Abstract
Respiratory viruses are difficult to characterize in the airborne environment due to their low concentration and the presence of a wide range of inhibitors. As a first step in studying airborne viruses, we optimized molecular biology methods to quantify influenza viruses and human rhinovirus. Quantitative PCR was used as an endpoint to evaluate RNA extraction techniques and reverse transcription protocols. We found that a Trizol-chloroform extraction and MultiScribe™ RT increased virus detection 10-fold compared to methods used in published field studies of airborne respiratory viruses. Virus was recovered without inhibition from samples contaminated with up to 50 μg/sample of particulate matter. The methods developed can be used in studies of airborne respiratory viruses.
Introduction
Human respiratory viruses are difficult to characterize in the airborne environment due to their low concentration and the presence of a wide range of contaminants which can inhibit laboratory assays. Molecular methods such as quantitative polymerase chain reaction (qPCR) offer increasingly sensitive techniques to detect small numbers of viruses but can be inhibited or enhanced by airborne contaminants such as humic acids, metals, proteins and other free nucleic acids[1]. Methods developed to overcome inhibitor effects include increasing template concentrations[2], adding internal standards[3], diluting environmental samples[4, 5], using alternative DNA polymerases[4, 6], and adding proteins post-extraction[7]. However, due to the complexity of the contaminant mixtures and their degree of influence on microorganisms, there is no general method that can be used to eliminate their effects in all cases[1, 5, 8] and optimized protocols have to be evaluated for each application.
Viruses are entrained in the air when infected humans cough, sneeze, talk, and breathe[9]. They are quickly diluted when released into the airborne environment, necessitating large sample collection volumes in order to achieve numbers detectable by laboratory assays[10]. As a first step in studying airborne human respiratory viruses, we optimized molecular biology methods to quantify influenza viruses and human rhinovirus in bench experiments. Both viruses are important agents of respiratory disease, believed to be transmitted at least in part via the airborne route[11–14]. We first used quantitative PCR as an endpoint to evaluate RNA extraction techniques and reverse transcription protocols for influenza virus. We then evaluated the degree of inhibition by contaminating influenza virus and human rhinovirus samples with a range of airborne particulate matter.
Materials and methods
Influenza virus optimization study
Virus
Influenza A/PR8/34 (H1N1) virus was obtained from Advanced Biotechnologies Incorporated (Columbia, MD) with a concentration of 1.9×1011 virus particles per ml as determined by electron microscopy. Influenza A virus was diluted 10−1 in phosphate buffered saline with calcium and magnesium (PBS++) and 0.1% bovine serum albumin (BSA) (HyClone, Logan, UT) and stored at −80°C in 15 μl single-use aliquots.
RNA extraction
Two viral RNA extraction methods were tested: a Trizol-chloroform based method modified from a protocol developed for extraction of nasal swab and lavage samples[15] and QIAamp viral RNA columns (Qiagen, Valencia, CA). The Qiagen columns were selected because they have been used previously to isolate human rhinovirus and SARS from air samples [16, 17]. A third method using magnetic beads (Agencourt, Beverly, MA) was tested and eliminated in early experiments due to low viral RNA recovery (data not shown). In all methods tested, RNA was extracted from a 400 μl aliquot of influenza virus A, suspended in 60 μl of nuclease-free water (Promega, Fitchburg, WI), and immediately reverse-transcribed to cDNA. Unused RNA was stored at −80°C.
Reverse-transcription
Three reverse transcription (RT) kits were tested with influenza virus RNA extracted using the methods described above: 1) Omniscript RT kit (Qiagen, Valencia, CA), 2) MultiScribe™ High capacity cDNA RT kit (Applied Biosystem, Foster City, CA) and 3) AMV RT kit (Promega, Fitchburg, WI). cDNA was synthesized according to each kit manufacturer’s instructions in a PTC-200 Peltier Thermal Cycler (BioRad, Hercules, CA). The AMV kit used 12 μl of RNA and yielded a cDNA volume of 30 μl, the MultiScribe™ and Omniscript kits used 10 μl of RNA and produced 20 μl of cDNA. All results were normalized based on sample volumes and concentration factors.
