Summary
During spermatogenesis, global nucleosome removal occurs where histones are initially replaced by transition proteins and subsequently by protamines. This chromatin reorganization is thought to facilitate the compaction of the paternal genome into the sperm head and to protect the DNA from damaging agents. Histone ubiquitination has been suggested to be important for sex chromosome inactivation during meiotic prophase and nucleosome removal at post-meiotic stages. However, the mechanisms regulating these ubiquitin-mediated processes are unknown. In this study, we investigate the role of the ubiquitin ligase RNF8 during spermatogenesis and find that RNF8-deficient mice are proficient in meiotic sex chromosome inactivation (MSCI), but deficient in global nucleosome removal. Moreover, we show that RNF8-dependent histone ubiquitination induces H4K16 acetylation, which may be an initial step in nucleosome removal. Thus, our results show that RNF8 plays an important role during spermatogenesis through histone ubiquitination, resulting in trans-histone acetylation and global nucleosome removal.
Introduction
The basic subunit of chromatin are nucleosomes which 147 bp of DNA wrapped around a histone octamers each containing of two copies of histones H2A, H2B, H3 and H4 (Kornberg and Thomas, 1974). Posttranslational modifications of histone tails have been tightly coupled with many nuclear activities such as replication, transcription and DNA repair (Li et al., 2007; Martin and Zhang, 2007; Vidanes et al., 2005). These modifications may directly affect the electrical charge of histone octamers and subsequently their affinity to DNA (Shahbazian and Grunstein, 2007). Histone modifications may also be recognized by “reader” proteins which may recruit other “effector” proteins for the regulation of various processes relating to DNA metabolism (Ruthenburg et al., 2007). Due to the importance of histone modifications in the regulation of DNA metabolism, perturbations of proteins, or pathways, responsible for these modifications are linked to various developmental defects and human diseases (Bhaumik et al., 2007).
Histone ubiquitination is unique compared to other histone modifications, because of the relatively bulky size of ubiquitin. Histone ubiquitination is predominantly monomeric on histone H2A at lysine 119 and H2B at lysine 120. Histone ubiquitination can both stimulate or repress various cellular processes (Weake and Workman, 2008). For example, ubiquitination of H2A at gene promoter regions suppresses gene transcription (Cao et al., 2005; de Napoles et al., 2004; Wang et al., 2004), while intragenic ubiquitination of H2B facilitates transcription elongation (Fleming et al., 2008; Minsky et al., 2008; Pavri et al., 2006; Shema et al., 2008; Zhu et al., 2005). Histone ubiquitination is also associated with DNA damage responses where H2A ubiquitination is enriched at sites of DNA damage (Bergink et al., 2006; Doil et al., 2009; Huen et al., 2007; Mailand et al., 2007; Stewart et al., 2009; Zhao et al., 2007). H2B ubiquitination is also induced by DNA damage (Wu et al., 2009).
In addition to playing important roles in DNA damage responses, histone ubiquitination is important for normal development and spermatogenesis. During the pachytene stage of meiotic prophase I, ubiquitinated H2A is highly enriched in the XY body (Baarends et al., 1999), where X and Y chromosomes has become partially synapsed through pseudo-autosomal regions and silenced transcriptionally. This phenomena is known as meiotic sex chromosome inactivation (MSCI) (Turner, 2007). Consistent with its transcriptionally silenced status, the XY body contains a unique combination of histone modification markers associated with gene silencing including dimethylation of H3K9 and deacetylation of histone H3 and H4 (Khalil et al., 2004). MSCI is important for proper meiosis and disruption of MSCI leads to the arrest of spermatocytes at the pachytene stage of meiotic prophase (Fernandez-Capetillo et al., 2003). Because of its role in gene silencing, ubiquitinated H2A is believed to be important for the silencing of sex chromosomes in the XY body.
Besides meiosis, ubiquitinated H2A and H2B are also enriched in elongating spermatids (Baarends et al., 1999; Chen et al., 1998), indicating that ubiquitinated H2A and H2B play roles in chromatin remodeling during spermiogenesis. Unlike somatic DNA, sperm DNA is highly condensed and tightly wrapped around protamines instead of histone octamers (Oliva, 2006). The transition from nucleosomes to protamines occurs during the post-meiotic stages of spermatogenesis, termed spermiogenesis, when round haploid spermatids elongate and transform into mature sperm. During this process, most nucleosomal histones are initially replaced by two transition proteins, transition protein 1 and 2, and subsequently by two protamines, protamine 1 and 2 (Meistrich et al., 2003; Oliva, 2006). Although the biological function of these massive chromatin remodeling events are not clear, it is hypothesized that protamine promotes further DNA condensation facilitating the packaging of DNA into tiny sperm heads (Oliva, 2006). Failure to accomplish this global chromatin restructuring causes male sterility (Cho et al., 2001; Shirley et al., 2004; Zhao et al., 2004).
RNF8 is a 485-residue nuclear polypeptide with an N-terminal FHA domain and a C-terminal RING domain. Like other RING domain proteins, RNF8 has ubiquitin E3 ligase activity (Ito et al., 2001). RNF8 has been shown to participate in the DNA damage response by interacting with MDC1, which is rapidly recruited to DNA damage sites through its interaction with γH2AX. At sites of DNA damage, RNF8 ubiquitinates histones, promoting the recruitment of downstream DNA damage response factors, such as 53BP1, BRCA1 and Rad51 (Huen et al., 2007; Kolas et al., 2007; Mailand et al., 2007; Wang and Elledge, 2007). Although the molecular mechanism remains elusive, it has been shown that the RING domain of RNF8 regulates both H2A and H2B ubiquitination at DNA damage sites (Huen et al., 2007; Mailand et al., 2007; Wu et al., 2009), indicating that RNF8 could play an important role in chromatin remodeling.
In this study, we used RNF8-deficient mice and demonstrated that RNF8 regulates both H2A and H2B ubiquitinations in cells of the testes. Surprisingly, RNF8-mediated histone ubiquitination was not important for meiotic sex chromosome inactivation in the XY body prior to meiosis. Instead, RNF8-dependent histone ubiquitination appears to promote H4 K16 acetylation, which is a critical modification for the replacement of histones by protamines during spermiogenesis.
Results
RNF8-deficiency results in male infertility in mice
To examine the functions of RNF8-dependent ubiquitination in vivo, we have examined RNF8-deficient mice generated by using the gene trap ES cell clone RRR260. The gene trap vector was inserted after exon 4, and the exact genomic location was mapped using genomic PCR and DNA sequencing (Supplementary Figure S1a, b). The gene trap allele disrupted proper transcription of the RNF8 gene and generates a fusion product of β-geo and the N-terminal part of RNF8 without its C-terminal RING domain critical for RNF8’s function (Supplementary Figure S1c, d) (Huen et al., 2007; Kolas et al., 2007; Mailand et al., 2007; Sakasai and Tibbetts, 2008; Wang and Elledge, 2007). Although no obvious defect was observed in the germ cells and reproductive system in female RNF8-deficient mice, RNF8-deficient male mice were infertile. The average weight of the testes from 3-month old RNF8-deficient mice was only 60% of that of wild type of the same age (Supplementary Figure S2a). The infertility was unlikely due to changes of hormone levels in RNF8-deficient mice, as FSH, LH, and testosterone levels remained unaltered (Supplementary Figure S2b).
