Abstract
Cancer stem cells, which share many common properties and regulatory machineries with normal stem cells, have recently been proposed to be responsible for tumorigenesis and to contribute to cancer resistance1. The main challenges in cancer biology are to identify cancer stem cells and to define the molecular events required for transforming normal cells to cancer stem cells. Here we show that Pten deletion in mouse haematopoietic stem cells leads to a myeloproliferative disorder, followed by acute T-lymphoblastic leukaemia (T-ALL). Self-renewable leukaemia stem cells (LSCs) are enriched in the c-KitmidCD3+Lin− compartment, where unphosphorylated β-catenin is significantly increased. Conditional ablation of one allele of the β-catenin gene substantially decreases the incidence and delays the occurrence of T-ALL caused by Pten loss, indicating that activation of the β-catenin pathway may contribute to the formation or expansion of the LSC population. Moreover, a recurring chromosomal translocation, T(14;15), results in aberrant overexpression of the c-myc oncogene in c-KitmidCD3+Lin− LSCs and CD3+ leukaemic blasts, recapitulating a subset of human T-ALL. No alterations in Notch1 signalling are detected in this model, suggesting that Pten inactivation and c-myc overexpression may substitute functionally for Notch1 abnormalities2,3, leading to T-ALL development. Our study indicates that multiple genetic or molecular alterations contribute cooperatively to LSC transformation.
The PTEN–phosphatidylinositol-3-OH kinase (PI(3)K) pathway has been implicated in human leukaemogenesis2,4. Although acute deletion of the murine Pten gene in adult haematopoietic stem cells (HSCs) leads to defects in HSC self-renewal, it also causes a brief myeloproliferative disorder (MPD), followed by the development of acute leukaemia5,6. However, LSCs responsible for this transplantable disease have not been identified and, more significantly, the molecular mechanisms responsible for LSC formation remain to be elucidated.
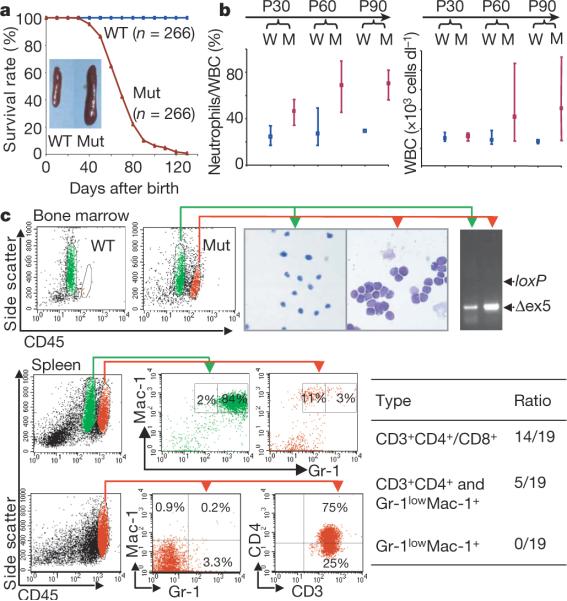
Here we report the establishment of a new leukaemia model. In contrast to the Mx-1–Cre inducible system, in which Pten is deleted in nearly 100% of adult HSCs5,6, Pten in this new model is conditionally deleted in 40% of fetal liver HSCs and their differentiated progeny (W.G. and H.W., unpublished observations) by the expression of the VE-cadherin-Cre (VEC-Cre) transgene7. VEC-Cre-mediated PTEN loss leads to a progressive development of MPD in the chronic phase followed by blast crisis. As early as one month after birth, mutant mice developed a myeloid shift with increased neutrophil counts (Fig. 1b, P30, and Supplementary Fig. 1, CP). One to two months later, mutant mice showed a marked increase in circulating neutrophils and white blood cells (Fig. 1b, P60–P90) and leukaemic blast invasion into haematopoietic and non-haematopoietic organs (Supplementary Fig. 1, BC). All mutant mice (n = 266) died with hepatomegaly and splenomegaly, and 70% of them suffered from enlarged thymus and lymph nodes (Fig. 1a and data not shown). By adopting CD45/side-scatter (CD45/SSC) fluorescence-activated cell sorting (FACS) analysis, a methodology used for the characterization of human leukaemic blasts8, we further identified two leukaemia subtypes (Fig. 1c): T-ALL (CD3+CD4+/CD8+) in 74% of the mice and acute myeloid leukaemia (AML)/T-ALL (Gr-1lowMac-1+ and CD3+CD4+) in the remaining 26%. This new model therefore shares a similar phenotype with the Mx-1-inducible model5,6 but progresses at a much slower pace (three to four months instead of three weeks) and develops a predominant T-ALL phenotype.