Quantitative PCR
Quantitative PCR was performed using an Applied Biosystems Prism 7300 detection system (Foster City, CA). Triplicate samples were analyzed in a 96-well plate with optical caps (Applied Biosystems, Foster City, CA) according to methods described by van Elden et al[18]. Influenza virus was quantified using two forward primers (INFA-1 and INFA-2) with sequences 5′-GGA CTG CAG CGT AGA CGC TT - 3′ and 5′-CAT CCT GTT GTA TAT GAG GCC CAT - 3′; reverse primer (INFA-3) with sequence 5′-CAT TCT GTT GTA TAT GAG GCC CAT - 3′ and probe (INFAprobe) sequence 5′-CTC AGT TAT TCT GCT GGT GCA CTT GCC A -3′. We constructed standard curves for the qPCR by making 1:10 dilutions of cDNA made from the virus stock. The virus was extracted using the Trizol-chloroform method and cDNA synthesized using the MultiScribe™ RT kit. The limit of quantification for the qPCR was 6 influenza virus A particles per PCR well, with all replicates crossing the qPCR fluorescence threshold within 37 cycles.
Inhibition characterization of human rhinovirus and influenza viruses
Virus
Human rhinovirus 16 strain 11757 (HRV) was obtained from ATCC (Manassas, VA) with a concentration of 1.14×1011 virus particles per ml as determined by qPCR. Human rhinovirus was diluted in phosphate buffered saline (PBS) (HyClone, Logan, UT) and stored at −80°C in single-use aliquots. For this experiment HRV RNA was extracted from 100 μl aliquots of a 3×10 −4 dilution of the original stock. The influenza A PR8 virus stock was described above. Influenza virus RNA was extracted from 100 μl aliquots of a 5×10−4 dilution of the original stock.
RNA extraction & reverse transcription
Influenza virus and human rhinovirus were extracted using QIAamp viral RNA columns and Trizol-chloroform extraction as described above. cDNA synthesis was carried out using the MultiScribe™ RT kit (Applied Biosystems, Foster City, CA) as described above.
Quantitative PCR
Quantitative PCR for influenza virus was carried out as described by van Elden et al[18]. HRV qPCR was performed as described by Hayden et al[19] with the following sequences: 5′-GTG AAG AGC CSC RTG TGC T-3′ for the forward primer, 5′-GCT SCA GGG TTA AGG TTA GCC-3′ for the reverse primer and FAM-TGA GTC CTC CGG CCC CTG AAT G-TAMRA for the probe. The limits of quantification for the qPCR were 6 influenza virus A particles per PCR well, and 24 HRV particles per PCR well, with all replicates crossing the qPCR fluorescence threshold within 37 cycles.
Particulate matter
Virus inhibition was tested by spiking samples with fine Arizona road dust (ARD) (AC Spark Plug, General Motors, Detroit, MI) and indoor air dust (IAD) collected at a residence. ARD was weighed out and suspended in sterile filtered water to make stock solutions of 5 mg/ml, 0.5 mg/ml and 0.05 mg/ml. IAD was collected on three Teflon filters from air sampled at 4 liters per minute over a period of two months at a house in Boston, MA. Three people inhabited the residence which was heated by baseboard heat. The room sampled was carpeted, and the windows remained closed over the sampling period. No pets were in residence, and no water damage was noted over the sampling period. The Teflon filters were scraped, the IAD weighed and suspended in water to make stock solutions of 0.2 mg/ml, 0.02 mg/ml and 0.002 mg/ml. Stocks were stored at −20°C.
Triplicate 100 μl aliquots of influenza virus and human rhinovirus were spiked with 100 μl of 5 mg/ml, 0.5 mg/ml and 0.05 mg/ml each of Arizona road dust and with 100 μl of 0.2 mg/ml, 0.02 mg/ml and 0.002 mg/ml each of indoor air dust. Positive controls were virus samples free of dusts. Water blanks as well as Arizona road dust and indoor air dust blanks were analyzed.