Absence of ubiquitinated H2A in the XY body did not affect meiosis in RNF8-deficient mice
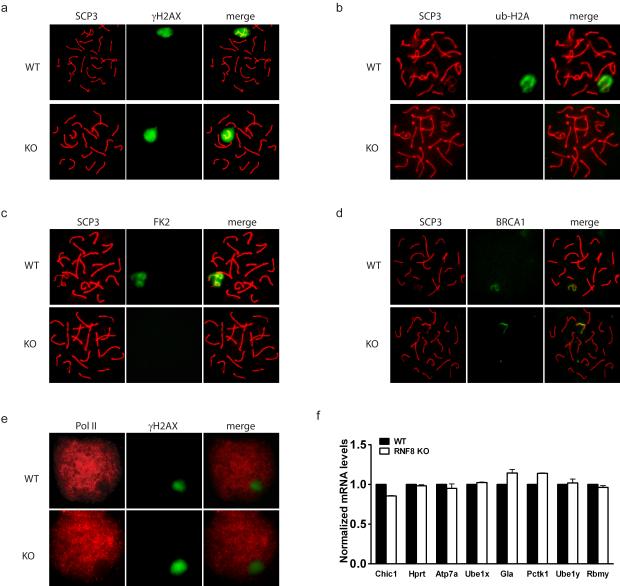
RNF8 regulates H2A ubiquitination (Huen et al., 2007; Mailand et al., 2007) and ubiquitinated H2A is highly enriched in the XY body during the pachytene stage of meiotic prophase I (Baarends et al., 1999). To investigate whether RNF8 plays a role in the ubiquitination of H2A in the XY body, we investigated the level of ubiquitination of H2A in RNF8-deficient mice. It was found that, although the XY body still formed and was clearly marked by γH2AX essential for XY body formation (Fernandez-Capetillo et al., 2003) (Figure 1a), little ubiquitination of H2A was observed in the XY body in RNF8-deficient mice (Figure 1b, Supplementary Figure S3a). Ubiquitinated proteins, detected by the monoclonal antibody FK2, were similarly absent from the XY body of RNF8-deficient mice (Figure 1c). Since histone H2A is the most abundant ubiquitinated protein in cells (Jason et al., 2002), absence of staining for ubiquitinated proteins further supported a role of RNF8 in that H2A ubiquitination. It has been shown that RNF8 regulates the function and localization of BRCA1 following DNA damage (Huen et al., 2007; Kolas et al., 2007; Mailand et al., 2007; Wu et al., 2009). Since BRCA1 is important for sex chromosome inactivation (Turner et al., 2004) and localizes to the unsynapsed axial elements of sex chromosomes in the XY body, we examined BRCA1 localization in the XY body of RNF8-deficient mice. To our surprise, BRCA1 localization appeared normal in the XY body with a string-like shape (Figure 1d), suggesting that BRCA1 might have RNF8-independent roles in the XY body which are distinct from those in DNA damage response in somatic cells. This hypothesis is consistent with a previous report showing that BRCA1 associates with sex chromosomes even when the XY body does not form (Fernandez-Capetillo et al., 2003). The specific role of ubiquitinated H2A in the XY body is not clear but it has been suggested to be important for inactivation of sex chromosomes (Baarends et al., 2005). To investigate whether RNF8 plays a role in sex chromosome inactivation by ubiquitinating H2A, we examined whether RNF8-deficient mice showed evidence of any defect in sex chromosome inactivation. One of the hallmarks of MSCI is that the transcriptional machinery is excluded from the XY body (Richler et al., 1994). Interestingly, we found that RNA polymerase was similarly absent from the XY body in RNF8-deficient mice as in wild type mice, suggesting that transcriptional activity was low in either case (Figure 1e). Consistently, the level of Cot-1 RNA FISH signals, which represent newly synthesized RNA (Namekawa et al., 2006; Turner et al., 2005), was found to be low in leptotene stage spermatocytes, increased in pachytene stage, and remained excluded from the XY body in RNF8-deficient mice, further suggesting that overall sex gene expressions were not altered (Supplementary Figure S3b,c). To confirm these observations, we examined the expression levels of several sex chromosome genes (Chic1, Hprt, Atp7a, Ube1x, Gla and Pctk1 on X chromosome; Ube1y and Rbmy on Y chromosome) using purified spermatocytes and found that their RNA levels were indeed not elevated (Figure 1f). These results suggest that the process of sex chromosome inactivation is independent of RNF8 ubiquitin ligase activity, and that ubiquitination of H2A is not required for this process. Furthermore, the induction of meiotic synapsis was unaffected (Supplementary Figure S3d), as well as crossover events (marked by MLH1), which occur after synapsis (Supplementary Figure S3e). In addition, metaphase I spermatocytes and haploid round spermatids (identified by centromere and SCP3 staining) could be readily observed in the testes of RNF8-deficient mice (Supplementary Figure S3f). Taken together, these observations suggest that the spermatogenic defect in RNF8-deficient mice occurs after meiosis.
Figure 1. Absence of ubiquitinated H2A in the XY body did not affect meiosis in RNF8-deficient testes.
a.-d. Ubiquitinated H2A and ubiquitinated proteins (FK2) were absent in the XY body of RNF8-deficient spermatocytes, while γH2AX and BRCA1 were intact. Typical immunostaining images of spermatocytes at pachytene stage are shown. SCP3 represents synapsed chromosomes and γH2AX marks the XY body. e. RNA polymerase II remained excluded from the XY body in RNF8-deficient spermatocytes. Typical immunostaining images of spermatocytes at pachytene stage are shown. f. Sex chromosome inactivation was maintained in RNF8-deficient mice. Real time PCR results of expression levels of several genes on sex chromosomes in purified spermatocytes are shown. The levels in WT mice were arbitrarily set to 1 and those in RNF8-deficient mice were normalized accordingly. See also Supplementary Figure S3.
Spermiogenesis is defective in RNF8-deficient mice
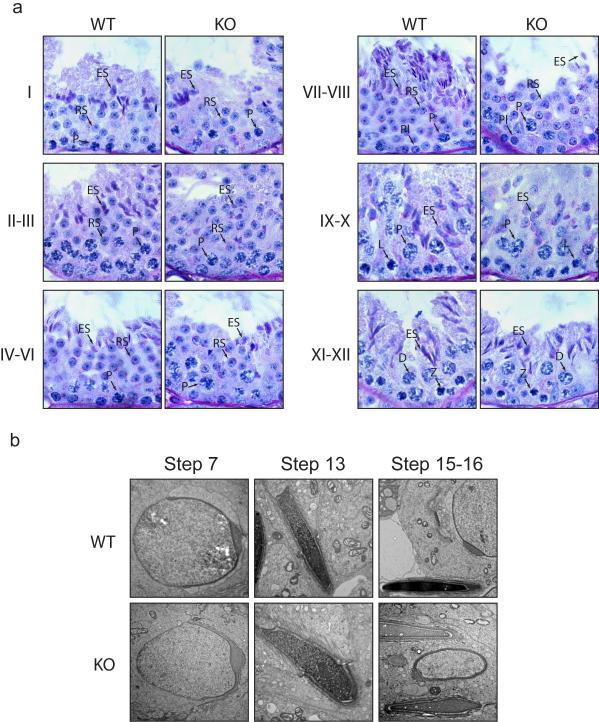
Next we analyzed the post-meiotic development in the testes of RNF8-defecient mice. Histological analysis of the testes revealed that RNF8-deficient testes contained similar numbers of tubule cross sections as in wild type testes (Supplementary Figure S4a). Although there was a slight decrease of the cellularity of Sertoli cells in each tubule cross section (Supplementary Figure S4b), RNF8-deficient male germ cells progressed through meiosis normally. Compared with Sertoli cells, there was little reduction of haploid round spermatids that just finished meiosis and started acrosomes development (Figure 2a, Supplementary Figure S4c) However, late spermatid development was compromised. Fewer condensing spermatids were identified (stage I, II-III) and the number of condensed mature spermatids was drastically reduced (stage IV-VI, VII-VIII) (Figure 2a, Supplementary Figure S4c). Inspection with electron microscopy suggested that spermatid elongation occur normally and acrosomes were formed. DNA condensation in step 13 spermatids could be observed with anterior-to-posterior thickening of chromatin fibers, however, the extent of condensation was slightly lower than that in wild type mice. Strikingly, DNA condensation in stage V-VIII spermatids, which have been expected to mature to step 15-16 with highly compacted DNA, was severely abrogated in RNF8-deficient mice (Figure 2b). Consistently, dramatically fewer sperm were harvested from cauda epididymis in RNF8-deficient mice than from wild type mice (Figure 3a). Most of these sperm displayed abnormal rounded heads (Figure 3b). Hematoxylin staining revealed that most of these sperm still contained residue cytoplasm (Figure 3b). Although these sperm had tails, the majority of them (>95%) were immotile (Figure 3c). In vitro fertilization using these abnormal sperm revealed that they could not fertilize wild type oocytes (Supplementary Figure 5a). Sections of cauda epididymis also showed a similar dramatic decrease of sperm. Consistently, we observed the presence of rounded sperm heads in more than 95% of the remaining specimens from RNF8-deficient mice (Figure 3d). Electron microscopy inspection of epididymal sperm further confirmed that the sperm heads were much less condensed and residual cytoplasm had not been completely removed in most of these sperm (Figure 3e). Notably, a slight elevation of apoptosis was observed in both testes and epididymis, but this elevated level of apoptosis was not associated with a specific cell type (Supplementary Figure S5c). To test if nuclei of sperm from RNF8-deficient mice could fertilize oocytes and support embryonic development, we performed intracytoplasmic sperm injection (ICSI) using sperm derived from RNF8-deficient mice and oocytes from wild type mice. Interestingly, 5 out of 28 oocytes that survived after injection developed into blastocysts and hatched out of the zona pellucida (Supplementary Figure S5a). Genotyping using these hatched blastocysts confirmed that they were all heterozygous for the RNF8-deficient allele (Supplementary Figure S5b), suggesting that chromatin integrity was maintained in the sperm from RNF8-deficient mice. Collectively, these observations suggested that the infertility of RNF8-deficient male mice was likely due to defects at post-meiotic stages of spermatogenesis, known as spermiogenesis.