Figure 1. VEC-Cre-mediated Pten loss leads to MPD and leukaemogenesis.
a, Survival curve for mouse littermate pairs. Inset: representative spleens from mice at postnatal day 60 (P60). Mut, mutant; WT, wild type. b, Progressive alterations in PB: 30 littermate pairs (W, wild-type; M, mutant) per time point. Bars show the data range; points show averages. WBC, white blood cells. c, Identification of blast population by CD45/SSC plot in BM and spleen. The blasts were sorted for Giemsa–Wright staining, PCR genotyping and lineage analyses. Δex5, exon-5-deleted Pten allele.
To understand the molecular and genetic mechanisms involved in LSC formation, we sought to identify the LSC population in our model. We first determined whether leukaemia development was transplantable and autonomous for Pten-null cells by transplanting 2 × 106 cells isolated from the bone marrow (BM), spleen or thymus of chronic-phase mutant male mice with the ROSA26-LacZ reporter gene9 into sublethally irradiated mice with severe combined immunodeficiency disease (SCID) (Supplementary Fig. 2a). Because both LacZ reporter activation and the Pten gene deletion are controlled by the same Cre transgene, LacZ expression, measurable by a β-galactosidase fluorescent substrate and FACS analysis (FACS-Gal), was used to mark Pten-null cells in this and subsequent experiments. Our results demonstrated that 90% of the recipient mice (n = 22) developed leukaemia within three months after transplantation. Strikingly, more than 95% of leukaemic blasts in SCID recipients were LacZ+ and Pten-null, and infiltrated into non-haematopoietic organs (Supplementary Fig. 2b–d), in a similar manner to the primary disease (Supplementary Fig. 1). This result suggests that the leukaemia is initiated by donor-derived Pten-null LSCs.
LSCs have been reported to arise from HSCs10,11 or from myeloid and B-lineage progenitors12,13. Our initial screening of several stem-cell and progenitor markers identified a small c-Kitmid subgroup within the blast compartment (Supplementary Fig. 2e). When combined with CD3, a pan-T-cell marker, we further identified three major populations: CD3−, c-Kit−CD3+ and c-KitmidCD3+ (Supplementary Fig. 3). We then sorted these three subpopulations from an individual leukaemic mouse and performed transplantation experiments with various numbers of sorted cells (Fig. 2a and Supplementary Fig. 3). When 104 cKit−CD3+ cells were transplanted into sublethally irradiated SCID mice, no leukaemia development was observed within 100 days, the longest time point we have followed for transplantation experiments. This is in sharp contrast with T-ALL development in those recipients transplanted with 102–103 cKitmidCD3+ cells. These results suggest that LSCs are enriched in a cKitmidCD3+ subpopulation. Therefore, in contrast with the recent report14, leukaemia development in our Pten-null leukaemia model is driven by rare LSCs.
Figure 2. Self-renewing LSCs are enriched in the c-KitmidCD3+ compartment.
a, LSC identification. The experimental design is illustrated in Supplementary Fig. 3. Top: illustration of the three subpopulations that were sorted from BM of five independent leukaemic donor mice (cell fractions are denoted on each FACS plot). Bottom: summary of independent transplantation experiments with sorted and serially diluted cells. +, leukaemia development; −, leukaemia-free for more than 100 days; n.a., viable cells after sorting were not enough for transplantation. b, LSCs are self-renewing and lead to accelerated leukaemogenesis during serial transplantations. Top: illustration of the experimental procedure. Bottom: summary of the results. Red lines in the lower chart represent the means of leukaemia latencies.
LSCs should be able to self-renew and initiate leukaemogenesis through serial transplantations. To test the self-renewal capability of the putative c-KitmidCD3+ LSCs, we transplanted 1,000 c-KitmidCD3+Lin− cells sorted from primary mice and then from each passage. The same immunophenotypic c-KitmidCD3+Lin− cells were able to mediate 100% leukaemia development in the quaternary transplantation (Fig. 2b and Supplementary Fig. 4c). Although the same numbers of c-KitmidCD3+Lin− cells were serially transplanted, disease latencies were significantly shortened from 64 days to 20 days (Fig. 2b). Similar results were obtained when LacZ+c-KitmidCD3+-derived BM cells were serially transplanted (Supplementary Fig. 4a, b). Thus, similarly to a previous study of human LSCs10, LSCs in our model are self-renewable and give rise to a more accelerated disease through serial transplantations.