Data analysis
Regression models were built using the proc MIXED procedure in the SAS System for Windows 9.13 (Cary, North Carolina). In the optimization study the dependent variable was the log of the number of influenza virus particles per PCR well and the covariates were experiment replicates, RNA extraction method (Trizol-chloroform or QIAamp), and reverse transcription kit (AMV, MultiScribe™ or Omniscript). In the validation study, the dependent variable was the log of the number of influenza virus or human rhinovirus particles per PCR well and the covariates were the extraction method (Trizol-chloroform or QIAamp) and the particulate matter dose (Indoor air dust or Arizona road dust). The mixed model adjusted for the correlation among PCR wells for each sample and calculated a different effect for each dust dose-kit combination. Excel 2002 (Microsoft Corporation, Redmond, WA) was used for t-tests and plotting.
Results
Influenza virus recovery
RNA extraction
The Trizol-chloroform extraction recovered 2.5 times more influenza virus RNA than the QIAamp columns, significant at the 0.05 level. We quantified 8.8×105 (95% CI: 6.2×105–1.3×106) and 3.4×105 (95% CI: 2.4×105–5.2×105) influenza virus particles per PCR well from samples extracted using Trizol-chloroform and QIAamp columns respectively (n=18 [6 samples × 3 PCR wells]).
Reverse transcription
The MultiScribe™ RT kit yielded 2.6-times more influenza virus cDNA compared to that synthesized with the AMV kit and 3.7-times more than that synthesized with the Omniscript kit. Differences were significant at the 0.05 level. From influenza virus samples extracted using Trizol-chloroform, we recovered 2.7×105 (95% CI: 2.0×105–3.6×105), 1.9×105 (95% CI: 1.6×105–2.1×105), and 7.0×105 (95% CI: 5.1×105–9.3×105) from RNA synthesized using the AMV RT, Omniscript RT and MultiScribe™ RT kits respectively (n=8 [2 samples × 2 dilutions × 2 PCR wells]).
Inhibition characterization of human rhinovirus and influenza viruses
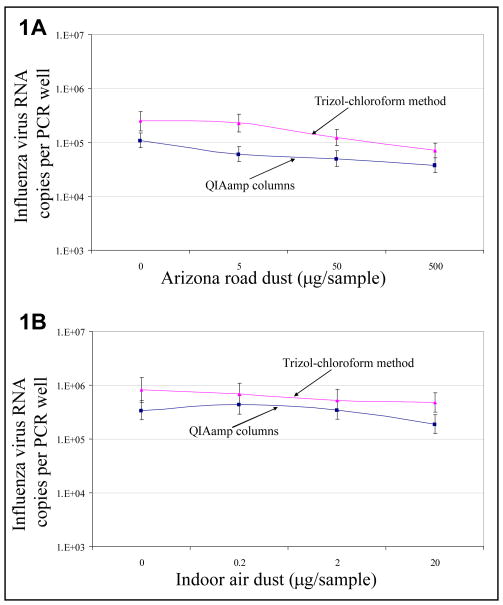
Figures 1 and 2 show the recovered influenza virus and human rhinovirus concentrations from samples contaminated with Arizona road dust (ARD) and indoor air dust (IAD), extracted with QIAamp columns and Trizol-chloroform. At all ARD doses (Figure 1A), Trizol-chloroform extraction yielded 1.9 to 3.5-fold more influenza virus concentration compared to QIAamp columns. At all IAD concentrations (Figure 1B) Trizol-chloroform extraction yielded 1.5 to 3.3-fold more concentration of influenza virus compared to QIAamp columns.
Fig 1.
Total influenza virus recovered from virus samples contaminated with A) Arizona road dust and B) indoor air dust, extracted using 2 RNA extraction methods (n=9 [3 samples × 3 PCR wells]). Concentrations and 95% confidence intervals were estimated from a mixed regression model and controlled for correlation among wells.