Figure 2. Late steps of spermatid development were abrogated in RNF8-deficient testes.
a. RNF8-deficient testes contained less condensed late spermatids. PAS-hematoxylin staining of testes from 3-month old wild type (WT) and RNF8-deficient (KO) mice were performed. Stages of seminiferous epithelium cycle were determined according to the morphology of spermatocytes and round spermatids. Spermatids at different stages are shown. Pl, preleptotene; L, leptotene; Z, zygotene; P, packytene; D, diplotene; RS, round spermatids; ES, elongating spermatids. b. Defect of DNA condensation during step 13-16 of spermatid development was observed in RNF8-deficient testes. Typical electron microscopic images of spermatids at different steps are shown. See also Supplementary Figure S4.
Figure 3. Spermiogenesis was defective in RNF8-deficient mice.
a. Number of sperm was decreased in RNF8-deficient mice. Sperm from 3-month old wild type (WT) and RNF8-deficient (KO) mice were counted. Sperm from both cauda epididymis were included. 6 mice of each genotype were used, p<0.001. b. Abnormal rounded sperm heads were observed in RNF8-deficient mice. Upper, typical morphologies of sperm from wild type (WT) and RNF8-deficient (KO) mice with hematoxylin staining are shown; Lower, quantification of sperm with abnormal rounded head is shown. c. Majorities of sperm from RNF8-deficient mice were immotile. Sperm mobility of wild type (WT) and RNF8-deficient (KO) mice are compared. d. H&E staining of paraffin sections of cauda epididymis from wild type (WT) and RNF8-deficient (KO) mice are shown. On right are images with higher magnification showing details of sperm heads. e. Sperm were less condensed and contained residue cytoplasm. Typical electron microscopic images of sperm heads from wild type (WT) and RNF8-deficient (KO) are shown. See also Supplementary Figure S5.
Histones failed to be replaced in sperm from RNF8-deficient mice
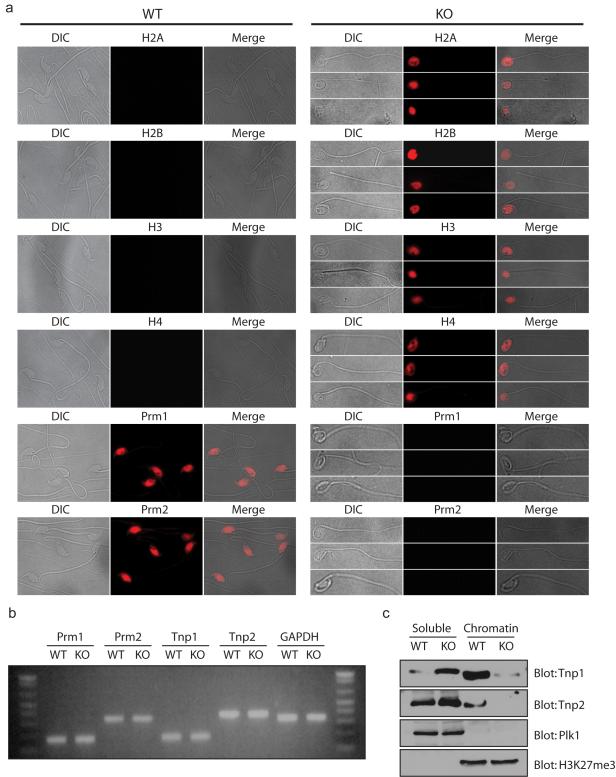
During spermiogenesis, histones are first replaced by transition proteins, which are subsequently replaced by protamines (Meistrich et al., 2003; Oliva, 2006). DNA is tightly wrapped around these basic proteins and packaged into sperm head. In order to check if histone replacement was affected in RNF8-deficient mice, we stained histones and protamines in the sperm. Consistent with previous reports showing that more than 85% of histones are replaced by protamines in sperm (Tanphaichitr et al., 1978), sperm from wild type mice showed strong staining for both Prm1 and Prm2 but not histones. On the contrary, sperm from RNF8-deficient mice contained histones but not protamines in their rounded heads (Figure 4a). This suggests that histones are not properly replaced by protamines in sperm from RNF8-deficient mice. Previous reports have found that disrupted expression of transition proteins or protamines could lead to a defect in histone replacement (Nair et al., 2008; Okada et al., 2007; Wu et al., 2000). This prompted us to examine whether the level of expression of two transition proteins (Tnp1 and Tnp2) and two protamines (Prm1 and Prm2) differed between wild type and RNF8-deficient testes using semi-quantitative RT-PCR. Our results show no significant difference in the RNA expression pattern of these genes in wild-type and RNF8-deficient testes (Figure 4b). Transition proteins were intermediates between histones and protamines, and the presence of histones in the sperm from RNF8-deficient mice suggested that the defect lies at the replacement of histones by transition proteins. As transition proteins are only expressed at step 9-12 during spermatid development, we analyzed the levels of chromatin-bound Tnp1 and Tnp2 in the testes of wild type and RNF8-defective mice. In the testes of wild type mice, a portion of both Tnp1 and Tnp2 were chromatin-bound, suggesting that some of the histones had been replaced by these proteins. However, we found that chromatin-associated Tnp1 and Tnp2 levels were dramatically reduced in RNF8-deficient testes while they were significantly increased in the chromatin-free fraction (Figure 4c). This suggested that most transition proteins failed to replace the histones and to become incorporated into the sperm chromatin in the testes of RNF8-deficient mice. Alternatively, changes in chromatin structure in RNF8-deficient mice rendered transition proteins loosely associated with chromatin and failed to pellet during chromatin extraction. Consistently, we did not detect any transition proteins in RNF8-deficient sperm (data not shown), which was likely due to proteolysis or other types of degradation of non-chromatin-associated transition proteins, together with many other non-chromatin-associated proteins, in the final stages of spermatid development. Very little protamine could be detected in testes of RNF8-deficient mice (Supplementary Figure S6), probably due to developmental defect at a stage prior to the protein synthesis of protamines (Braun, 1998). This hypothesis is consistent with the observation that spermatids of steps13-16 were lost. Alternatively, it could be due to a rapid proteolysis of protamines unable to be deposited on DNA.
Figure 4. Histones failed to be replaced in sperm from RNF8-deficient mice.
a. Protamines failed to replace histones in RNF8-deficient sperm. Typical immunostaining images of sperm from wild type (WT) and RNF8-deficient (KO) mice are shown. Antibodies used are indicated. Prm1: protamine 1; Prm2: protamine 2. b. mRNA levels of transition proteins and protamines were similar in wild type and RNF8-deficient mice. Semi-quantitative RT-PCR was performed using testes cDNA from wild type (WT) and RNF8-deficient (KO) mice. Tnp1: transition protein 1; Tnp2: transition protein 2. GAPDH was used as internal control. c. Incorporation of transition proteins into chromatin was defective in RNF8-deficient mice. Western blots using testes from wild type (WT) and RNF8-deficient (KO) mice are shown. Both NETN-soluble and HCl-released chromatin-bound fractions were used. Antibodies used are indicated, and Plk1 and H3K27me3 were used as soluble and chromatin-bound fraction loading controls respectively. See also Supplementary Figure S6.