Because Pten deletion in HSCs results in a self-renewal defect and stem cell exhaustion5,6 (W.G., J.L. and H.W., unpublished observations), understanding the molecular mechanism involved in the self-renewal acquisition of the c-KitmidCD3+ LSCs became the central focus of our study. The Wnt/β-catenin signalling pathway is known to be involved in the regulation of HSC self-renewal15, and its activation is required for the in vitro replating activity of the LSCs from myeloid blast crisis of human chronic myeloid leukaemia13. To assess β-catenin activation in our model, we employed a monoclonal antibody (8E4) that specifically recognizes the unphosphorylated and thus activated form of β-catenin. When making a comparison with the same immunophenotypic populations from wild-type (WT) mice, we detected a marked increase in unphosphorylated β-catenin in more than 90% of c-KitmidCD3+ LSCs and c-Kit−CD3+ blasts (Fig. 3a, b, Supplementary Figs 5 and 6c, and data not shown) and modest increases in the c-KithighLin− HSC/progenitor population in the blast-crisis mice (Fig. 3b) and the CD3−CD4−CD8− T progenitors in the chronic-phase mice (Supplementary Fig. 6b). This result suggests that activation of β-catenin may be associated with LSC formation and T-ALL development.
Figure 3. β-Catenin activation in LSCs and its role in leukaemogenesis.
a, Accumulation of unphosphorylated β-catenin in blasts. Cytospin slides with thymic cells were stained with the monoclonal antibody 8E4, which recognizes unphosphorylated β-catenin. Top: representative fluorescent images. CP, chronic phase; BC, blast crisis. Scale bars, 25 μm. Bottom: representative confocal images. DAPI, 4′,6-diamidino-2-phenylindole. Original magnification ×100. b, Marked increase in unphosphorylated β-catenin levels in the LSC and blast populations. BM cells were pooled from two blast-crisis or WT littermates and were lineage (Lin)-depleted before FACS analysis. c, Decreased and delayed leukaemogenesis after ablation of one allele of Ctnnb1. Kaplan–Meier survival curves with Logbank statistical analysis (denoted on the curves) summarize leukaemia development in transplantation experiments. Blue line, PtenloxP/loxP;Ctnnb1loxP/+;VEC-Cre+; green line, PtenloxP/loxP;VEC-Cre+.
To improve our understanding of the role of β-catenin in LSC formation, we took a genetic approach by simultaneously deleting the genes encoding PTEN and β-catenin (Pten and Ctnnb1, respectively) in HSCs. The resulting PtenloxP/loxP;Ctnnb1loxP/loxP;VEC-Cre+ mice are embryonic lethal. PtenloxP/loxP;Ctnnb1loxP/+;VEC-Cre+ mice developed MPD with 100% penetrance at about 30 postnatal days (Supplementary Fig. 7a, b) but died with vascular complication before 68 postnatal days (data not shown). This prompted us to use the transplantation assay to compare the leukaemogenic potential between PtenloxP/loxP;Ctnnb1loxP/+;VEC-Cre+ (blue line; three independent experiments) and PtenloxP/loxP;VEC-Cre+ (green line; four independent experiments) mice. A gradient of leukaemogenic potential was observed in cells derived from different mutant haematopoietic organs (thymus > spleen > BM), which was consistent with the relative enrichment of LSCs in these organs. The occurrence of acute leukaemia and death were markedly decreased and delayed when one allele of Ctnnb1 was deleted from donor cells (Fig. 3c and Supplementary Fig. 7c). These results suggest that dysregulated β-catenin activity may contribute to LSC formation and leukaemogenesis in our model.
Chromosomal translocations are frequently associated with human leukaemia and lymphoma. To investigate chromosomal abnormalities in our model, we employed cytogenetic approaches, including spectral karyotyping (SKY) and fluorescence in situ hybridization (FISH) analyses. SKY analysis revealed no recurrent structural abnormalities in BM cells from chronic-phase mutant or WT mice (Fig. 4d). In contrast, all three blast-crisis samples we initially analysed showed the same T(14;15) chromosomal translocation in 9–61% of analysed metaphases (Fig. 4a, boxed chromosomes, and Fig. 4d), which was reminiscent of leukaemic clonal expansion in humans. Of the cells carrying T(14;15), 53% contained two or even three copies of Der(15)T(14;15) (Fig. 4a, b), suggesting a strong selective pressure in cells with this translocation and implying a possible role of translocation-associated genes in leukaemic transformation.