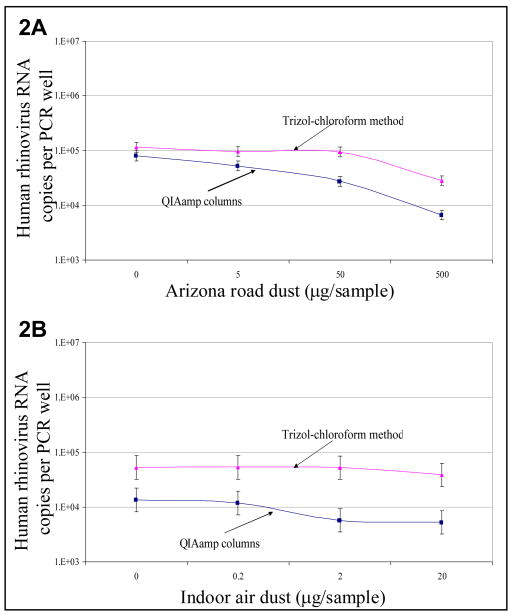
Fig 2.
Total human rhinovirus recovered from samples contaminated with A) Arizona road dust and B) indoor air dust, extracted using 2 RNA extraction methods (n=9 [3 samples × 3 PCR wells]). Concentrations and 95% confidence intervals were estimated from a mixed regression model and controlled for correlation among wells.
For HRV recovery, at all ARD doses (Figure 2A) Trizol-chloroform extraction yielded 1.4 to 4.2-fold more HRV concentrations compared to QIAamp columns. At all IAD concentrations (Figure 2B) Trizol-chloroform extraction yielded 3.9 to 9.0-fold more concentration of HRV compared to QIAamp columns.
Discussion
The methods presented are the first step in the development of protocols to collect and quantify airborne respiratory viruses. The combined optimized methods recovered 10-fold more viruses compared to methods used previously to isolate human rhinovirus from air samples[16]. This increased sensitivity was observed for influenza virus and human rhinovirus contaminated with ARD and IAD. In contrast to previous studies of airborne respiratory viruses which used standard PCR[16, 17, 20, 21], we analyzed viruses via quantitative PCR, a method that is more sensitive and allows us to measure the number of viruses in the sample. Quantification is also essential for exposure assessment and dose response studies.
Samples in the field are collected via air sampling onto media, requiring extra steps to suspend the virus in a solution ready to be extracted. In the case of air samples collected on filter media, the filter material has be washed by vortexing or sonication to suspend the virus in solution. Air samples collected with liquid impingers have to be concentrated using ultracentrifugation columns prior to extraction.
The RNA extraction method we selected was robust with human rhinovirus and influenza virus contaminated with typical air particulate matter up to 50 μg per sample. ARD represented outdoor airborne particulate matter that was well characterized both in composition and size. It is composed of 40% clay material with nominal particle sizes between <1 and 80 μm[22]. The IAD is a more complex dust matrix representative of dust found in an urban residence, and likely contained a mixture of microorganisms, outdoor pollutants and indoor pollutants. The range of dust concentrations was estimated from studies of airborne particulate matter in schools and residences. In one study, 1 milligram of dust was collected at an elementary school over a 2 day sampling period at an airflow of 36.5 lpm (~29 μg/m 3)[23]. In a study of residential indoor air particulate matter, concentration of PM10 in houses of healthy adults was 12.6 μg/m3 air[24], corresponding to a total of 46 μg of dust collected if the air is sampled for 2 hours at a rate of 30 lpm. For our reported study dust concentrations simulated particulate matter between 0.2 and 500 μg per sample, and no inhibitory effect was noted for neither influenza virus nor human rhinovirus at concentrations under 50 μg per sample.
Although assay methods can reduce the effect of inhibition, one approach to reducing inhibitors is to restrict aerosol collection to particle sizes that are relevant and avoid collecting particles that may not contain viruses but probably contain inhibitors. Studies of airborne virus detection have used open-faced filters which do not restrict the particle size they collect[16, 17, 20, 21]. The use of a particle collector that reduces the particle cutoff size of a sampler would eliminate large particles that aren’t likely to contain viruses of interest but may be a major source of inhibitors. Mass is a cubed function of particle size, thus removing a small number of large particles has a large impact on the total mass in a sample.
In contrast with clinical samples which contain high concentration of a single virus, air samples typically contain low numbers of viruses. Air samples also typically contain a wide array of contaminants which can inhibit molecular biology assays. These factors make the likelihood of detecting viruses in air samples very low and can explain the small number of field studies that have detected airborne respiratory viruses as well as the larger number studies with negative findings. After the optimization work was completed, the methods presented in this paper were used in the field to detect low levels of influenza virus in the exhaled breath of infected patients[25].