Decreased ubiquitination of H2A, H2B and acetylation of H4 K16 in sperm from RNF8-deficient mice
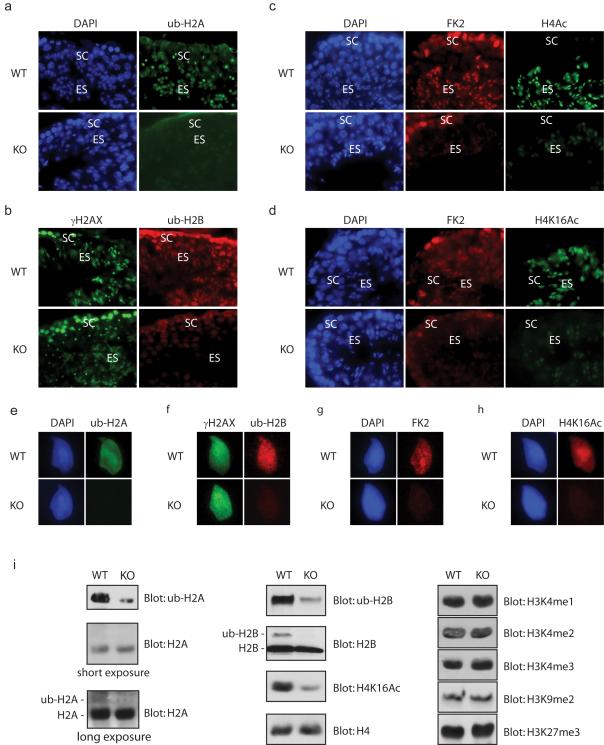
We next explore the potential mechanism by which RNF8 participates in the replacement of histone in sperm nuclei. Interestingly the substrate of RNF8, ubiquitinated H2A, was enriched not only in the XY body, but also in elongating spermatids before histone replacement (Baarends et al., 1999). We therefore hypothesized that RNF8 may be required for H2A ubiquitination in elongating spermatids, which might be important for the incorporation of transition proteins in sperm chromatin. We compared the ub-H2A levels between wild type and RNF8-deficient testes and found that ub-H2A staining was significantly decreased in elongating spermatids in the tesis of RNF8-deficient mice (Figure 5a). Consistent with a previous report (Chen et al., 1998), we found that ubiquitinated H2B was present in elongating spermatids and colocalized with γH2AX staining, and the level of ub-H2B was decreased in RNF8-deficient mice (Figure 5b). We also used the FK2 antibody, which recognizes ubiquitinated proteins, and we confirmed that FK2 signal was decreased in elongating spermatids as well (Figure 5c, d). It has also been suggested that the N-terminal tail of histone H4 is highly acetylated in elongating spermatids before histone replacement (Meistrich et al., 1992). Since acetylation adds negative charges to nucleosomes, it has been hypothesized that acetylation of H4 could loose chromatin fibers which would enhance histone replacement (Sonnack et al., 2002). Consistently, we observed the presence of H4 acetylation in elongating spermatids, and interestingly this was significantly decreased in the testes of RNF8-deficient mice (Figure 5c). Although several lysine residues could be acetylated at the N-terminus of H4 (Allfrey et al., 1968; Shahbazian and Grunstein, 2007), H4 K16 acetylation has been shown to be a key step in chromatin remodling to a more open conformation (Dion et al., 2005; Shogren-Knaak et al., 2006). Since acetylation of H4 K16 is prevelent in elongating spermatids (van der Heijden et al., 2006), we examined the levels of H4 K16 acetylation in the testes of RNF8-deficient mice and found that it was significantly decreased compared with that in testes from wild type mice (Figure 5d). The decrease of these histone modification markers was further confirmed by staining individual spermatids (Figure 5e-h). In order to quantitatively assess histone modifications, we used Western blot and found that the levels of both ubiquitinated H2A and H2B were decreased in testes from RNF8-deficient mice, and that acetylation of H4 K16 was dramatically decreased as well (Figure 5i). These results suggest that RNF8-dependent H2A and H2B ubiquitinations are likely to induce H4 K16 acetylation which facilitates global removal of histones from sperm chromatin. Since histone ubiquitination has been shown to associate with histone methylation (Briggs et al., 2002; Lee et al., 2007a; Lee et al., 2007b; McGinty et al., 2008; Sun and Allis, 2002), we also examined several histone methylation markers in the testes of RNF8-deficient mice. No obvious defects of histone methylation markers were observed in RNF8-deficient mice (Figure 5i).
Figure 5. Ubiquitinated H2A, H2B and H4 K16 acetylation were decreased in RNF8-deficient testes.
a.-d. ub-H2A, ub-H2B and K4 K16 acetylation were decreased in elongating spermatids of RNF8-deficient mice. Typical immunostaining images of testes seminiferous tubules at similar stages from wild type (WT) and RNF8-deficient (KO) mice are shown. SC: spermatocytes; ES: elongating spermatids. Antibodies used are indicated, FK2: antimono/poly-ubiquitinated proteins. e.-h. Typical immunostaining images of individual elongating spermatids from wild type (WT) and RNF8-deficient (KO) mice are shown. i. ub-H2A, ub-H2B and H4 K16 acetylation were decreased in testes from RNF8-deficient mice. Western blots of chromatin-bound proteins using testes from wild type (WT) and RNF8-deficient (KO) mice are shown. Antibodies used are indicated, and H4 was used as loading control.
RNF8 controls the association of H4K16 acetyltransferase MOF with chromatin
To confirm our observation on the changes of histone modifications in the testes of RNF8-deficient mice, we examined these histone markers again by analyzing RNF8-deficient mouse embryonic fibroblasts (MEFs). As was found in the testes of RNF8-defective mice, the ubiquitnation of H2A and H2B as well as the acetylation of H4 K16 were all decreased in RNF8-deficient MEFs (Figure 6a). Furthermore, depleting RNF8 in HeLa cells suppressed not only H2A and H2B ubiquitinations, but also H4 K16 acetylation (Figure 6b). RNF8 is an E3 ubiquitin ligase and has been reported to directly ubiquitinate H2A in vitro (Mailand et al., 2007), It can also ubiquitinate H2B in vitro, but neither contain any acetyltransferase activity to directly acetylates H4 K16 nor associate with any known acetyltransferases (data not shown). Thus, it is likely that the decrease of H4 K16 acetylation is due to loss of RNF8-mediated ubiquitinations of H2A and H2B. The major acetyltransferase responsible for H4 K16 acetylation is MOF (males absent of the first). It was initially identified as a key component of the male specific lethal (MSL) complex in drosophila, which binds to male X chromosome and maintains dosage compensation of male X-linked genes (Akhtar and Becker, 2000; Hilfiker et al., 1997). Mammalian MOF is also required for H4 K16 acetylation (Gupta et al., 2008; Smith et al., 2005; Taipale et al., 2005; Thomas et al., 2008). Although MOF is highly expressed in the male reproduction system (Thomas et al., 2007), the role of MOF in spermatogenesis is not clear. We examined the expression pattern of MOF proteins in the testes of wild type mice and found that MOF was highly enriched in the elongating spermatids (Figure 6c), which correlated well with the distribution of H4 K16 acetylation in the testes. We went on to check the expression level of MOF in RNF8-deficient testes, but did not find any significant alterations (Figure 6c). However, Western blot analysis revealed that chromatin-bound MOF was significantly reduced in the testes of RNF8-deficient mice while MOF levels were elevated in the chromatin-free fraction (Figure 6d). Similar results were also obtained from wild type and RNF8-deficient MEFs (Figure 6e), suggesting that ubiquitinated H2A and H2B are needed for the efficient association of MOF to the chromatin in order to acetylate H4 K16. Depletion of MOF from HeLa cells abolished more than 90% of H4 K16 acetylation, while ubiquitinations of H2A and H2B were normal (Figure 6f), indicating that H4 K16 acetylation is a downstream event of H2A and H2B ubiquitination during histone replacement in the elongating spermatids. Since H4 K16 has been shown to control chromatin relaxation in vitro (Shogren-Knaak et al., 2006), H4 K16 acetylation likely promotes histone removal in elongating spermatids.
Figure 6. RNF8 controlled chromatin association of H4K16 acetyltransferase MOF.
a. ub-H2A, ub-H2B and H4 K16 acetylation were decreased in RNF8-deficient MEFs. Western blots of chromatin-bound proteins from wild type (WT) and RNF8-deficient (KO) MEFs are shown. Antibodies used are indicated, and H4 was used as loading control. b. Depletion of RNF8 affects not only H2A and H2B ubiquitination, but also H4K16 acetylation. Western blots of chromatin-bound proteins from control siRNA or RNF8 siRNA-treated HeLa cells are shown. c. MOF is enriched in elongating spermatids. Typical immunostaining images of seminiferous tubules at similar stages from wild type (WT) and RNF8-deficient (KO) mice are shown. SC: spermatocytes; ES: elongating spermatids. Antibodies used are indicated. d.-e. Chromatin-associated MOF was decreased in both testes and MEFs from RNF8-defiecient mice. Western blot using wild type (WT) and RNF8-deficient (KO) testes (d) and MEFs (e) are shown. Both NETN-soluble and chromatin-bound fractions were used. Antibodies used are indicated. Plk1 and H3K27me3 were used as soluble and chromatin-bound fraction loading controls, respectively. f. Depletion of MOF affects H4K16 acetylation only, but not H2A and H2B ubiquitination. Western blots of chromatin-bound proteins from control siRNA or MOF siRNA-treated HeLa cells are shown.