Figure 4. The recurring translocation T(14;15) involves the Tcra/Tcrd cluster and the c-myc gene and results in aberrant overexpression of c-myc in LSCs and T-ALL blasts.
a, Detection of T(14;15) by SKY analysis in metaphases prepared directly from BM of colcemid-treated WT or mutant mice. b, Breakpoint (BP) identification by two-colour FISH analysis. The results with green-labelled and red-labelled BAC probes are illustrated in the lower images and explained in the upper diagram. c, A schematic mapping of the T(14;15) translocation. BP, breakpoint. mChr, mouse chromosome. d, Recurring T(14;15) in primary T-ALL mutants and transplants with chronic-phase BM (BMT), splenic (SpT) or LSC cells was analysed by metaphase SKY (mSKY), metaphase FISH (mFISH) and interphase FISH (iFISH). Values shown are percentages. e, c-myc messenger RNA overexpression (means ± s.d.) in both primary blast-crisis (BC) mice and leukaemia transplants was detected by RT–PCR and analysed by Student's t-test. CP, chronic phase; Ct control, WT littermates (for primary mice) or an unrelated SCID mouse (for WT and leukaemic transplants). f, Overexpression of c-myc only in CD3+ splenic cells of T-ALL (n = 3). The percentage with c-myc overexpression in the CD3− or CD3+ compartment is denoted in parentheses. Grey, control (a mixture of WT and T-ALL cells); blue, WT; red, T-ALL.
Because breakpoints in chromosomes 14 and 15 seemed to be located at 14qC2–C3 and 15qD3 (Fig. 4a), we searched for translocation targets in the public domain and identified the T-cell antigen receptor (TCR)-α/δ (Tcra/Tcrd) cluster on chromosome 14qC2 and c-myc on chromosome 15qD3 as potential candidates. A similar translocation of t(8;14) is known to be associated with a subset of human T-ALL16. Using two-colour FISH analysis with validated bacterial artificial chromosome (BAC) clones (Supplementary Fig. 8a), we confirmed the translocation between Tcra/Tcrd and c-myc and narrowed down the breakpoints to two minimal regions (Fig. 4b, c, and Supplementary Fig. 8b): a roughly 209,000-base-pair (209-kb) fragment containing the TRAC gene and the TRA enhancer and 680 kb between the genes Pvt1 and Tsg101-ps. The same translocation was found in four additional leukaemic recipients transplanted with primary chronic mutant cells or sorted LSCs (Fig. 4d and Supplementary Fig. 8c), suggesting that T-ALL can be recapitulated at the genetic level and that the genomic abnormality is intrinsic to LSCs.
To determine whether c-myc expression was indeed altered by Tcra regulatory machinery, as reported for human T-ALL17, we quantified c-myc expression in BM and thymic cells with the use of quantitative RT–PCR and FACS analysis. Strikingly, c-myc expression was not changed in the chronic phase but was markedly increased in BM or thymic cells isolated from primary or transplanted blast-crisis mice (Fig. 4e). Furthermore, c-myc overexpression was detected only in LSCs and CD3+ blast cells but not in HSCs or CD3− cells (Fig. 4f and Supplementary Fig. 9a, b). These results rule out a predominant role of the deregulated PTEN–PI(3)K–AKT pathway in c-myc overexpression and highlight the importance of overexpressed C-myc in the formation of CD3+ LSCs and T-ALL development in our model.
Our study provides strong evidence that the molecular and genetic events involved in `multiple-hit' leukaemogenesis are likely to take place at the levels of HSCs and LSCs18. In this model, Pten inactivation in HSCs serves as the first hit to activate the PI(3)K–AKT pathway, conferring survival and proliferative advantages, and to promote genomic instability19, leading to additional alterations. Among them, activation of β-catenin may contribute to the acquisition of self-renewal capacity of LSCs, while the T(14;15) chromosomal translocation results in T-lineage-specific overexpression of c-myc, which may promote Pten-null LSC self-renewal and lead to T-ALL development. The sequential order of β-catenin activation and c-myc overexpression is currently unknown but it does not seem that c-Myc function is mediated by β-catenin signalling (Supplementary Fig. 9c, d).