We studied influenza virus and human rhinovirus because they are both important causes of respiratory disease human rhinovirus causes 40–65% of common colds annually[26, 27] and influenza virus causes 36,000 deaths yearly in the US[28]. Their predominant mode of transmission remains controversial in spite of the numerous studies supporting either airborne transmission[14, 29, 30] and other transmission modes[31, 32]. The protocols we propose can be used in future studies designed to answer transmission questions.
Conclusions
In this study of respiratory virus quantification from samples contaminated with airborne particulate matter, we found that we could improve virus detection 10-fold compared to methods published in field studies of airborne respiratory viruses. We increased recovery by using the optimal molecular biology methods to extract nucleic acid (RNA), reverse transcribe RNA and quantify cDNA using quantitative PCR. The improvement in detection is critical for studies of the environment, where virus concentrations are low and many inhibitory substances are collected along with the microorganisms under study. When tested in a controlled environment, quantification of human rhinovirus and influenza virus was unaffected by particulate matter concentrations up to 50 μg per sample. In conjunction with efficient air sampling techniques, these methods can be used to improve recovery of respiratory viruses from the airborne environment.
Acknowledgments
This work has received financial support from the US Centers for Disease Control and Prevention (CDC) (cooperative grant #1U01CI000446-01), the Natonal Institute of Health (training grants # HL07118 and #AI061884), the Alfred P. Sloan Foundation, and the Federal Aviation Administration (FAA) Office of Aerospace Medicine through the Air Transportation Center of Excellence for Airliner Cabin Environment Research (ACER). Funding was also provided by a cooperative agreement from CDC through the Association of Schools of Public Health (#S2238-22/22).
Notes and references
- 1.Chandler D. J Ind Microbiol Biot. 1998b;21:128–140. [Google Scholar]
- 2.Chandler D, Wagnon C, Bolton H. Appl Environ Microb. 1998a;64:669–677. doi: 10.1128/aem.64.2.669-677.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McDevitt JJ, Lees PSJ, Merz WG, Schwab KJ. Aerobiologia. 2007;23:35–45. [Google Scholar]
- 4.Wiedbrauk DL, Werner JC, Drevon AM. J Clin Microbiol. 1995;33:2643–2646. doi: 10.1128/jcm.33.10.2643-2646.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alvarez AJ, Buttner MP, Stetzenbach LD. Appl Environ Microb. 1995;61:3639–3644. doi: 10.1128/aem.61.10.3639-3644.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poddar SK, Sawyer MH, Connor JD. J Med Microbiol. 1998;47:1131–1135. doi: 10.1099/00222615-47-12-1131. [DOI] [PubMed] [Google Scholar]
- 7.Moppett J, van der Velden V, Wikhuijs A, Hancock J, van Dongen J, Goulden N. Leukemia. 2003:17. doi: 10.1038/sj.leu.2402751. [DOI] [PubMed] [Google Scholar]
- 8.Wilson IG. Appl Environ Microb. 1997;63:3741–3751. doi: 10.1128/aem.63.10.3741-3751.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roy CJ, Milton DK. N Engl J Med. 2004;350:1710–1712. doi: 10.1056/NEJMp048051. [DOI] [PubMed] [Google Scholar]
- 10.Payment P. In: Cultivation and Assay of Viruses. Hurst CJ, Knudsen GR, McInerney MJ, Stetzenbach LD, Walter MV, Knudsen GR, editors. ASM Press; Washington, USA: 1997. pp. 72–78. [Google Scholar]