Collectively, our results demonstrate that RNF8-dependent histone ubiquitination is not essential for meiotic sex chromosome inactivation, but is important for global histone removal at post-meiotic stages. Furthermore, our results suggest that RNF8-dependent histone ubiquitination controls H4 K16 acetylation by regulating association of MOF on the chromatin, which could be a crucial step for histone removal in the elongating spermatids.
Discussion
Global histone removal from the DNA of spermatids and replacement by transition proteins and protamines are critical for proper spermiogenesis. In this study, we demonstrate that histone ubiquitination and histone H4 K16 acetylation play important roles in global histone replacement in spermatids. Transition proteins and protamines are very basic proteins that help generate an extremely condensed chromatin that is packaged into the sperm head. This process is proposed to protect the paternal genome and make it inaccessible to nucleases and mutagens (Oliva and Dixon, 1991). It is thus not surprising that several studies have shown that mice with reduced levels of transition proteins or protamine displayed defects in spermiogenesis (Nair et al., 2008; Okada et al., 2007; Wu et al., 2000). In our study, we found that RNF8-dependent histone ubiquitination and histone H4 K16 acetylation do not affect the mRNA levels of transition proteins or protamines. Instead, these histone modifications appear to control histone removal and transition protein incorporation into the chromatin.
H2A ubiquitination has been shown to be important for spermatogenesis. During pachytene stage of meiosis I, ubiquitinated H2A is accumulated in the XY body and enriched again in elongating spermatids after meiosis (Baarends et al., 1999). The role of ubiquitinated H2A in the XY body is not clear, but it is thought that these modifications may mediate MSCI (Baarends et al., 2005). Interestingly, absence of ubiquitinated H2A in the XY body does not affect XY body formation, sex chromosome inactivation or meiotic progression, indicating that H2A ubiquitination is not essential for these processes. The ubiquitination of H2A was significantly suppressed in elongating spermatids in the testes of RNF8-deficient mice (Figure 5i) suggesting that H2A ubiquitination may play an essential role for nucleosome removal during post-meiotic stages. Besides H2A ubiquitination, we provide evidence that H2B ubiquitination is also present in elongating spermatids and it is decreased in the spermatids of RNF8-deficient mice.
Yeast Rad6 is an E2 ubiquitin conjugase for H2B. Similar to a mutation of the Rad6-directed ubiquitination site of H2B, Rad6 mutants are defective in sporulation (Lawrence, 1994; Robzyk et al., 2000). Genetic deletion of its mouse homolog HR6B causes male sterility due to defects at post-meiotic stages of spermiogenesis (Roest et al., 1996). It is possible that RNF8 and HR6B work in concert to regulate H2B ubiquitination in the elongating spermatids, which could be essential for nucleosome removal. Protein structure analysis of recombinant nucleosomes indicates that H2B ubiquitination may be involved in decondensation of chromatin (Luger et al., 1997). In addition, ubiquitinated H2B, which are usually associated with active genes (Minsky et al., 2008; Pavri et al., 2006), may promote local nucleosome removal during transcription.
We and others have previously demonstrated that RNF8 regulates the ubiquitination of H2A following DNA damage (Huen et al., 2007; Mailand et al., 2007). Here we show that RNF8 also affects the basal levels of H2A and H2B ubiquitination in the absence of DNA damage. Previous reports have suggested that the PRC1 complex, containing RING1A/RING1B, is responsible for H2A ubiquitination while the RNF20/RNF40 complex is responsible for H2B ubiquitination (Wang et al., 2004; Zhu et al., 2005). It is possible that RNF8 is in a large complex together with PRC1 and other proteins or that RNF8 is a tissue-specific E3 for histone H2A/H2B. Although topoisomerase II-induced DNA decatenation occurs in the elongating spermatids (Leduc et al., 2008), it is unlikely that RNF8 participates in the global DNA damage response in elongating spermatids since key DNA damage repair factors, such as MDC1, Rad51 and DNAPK, are not expressed in the elongating spermatids (data not shown).
Trans-histone modification has been suggested to occur in several organisms (Briggs et al., 2002; Kirmizis et al., 2007; Lee et al., 2007b; Sun and Allis, 2002; Weake and Workman, 2008; Wood et al., 2005). In yeast, ubiquitinated H2B has been shown to regulate methylation of H3 K4 and H3 K79 (Briggs et al., 2002; Lee et al., 2007a; McGinty et al., 2008; Sun and Allis, 2002). In our study, we did not observe any dramatic alterations in the levels of in H3 K4 methylation in RNF8-deficient mice (Figure 3e). It is likely that histone modifications in yeast and mammals are regulated by different mechanisms. Alternatively, since H2B ubiquitination only occurs in about 1.5 % of the total pool of H2B proteins in mammalian cells (West and Bonner, 1980), this ubiquitination event may not affect the global level of H3 K4 methylation. Nevertheless, we here found evidence that H2A and H2B ubiquitination may control H4 K16 acetylation, which represents a novel trans-histone modification in mammalian cells. We have further demonstrated ub-H2A and ub-H2B regulates chromatin-association of MOF, the acetyltransferase responsible for H4 K16 acetylation. It has been shown that the N-terminal tail of H4, including H4 K16 site, from one nucleosome interacts with a H2A/H2B heterodimer on an adjacent nucleosome (Luger et al., 1997). Thus, it is possible that the bulky ubiquitin group may simply change the internucleosomal space to expose the H4 K16 site or to accommodate for the MOF complex to associate with chromatin. A similar mechanism has been recently suggested for the recruitment of the Dot1 methyltransferase where the ubiquitination of H2B promotes Dot1-mediated H3 K79 methylation possibly by facilitating allosteric binding of Dot1 to the ubiquitinated H2B site (McGinty et al., 2008).
In this study, we provided the first direct evidence in vivo that H4 K16 acetylation is critical for histone removal during spermatogenesis. H4 K16 acetylation has been shown to change the higher-order chromatin structure and prevent the formation of 30 nm fibers in vitro, suggesting that this modification prevents chromatin condensation (Shogren-Knaak et al., 2006). H4 K16 acetylation is indeed correlated with decondensed chromatin structure in vivo. In yeast, over 80% of H4 proteins are acetylated at K16 and the chromatin is in a more decondensed state than in higher eukaryotes (Lohr et al., 1977; Smith et al., 2003). In Drosophila, the male X chromosome is also highly enriched for H4 K16 acetylation and is highly decondensed compared to the other chromosomes (Turner et al., 1992). Although no chromosome locus that is highly enriched for H4 K16 acetylation have been identified in human or mice, we have shown here that H4 K16 acetylation is dramatically increased globally in elongating spermatids and this acetylation event precedes histone replacement during spermatogenesis. Such global H4 K16 acetylation correlates well with the global removal of histones and incorporation of transition proteins, suggesting that acetylation of H4 K16 induces global chromatin decondensation and triggers histone removal. Consistently, we have demonstrated that a defect in H4 K16 acetylation indeed significantly suppresses histone removal and transition protein incorporation. It has also been noticed that in addition to acetylation of H4 K16, the K5, K8 and K12 sites of H4 are also hyperacetylated during spermatogenesis (Govin et al., 2007). The functional significance of these acetylations needs to be further examined both in vitro and in vivo.
The mechanisms responsible for global nucleosome removal may be similar to mechanisms regulating local removal of nucleosome during transcription and DNA repair. It has been reported that both ubiquitination of H2A and acetylation of H4 K16 are induced by DNA damage (Gupta et al., 2005; Huen et al., 2007; Mailand et al., 2007; Nicassio et al., 2007). It is conceivable that these histone modifications may render the structure of chromatin more open and trigger removal of histones surrounding the DNA damage sites to provide accessibility for DNA repair proteins. Similarly, acetylation of histones is also required for efficient histone eviction during transcription (Reinke and Horz, 2003; Williams et al., 2008).
Taken together, our results reveal a novel role of the ubiquitin ligase RNF8 in the nucleosome removal during spermiogenesis. RNF8 ubiquitinates histones H2A and H2B promoting the recruitmant of the MOF acetyltransferase by increased accessibility of H4 histones resulting in trans-histone acetylation of H4 K16 by MOF. The MOF-mediated acetylation of H4 K16 initiates a global histone removal leading to the restructuring of the sperm chromatin into a compact structure, impervious to DNA-damaging agents and malleable to the tight packaging of the paternal genome into the sperm head.
Experimental Procedures
Animals
Gene trap ES cell RRR260 was purchased from Bay Genomics. After germline transmission of gene trap ES cell RRR260, the chimeras were backcrossed with C57BL/6J mice. All of the mice were housed in an AAALAC accredited facility in accordance with the National Research Council’s guide for the care and use of laboratory animals. Genotyping primer sequences were listed in Supplementary Table S1.