Dysregulated NOTCH1 signalling is involved in about 56% of human T-ALL3. However, in our leukaemia samples we detected no mutations in Notch1 heterodimerization (HD) domain and PEST domain, two mutation hot-spots associated with human T-ALL (Supplementary Fig. 10a). Furthermore, neither a consistent decrease in Fbxw7 expression, a negative regulator of Notch signalling that is frequently altered in human T-ALL2, nor an increase in the expression of the NOTCH1 target gene Hes1 was detected in this model (Supplementary Fig. 10b). Given recent reports that the activated Notch1 signalling directly induces c-myc overexpression20–22 and modulates Pten expression23, the T(14,15)-mediated c-myc overexpression and Pten deletion together may substitute functionally for Notch1 mutations, leading to T-ALL development in our model.
METHODS SUMMARY
Mice
Mice were maintained in the animal facility of University of California Los Angeles in accordance with federal and institutional guidelines and were used between 1 and 5 months of age unless otherwise noted. PtenloxP/loxP mice have been deposited to the Jackson Laboratory.
Transplantation assay
Cells were directly collected from or sorted from BM, spleen or thymus, balanced with SCID carrier cells in some cases and transplanted into sublethally irradiated SCID recipients. Leukaemia was determined by CD45/SSC FACS, FACS-Gal and histological analyses.
Flow cytometric (FACS) analysis
Haematopoietic cells were obtained by flushing femurs or smashing the spleen and thymus, and were then stained with antibodies. These cells were either analysed or sorted by flow cytometry. Lineage depletion was performed with microbeads (Miltenyi Biotec). FACS-Gal analysis was performed by fluorescein di-β-D-galactopyranoside loading and FACS analysis. To analyse activated β-catenin and c-Myc, cells were stained with surface markers, fixed, permeabilized and stained with corresponding antibodies for FACS analysis.
Profiling and histology analysis of peripheral blood
Peripheral blood (PB) was collected, analysed for PB profile, spread for blood smears and stained with Giemsa stain or Giemsa–Wright (Fisher). Tissues dissected from killed mice were fixed, embedded in paraffin and sectioned for haematoxylin/eosin staining. Cytospin slides with single suspension cells were prepared for immunochemistry with antibodies.
SKY and FISH analysis
Metaphases were prepared directly from BM cells in colcemid-treated mice for SKY assay and FISH analysis. Six murine DNA BAC clones were obtained, prepared, and validated for FISH analysis.
RT–PCR
Total RNA was extracted and reverse-transcribed into complementary DNAs. These cDNAs were diluted for real-time RT–PCR with primers for the genes of interest and reference. Ct was obtained for a statistical calculation of relative fold changes.
METHODS
Mice
VEC-Cre+ transgenic mice (129/C57 mixed background)7 were carefully analysed and confirmed without any aberrant phenotypes before being crossed to PtenloxP/loxP mice (129/Balb/c mixed background)24 to obtain PtenloxP/+;VEC-Cre+ mice, which were backcrossed to PtenloxP/loxP mice to obtain PtenloxP/loxP;VEC-Cre+ mutant mice for this study. We also crossed PtenloxP/loxP;VEC-Cre+ mice with ROSA26-LacZ+ reporter transgenic mice9, to obtain mice of PtenloxP/loxP;VEC-Cre+;LacZ+, Pten+/+;VEC-Cre+;LacZ+ and PtenloxP/loxP;VEC-Cre−;LacZ+ for transplantation and FACS-Gal analysis. Ctnnb1loxP/loxP mice25 were crossed with PtenloxP/loxP;VEC-Cre+ mice to generate PtenloxP/loxP;Ctnnb1loxP/+;VEC-Cre+ mice. Mouse genotypes were verified by PCR analysis with the primer sets for Pten, Ctnnb1, Cre and LacZ (Supplementary Fig. 11).