- 11.Barker J, Stevens D, Bloomfield SF. J Appl Microbiol. 2001;91:7–21. doi: 10.1046/j.1365-2672.2001.01364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riley RL. Am J Med. 1974;57:466–475. doi: 10.1016/0002-9343(74)90140-5. [DOI] [PubMed] [Google Scholar]
- 13.Goldmann DA. Pediatr Infect Dis J. 2000;19:S97–102. doi: 10.1097/00006454-200010001-00002. [DOI] [PubMed] [Google Scholar]
- 14.Tellier R. Emerg Infect Dis. 2006;12:1657–1662. doi: 10.3201/eid1211.060426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee WM, Grindle K, Pappas T, Marshall DJ, Moser MJ, Beaty EL, Shult PA, Prudent JR, Gern JE. J Clin Microbiol. 2007;45:2626–2634. doi: 10.1128/JCM.02501-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Myatt TA, Johnston SL, Zuo Z, Wand M, Kebadze T, Rudnick S, Milton DK. AJRCCM. 2004;169:1187–1190. doi: 10.1164/rccm.200306-760OC. [DOI] [PubMed] [Google Scholar]
- 17.Booth TF, Kournikakis B, Bastien N, Ho J, Kobasa D, Stadnyk L, Li Y, Spence M, Paton S, Henry B, Mederski B, White D, Low DE, McGeer A, Simor A, Vearncombe M, Downey J, Jamieson FB, Tang P, Plummer F. J Infect Dis. 2005;191:1472–1477. doi: 10.1086/429634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Elden LJ, Nijhuis M, Schipper P, Schuurman R, van Loon AM. J Clin Microbiol. 2001;39:196–200. doi: 10.1128/JCM.39.1.196-200.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayden FG, Herrington DT, Coats TL, Kim K, Cooper EC, Villano SA, Liu S, Hudson S, Pevear DC, Collett M, McKinlay M, Group PRIS. Clin Infect Dis. 2003;36:1523–1532. doi: 10.1086/375069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aintablian N, Walpita P, Sawyer MH. Infect Control Hosp Epidemiol. 1998;19:918–923. doi: 10.1086/647764. [DOI] [PubMed] [Google Scholar]
- 21.Sawyer MH, Chamberlin CJ, Wu YN, Aintablian N, Wallace MR. J Infect Dis. 1994;169:91–94. doi: 10.1093/infdis/169.1.91. [DOI] [PubMed] [Google Scholar]
- 22.PTI. ISO STANDARD 12103-1 Test Dust. Vol. 2007. P. T. Incorporated; 2007. [Google Scholar]
- 23.Fox A, Harley W, Feigley C, Salzberg D, Toole C, Sebastian A, Larsson L. J Environ Monit. 2005;7:450–456. doi: 10.1039/b418038k. [DOI] [PubMed] [Google Scholar]
- 24.Liu LJ, Box M, Kalman D, Kaufman J, Koenig J, Larson T, Lumley T, Sheppard L, Wallace L. Environ Health Perspect. 2003;111:909–918. doi: 10.1289/ehp.6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fabian P, McDevitt JJ, DeHaan WH, Fung RO, Cowling BJ, Chan KH, Leung GM, Milton DK. PLoS ONE. 2008;3:e2691. doi: 10.1371/journal.pone.0002691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gwaltney J. Trans Am Clin Climatol Assoc. 1984;96:159–175. [PMC free article] [PubMed] [Google Scholar]
- 27.Hayden F. Am J Med. 2002;112:1S–3S. [Google Scholar]
- 28.CDC. Key Facts About Influenza and Influenza Vaccine. Centers for Disease Control and Prevention; Atlanta: 2006. [Google Scholar]
- 29.Meschievitz CK, Schultz SB, Dick EC. J Infect Dis. 1984;150:195–201. doi: 10.1093/infdis/150.2.195. [DOI] [PubMed] [Google Scholar]
- 30.Dick EC, Jennings LC, Mink KA, Wartgow CD, Inhorn SL. J Infect Dis. 1987;156:442–448. doi: 10.1093/infdis/156.3.442. [DOI] [PubMed] [Google Scholar]
- 31.Brankston G, Gitterman L, Hirji Z, Lemieux C, Gardam M. Lancet Infect Dis. 2007;7:257–265. doi: 10.1016/S1473-3099(07)70029-4. [DOI] [PubMed] [Google Scholar]
- 32.Gwaltney JM, Jr, Moskalski PB, Hendley JO. Ann Intern Med. 1978;88:463–467. doi: 10.7326/0003-4819-88-4-463. [DOI] [PubMed] [Google Scholar]