res. 2000;6:1833–1839. [PubMed] [Google Scholar]
- 11.Mahlamaki EH, Barlund M, Tanner M, Gorunova L, Hoglund M, et al. Frequent amplification of 8q24, 11q, 17q, and 20q-specific genes in pancreatic cancer. Genes Chromosomes Cancer. 2002;35:353–358. doi: 10.1002/gcc.10122. [DOI] [PubMed] [Google Scholar]
- 12.D'Arcy V, Abdullaev ZK, Pore N, Docquier F, Torrano V, et al. The potential of BORIS detected in the leukocytes of breast cancer patients as an early marker of tumorigenesis. Clin Cancer Res. 2006;12:5978–5986. doi: 10.1158/1078-0432.CCR-05-2731. [DOI] [PubMed] [Google Scholar]
- 13.Dougherty CJ, Ichim TE, Liu L, Reznik G, Min WP, et al. Selective apoptosis of breast cancer cells by siRNA targeting of BORIS. Biochem Biophys Res Commun. 2008;370:109–112. doi: 10.1016/j.bbrc.2008.03.040. [DOI] [PubMed] [Google Scholar]
- 14.Hoffmann MJ, Muller M, Engers R, Schulz WA. Epigenetic control of CTCFL/BORIS and OCT4 expression in urogenital malignancies. Biochem Pharmacol. 2006;72:1577–1588. doi: 10.1016/j.bcp.2006.06.020. [DOI] [PubMed] [Google Scholar]
- 15.Hong JA, Kang Y, Abdullaev Z, Flanagan PT, Pack SD, et al. Reciprocal binding of CTCF and BORIS to the NY-ESO-1 promoter coincides with derepression of this cancer-testis gene in lung cancer cells. Cancer Res. 2005;65:7763–7774. doi: 10.1158/0008-5472.CAN-05-0823. [DOI] [PubMed] [Google Scholar]
- 16.Looijenga LH, Hersmus R, Gillis AJ, Pfundt R, Stoop HJ, et al. Genomic and expression profiling of human spermatocytic seminomas: primary spermatocyte as tumorigenic precursor and DMRT1 as candidate chromosome 9 gene. Cancer Res. 2006;66:290–302. doi: 10.1158/0008-5472.CAN-05-2936. [DOI] [PubMed] [Google Scholar]
- 17.Risinger JI, Chandramouli GV, Maxwell GL, Custer M, Pack S, et al. Global expression analysis of cancer/testis genes in uterine cancers reveals a high incidence of BORIS expression. Clin Cancer Res. 2007;13:1713–1719. doi: 10.1158/1078-0432.CCR-05-2569. [DOI] [PubMed] [Google Scholar]
- 18.Ulaner GA, Vu TH, Li T, Hu JF, Yao XM, et al. Loss of imprinting of IGF2 and H19 in osteosarcoma is accompanied by reciprocal methylation changes of a CTCF-binding site. Hum Mol Genet. 2003;12:535–549. doi: 10.1093/hmg/ddg034. [DOI] [PubMed] [Google Scholar]
- 19.Vatolin S, Abdullaev Z, Pack SD, Flanagan PT, Custer M, et al. Conditional expression of the CTCF-paralogous transcriptional factor BORIS in normal cells results in demethylation and derepression of MAGE-A1 and reactivation of other cancer-testis genes. Cancer Res. 2005;65:7751–7762. doi: 10.1158/0008-5472.CAN-05-0858. [DOI] [PubMed] [Google Scholar]
- 20.Woloszynska-Read A, James SR, Link PA, Yu J, Odunsi K, et al. DNA methylation-dependent regulation of BORIS/CTCFL expression in ovarian cancer. Cancer Immun. 2007;7:21. [PMC free article] [PubMed] [Google Scholar]
- 21.D'Arcy V, Pore N, Docquier F, Abdullaev ZK, Chernukhin I, et al. BORIS, a paralogue of the transcription factor, CTCF, is aberrantly expressed in breast tumours. Br J Cancer. 2008;98:571–579. doi: 10.1038/sj.bjc.6604181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kholmanskikh O, Loriot A, Brasseur F, De Plaen E, De Smet C. Expression of BORIS in melanoma: lack of association with MAGE-A1 activation. Int J Cancer. 2008;122:777–784. doi: 10.1002/ijc.23140. [DOI] [PubMed] [Google Scholar]
- 23.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Renaud S, Pugacheva EM, Delgado MD, Braunschweig R, Abdullaev Z, et al. Expression of the CTCF-paralogous cancer-testis gene, brother of the regulator of imprinted sites (BORIS), is regulated by three alternative promoters modulated by CpG methylation and by CTCF and p53 transcription factors. Nucleic Acids Res. 2007;35:7372–7388. doi: 10.1093/nar/gkm896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rae JM, Creighton CJ, Meck JM, Haddad BR, Johnson MD. MDA-MB-435 cells are derived from M14 Melanoma cells–a loss for breast cancer, but a boon for melanoma research. Breast Cancer Res Treat. 2007;104:13–19. doi: 10.1007/s10549-006-9392-8. [DOI] [PubMed] [Google Scholar]
- 26.Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, et al. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- 27.Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer. 2005;5:615–625. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- 28.Yao J, Weremowicz S, Feng B, Gentleman RC, Marks JR, et al. Combined cDNA array comparative genomic hybridization and serial analysis of gene expression analysis of breast tumor progression. Cancer Res. 2006;66:4065–4078. doi: 10.1158/0008-5472.CAN-05-4083. [DOI] [PubMed] [Google Scholar]
- 29.Allinen M, Beroukhim R, Cai L, Brennan C, Lahti-Domenici J, et al. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell. 2004;6:17–32. doi: 10.1016/j.ccr.2004.06.010. [DOI] [PubMed] [Google Scholar]