Transplantation assay
As illustrated in Supplementary Fig. 2a, cells harvested from BM, spleen or thymus of P30–P60 Pten+/+;VEC-Cre+;LacZ+ WT, PtenloxP/loxP;VEC-Cre+;LacZ+ or PtenloxP/loxP;Ctnnb1loxP/+;VEC-Cre+ chronic mutant mice were injected into tail veins of 2–4-month-old CBySmn.CB17-Prkdcscid SCID female mice (Taconic), which had been sublethally irradiated at a dose of 180–200 rad on a 137Cs Mark I irradiator (J. L. Shepherd & Associates). Development of MPD and leukaemia was monitored by FACS-Gal analysis on PB every 2 weeks.
For LSC identification, each of the three populations, as shown in Fig. 2b, was sorted from BM of blast-crisis mutants, serially diluted and balanced with Pten+/+ carrier cells from SCID mice before being transplanted into sublethally irradiated SCID recipients. The relative fractions of these three populations varied from mouse to mouse. Leukaemia was determined by CD45/SSC FACS, FACS-Gal and histological analyses.
In serial transplantation assays, when a transplant recipient with 900 or 1,000 candidate LSCs (c-KitmidCD3+Lin−) developed leukaemia, the same number of LSCs sorted (Fig. 2c) or various numbers of BM cells harvested (Supplementary Fig. 4c) from this recipient mouse were balanced with Pten+/+ SCID carrier cells in most cases (indicated in the figure legends) and transplanted into secondary SCID recipients. The same procedure was repeated up to the fourth passage.
Flow cytometric (FACS) analysis
The cells stained with antibodies were either analysed by flow cytometry on a BD FACSCalibur or FACScan flow cytometer (BD Biosciences) with modifications by Cytek or sorted on a BD FACSV antage s.e. sorter (BD Biosciences). Cells were stained with fluorescein isothiocyanate (FITC)-conjugated, R-phycoerythrin (PE)-conjugated, allophycocyanin (APC)-conjugated or APC-Cy7-conjugated antibodies, including Ter119, Gr-1 (RB6-8C5), Mac-1 (M1/70), CD3 (145-2C11), CD4 (GK1.5), CD8 (53-6.7), B220 (RA3-6B2), CD19 (1D3), c-Kit (2B8), Sca-1(E13-161.7) or CD45 (30-F11) (from BD Pharmingen or eBioscience). For partial lineage depletion, cells were stained with lineage markers (PE–Ter119 and B220), followed by anti-PE microbeads (Miltenyi Biotec) and sorted for Linlow/− population in accordance with the manufacturer's instruction. Fluorescein di(β-D-galactopyranoside) (FDG) was obtained from Invitrogen or Sigma and used for FACS-Gal analysis in accordance with the manufacturer's instructions.
Leukaemic blasts were analysed on CD45/SSC plots as described previously8. An abnormal blast population was detected in all mutant animals at blast crisis and constituted more than 20% of the total leukocytes (Fig. 1c), satisfying the French–American–British (FAB) criteria for human acute leukaemia. The blast cells were Pten-null, large, immature and morphologically distinct from normal lineage cells but similar to human leukaemic blasts (Fig. 1c). Leukaemia with blasts positive for CD3, CD4 and/or CD8 was considered T-ALL, whereas leukaemia with more than 3% of Gr-1lowMac-1+ blasts was AML. Blasts with both CD3+CD4+ and Gr-1lowMac-1+ may be either a mixture of T-ALL and AML or T-ALL bearing myeloid markers26.
Intracellular staining with an anti-β-catenin (unphosphorylated) mouse monoclonal antibody (clone 8E4; AXXORA or Millipore/Upstate), anti-c-myc rabbit polyclonal antibody (Cell Signaling Technology) or control mouse or rabbit IgG, followed by staining with donkey anti-mouse-FITC or anti-rabbit-FITC antibody (Jackson Immunoresearch Laboratories), was performed with the Fix & Perm Kit (Caltag) in accordance with the manufacturer's instructions.
PB profiling and histology analysis
For PB profile analysis, 200 μl of PB was collected by retro-orbital bleeding and analysed on a HemaVet HV950FS (Drew Scientific). Blood smears with eye or tail PB were subjected to Giemsa (Sigma) or Giemsa–Wright (Fisher) staining, in accordance with the manufacturers' instructions.
For histological analysis, tissues were fixed with Z-Fix (Anatech) or 10% formalin (Fisher) for 12 h, embedded with paraffin and sectioned for haematoxylin and eosin staining at the Tissue Procurement and Histology Core Laboratory at UCLA. Cytospin slides prepared with single suspension cells on a Shandon Cytospin 2 (Thermo) were fixed in 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, blocked with Mouse-on-Mouse blocking solution (Vector Laboratories) and immunostained with anti-un-phosphorylated β-catenin antibody (clone 8E4), anti-mouse IgG-biotin, and streptavidin-FITC (Vector Laboratories). Images were taken with a Macrofire charge-coupled device camera (Optronics) under a BX60 microscope (Olympus).