Sperm analysis
Sperm were harvested by dissecting and cutting cauda epididymis in Research Vitro Fert medium (Cook, Australia). Sperm counting and sperm activity analysis were performed as described previously (Shirley et al., 2004).
Transmission electron microscopy
For transmission electron microscopy, mice were perfused with 1% glutaraldehyde and 4% PFA in PBS. Testes and Epididymis were quickly harvested after perfusion and post-fixed with1% osmium tetroxide. The tissues were then dehydrated, embedded, and thick sections were cut. After toluidine blue staining, ultra thin sections at75 nm thickness were cut, stained by lead citrate, and observed using Philips CM-100 transmission electron microscope.
In vitro fertilization (IVF) and intracytoplasmic sperm injection (ISCI)
IVF was preformed according to protocol from Jackson Laboratory (http://cryo.jax.org/ivf.html) and ISCI was performed as described (Kimura and Yanagimachi, 1995). Genotyping of blastocysts were performed using nested PCR. All primers sequences are listed in Supplementary Table S1.
Germ cell spread and fractionation
Germ cell spread was performed as described with modifications (Peters et al., 1997). Germ cells fractionation was performed using sedimentation in 2-4% BSA gradient following established protocols (Pivot-Pajot et al., 2003). Fractions enriched with certain populations were indentified under a light microscope, and fractions of similar populations were pooled.
Cell culture, siRNA and Western blots
Cells were cultured in DMEM supplemented with 10% FBS. For siRNA experiments, HeLa cells were seeded in 6-well plates with 5×105 cells per well, and two sequential siRNA transfections were performed after 24 and 48 hours respectively. Lysates were prepared 36 hours post-transfection and subjected for Western blots. All siRNAs were purchased from Dharmacon and individual siRNAs for the same gene were pooled for optimal effects. siRNA sequences for RNF8 were AGAAUGAGCUCCAAUGUAUdTdT and GAUGGGUGCGAGGUGACUGdTdT, and those for MOF were GCACGAAGUUGAUGGCAAAdTdT and GGAAAGAGAUCUACCGCAAdTdT. For Western blot, cell pellet was lysed with NETN (50mM Tris-HCl pH8.0, 100mM NaCl, 2 mM EDTA and 0.5% NP-40). Soluble fraction was collected and the pellet was treated with micrococcal nuclease or 0.2 M HCl to release chromatin-bound proteins, which were then neutralized using 1 M Tris-HCl pH 8.5. Both fractions were subjected to electrophoresis, Western blot, and then probed with antibodies as indicated. Acid urea gel was used for detecting protamine 1 and 2. Anti-RNF8 N and C terminus antibodies were generated against a.a. 1-324 and 380-488 respectively. Myc antibody was from Covance. Plk1 antibody was generated as described previously (Lu et al., 2008). Antibodies for transition proteins, protamines and MOF were kind gifts from Dr. Stephen Kistler, Dr. Rod Balhorn and Dr. Yali Dou respectively.
Histology, Immunostaining, TUNEL assay, and RNA FISH
For histology, testes were fixed by perfusing mice with Bouin’s fixatives (Polysciences). 5 μm paraffin sections were cut and the stained with periodic acid schiff (PAS)-hematoxylin. Stages of seminiferous epithelium cycle and steps of spermatid development were determined described (Russell et al., 1990). For immunostaining, 5 μm paraffin-embedded sections of testis were used for staining FK2, H4 acetylation and H4 K16 acetylation. After deparaffinization and rehydration, slides were incubated with boiling 0.01 M sodium citrate, pH 6.0 for 10 minutes to retrieve the antigens before immunostaining. For ub-H2A, ub-H2B and MOF staining, testes were fixed in fast fix (Polysciences) and 8 m frozen sections were taken. Each section was treated with 2 M HCl for 30 minutes before staining ub-H2B. Standard immunostaining procedures were used. All histone antibodies were from Millipore except anti ub-H2B antibody, which was from Medimabs. FK2 antibody was from Biomol. Anti-RNA polymerase II antibody was from Santa Cruz. Centromere antibody was from Antibodies Incorporated. MLH1 antibody was from BD. BRCA1 antibody was generated as described previously (Wu et al., 2009). SCP3 antibodies were from Genetex (rabbit polyclonal) and Dr. Stephen West (mouse monoclonal). Apoptosis analysis was performed using ApopTag Plus Peroxidase In Situ Apoptosis Kit (Millipore). Cot-1 RNA FISH was performed as described (Turner et al., 2005) besides that biotinylated Cot-1 probes were prepared using mouse Cot-1 DNA and BioNick DNA Labeling System (Invitrogen) and incubated overnight.
Statistical methods
Student t-tests were used for all statistical analyses. Mean and standard deviation were plotted.
Supplementary Material
Acknowledgements
We thank Drs. Eric Fearon, Kathleen Cho, and Benjamin Margolis for their generous sharing of experimental equipments. We thank Dr. Mats Ljungman for invaluable suggestions and editing the manuscript. We are grateful to Sasha Meshinchi and Dorothy Sorenson for help on electron microscopy. We thank Drs. Rod Balhorn, Stephen Kistler, Stephen West, and Yali Dou for reagents. L.L. is a recipient of postdoctoral fellowship from the Center for Genetics in Health and Medicine in the University of Michigan. This work was supported in part by grants from the National Institutes of Health (CA130899 to XY) and the Developmental fund from the University of Michigan Cancer Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akhtar A, Becker PB. Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Molecular cell. 2000;5:367–375. doi: 10.1016/s1097-2765(00)80431-1. [DOI] [PubMed] [Google Scholar]
- Allfrey VG, Pogo BG, Littau VC, Gershey EL, Mirsky AE. Histone acetylation in insect chromosomes. Science (New York, NY. 1968;159:314–316. doi: 10.1126/science.159.3812.314. [DOI] [PubMed] [Google Scholar]
- Baarends WM, Hoogerbrugge JW, Roest HP, Ooms M, Vreeburg J, Hoeijmakers JH, Grootegoed JA. Histone ubiquitination and chromatin remodeling in mouse spermatogenesis. Developmental biology. 1999;207:322–333. doi: 10.1006/dbio.1998.9155. [DOI] [PubMed] [Google Scholar]
- Baarends WM, Wassenaar E, van der Laan R, Hoogerbrugge J, Sleddens-Linkels E, Hoeijmakers JH, de Boer P, Grootegoed JA. Silencing of unpaired chromatin and histone H2A ubiquitination in mammalian meiosis. Molecular and cellular biology. 2005;25:1041–1053. doi: 10.1128/MCB.25.3.1041-1053.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergink S, Salomons FA, Hoogstraten D, Groothuis TA, de Waard H, Wu J, Yuan L, Citterio E, Houtsmuller AB, Neefjes J, et al. DNA damage triggers nucleotide excision repair-dependent monoubiquitylation of histone H2A. Genes & development. 2006;20:1343–1352. doi: 10.1101/gad.373706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik SR, Smith E, Shilatifard A. Covalent modifications of histones during development and disease pathogenesis. Nature structural & molecular biology. 2007;14:1008–1016. doi: 10.1038/nsmb1337. [DOI] [PubMed] [Google Scholar]
- Braun RE. Post-transcriptional control of gene expression during spermatogenesis. Seminars in cell & developmental biology. 1998;9:483–489. doi: 10.1006/scdb.1998.0226. [DOI] [PubMed] [Google Scholar]
- Briggs SD, Xiao T, Sun ZW, Caldwell JA, Shabanowitz J, Hunt DF, Allis CD, Strahl BD. Gene silencing: trans-histone regulatory pathway in chromatin. Nature. 2002;418:498. doi: 10.1038/nature00970. [DOI] [PubMed] [Google Scholar]
- Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Molecular cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Chen HY, Sun JM, Zhang Y, Davie JR, Meistrich ML. Ubiquitination of histone H3 in elongating spermatids of rat testes. The Journal of biological chemistry. 1998;273:13165–13169. doi: 10.1074/jbc.273.21.13165. [DOI] [PubMed] [Google Scholar]
- Cho C, Willis WD, Goulding EH, Jung-Ha H, Choi YC, Hecht NB, Eddy EM. Haploinsufficiency of protamine-1 or -2 causes infertility in mice. Nature genetics. 2001;28:82–86. doi: 10.1038/ng0501-82. [DOI] [PubMed] [Google Scholar]