SKY and FISH analysis
Metaphases of BM cells were prepared directly from colcemid-treated mice to avoid any artificial chromosomal abnormality introduced by in vitro cell culture. To prepare metaphases, 250 μl of 200 μgml−1 colcemid (Sigma) was injected intraperitoneally into mice. After 30 min, BM cells were flushed out with 30 ml of 0.06M potassium chloride, lysed at 37 °C for 20 min and fixed with 20 ml of fixative (3:1 methanol/acetic acid) for 10 min at 22–25 °C. The SKY assay was performed in accordance with the manufacturer's instructions (Applied Spectral Imaging) and images were acquired using a SD300 Spectracube system (Applied Spectral Imaging) mounted in an Olympus BX60 microscope equipped with a custom-designed optical filter (SKY-1; Chroma Technology). The conversion of the emission spectra to the display colours was achieved by assigning blue, green and red colours to specific sections of the emission spectrum. For FISH analysis, murine DNA BAC clones RP23-6A14, RP23-357G5, RP23-98D8, RP24-194H23 and RP23-55P19 were obtained from the Children's Hospital Oakland Research Institute, and RG331N4 was obtained from Invitrogen. Their genomic sequences are available in the National Center for Biotechnology Information mouse genome resources (http://www.ncbi.nlm.nih.gov/genome/guide/mouse/). The BAC DNAs were prepared with a Qiagen Large Construction Kit (Qiagen) and labelled with SpectrumGreen-conjugated dUTPs (RG331N4, RP23-6A14 and RP23-357G5 on chromosome 14) or SpectrumRed-conjugated dUTPs (RP23-98D8, RP24-194H23 and RP23-55P19 on chromosome 15) using the Vysis Nick Translation Kit (Abbott Molecular), in accordance with the manufacturers' instructions. Single-colour and dual-colour FISH assays were performed as described previously27. FISH images were captured with a charge-coupled device camera under a Zeiss Axio Imager Z1 fluorescence microscope equipped with proper filters (Zeiss).
Quantitative RT–PCR
Total RNA was extracted with a Qiagen RNeasy Micro RNA Kit (Qiagen) from mouse BM, splenic cells and thymic cells, or FACS-sorted cells from BM, and reverse-transcribed into cDNA with Superscript III Reverse Transcriptase (Invitrogen), in accordance with the manufacturers' instructions. cDNA was diluted 1:20 and mixed with the SYBR Green I mix (Bio-Rad) to perform RT–PCR with primers for the target genes c-myc, Fbxw7 or Hes1 or the reference gene for β-actin (Actb; sequences are given in Supplementary Fig. 11) in an iCycler (Bio-Rad). Amplification of correct products with no genomic or non-specific noise was verified on agarose gels. Each reaction was repeated three or four times to ensure Ct consistency (a Ct variation of one cycle or less), and the mean Ct was used for statistical calculation of relative fold changes. The relative fold change for each target mRNA was calculated as 2ΔCt,target(control−sample)−ΔCt,reference(control−sample). The control samples were obtained from either unrelated WT mice or WT littermates (see figure legends).
Supplementary Material
Acknowledgements
We thank O. Witte, K. Dorshkind, J. Said, R. Gatti, K. Sakamoto, S. Schubbert, R. Hill and B. Valamehr for helpful comments; D. Cheng from O. Witte's laboratory for cell sorting; J. Gao for retro-orbital bleeding and genotyping; and the Department of Pathology's Tissue Procurement Core Laboratory (partly supported by National Cancer Institute grant CA16042) for tissue procurement. W.G. is supported by a California Institute of Regenerative Medicine (CIRM) training fellowship. J.E.Y. was supported by the University of California Los Angeles Amgen Scholar Program. J.Y.C. is supported by the Beckman Undergraduate Research Program.