- de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Developmental cell. 2004;7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Dion MF, Altschuler SJ, Wu LF, Rando OJ. Genomic characterization reveals a simple histone H4 acetylation code. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:5501–5506. doi: 10.1073/pnas.0500136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136:435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- Fernandez-Capetillo O, Mahadevaiah SK, Celeste A, Romanienko PJ, Camerini-Otero RD, Bonner WM, Manova K, Burgoyne P, Nussenzweig A. H2AX is required for chromatin remodeling and inactivation of sex chromosomes in male mouse meiosis. Developmental cell. 2003;4:497–508. doi: 10.1016/s1534-5807(03)00093-5. [DOI] [PubMed] [Google Scholar]
- Fleming AB, Kao CF, Hillyer C, Pikaart M, Osley MA. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Molecular cell. 2008;31:57–66. doi: 10.1016/j.molcel.2008.04.025. [DOI] [PubMed] [Google Scholar]
- Govin J, Escoffier E, Rousseaux S, Kuhn L, Ferro M, Thevenon J, Catena R, Davidson I, Garin J, Khochbin S, et al. Pericentric heterochromatin reprogramming by new histone variants during mouse spermiogenesis. The Journal of cell biology. 2007;176:283–294. doi: 10.1083/jcb.200604141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Guerin-Peyrou TG, Sharma GG, Park C, Agarwal M, Ganju RK, Pandita S, Choi K, Sukumar S, Pandita RK, et al. The mammalian ortholog of Drosophila MOF that acetylates histone H4 lysine 16 is essential for embryogenesis and oncogenesis. Molecular and cellular biology. 2008;28:397–409. doi: 10.1128/MCB.01045-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Sharma GG, Young CS, Agarwal M, Smith ER, Paull TT, Lucchesi JC, Khanna KK, Ludwig T, Pandita TK. Involvement of human MOF in ATM function. Molecular and cellular biology. 2005;25:5292–5305. doi: 10.1128/MCB.25.12.5292-5305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilfiker A, Hilfiker-Kleiner D, Pannuti A, Lucchesi JC. mof, a putative acetyl transferase gene related to the Tip60 and MOZ human genes and to the SAS genes of yeast, is required for dosage compensation in Drosophila. The EMBO journal. 1997;16:2054–2060. doi: 10.1093/emboj/16.8.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Adachi S, Iwakami R, Yasuda H, Muto Y, Seki N, Okano Y. N-Terminally extended human ubiquitin-conjugating enzymes (E2s) mediate the ubiquitination of RING-finger proteins, ARA54 and RNF8. European journal of biochemistry / FEBS. 2001;268:2725–2732. doi: 10.1046/j.1432-1327.2001.02169.x. [DOI] [PubMed] [Google Scholar]
- Jason LJ, Moore SC, Lewis JD, Lindsey G, Ausio J. Histone ubiquitination: a tagging tail unfolds? Bioessays. 2002;24:166–174. doi: 10.1002/bies.10038. [DOI] [PubMed] [Google Scholar]
- Khalil AM, Boyar FZ, Driscoll DJ. Dynamic histone modifications mark sex chromosome inactivation and reactivation during mammalian spermatogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:16583–16587. doi: 10.1073/pnas.0406325101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y, Yanagimachi R. Intracytoplasmic sperm injection in the mouse. Biology of reproduction. 1995;52:709–720. doi: 10.1095/biolreprod52.4.709. [DOI] [PubMed] [Google Scholar]
- Kirmizis A, Santos-Rosa H, Penkett CJ, Singer MA, Vermeulen M, Mann M, Bahler J, Green RD, Kouzarides T. Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature. 2007;449:928–932. doi: 10.1038/nature06160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science (New York, NY. 2007;318:1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD, Thomas JO. Chromatin structure; oligomers of the histones. Science (New York, NY. 1974;184:865–868. doi: 10.1126/science.184.4139.865. [DOI] [PubMed] [Google Scholar]
- Lawrence C. The RAD6 DNA repair pathway in Saccharomyces cerevisiae: what does it do, and how does it do it? Bioessays. 1994;16:253–258. doi: 10.1002/bies.950160408. [DOI] [PubMed] [Google Scholar]
- Leduc F, Maquennehan V, Nkoma GB, Boissonneault G. DNA damage response during chromatin remodeling in elongating spermatids of mice. Biology of reproduction. 2008;78:324–332. doi: 10.1095/biolreprod.107.064162. [DOI] [PubMed] [Google Scholar]
- Lee JS, Shukla A, Schneider J, Swanson SK, Washburn MP, Florens L, Bhaumik SR, Shilatifard A. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell. 2007a;131:1084–1096. doi: 10.1016/j.cell.2007.09.046. [DOI] [PubMed] [Google Scholar]
- Lee MG, Villa R, Trojer P, Norman J, Yan KP, Reinberg D, Di Croce L, Shiekhattar R. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science (New York, NY. 2007b;318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Lohr D, Kovacic RT, Van Holde KE. Quantitative analysis of the digestion of yeast chromatin by staphylococcal nuclease. Biochemistry. 1977;16:463–471. doi: 10.1021/bi00622a020. [DOI] [PubMed] [Google Scholar]
- Lu LY, Wood JL, Minter-Dykhouse K, Ye L, Saunders TL, Yu X, Chen J. Polo-like kinase 1 is essential for early embryonic development and tumor suppression. Molecular and cellular biology. 2008;28:6870–6876. doi: 10.1128/MCB.00392-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- Martin C, Zhang Y. Mechanisms of epigenetic inheritance. Current opinion in cell biology. 2007;19:266–272. doi: 10.1016/j.ceb.2007.04.002. [DOI] [PubMed] [Google Scholar]
- McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature. 2008;453:812–816. doi: 10.1038/nature06906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meistrich ML, Mohapatra B, Shirley CR, Zhao M. Roles of transition nuclear proteins in spermiogenesis. Chromosoma. 2003;111:483–488. doi: 10.1007/s00412-002-0227-z. [DOI] [PubMed] [Google Scholar]
- Meistrich ML, Trostle-Weige PK, Lin R, Bhatnagar YM, Allis CD. Highly acetylated H4 is associated with histone displacement in rat spermatids. Molecular reproduction and development. 1992;31:170–181. doi: 10.1002/mrd.1080310303. [DOI] [PubMed] [Google Scholar]
- Minsky N, Shema E, Field Y, Schuster M, Segal E, Oren M. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nature cell biology. 2008;10:483–488. doi: 10.1038/ncb1712. [DOI] [PubMed] [Google Scholar]
- Nair M, Nagamori I, Sun P, Mishra DP, Rheaume C, Li B, Sassone-Corsi P, Dai X. Nuclear regulator Pygo2 controls spermiogenesis and histone H3 acetylation. Developmental biology. 2008;320:446–455. doi: 10.1016/j.ydbio.2008.05.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namekawa SH, Park PJ, Zhang LF, Shima JE, McCarrey JR, Griswold MD, Lee JT. Postmeiotic sex chromatin in the male germline of mice. Curr Biol. 2006;16:660–667. doi: 10.1016/j.cub.2006.01.066. [DOI] [PubMed] [Google Scholar]
- Nicassio F, Corrado N, Vissers JH, Areces LB, Bergink S, Marteijn JA, Geverts B, Houtsmuller AB, Vermeulen W, Di Fiore PP, et al. Human USP3 is a chromatin modifier required for S phase progression and genome stability. Curr Biol. 2007;17:1972–1977. doi: 10.1016/j.cub.2007.10.034. [DOI] [PubMed] [Google Scholar]
- Okada Y, Scott G, Ray MK, Mishina Y, Zhang Y. Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature. 2007;450:119–123. doi: 10.1038/nature06236. [DOI] [PubMed] [Google Scholar]
- Oliva R. Protamines and male infertility. Human reproduction update. 2006;12:417–435. doi: 10.1093/humupd/dml009. [DOI] [PubMed] [Google Scholar]
- Oliva R, Dixon GH. Vertebrate protamine genes and the histone-to-protamine replacement reaction. Progress in nucleic acid research and molecular biology. 1991;40:25–94. doi: 10.1016/s0079-6603(08)60839-9. [DOI] [PubMed] [Google Scholar]
- Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- Peters AH, Plug AW, van Vugt MJ, de Boer P. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res. 1997;5:66–68. doi: 10.1023/a:1018445520117. [DOI] [PubMed] [Google Scholar]
- Pivot-Pajot C, Caron C, Govin J, Vion A, Rousseaux S, Khochbin S. Acetylation-dependent chromatin reorganization by BRDT, a testis-specific bromodomain-containing protein. Molecular and cellular biology. 2003;23:5354–5365. doi: 10.1128/MCB.23.15.5354-5365.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke H, Horz W. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Molecular cell. 2003;11:1599–1607. doi: 10.1016/s1097-2765(03)00186-2. [DOI] [PubMed] [Google Scholar]