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 2.Maser RS, et al. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature. 2007;447:966–971. doi: 10.1038/nature05886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weng AP, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 4.Fukuda R, et al. Alteration of phosphatidylinositol 3-kinase cascade in the multilobulated nuclear formation of adult T cell leukemia/lymphoma (ATLL) Proc. Natl Acad. Sci. USA. 2005;102:15213–15218. doi: 10.1073/pnas.0507184102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yilmaz OH, et al. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 2006;441:475–482. doi: 10.1038/nature04703. [DOI] [PubMed] [Google Scholar]
- 6.Zhang J, et al. PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature. 2006;441:518–522. doi: 10.1038/nature04747. [DOI] [PubMed] [Google Scholar]
- 7.Alva JA, et al. VE-Cadherin-Cre-recombinase transgenic mouse: a tool for lineage analysis and gene deletion in endothelial cells. Dev. Dyn. 2006;235:759–767. doi: 10.1002/dvdy.20643. [DOI] [PubMed] [Google Scholar]
- 8.Borowitz MJ, Guenther KL, Shults KE, Stelzer GT. Immunophenotyping of acute leukemia by flow cytometric analysis. Use of CD45 and right-angle light scatter to gate on leukemic blasts in three-color analysis. Am. J. Clin. Pathol. 1993;100:534–540. doi: 10.1093/ajcp/100.5.534. [DOI] [PubMed] [Google Scholar]
- 9.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nature Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 10.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nature Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 11.Passegue E, Wagner EF, Weissman Jun IL. B deficiency leads to a myeloproliferative disorder arising from hematopoietic stem cells. Cell. 2004;119:431–443. doi: 10.1016/j.cell.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 12.Castor A, et al. Distinct patterns of hematopoietic stem cell involvement in acute lymphoblastic leukemia. Nature Med. 2005;11:630–637. doi: 10.1038/nm1253. [DOI] [PubMed] [Google Scholar]
- 13.Jamieson CH, et al. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N. Engl. J. Med. 2004;351:657–667. doi: 10.1056/NEJMoa040258. [DOI] [PubMed] [Google Scholar]
- 14.Kelly PN, et al. Tumor growth need not be driven by rare cancer stem cells. Science. 2007;317:337. doi: 10.1126/science.1142596. [DOI] [PubMed] [Google Scholar]
- 15.Reya T, et al. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature. 2003;423:409–414. doi: 10.1038/nature01593. [DOI] [PubMed] [Google Scholar]
- 16.Erikson J, et al. Deregulation of c-myc by translocation of the alpha-locus of the T-cell receptor in T-cell leukemias. Science. 1986;232:884–886. doi: 10.1126/science.3486470. [DOI] [PubMed] [Google Scholar]
- 17.Boxer LM, Dang CV. Translocations involving c-myc and c-myc function. Oncogene. 2001;20:5595–5610. doi: 10.1038/sj.onc.1204595. [DOI] [PubMed] [Google Scholar]
- 18.Weissman I. Stem cell research: paths to cancer therapies and regenerative medicine. J. Am. Med. Assoc. 2005;294:1359–1366. doi: 10.1001/jama.294.11.1359. [DOI] [PubMed] [Google Scholar]
- 19.Shen WH, et al. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell. 2007;128:157–170. doi: 10.1016/j.cell.2006.11.042. [DOI] [PubMed] [Google Scholar]
- 20.Palomero T, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc. Natl Acad. Sci. USA. 2006;103:18261–18266. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma VM, et al. Notch1 contributes to mouse T-cell leukemia by directly inducing the expression of c-myc. Mol. Cell. Biol. 2006;26:8022–8031. doi: 10.1128/MCB.01091-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weng AP, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20:2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palomero T, et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nature Med. 2007;13:1203–1210. doi: 10.1038/nm1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lesche R, et al. Cre/loxP-mediated inactivation of the murine Pten tumor suppressor gene. Genesis. 2002;32:148–149. doi: 10.1002/gene.10036. [DOI] [PubMed] [Google Scholar]
- 25.Brault V, et al. Inactivation of the β-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development. 2001;128:1253–1264. doi: 10.1242/dev.128.8.1253. [DOI] [PubMed] [Google Scholar]
- 26.Putti MC, et al. Expression of myeloid markers lacks prognostic impact in children treated for acute lymphoblastic leukemia: Italian experience in AIEOP-ALL 88–91 studies. Blood. 1998;92:795–801. [PubMed] [Google Scholar]
- 27.Boomer T, et al. Detection of E2A translocations in leukemias via fluorescence in situ hybridization. Leukemia. 2001;15:95–102. doi: 10.1038/sj.leu.2401988. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.