- Richler C, Ast G, Goitein R, Wahrman J, Sperling R, Sperling J. Splicing components are excluded from the transcriptionally inactive XY body in male meiotic nuclei. Molecular biology of the cell. 1994;5:1341–1352. doi: 10.1091/mbc.5.12.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robzyk K, Recht J, Osley MA. Rad6-dependent ubiquitination of histone H2B in yeast. Science (New York, NY. 2000;287:501–504. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- Roest HP, van Klaveren J, de Wit J, van Gurp CG, Koken MH, Vermey M, van Roijen JH, Hoogerbrugge JW, Vreeburg JT, Baarends WM, et al. Inactivation of the HR6B ubiquitin-conjugating DNA repair enzyme in mice causes male sterility associated with chromatin modification. Cell. 1996;86:799–810. doi: 10.1016/s0092-8674(00)80154-3. [DOI] [PubMed] [Google Scholar]
- Russell LD, Ettlin RA, Sinha Hikim AP, Clegg ED. Histological and histopathological evaluation of the testis. Cache River Press; 1990. [Google Scholar]
- Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nature reviews. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakasai R, Tibbetts R. RNF8-dependent and RNF8-independent regulation of 53BP1 in response to DNA damage. The Journal of biological chemistry. 2008;283:13549–13555. doi: 10.1074/jbc.M710197200. [DOI] [PubMed] [Google Scholar]
- Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annual review of biochemistry. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- Shema E, Tirosh I, Aylon Y, Huang J, Ye C, Moskovits N, Raver-Shapira N, Minsky N, Pirngruber J, Tarcic G, et al. The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes & development. 2008;22:2664–2676. doi: 10.1101/gad.1703008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley CR, Hayashi S, Mounsey S, Yanagimachi R, Meistrich ML. Abnormalities and reduced reproductive potential of sperm from Tnp1- and Tnp2-null double mutant mice. Biology of reproduction. 2004;71:1220–1229. doi: 10.1095/biolreprod.104.029363. [DOI] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science (New York, NY. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- Smith CM, Gafken PR, Zhang Z, Gottschling DE, Smith JB, Smith DL. Mass spectrometric quantification of acetylation at specific lysines within the amino-terminal tail of histone H4. Analytical biochemistry. 2003;316:23–33. doi: 10.1016/s0003-2697(03)00032-0. [DOI] [PubMed] [Google Scholar]
- Smith ER, Cayrou C, Huang R, Lane WS, Cote J, Lucchesi JC. A human protein complex homologous to the Drosophila MSL complex is responsible for the majority of histone H4 acetylation at lysine 16. Molecular and cellular biology. 2005;25:9175–9188. doi: 10.1128/MCB.25.21.9175-9188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnack V, Failing K, Bergmann M, Steger K. Expression of hyperacetylated histone H4 during normal and impaired human spermatogenesis. Andrologia. 2002;34:384–390. doi: 10.1046/j.1439-0272.2002.00524.x. [DOI] [PubMed] [Google Scholar]
- Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, Nakada S, Ylanko J, Olivarius S, Mendez M, et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136:420–434. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- Taipale M, Rea S, Richter K, Vilar A, Lichter P, Imhof A, Akhtar A. hMOF histone acetyltransferase is required for histone H4 lysine 16 acetylation in mammalian cells. Molecular and cellular biology. 2005;25:6798–6810. doi: 10.1128/MCB.25.15.6798-6810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanphaichitr N, Sobhon P, Taluppeth N, Chalermisarachai P. Basic nuclear proteins in testicular cells and ejaculated spermatozoa in man. Experimental cell research. 1978;117:347–356. doi: 10.1016/0014-4827(78)90148-9. [DOI] [PubMed] [Google Scholar]
- Thomas T, Dixon MP, Kueh AJ, Voss AK. Mof (MYST1 or KAT8) is essential for progression of embryonic development past the blastocyst stage and required for normal chromatin architecture. Molecular and cellular biology. 2008;28:5093–5105. doi: 10.1128/MCB.02202-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas T, Loveland KL, Voss AK. The genes coding for the MYST family histone acetyltransferases, Tip60 and Mof, are expressed at high levels during sperm development. Gene Expr Patterns. 2007;7:657–665. doi: 10.1016/j.modgep.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Turner BM, Birley AJ, Lavender J. Histone H4 isoforms acetylated at specific lysine residues define individual chromosomes and chromatin domains in Drosophila polytene nuclei. Cell. 1992;69:375–384. doi: 10.1016/0092-8674(92)90417-b. [DOI] [PubMed] [Google Scholar]
- Turner JM. Meiotic sex chromosome inactivation. Development (Cambridge, England) 2007;134:1823–1831. doi: 10.1242/dev.000018. [DOI] [PubMed] [Google Scholar]
- Turner JM, Aprelikova O, Xu X, Wang R, Kim S, Chandramouli GV, Barrett JC, Burgoyne PS, Deng CX. BRCA1, histone H2AX phosphorylation, and male meiotic sex chromosome inactivation. Curr Biol. 2004;14:2135–2142. doi: 10.1016/j.cub.2004.11.032. [DOI] [PubMed] [Google Scholar]
- Turner JM, Mahadevaiah SK, Fernandez-Capetillo O, Nussenzweig A, Xu X, Deng CX, Burgoyne PS. Silencing of unsynapsed meiotic chromosomes in the mouse. Nature genetics. 2005;37:41–47. doi: 10.1038/ng1484. [DOI] [PubMed] [Google Scholar]
- van der Heijden GW, Derijck AA, Ramos L, Giele M, van der Vlag J, de Boer P. Transmission of modified nucleosomes from the mouse male germline to the zygote and subsequent remodeling of paternal chromatin. Developmental biology. 2006;298:458–469. doi: 10.1016/j.ydbio.2006.06.051. [DOI] [PubMed] [Google Scholar]
- Vidanes GM, Bonilla CY, Toczyski DP. Complicated tails: histone modifications and the DNA damage response. Cell. 2005;121:973–976. doi: 10.1016/j.cell.2005.06.013. [DOI] [PubMed] [Google Scholar]
- Wang B, Elledge SJ. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:20759–20763. doi: 10.1073/pnas.0710061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Molecular cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- West MH, Bonner WM. Histone 2B can be modified by the attachment of ubiquitin. Nucleic acids research. 1980;8:4671–4680. doi: 10.1093/nar/8.20.4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SK, Truong D, Tyler JK. Acetylation in the globular core of histone H3 on lysine-56 promotes chromatin disassembly during transcriptional activation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:9000–9005. doi: 10.1073/pnas.0800057105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A, Schneider J, Shilatifard A. Cross-talking histones: implications for the regulation of gene expression and DNA repair. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2005;83:460–467. doi: 10.1139/o05-116. [DOI] [PubMed] [Google Scholar]
- Wu J, Huen MS, Lu LY, Ye L, Dou Y, Ljungman M, Chen J, Yu X. Histone ubiquitination associates with BRCA1-dependent DNA damage response. Molecular and cellular biology. 2009;29:849–860. doi: 10.1128/MCB.01302-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JY, Ribar TJ, Cummings DE, Burton KA, McKnight GS, Means AR. Spermiogenesis and exchange of basic nuclear proteins are impaired in male germ cells lacking Camk4. Nature genetics. 2000;25:448–452. doi: 10.1038/78153. [DOI] [PubMed] [Google Scholar]
- Zhao GY, Sonoda E, Barber LJ, Oka H, Murakawa Y, Yamada K, Ikura T, Wang X, Kobayashi M, Yamamoto K, et al. A critical role for the ubiquitin-conjugating enzyme Ubc13 in initiating homologous recombination. Molecular cell. 2007;25:663–675. doi: 10.1016/j.molcel.2007.01.029. [DOI] [PubMed] [Google Scholar]
- Zhao M, Shirley CR, Hayashi S, Marcon L, Mohapatra B, Suganuma R, Behringer RR, Boissonneault G, Yanagimachi R, Meistrich ML. Transition nuclear proteins are required for normal chromatin condensation and functional sperm development. Genesis. 2004;38:200–213. doi: 10.1002/gene.20019. [DOI] [PubMed] [Google Scholar]
- Zhu B, Zheng Y, Pham AD, Mandal SS, Erdjument-Bromage H, Tempst P, Reinberg D. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Molecular cell. 2005;20:601–611. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.